Summary
Hepatitis viruses cause mitotic error-induced chromosome instability (ME-CIN). Since liver is naturally aneuploidgenic, the effect of additional ME-CIN on carcinogenesis was hard to dissect. This study using ME-CIN model mice (Sgo1−/+ mice) demonstrates additional ME-CIN can be tumorigenic in liver.
Abstract
A major etiological risk factor for hepatocellular carcinoma (HCC) is infection by Hepatitis viruses, especially hepatitis B virus and hepatitis C virus. Hepatitis B virus and hepatitis C virus do not cause aggressive activation of an oncogenic pathway, but they transactivate a broad array of genes, cause chronic inflammation, and, through interference with mitotic processes, lead to mitotic error-induced chromosome instability (ME-CIN). However, how ME-CIN is involved in the development of HCC remains unclear. Delineating the effect of ME-CIN on HCC development should help in identifying measures to combat HCC. In this study, we used ME-CIN model mice haploinsufficient in Shugoshin 1 (Sgo1−/+) to assess the role of ME-CIN in HCC development. Treatment with the carcinogen azoxymethane caused Sgo1−/+ ME-CIN model mice to develop HCCs within 6 months, whereas control mice developed no HCC (P < 0.003). The HCC development was associated with expression of early HCC markers (glutamine synthetase, glypican 3, heat shock protein 70, and the serum marker alpha fetoprotein), although without fibrosis. ME-CIN preceded the expression of HCC markers, suggesting that ME-CIN is an important early event in HCC development. In 12-month-old untreated Sgo1 mice, persistent DNA damage, altered gene expression, and spontaneous HCCs were observed. Sgo1 protein accumulated in response to DNA damage in vitro. Overall, Sgo1−/+-mediated ME-CIN strongly promoted/progressed development of HCC in the presence of an initiator carcinogen, and it had a mild initiator effect by itself. Use of the ME-CIN model mice should help in identifying drugs to counteract the effects of ME-CIN and should accelerate anti-HCC drug development.
Introduction
Hepatocellular carcinomas (HCCs) remain a major health threat with 23000 predicted deaths in the US and 600000+ worldwide in 2014 (1,2). The 5-year survival rate for HCC is 15% for all stages combined (1,2). There is an unmet need for intervention strategies for HCC. A major etiological risk factor for HCC is infection by a hepatitis virus [especially hepatitis B virus (HBV) and hepatitis C virus (HCV)] (3,4). High viral infection rate in Asia and West Africa is leading to endemic HCC, accounting for 80% of HCC deaths worldwide. Although vaccination against HBV is readily available for preventing the infection in Western countries, HCV vaccine has not been available.
For translational strategies to prevent and/or treat HCC, animal models that reflect cancer-specific etiology play a major role (5). An issue in translational HCC study is the limited number of practical animal models that reflect HCC-specific etiology, especially that involving Hepatitis virus infection (6–9). HBV and HCV are BSL-2 biohazards, thus limiting the number of capable animal-based research environments. In addition, common laboratory rodents cannot be infected by HBV or HCV due to species barriers. The only two known rodents that can host the viruses are Woodchuck and tupaia, and they are uncommon as lab animals (10). To investigate immuno-oncological effects of HBV or HCV infection, several models have been generated, including a human hepatocyte-transplanted immunodeficient mouse model (chimera) and cre-lox-based viral protein expressing transgenic mice models (6–9,11–14). However, these mice are hard to generate or maintain for regular drug testing purposes. Mice treated with the hepatocarcinogen diethylnitrosamine at the neonatal stage and subsequently fed a high fat diet develop HCC rapidly, yet the model more likely reflects NonAlcoholic SteatoHepatitis-HCC (15,16). It is imperative to have a practical model that reflects HCCs with a viral etiology/oncogenic events.
Unlike certain oncogenic viruses, hepatitis viruses do not immediately induce HCC through an oncogenic pathway. The infection is theorized to lead to carcinogenesis via multiple pathways; transactivation of a broad array of genes, chronic infection and lingering immune attack on hepatocytes, direct genomic alteration via viral genome integration, and an immediate increase in chromosome instability (CIN) that leads to further accumulation of mutations (2–4). The HBV viral protein HBx binds to and interferes with functions of cellular HBx Interacting Protein, which regulates centrosomal function, and BubR1, a mitotic checkpoint protein (17–21). HCV infection leads to expression of viral NS5A protein, which can induce CIN via mitotic cell cycle dysregulation (22). These results link HBV and HCV infection to CIN through mitotic errors (ME-CIN). Since HBV or HCV infection induces ME-CIN in liver and HCC, the influence of high ME-CIN on HCC development has been suspected; but the role of ME-CIN effects in HCC carcinogenesis have not been distinguished from other effects of HBV or HCV infection.
Furthermore, liver is naturally aneuploidgenic (23), leading to an assumption that the effect of ME-CIN may be well tolerated. Here, we questioned whether additional ME-CIN in liver manifests as carcinogenesis.
CIN is widespread among cancers, is a poor prognostic marker, and is deeply involved in carcinogenesis and recurrence (24–28). For oncological study purposes, several high ME-CIN mouse models have been generated (24,29–32). Previously, we used haploinsufficiency in BubR1, a spindle checkpoint component (33,34). Haploinsufficiency (−/+) in the gene resulted in 40–60% reduction in gene expression and in protein amount, causing insufficient functions in the corresponding protein. Mouse Embryonic Fibroblasts (MEFs) from BubR1−/+ mice showed chromosome mis-segregation due to spindle checkpoint impairment, and the mice showed enhanced carcinogenesis in liver, lung, and colon upon treatment with the carcinogen azoxymethane (AOM) (33). In this study, we used another ME-CIN model: haploinsufficiency in Sgo1, a protector of chromosome cohesion and centrosome integrity (35–37). The Sgo1 defect interferes with the same two pathways that the HBV protein HBx targets, namely centrosomal functions and mitotic processes, thus Sgo1 mice may represent a model mimicking ME-CIN that is caused by viral infection and that occurs in addition to natural aneuploidy in liver (17–22). Although Sgo1 has two functions (in centromeric cohesion and centrosome integrity), defect in either one pathway can lead to ME-CIN. MEFs from Sgo1−/+ mice showed chromosome mis-segregation and multiple centrosomes, consistent with the dual functions of Sgo1 in maintenance of mitotic chromosome cohesion and centrosome integrity (38). AOM treatment resulted in five-times more colonic precancerous aberrant crypt foci lesions in Sgo1 mice (38). Here, we analyzed the role of ME-CIN in HCC development with the Sgo1−/+ ME-CIN model mice. Gross HCCs have been reported in other ME-CIN mice models (32,33), and this report is the first in-detail HCC study in an ME-CIN model mice with histological characterization.
Materials and Methods
Animals
Generation, genotyping, and characterization of MEF and the colonic carcinogenesis assay with AOM injections in haploinsufficient mice have been described previously (33,38). Briefly, after genotyping and grouping, we treated 8-week-old female mice (9 wild type, 10 Sgo1−/+) with AOM (4-mg/kg body weight, i.p. injection) twice per week for 4 weeks, maintained them for an additional 12 weeks without further treatment, then collected samples at the endpoint after euthanizing. All mice were generated with the non-cancer-prone C57/BL6 background.
For the spontaneous HCC assay, all Sgo1−/+ and control wild-type male mice were maintained in the OUHSC rodent barrier facility with regular diet (Purina, St. Louis, MO) without any experimental treatment. At 12 months of age, they were euthanized with CO2, and tissue samples were collected promptly and examined for presence of tumor. N = 16 (wild type) and 15 (Sgo1−/+). All treatments were in compliance with protocols approved by the OUHSC institutional animal care and use committee.
