Summary
This study describes a novel mechanism of the inflammatory cytokine IL-6 induced Fra-1 upregulation through activating STAT3 by phosphorylation and acetylation, and demonstrates that this signaling pathway plays a critical role in promoting epithelial–mesenchymal transition and aggressiveness of colorectal cancer.
Abstract
The pro-inflammatory cytokine interleukin-6 (IL-6) in tumor microenvironment has been suggested to promote development and progression of colorectal cancer (CRC). However, the underlying molecular mechanisms remain elusive. In this study, we demonstrate that fos-related antigen-1 (Fra-1) plays a critical role in IL-6 induced CRC aggressiveness and epithelial–mesenchymal transition (EMT). In CRC cell lines, the expression of Fra-1 gene was found significantly upregulated during IL-6-driven EMT process. The Fra-1 induction occurred at transcriptional level in a manner dependent on signal transducer and activator of transcription 3 (STAT3), during which both phosphorylated and acetylated post-translational modifications were required for STAT3 activation to directly bind to the Fra-1 promoter. Importantly, RNA interference-based attenuation of either STAT3 or Fra-1 prevented IL-6-induced EMT, cell migration and invasion, whereas ectopic expression of Fra-1 markedly reversed the STAT3-knockdown effect and enhanced CRC cell aggressiveness by regulating the expression of EMT-promoting factors (ZEB1, Snail, Slug, MMP-2 and MMP-9). Furthermore, Fra-1 levels were positively correlated with the local invasion depth as well as lymph node and liver metastasis in a total of 229 CRC patients. Intense immunohistochemical staining of Fra-1 was observed at the tumor marginal area adjacent to inflammatory cells and in parallel with IL-6 secretion and STAT3 activation in CRC tissues. Together, this study proposes the existence of an aberrant IL-6/STAT3/Fra-1 signaling axis leading to CRC aggressiveness through EMT induction, which suggests novel therapeutic opportunities for the malignant disease.
Introduction
Colorectal cancer (CRC) is one of the most frequent causes of malignant morbidity and mortality worldwide, and in most cases, lethality in CRC patients is caused by metastasis that results in tumor resistance to conventional therapies and an overall poor prognosis (1,2). Although vigorous studies have greatly improved the knowledge of colorectal tumorigenesis, the relevant factors that contribute to metastasis are still not well determined, which is urgently required for the early detection and treatment of metastatic CRC.
Chronic intestinal inflammation has been closely linked to CRC risk in epidemiological studies, and several pro-inflammatory cytokines released by infiltrating immune cells and other cells in the microenvironment are suggested to regulate tumor initiation and progression (3). In particular, interleukin-6 (IL-6) and its intracellular signaling molecule signal transducer and activator of transcription 3 (STAT3) seem to take center stage in bridging chronic inflammation to CRC promotion (4–6). Even the cells that do not harbor the membrane-bound IL-6 receptor can be activated by IL-6 via a soluble form of the IL-6 receptor (7). Moreover, serum and cancer tissue IL-6 levels are elevated in CRC patients, and the concentration is correlated with tumor size, metastasis and reduced survival (8,9).
Fos-related antigen-1 (Fra-1), an important member of the Fos family, is frequently elevated by oncogenic signaling in a variety of human cancers and is strongly implicated in metastasis and poor prognosis. In contrast to the tumorigenic activity of c-Fos, Fra-1 seems to play a role in the motile and invasive phenotypes of cancer cells (10). Overexpression of Fra-1 results in fibroblastic morphological changes and correlates with mesenchymal characteristics and E-cadherin downregulation in carcinoma cells (11,12). More recently, Fra-1 has been proposed as a gatekeeper of the epithelial–mesenchymal transition (EMT) program during cancer progression (13–15). However, the stimulating signal and regulatory mechanism of Fra-1 in cancer EMT and aggressiveness are still not well understood.
In the present study to investigate the expression changes of key EMT regulators in IL-6-mediated CRC progression, we provided evidence that Fra-1 is significantly upregulated in response to IL-6 stimulation and plays a central role in IL-6 induced EMT process of CRC cells. The regulatory mechanism investigation demonstrated that IL-6 stimulated Fra-1 transcription though the direct binding of STAT3 to the Fra-1 gene promoter, and the activity of STAT3 for Fra-1 transactivation was controlled by tyrosine phosphorylation and lysine acetylation simultaneously. Further clinical specimens analyses showed that increased Fra-1 expression was positively correlated with IL-6 secretion, STAT3 activation and cancer progression in CRC tissues. Thus, we propose the existence of an aberrant IL-6/STAT3/Fra-1 signaling axis through which pro-inflammatory cytokine IL-6 in the tumor microenvironment promotes EMT and aggressiveness of CRC.