Immunohistochemistry (IHC) and immunofluorescence (IF)
Liver tissues were fixed in 10% formalin, paraffin-embedded and subjected to immunohistochemistry (IHC; Histostain SP kit or SuperPicture 3rd Gen IHC kit, Invitrogen/LifeTechnologies, Grand Island, NY). The following primary antibodies were used at 1.0 μg/ml: antiphospho-H2AX (gammaH2AX, Novus Biologicals, Littleton, CO; Catalog No. NBP-1-19931), anti-p53 (Santa Cruz Biotechnology, Dallas, TX; SC-6243), anti-p16INK4A (Lifespan Biosciences, Seattle, WA; LS-B1347), anti-Bcl2 (Santa Cruz Biotechnology, SC-492), anti-alpha fetoprotein (AFP) (biorbyt, San Francisco, CA; orb129505), anti-heat shock protein 70 (HSP70) (biorbyt, orb10848), anti-glutamine synthetase (GS)/GLUL (biorbyt, orb4662), anti-glypican 3 (GPC3) (biorbyt, orb10735), antigamma-Tubulin (Novus, NB100-92115; Santa Cruz Biotechnology, SC-17787; Abcam ab27074), and anti-Sgo1 (SGOL1; Proteintech, Chicago, IL; 16977-1-AP). IHC experiments were repeated at least twice with appropriate controls.
For immunofluorescence, after de-paraffinization and re-hydration, we performed antigen-retrieval with 20-min incubation in boiling 10-mM citrate buffer (pH 6.0). When antimouse primary antibody was used, after cooling, the slides were rinsed twice in PBS-tween20 (0.05%), and incubated with blocking buffer with 1% bovine serum albumin and antimouse IgG (0.1mg/ml) (AffiPure Donkey antimouse IgG, Jackson Immuno Research, cat. 715-005-151) for 1h in room temperature. Then, we applied primary antibodies and incubated overnight in 4°C. The next day after three times 3-min rinse in PBS-tween20, secondary fluorescent antibodies were applied [green and far-red combination; Alexa 488- and Alexa 680-conjugated (Invitrogen), alternatively cy2- and cy5-conjugated (Jackson Immuno Research)], incubated for 1–2h in the dark condition, rinsed the slides three times with PBS, then stained nuclei with DAPI (4′,6-diamidino-2-phenylindole) (20 µg/ml) and mounted with ProLong Gold antifade/mounting medium (Molecular Probes). IF images were taken with confocal microscope (Leica SP2 using LCS software, Mannheim, Germany) in the OUHSC Laboratory for Molecular Biology and Cytometry Research.
Statistics
Four to six mice per strain were analyzed, at least 10 IHC images were captured from each liver, and percentages of IHC-positive cells were calculated. Data were expressed as means ± SD, or as variances. The differences between groups were analyzed using Student’s t-test with Graphpad Prism5 software (La Jolla, CA).
Immunoblots
Our standard procedures were followed (38). For serum samples, 2× sodium dodecyl sulfate sample buffer (Sigma, St. Louis, MO; S3401-10VL) was added (1:1 volume), and samples were boiled for 5min and subjected to immunoblotting. Equal volumes (sera) or protein amount (tissue) were loaded per lane.
Cell culture and in vitro treatments
The human HCC cell line Huh7 was obtained from Dr. Naushad Ali (OUHSC), originally from Apath LLC (St. Louis); HepG2 from Dr. Jian-Xing Ma (OUHSC) originally from American Type Culture Collection (ATCC); cervical cancer-origin HeLa cells from ATCC. The cells were maintained in Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum and 1% penicillin/streptomycin in a 37°C, 5% CO2 incubator. The cells were plated on coverslips in 6 well plates and treated for 72h with one of the following reagents from R&D systems (Minneapolis, MN) or Sigma: interleukin 2 (IL2) (4ng/ml), lipopolysaccharide (80ng/ml), interleukin 10 (IL10) (10ng/ml), interleukin 22 (IL22) (10ng/ml), 5-fluorouracil (5-FU) (2 µg/ml), etoposide (10 µM), cisplatin (5 µg/ml), MG132 (1 µg/ml), and taxol (20nM). The cells were fixed with 4% paraformaldehyde for 20min, permeabilized with 100% methanol (−20°C) for 5min, rehydrated in phosphate-buffered saline, then subjected to IHC.
Primary fibroblast cultures of wild type and Sgo1−/+ mice were established from neonate tail with collagenase XI-S treatment (Sigma, 0.5mg/ml), and used for experiments within four passages. Drug treatments followed the same procedure as huh7 cells.
Cell line authentication
Cell lines used in this study were originally purchased from ATCC [HepG2 (ATCC-HB-8065TM) and HeLa (ATCC-CCL-2TM)] or distributed from Apath LLC [huh7] and immediately frozen on arrival. The cell lines have been tested and authenticated by DNA fingerprinting (Short Tandem Repeat profiling) by the ATCC or by the Apath LLC before original shipment. The cells were used in experiments within 2 weeks after reactivation.
Results
ME-CIN promotes carcinogen-initiated HCC
HCC development often is discussed in the context of a classical three-step model (i.e. initiation, promotion, and progression) (39). We hypothesized that additional ME-CIN in liver would serve as either an initiator or a promoter of HCC, or both, leading to rapid progression. (Note that in this genetic model, unlike chemical carcinogenesis, making distinction between promotion and progression may be difficult.) To test whether ME-CIN is involved at a carcinogenesis stage, we examined livers of AOM-treated 6-month-old Sgo1−/+ and control wild-type mice available from a previous colon cancer study (38). AOM creates DNA adducts and is a colon carcinogen and a hepatotoxin, and can serve as an initiator for HCC possibly through mutating various genes such as c-fos, c-myc, or k-ras (40,41). We reasoned that the AOM treatments would serve as an initiator for HCC development, and that HCC development specifically in a Sgo1−/+ high-ME-CIN background mice would indicate a promoter role of ME-CIN in HCC.
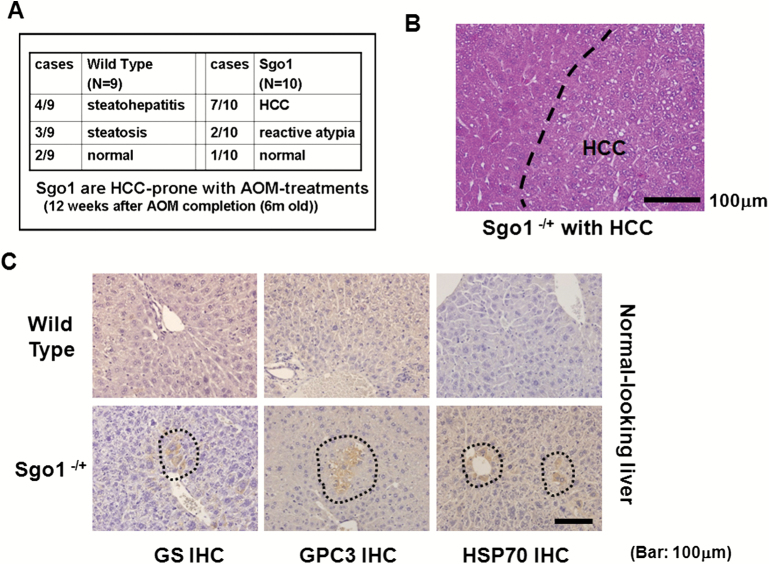
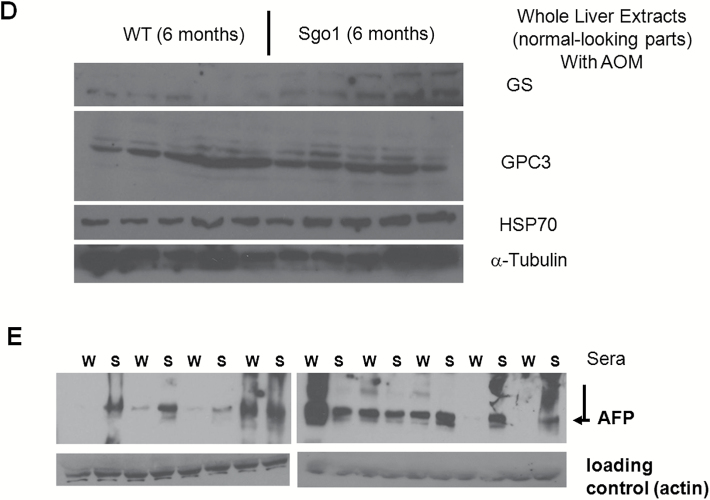
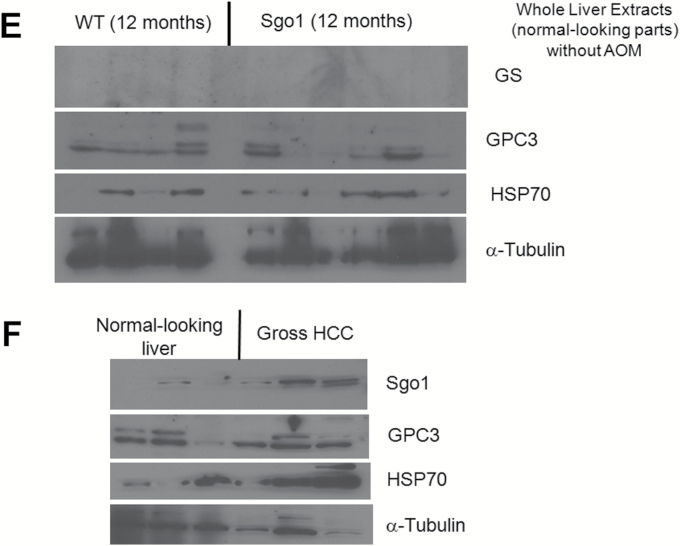
With the dosing of AOM treatments employed, control wild-type mice showed only mild histopathological changes (i.e. steatosis, steatohepatitis) in the liver and developed no HCC (0/9 animals). In contrast, Sgo1−/+ mice developed HCC in 7 out of 10 animals (P < 0.0031) at the 6 months of age endpoint (Figure 1A and B). The HCC was associated with steatohepatitis and localized expression of three early HCC markers (HSP70, GPC3, and GS) (Figure 1C and D). These three molecular markers are able to detect human HCC with 50–95% probability (42–44). These Sgo1−/+, high ME-CIN mice also were tested for an HCC serum marker, AFP (Figure 1E). Sgo1−/+ mice were 100% AFP-positive (9/9 tested). Among nine wild type mice, only the four with steatohepatitis were strongly AFP-positive and the two with steatosis were weakly positive. Thus, AFP expression was associated strongly with histological progression toward HCC in the AOM-treated mice. AFP is conventionally considered a tumor marker. Our results suggest that AFP may also serve as a marker for (tumor induced) inflammation.
Figure 1.
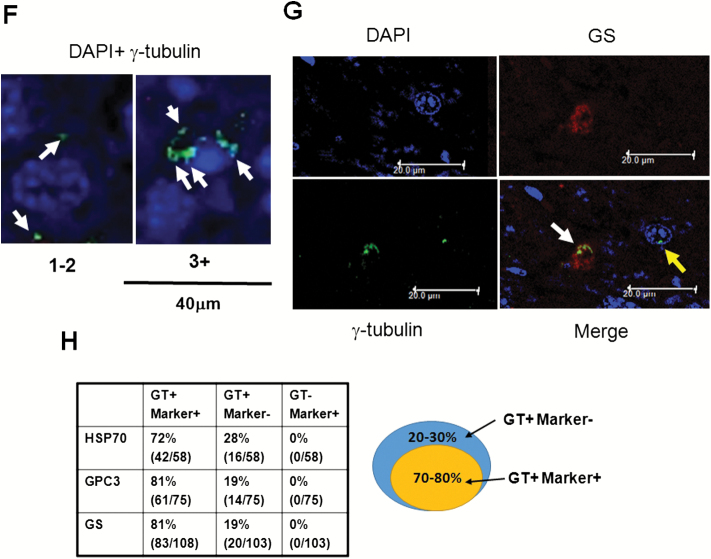
Sgo1 haplo-insufficiency promotes development of HCC with expression of early HCC markers after initiation with AOM. (A) With AOM treatments, wild type mice showed only modest histopathological change in the liver with no HCC, but 7 out of 10 Sgo1−/+ mice rapidly developed HCC in the 6-month period (P < 0.003 by two-tailed Fisher’s exact test). (B) An example of histologically identified HCC in Sgo1−/+ mice (H/E staining). (C) Normal-looking livers of Sgo1−/+ mice showed sporadic positive staining for early HCC markers (marked in dashed circle). (D) Immunoblots for whole extracts of normal-looking part of liver. Increase in GS in Sgo1 liver was notable, whereas increases in GPC3 and HSP70 total amounts appeared subtle in wild type and Sgo1, likely because of highly localized expressions of the markers and differences in basal expression for each marker. Since all mice were treated with AOM, GPC3, and HSP70 may have been up-regulated in wild type than untreated condition. Note: GPC3 is a proteoglycan and produces multiple bands in immunoblots. (E) The serum HCC marker AFP was elevated in nine out of nine Sgo1−/+ mice (marked S), whereas only four out of nine wild type (marked W) showed AFP elevation and these all had steatohepatitis. (F) Examples of normal cells that carry 1–2 centrosomes (left panel, green dots marked by white arrows), and of GT+ cells that carry multiple (3+) centrosomes (right panel) in Sgo1 liver. Blue: nuclear staining with DAPI. Possibly through disintegration of centrosome structure, through abnormal centrosome replication, or through failed centrosomal distribution, up to several GT+ signals could be observed. (G) Early HCC marker expression coincided with multiple GT staining, a consequence of the Sgo1 haploinsufficiency with partial loss of function. In this example, left cell had multiple GT-positive immunofluorescence signals and also was GS-positive (white arrow). The cell on the right was an example of normal cell; GS (red)-negative and showing one GT signal (green, indicated with yellow arrow). (H) Expression of multiple GT foci per cell precedes early expression of HCC markers. A majority (72–81%) of multiple GT+ cells was early HCC marker-positive. However, all cells positive for an HCC marker also were multiply GT-positive. The results indicate that appearance of multiple GT-positive foci per cell (Sgo1 defect) preceded early HCC marker expression, suggesting that early HCC marker-expressing cells arose from ME-CIN cells (diagram).
These results suggest that Sgo1 haploinsufficiency-mediated ME-CIN can serve as a tumor promoter in carcinogen-initiated HCC.