Materials and methods
Cell culture and transfection
HT-29, SW480, PC3 and 293T cells were obtained from the American Type Culture Collection (Manassas, VA), where the cell lines were authenticated by STR profiling before distribution. The cells were cultured and stored according to supplier’s instructions. After resuscitation, they were grown in DMEM with 10% FBS never passaged longer than 6 months and tested routinely by Hoechst DNA staining to ensure no mycoplasma contamination. Fra-1 or EV stably transfected HT-29 cells were derived from the parental cells by G418 (Sigma, St Louis, MO) selection. HT-29 cells were transfected with siRNAs or plasmids using X-tremeGENE siRNA Transfection Reagent (Roche, Basel, Switzerland), and 24h later, the cells were serum-starved overnight and stimulated with IL-6 (R&D Systems, Minneapolis, MN) as indicated.
Antibodies and reagents
Antibodies against E-cadherin, vimentin, fibronectin, ZO-1, Fra-1, Ku80 and GAPDH were from Santa Cruz Biotechnology (Santa Cruz, CA). The ZEB1, Snail, Slug, ERK1/2, p-ERK1/2, AKT, p-AKT, STAT3, p-STAT3 (Y705) and ac-STAT3 (K685) antibodies were from Cell Signaling Technology (Beverly, MA). H3 and c-Fos antibodies were from Abcam (Cambridge, UK). The siRNAs targeting STAT3, Fra-1 and c-Fos were from Santa Cruz. All the chemical inhibitors were from Calbiochem (San Diego, CA).
Quantitative real-time PCR analysis
Total RNA was isolated from cells using RNAiso Plus (TaKaRa, Kyoto, Japan). Reverse transcription was performed with the PrimeScript RT reagent Kit (TaKaRa). Quantitative real-time PCR (qRT-PCR) was achieved using the 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA). Target gene expression was normalized to GAPDH levels in respective samples as an internal control, and the results are representative of at least three independent experiments. The sequences of the qRT-PCR primers are listed in Supplementary Table S1, available at Carcinogenesis Online.
Western blot
Whole-cell extracts lysed from treated cells were clarified by centrifugation, and the protein concentration was determined by the Bradford method (Bio-Rad, Hercules, CA). The protein samples were separated on SDS–PAGE and transferred to nitrocellulose membranes (Whatman, Maidstone, UK). The membranes were probed with dilutions of primary antibodies followed by incubation with IRDye 800CW or IRDye 680-conjugated secondary antibodies, and then visualized by the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
Immunofluorescence
IL-6 treated cell layers on glass coverslips were fixed for 15min with 4% paraformaldehyde, permeabilized for 10min in PBS containing 0.2% Triton X-100, blocked for 1h with 1% BSA and 0.5% goat serum in PBS, and then probed with primary antibody at 4°C overnight. After rinsing in PBS, the cells were incubated with FITC- or TRITC-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) for 1h at room temperature and the nuclei were stained with DAPI (Sigma) for 5min. The slides were mounted and visualized using a fluorescence microscope (AX70, Olympus, Tokyo, Japan). The images are representative of three independent experiments.
Luciferase reporter assay
The genomic regions surrounding the promoter of human Fra-1 were amplified by PCR and inserted into the pGL3 vector. The reporter constructs containing various lengths of Fra-1 promoter or mutated STAT3 binding sites were generated by subsequent PCR-based cloning. The luciferase reporter assay was performed by transfecting the reporter construct with wild-type or mutant STAT3 expression vectors (kindly provided by Y. Eugene Chin, Brown University, Providence, RI) (16) into the indicated cell lines. The pRL-SV40 vector was co-transfected in each experiment as an internal control for transfection efficiency. At 24h post-transfection, cells were incubated with 50ng/ml IL-6 for 6h and the luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a luminometer (LB 9507, Berthold, Bad Wildbad, Germany). The firefly luciferase activity was corrected by the corresponding Renilla luciferase activity and all experiments were carried out in triplicate.