Early HCC marker-positive cells arise from ME-CIN cells
We questioned whether expression of these early HCC markers is linked to ME-CIN. Hepatocytes naturally become polyploid and then undergo reductive division, a process characterized by chromosome loss and the creation of near-diploid aneuploid cells (23). This naturally occurring polyploidization and aneuploidy in the liver complicates interpretation of conventional FISH results and other karyotyping analysis to detect CIN. Thus, we used a centrosome marker, gamma-tubulin (GT), to detect ME-CIN in this model as shown in Sgo1 MEF (38). Sgo1 dysfunction leads to multi-GT-positive (GT+) cells that carry multiple centrosomes (more than three per cell in contrast to normal 1 or 2), and we reasoned that GT+ cells are unique marker for Sgo1 defect and ME-CIN in this model (Figure 1F). Consistently, GT+ occurred in 16.4% −/+ 8.67% (SD) of hepatic cells from the Sgo1−/+ mice previously treated with AOM (n = 6), but rarely (less than 2%) in hepatocytes from the control wild-type mice treated with AOM. We tested with immunofluorescence and confocal microscopy whether the GT+ cells express the early HCC markers (HSP70, GPC3, GS) (Figure 1F and G). 70–80% of the GT+ cells co-expressed an early HCC marker (Example: Figure 1G left [white arrow]: GT+, GS+), yet the remaining 19–28% of GT+ cells were HCC marker-negative. This suggests that the Sgo1 defect and resulting ME-CIN is linked with or at least coincides with an early HCC marker expression. The fact that not all GT+ cells are HCC marker-positive suggests that an additional event may be required to trigger fully the HCC marker expression in the ME-CIN (GT+) cells. However, all HCC marker-positive cells were GT positive, suggesting that generation of ME-CIN cells with multiple centrosomes (GT+) precedes early HCC marker expression, or that HCC marker-positive cells arise from ME-CIN (GT+) cells (Figure 1H).
ME-CIN is a mild initiator of HCC
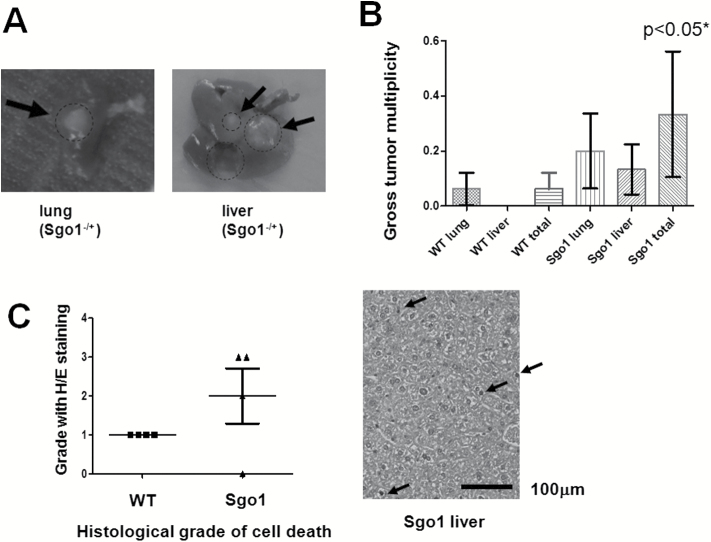
The HCC-promoting role of ME-CIN with an initiator carcinogen treatment prompted us to test spontaneous HCC carcinogenesis in the ME-CIN models to evaluate the role of ME-CIN as an HCC initiator. We maintained Sgo1−/+ mice and control wild type for 12 months with regular diet without other treatment. At the endpoint we observed by gross examination a mild yet significant increase of sporadic cancers, particularly in liver and lung (Figure 2); thus, ME-CIN appeared to be mildly tumorigenic in these organs. Note that the background C57BL/6 strain has a low rate of spontaneous liver tumors (in general only ~2.5% of animals in 2 years) (6,7).
Figure 2.
Increased spontaneous HCC in 12-month-old untreated Sgo1-/+ mice. (A) Examples of gross tumors in Sgo1−/+ mice. (B) Gross tumors in liver and lung were prominent in Sgo1−/+ mice. All gross liver tumors were HCC. “Gross lung tumors” in Sgo1 were one granuloma, one carcinoma, and one metastatic HCC. Gross tumor multiplicity is defined as the number of tumors per mouse observed through gross examination of a particular organ. (C) Livers from the Sgo1−/+ mice showed modestly higher levels of cell death (n = 4). Cells with dark nuclei (arrow) are hepatocytes undergoing apoptosis. Grading with H/E staining is as follows: 1 dead cell per 10 high power fields (HPF) is grade 1; 2–3 dead cells per 10 HPF is grade 2; and >2 dead cells per 10 HPF is grade 3; Zero dead cells is grade 0.
Persistent DNA damage and marker gene expression are associated with ME-CIN
Next, we questioned how ME-CIN leads to spontaneous HCC. In cultured cells, induced ME-CIN can cause DNA damage (45–47); however, induction of hepatic DNA damage had not been confirmed in vivo. We hypothesized that ME-CIN in the liver leads to an increase in DNA damage, and that the DNA damage would lead to further accumulation of mutations and HCC. We also reasoned that, if the hypothesis is correct, intrinsic cancer prevention mechanisms (e.g. apoptotic cell death or senescence) also should be activated. Consistent with the hypothesis, hematoxylin and eosin (H/E) staining for normal-looking parts of the Sgo1−/+ mice liver showed an increase in cell death (Figure 2C).
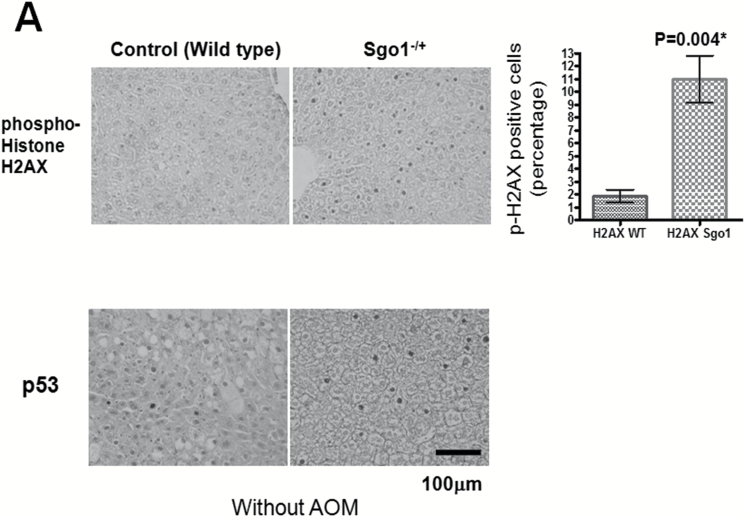
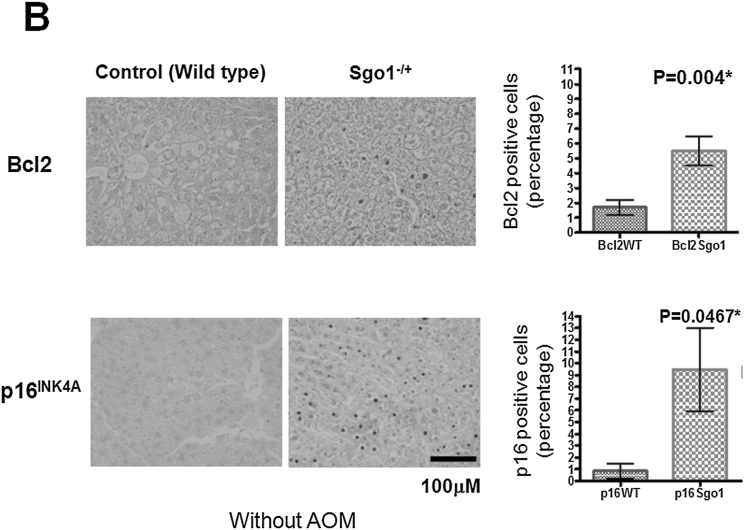
To further test the hypothesis, we analyzed expression of markers for DNA damage (phosphor-histone H2AX [p-H2AX] and p53), cell death pathways (Bcl2) and senescence (p16INK4A) in the liver. The normal-looking part of liver expressed significantly more of the DNA damage marker p-H2AX (P = 0.004) and of p53 (P = 0.0001), indicating ongoing DNA damage and subsequent p53-mediated repair/senescence response (Figure 3A). The cell death pathway marker Bcl2 was increased significantly in Sgo1−/+ mice (P = 0.004), as well as the senescence marker p16 INK4A (P = 0.0467) (Figure 3B).
Figure 3.
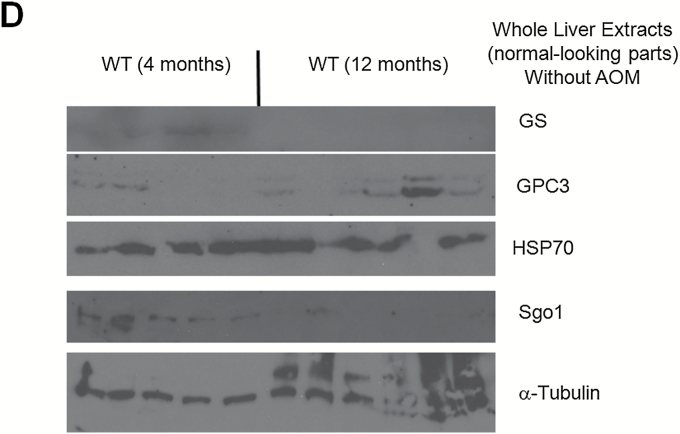
Persistent DNA damage in normal-looking part of the livers of the Sgo1−/+ mice. (A) The DNA damage marker phospho-gamma-H2AX and p53 were expressed at higher levels in livers of 12-month-old Sgo1−/+ mice. Based on IHC pictures, percentages of IHC-positive cells (black nuclear stain) are calculated. (*) indicates that the P value is statistically significant. (B) A senescence marker p16INK4A and an apoptosis pathway marker Bcl2 were expressed at higher in livers of Sgo1−/+ mice. (C) Expression of p53, p16INK4A, and Bcl2 were age-dependent. Livers from 4-month-old Sgo1−/+ mice were analyzed as in (A) and (B). Only the DNA damage marker p-H2AX showed a significant difference from the control in the younger Sgo1−/+ mice, suggesting that persistent DNA damage is an upstream event of the differential expression of other markers. (D) Age diminished Sgo1 protein expression level in untreated wild type. Younger wild type (4 months; left five lanes) expressed higher amount of Sgo1. In older mice (12 months; right six lanes), Sgo1 expression diminished. Modification of loading control α–tubulin was altered.
Marker expression is age-dependent
Since age is a major HCC risk factor (1,2), we questioned whether the marker expression profile is dependent on age. Instead of using 12-month-old mice that correspond to middle age in humans, we compared the expression of the same marker set between untreated 4-month-old (corresponds to human young adult) wild type and Sgo1−/+ mice. The 4-month-old Sgo1−/+ mice showed no histological HCC, yet had significantly higher p-H2AX expression compared with wild type (P < 0.0001), indicating the presence of persistent DNA damage. It also suggests that the DNA damage is a primary effect of ME-CIN. However, other markers tested (i.e. p53, Bcl2, p16 INK4A) showed no significant differences from control (Figure 3C). The average percentage of p53-positive cells was 5-fold higher in Sgo1−/+ mice, yet the wide variance resulted in a non-significant P value (P = 0.0966). Thus, expression of most of the markers examined in 12-month-old mice was age-dependent, except for the DNA damage marker p-H2AX that was consistently higher in Sgo1−/+ mice. The age-dependence in marker expression profiles suggests a progressive nature of the effect of ME-CIN and DNA damage in the liver.
Aging influences protein expressions. We tested effect of aging on Sgo1 protein using 4-month-old and 12-month-old untreated wild type (Figure 3D). Younger wild type mice (4 months) expressed overall higher amount of Sgo1 than older mice (12 months).
Spontaneous HCC in Sgo1−/+ mice express relevant HCC markers
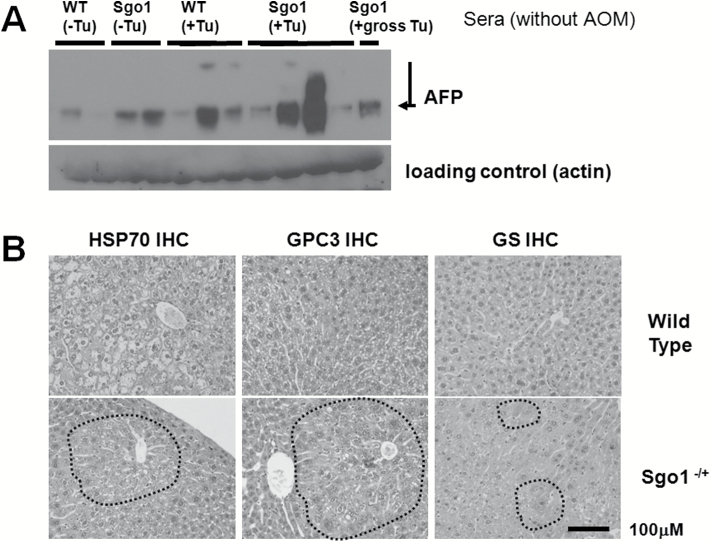
Next, we questioned that to what extent the HCCs in the Sgo1−/+ ME-CIN model mice mimic human HCC. We tested sera from the mice for human the HCC serum marker AFP (Figure 4A). In both 12-month-old wild type and Sgo1−/+, HCC-bearing mice (marked Tu+) showed high amounts of AFP in the sera, similar to the case with human HCC. We also tested expressions of the three early HCC markers in Sgo1−/+ and control mice (Figure 4B, IHC for HSP70 and GPC3. Typical IHC picture is shown). Three Sgo1−/+ mice out of four showed positive for two HCC markers, yet only one out of four control mice was positive for two markers. Additionally, the relationship between the HCC marker expression and steatosis was investigated, but no statistically significant correlation was demonstrated in Sgo1−/+ mice or in control wild-type.
Figure 4.
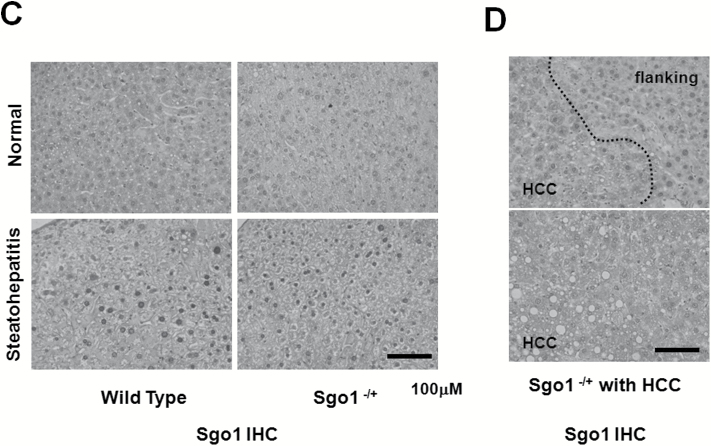
HCC-relevant marker expression in the 12-month-old untreated Sgo1−/+ mice. (A) The HCC marker AFP was elevated in the sera from Sgo1−/+ mice and HCC-bearing (Tu+) mice. (B) Early HCC markers HSP70, GPC3, and GS are elevated in the livers of Sgo1−/+ mice in a highly localized manner. The sporadic marker expression is not limited to the histologically cancerous area and can be observed in normal-looking parts of the liver. (C) Sgo1 protein level was correlated with disease progression. Consistent with haploinsufficiency, the IHC signal for Sgo1 protein (brown) was generally weaker in normal-looking parts of Sgo1−/+ mouse liver than in control wild type (top panels), but the IHC signal increased with steatohepatitis both in wild type and in Sgo1−/+ mice (bottom panels). Nuclear localization of Sgo1 protein became prominent with steatohepatitis. (D) Sgo1 protein level was higher in HCC than in normal-looking parts of liver in Sgo1−/+ mice. In HCC, nuclear localization of Sgo1 protein was not apparent. (E) Total HCC marker expression appeared comparable in whole liver (minus gross tumor) extracts, presumably due to the highly localized manner of the marker expression. (F) Sgo1 protein amount was higher in HCC than in normal-looking part of the liver. Extracts of normal-looking part of liver from 12-month-old AOM-treated Sgo1−/+ mice (left three lanes) and HCCs from 12-months-old AOM-treated Sgo1−/+ mice (right three lanes) were compared.
Human HCC commonly is associated with cirrhosis. In the 12-month-old mice and 4-month-old mice, however, there was no sign of fibrosis in the liver detectable with Trichrome staining (not shown).
Sgo1 protein accumulates in diseased liver
We also tested Sgo1 protein expression in the liver. In normal-looking tissue, Sgo1−/+ mice expressed only a nominal amount of Sgo1 protein, reflecting the predicted result of haploinsufficiency; a reduction that is presumed to cause diminished function and defect. The wild-type control expressed more Sgo1 in the normal-looking liver. Interestingly, liver segments with steatohepatitis were associated with overall high Sgo1 protein expression and, notably, with nuclear localized signals, both in Sgo1−/+ and control mice (Figure 4C). Immunoblots for whole liver extracts failed to reflect this difference in microlocalization in Sgo1 or HCC markers, presumably due to the highly localized nature (Figure 4E). HCCs observed in wild type and in Sgo1−/+ mice also expressed high amounts of Sgo1 protein in the cytoplasm, but nuclear localized Sgo1 was not apparent in HCC (Figure 4D). Gross HCCs expressed Sgo1 protein higher than normal-looking part of liver in immunoblots (Figure 4F).
DNA damage can cause nuclear accumulation of Sgo1 protein in vitro
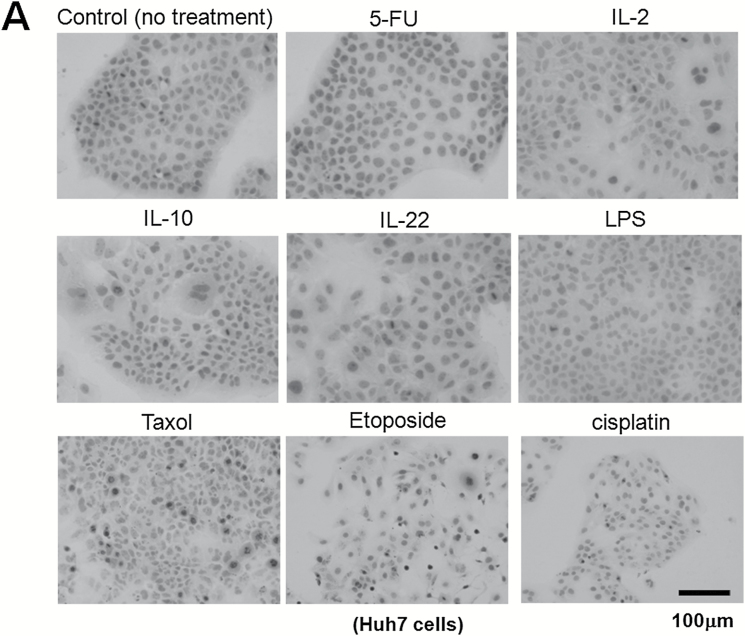
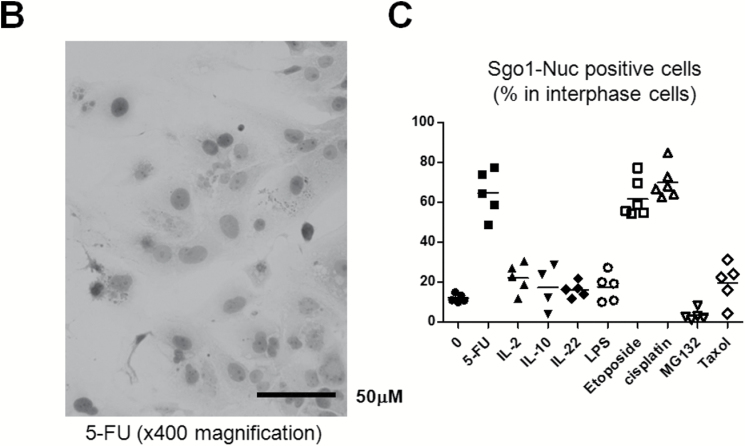
From the histological staining of Sgo1 protein shown in Figure 4, we hypothesized that the Sgo1 protein accumulation in the nucleus may be caused by inflammation or by DNA damage. To determine whether DNA damage or inflammation might contribute to nuclear accumulation of Sgo1, we treated the human HCC cell line Huh7 with a set of reagents (Figure 5A–C). Although treatment with the inflammatory cytokines IL2, IL22, or IL10, or with the bacterial cell wall compound lipopolysaccharide, or with proteasome inhibitor MG132 or spindle poison Taxol did not result in Sgo1 accumulation in the nucleus, all DNA damaging reagents tested (i.e. the uracil analog 5-FU, the topoisomerase inhibitor Etoposide, the platinum compound cisplatin) did. We concluded that the DNA damage can cause Sgo1 protein accumulation in nuclei of the human HCC cells.
Figure 5.
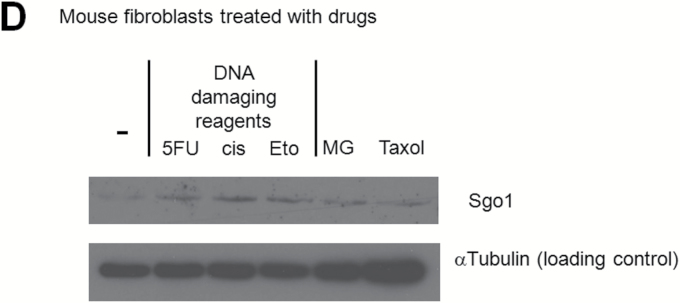
DNA damage can cause Sgo1 nuclear accumulation in vitro. (A) Human HCC Huh7 cells were treated with the indicated reagents. DNA damaging agents (5-FU, etoposide and cisplatin) led to nuclear accumulation of Sgo1 protein (brown). (B) Immunostain for Sgo1 protein in Huh7 cells treated with 5-FU, shown in a higher magnification (brown). (C) Quantification of the drug effect indicated that DNA damaging reagents, but not other reagents tested, increased nuclear Sgo1. Percentages of nuclear Sgo1-positive cells among all interphase cells are shown. Mitotic cells were omitted in this analysis because Sgo1 protein amount peaks during mitosis. (D) Total amount of Sgo1 protein increased in response to DNA damage in mouse primary fibroblasts compared with no drug treatment (-). Treatments with MG132 or Taxol enrich mitotic cells that express Sgo1 in higher amount. Due to the cell cycle effect, they show an increase in Sgo1 amount. (E) The localization change of Sgo1 in response to DNA damage occurs in Sgo1−/+ mouse primary fibroblasts as well. We treated Sgo1−/+ mouse primary fibroblasts with DNA damaging reagents cisplatin (cis) or 5-fluorouracil (5-FU), or proteasome inhibitor MG132 (MG). In untreated interphase cells (-), Sgo1 protein show diffused cytoplasmic stain (brown signal; white arrow). With DNA damaging reagents (cis, 5-FU), nuclear enrichment of Sgo1 signal was observed (black arrow), although it was not as clear as in Huh7 cells.
Since Huh7 is p53-mutated cell line, the process may not depend on p53 directly. Supporting the notion, the Sgo1 nuclear accumulation was observed in other cell lines both with mutated p53 (HeLa) and with wild type p53 (HepG2) (data not shown).
To test whether the nuclear accumulation of Sgo1 protein in response to DNA damage is limited to cancer-origin cell lines, we used mouse primary fibroblasts. Mouse primary fibroblasts also showed an increase in Sgo1 protein amount and its nuclear accumulation in response to DNA damaging reagents, although it was not as clear as in Huh7 cells (Figure 5D and E).
Discussion
Evaluation of drugs, diets, or other compounds for biological responses, including prevention, blockade, or reversal of carcinogenesis, requires appropriate animal models for translational studies prior to human clinical trials (5). Mouse models requiring minimal additional procedures (e.g. “breed and test”) are favored. Viral infection is observed in approximately 30–50% of all HCC. Yet a reliable test model for viral HCC comparable to the “apc” model studied in colonic carcinogenesis is yet to be developed due to the complex and variable HCC carcinogenesis processes. In this study, we focused on a common consequence of HBV/HCV infection, ME-CIN, and its effect on HCC development using a mitotic error-induced (ME-)CIN model mice (Sgo1−/+ mice) whose defect targets the same pathway as viral proteins HBx or NS5A (17–22).
Overall this study demonstrated an HCC-promoting/progressing role of ME-CIN in the Sgo1−/+ mouse model in the presence of a strong initiator. ME-CIN preceded early HCC marker expressions (Figure 1G and H), suggesting that ME-CIN may be an important early event in HCC development. Also suggested was a weak HCC-initiating role of ME-CIN through persistent generation of DNA damage. This study demonstrated that Sgo1−/+ ME-CIN model mice were prone to hepatic DNA damage and showed differential expression of tumor markers. Notably, a senescence marker p16INK4A was highly expressed in Sgo1−/+ mice. Since cellular senescence is a cancer risk factor, possibly through secretory factors or its association with accumulation of mutations, altered p16INK4A expression may be a part of the reason that Sgo1−/+ mice were prone to spontaneous HCC development (48,49).
In Sgo1−/+ mice, cirrhosis was not observed, at least up to 12 months of age. Development of cirrhosis may not be a primary effect of ME-CIN in this model. The lack of cirrhosis is reminiscent of HCV viral protein-expressing transgenic model mice, which also showed no sign of cirrhosis despite an increase in HCC (50).
We also discovered that the Sgo1 protein expression in liver is naturally diminished as mice age (Figure 3D [wild type], Supplementary Figure 1 [Sgo1]). This is reminiscent of a fact that BubR1, a mitotic checkpoint protein whose reduction also causes ME-CIN, also diminishes the expression with age. Severe reduction of BubR1 in hypomorph mice leads to premature aging (29,48). It is a plausible scenario that reduction of Sgo1 in older mice may have additional ME-CIN inducing effect, which can further accelerate aging and/or hepatic carcinogenesis. Testing whether the reduction of Sgo1 is functionally related to aging, or restoration of Sgo1 protects animals from aging and/or carcinogenesis, requires further study.
Another discovery is that, while overall protein expression level decreases, Sgo1 protein accumulates, particularly in the nuclei, as the liver develops steatohepatitis and HCC. The abnormal Sgo1 accumulation may indicate a need for tight control of Sgo1 expression in maintaining normal histology in the liver. Whether the increased or accumulated Sgo1 protein in HCC is functionally influencing HCC development, or is due to other mechanisms (e.g. misregulation of transcription, translation, or proteolysis of Sgo1 in HCC cells) and is only a result, requires further investigation. “In vitro experiments (Figure 5) suggested that nuclear accumulation can be caused by DNA damaging reagents. The nuclear accumulation of Sgo1 in the liver with steatohepatitis may indicate ongoing DNA damage in the tissue, and possibly may serve as a novel marker for DNA damage and HCC risk. Molecular mechanisms of the nuclear accumulation of Sgo1 and its function require further study. Although with further investigation Sgo1 protein may prove to have an additional and specific role in DNA damage, HCC proneness in other ME-CIN strains [e.g. BubR1−/+ (33), Mad1 (29,32)] would still suggest a common role of ME-CIN in HCC development.
Previously, the relationships among HCC risk factors were hard to dissect. Our results imply that viral infection and resulting ME-CIN can promote HCC initiated by other insults. This possibility of multiple initiating and promoting agents, though not yet validated in HCC, emphasizes the importance of minimizing additional insults in a ME-CIN-generating background for the prevention of HCC development. The ME-CIN mice may be used for screening for environmental or dietary factors that aggravate HCC in combination with ME-CIN. Alternatively, by generating double mutants with immune-modulated transgenic mice, the ME-CIN mice may provide mechanistic insights into immune surveillance against cells with ME-CIN.
In other ME-CIN models, both oncogenic and tumor-suppressing effects were observed in an organ-specific manner (29–32). We previously reported that in response to AOM treatments, Sgo1−/+ mice developed an increased size and number of precancerous lesions aberrant crypt foci and microadenomas in colon (38). Since AOM can induce tumors in colon, liver and kidney, we examined kidneys from the wild type and Sgo1−/+ mice (Supplementary Figure 2). In contrast to colon and liver, invasive transitional cell carcinoma developed in a higher frequency in wild type (6 among 9 mice) than in Sgo1−/+ mice (1 among 10). We cross-referenced all mice to see whether carcinogenesis is occurring only to particular mice or not. There was no correlation in the occurrence of HCC and transitional cell carcinoma (Supplementary Figure 2B). We interpret this as an indication of both oncogenic and tumor-suppressing effects of ME-CIN-causing mutation [as reviewed in Refs. (29–32)]. The paradoxical effects may be determined by organ-specific cell death rate or tissue turnover rate. Alternatively, organ-specific gene expression profile, or Sgo1−/+model-specific modulation of gene expression may have an effect on carcinogenesis response. The notable decrease in renal transitional cell carcinoma in Sgo1−/+ mice warrants further investigation.
Overall, the high ME-CIN mouse model may facilitate several aspects of studies for HCC, and also of cancers in colon and kidney.
Supplementary material
Supplementary Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
US National Institutes of Health (NCI R01CA094962) to C.V.R., (NCI RO3CA162538) to H.Y.Y., and (RO1CA090658) to W.D.; Chris4Life colon cancer foundation pilot study grant to H.Y.Y.
Supplementary Material
Acknowledgements
We thank Dr. Julie Sando for editing the manuscript; Mr. Jim Henthorn for providing help with confocal microscopy.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- 5-FU
5-fluorouracil
- AFP
alpha fetoprotein
- AOM
azoxymethane
- BubR1
Bub1-related 1
- CIN
chromosome instability
- GPC3
glypican 3
- GS
glutamine synthetase
- GT
gamma-tubulin
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- H/E
hematoxylin and eosin
- HSP70
heat shock protein 70
- IF
immunofluorescence
- IHC
immunohistochemistry
- IL2
interleukin 2
- IL10
interleukin 10
- IL22
interleukin 22
- LPS
lipopolysaccharide
- ME-CIN
mitotic error-induced chromosome instability
- MEFs
mouse embryonic fibroblasts
- p-H2AX
phosphor-histone H2AX
- Sgo1
shugoshin 1.
References
- 1. American Cancer Society statistics (2014). http://www.cancer.org/ cancer/livercancer/detailedguide/liver-cancer-what-is-key-statistics (16 December 2014, date last accessed).
- 2. El-Serag H.B. (2012). Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology, 142, 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bouchard M.J., et al. (2011). Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett., 305, 123–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fallot G., et al. (2012). Diverse roles of hepatitis B virus in liver cancer. Curr. Opin. Virol., 2, 467–473. [DOI] [PubMed] [Google Scholar]
- 5. Senderowicz A.M. (2010). Information needed to conduct first-in-human oncology trials in the United States: a view from a former FDA medical reviewer. Clin. Cancer Res., 16, 1719–1725. [DOI] [PubMed] [Google Scholar]
- 6. Fausto N., et al. (2010). Mouse models of hepatocellular carcinoma. Semin. Liver Dis., 30, 87–98. [DOI] [PubMed] [Google Scholar]
- 7. Li Y., et al. (2012). Hepatocellular carcinoma: insight from animal models. Nat. Rev. Gastroenterol. Hepatol., 9, 32–43. [DOI] [PubMed] [Google Scholar]
- 8. Chayama K., et al. (2011). Animal model for study of human hepatitis viruses. J. Gastroenterol. Hepatol., 26, 13–18. [DOI] [PubMed] [Google Scholar]
- 9. Bukh J. (2012). Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology, 142, 1279–1287.e3. [DOI] [PubMed] [Google Scholar]
- 10. Menne S., et al. (2007). The woodchuck as an animal model for pathogenesis and therapy of chronic hepatitis B virus infection. World J. Gastroenterol., 13, 104–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng Y., et al. (2007). Hepatitis B virus promotes hepatocarcinogenesis in transgenic mice. Hepatology, 45, 16–21. [DOI] [PubMed] [Google Scholar]
- 12. Mercer D.F., et al. (2001). Hepatitis C virus replication in mice with chimeric human livers. Nat. Med., 7, 927–933. [DOI] [PubMed] [Google Scholar]
- 13. Bissig K.D., et al. (2010). Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J. Clin. Invest., 120, 924–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Azuma H., et al. (2007). Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat. Biotechnol., 25, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vesselinovitch S.D. (1987). Certain aspects of hepatocarcinogenesis in the infant mouse model. Toxicol. Pathol., 15, 221–228. [DOI] [PubMed] [Google Scholar]
- 16. Aleksic K., et al. (2011). Evolution of genomic instability in diethylnitrosamine-induced hepatocarcinogenesis in mice. Hepatology, 53, 895–904. [DOI] [PubMed] [Google Scholar]
- 17. Forgues M., et al. (2003). Involvement of Crm1 in hepatitis B virus X protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol. Cell. Biol., 23, 5282–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yun C., et al. (2004). Mitotic aberration coupled with centrosome amplification is induced by hepatitis B virus X oncoprotein via the Ras-mitogen-activated protein/extracellular signal-regulated kinase-mitogen-activated protein pathway. Mol. Cancer Res., 2, 159–169. [PubMed] [Google Scholar]
- 19. Wen Y., et al. (2008). Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J. Biol. Chem., 283, 2793–2803. [DOI] [PubMed] [Google Scholar]
- 20. Kim S., et al. (2008). HBV X protein targets hBubR1, which induces dysregulation of the mitotic checkpoint. Oncogene, 27, 3457–3464. [DOI] [PubMed] [Google Scholar]
- 21. Wang L.H., et al. (2012). Aberrant cyclin A expression and centrosome overduplication induced by hepatitis B virus pre-S2 mutants and its implication in hepatocarcinogenesis. Carcinogenesis, 33, 466–472. [DOI] [PubMed] [Google Scholar]
- 22. Baek K.H., et al. (2006). Overexpression of hepatitis C virus NS5A protein induces chromosome instability via mitotic cell cycle dysregulation. J. Mol. Biol., 359, 22–34. [DOI] [PubMed] [Google Scholar]
- 23. Duncan A.W. (2013). Aneuploidy, polyploidy and ploidy reversal in the liver. Semin. Cell Dev. Biol., 24, 347–356. [DOI] [PubMed] [Google Scholar]
- 24. Rao C.V., et al. (2013). Genomic instability and colon carcinogenesis: from the perspective of genes. Front. Oncol., 3, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watanabe T., et al. (2012). Chromosomal instability (CIN) phenotype, CIN high or CIN low, predicts survival for colorectal cancer. J. Clin. Oncol., 30, 2256–2264. [DOI] [PubMed] [Google Scholar]
- 26. Nishida N., et al. (2003). Chromosomal instability and human hepatocarcinogenesis. Histol. Histopathol., 18, 897–909. [DOI] [PubMed] [Google Scholar]
- 27. Walther Z., et al. (2011). Molecular pathology of hepatic neoplasms: classification and clinical significance. Patholog. Res. Int., 2011, 403929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee Y.H., et al. (2009). Chromosomal instability, telomere shortening, and inactivation of p21(WAF1/CIP1) in dysplastic nodules of hepatitis B virus-associated multistep hepatocarcinogenesis. Mod. Pathol., 22, 1121–1131. [DOI] [PubMed] [Google Scholar]
- 29. Ricke R.M., et al. (2008). Whole chromosome instability and cancer: a complex relationship. Trends Genet., 24, 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foijer F., et al. (2008). Studying chromosome instability in the mouse. Biochim. Biophys. Acta, 1786, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rao C.V., et al. (2009). Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis, 30, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schvartzman J.M., et al. (2010). Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer, 10, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dai W., et al. (2004). Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res., 64, 440–445. [DOI] [PubMed] [Google Scholar]
- 34. Rao C.V., et al. (2005). Colonic tumorigenesis in BubR1+/-ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc. Natl. Acad. Sci. U. S. A., 102, 4365–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salic A., et al. (2004). Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell, 118, 567–578. [DOI] [PubMed] [Google Scholar]
- 36. Schöckel L., et al. (2011). Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol., 13, 966–972. [DOI] [PubMed] [Google Scholar]
- 37. Wang X., et al. (2008). sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev. Cell, 14, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamada H.Y., et al. (2012). Haploinsufficiency of SGO1 results in deregulated centrosome dynamics, enhanced chromosomal instability and colon tumorigenesis. Cell Cycle, 11, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weinberg R.A. (2013). The Biology of Cancer, 2nd edn Garland Science, New York, NY. [Google Scholar]
- 40. Matkowskyj K.A., et al. (1999). Azoxymethane-induced fulminant hepatic failure in C57BL/6J mice: characterization of a new animal model. Am. J. Physiol., 277(2 Pt 1), G455–G462. [DOI] [PubMed] [Google Scholar]
- 41. Nishihara T., et al. (2008). Adiponectin deficiency enhances colorectal carcinogenesis and liver tumor formation induced by azoxymethane in mice. World J. Gastroenterol., 14, 6473–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. The International Consensus Group for Hepatocellular Neoplasia (2009). Pathologic diagnosis of early hepatocellular carcinoma: a report of the international consensus group for hepatocellular neoplasia. Hepatology, 49, 658–664. [DOI] [PubMed] [Google Scholar]
- 43. Di Tommaso L., et al. (2009). The application of markers (HSP70 GPC3 and GS) in liver biopsies is useful for detection of hepatocellular carcinoma. J. Hepatol., 50, 746–754. [DOI] [PubMed] [Google Scholar]
- 44. Sturgeon C.M., et al. (2010). Laboratory Medicine Practice Guidelines for use of tumor markers in liver, bladder, cervical, and gastric cancers. The National Academy of Clinical Biochemistry, 56, e1–e48. [DOI] [PubMed] [Google Scholar]
- 45. Janssen A., et al. (2011). Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science, 333, 1895–1898. [DOI] [PubMed] [Google Scholar]
- 46. Orth J.D., et al. (2012). Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell, 23, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crasta K., et al. (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature, 482, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baker D.J., et al. (2011). Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature, 479, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. López-Otín C., et al. (2013). The hallmarks of aging. Cell, 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koike K., et al. (2002). Role of hepatitis C virus in the development of hepatocellular carcinoma: transgenic approach to viral hepatocarcinogenesis. J. Gastroenterol. Hepatol., 17, 394–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.