Chromatin immunoprecipitation
HT-29 cells were serum-starved overnight and treated with 50ng/ml IL-6 for 1h. Chromatin was cross-linked with 1% formaldehyde and sonicated to obtain a DNA smear of 200–1000bp. Following centrifugation, the supernatants were subjected to immunoprecipitation overnight at 4°C with antibodies against STAT3 or histone H3, or with normal rabbit IgG. Chromatin-antibody complexes were isolated using Protein A/G PLUS-Agarose (Santa Cruz). The crosslinking was reversed and genomic DNA fragments were purified and analyzed by PCR using the following primer pair for Fra-1 promoter: 5ʹ-TATCAGACCTCAGGACACCT-3ʹ (forward) and 5ʹ-CCAAGTAAGTGGGACTACAG-3ʹ (reverse). The results are representative of at least three independent experiments.
DNA pull-down assay
The DNA fragment of human Fra-1 promoter from −760 to −524 was amplified by PCR using a 5ʹ-biotin-labeled forward primer. A total of 100 μg nuclear proteins extracted from PC3 cells transiently transfected with wild-type or mutant STAT3 were mixed with 2 µg of biotinylated probes in 400 µl of binding buffer (10mM Tris–HCl, pH 7.5, 50mM NaCl, 1mM DTT, 1mM EDTA and 5% glycerol) for 1h at 4°C. Then 20 µl of streptavidin-agarose beads (Invitrogen, Carlsbad, CA) was added and incubated for an additional 1h at 4°C. The precipitates were washed three times with binding buffer, and the bound proteins were analyzed by SDS–PAGE and western blot with Ku80 as loading control. Unlabeled PCR product (cold probe) was added to the binding reaction for the competition assay.
Wound healing assay
Cell migration was determined by a scratch wound healing assay. HT-29 cells were allowed to reach confluence and a wound was created in the monolayer by scraping with a sterile pipette tip across the entire diameter of the well. The culture was then washed with medium to remove free-floating cells and debris and cultured in serum-free medium with or without 50ng/ml IL-6 for an additional 24h. To monitor the wound closure, images of the wound area were captured in six fields using an inverted microscope (Eclipse Ti, Nikon, Kyoto, Japan).
Cell invasion assay
The in vitro cell invasion assay was performed in 24-well transwell plates (Costar, Cambridge, MA) with 8 μm-pore inserts coated with Matrigel (BD Biosciences, San Jose, CA). HT-29 cells (1×105) were applied to a culture insert in serum-free medium, whereas complete medium with or without 50ng/ml IL-6 was applied to the lower compartment. After incubation for 48h, non-invaded cells on the upper surface of the filter were removed carefully with a cotton swab and the undersurface adherent cells that had invaded through the Matrigel were fixed in methanol and stained with 0.5% crystal violet. The air-dried filter membrane was viewed under a microscope and four random fields were selected for cell counting.
Gelatin zymography
Fra-1 or EV stably transfected HT-29 cells were incubated overnight in serum-free medium and the proteins in the conditioned medium were concentrated with Amicon Ultra-4 (Millipore, Billerica, MA). Proteins in non-reducing conditions were resolved on 8% polyacrylamide gel containing 1mg/ml gelatin (Sigma). The gel was rinsed in 2.5% Triton X-100 for 1h, developed overnight at 37°C in incubation buffer (50mM Tris–HCl, pH 7.6, 50mM NaCl and 5mM CaCl2), stained with Coomassie brilliant blue R250 for 2h, and de-stained to the desired degree. Gelatinolytic activity was shown as clear areas in the gel, and images were captured using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunohistochemistry
A total of 229 human CRC samples were collected at the First Affiliated Hospital of Zhejiang University School of Medicine after informed consent had been given by all patients. The immunohistochemistry was performed using an Envision Detection System (DAKO, Carpinteria, CA) according to the manufacturer’s instructions. To estimate the score for each slide, at least eight individual fields at 200× were chosen, and 100 cancer cells were counted in each field. Cells with nuclear Fra-1 immunoreactivity were considered positive. The score for each slide was calculated as the cross-product of the immunostaining intensity value and the proportion of positive-staining cells. The immunostaining intensity was divided into five grades: 0, negative; 1, weak; 2, moderate; 3, strong; and 4, very strong. The proportion of positive-staining cells was also divided into five grades: 0, <5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. The staining results were assessed and confirmed by two independent investigators blinded to the clinical data. To determine the correlation of Fra-1 expression with IL-6 secretion or with changes in STAT3 activation, consecutive sections and corresponding similar regions were used to score the immunostaining intensity. The clinico-pathological characteristics of the clinical specimens are summarized in Supplementary Table S2, available at Carcinogenesis Online.
Statistical analysis
Statistical data analysis was performed with the two-tailed Student’s t-test or Pearson’s correlation test, and the results were regarding as significant if the P value was <0.05.
Results
IL-6 induces EMT program and Fra-1 expression in CRC cells
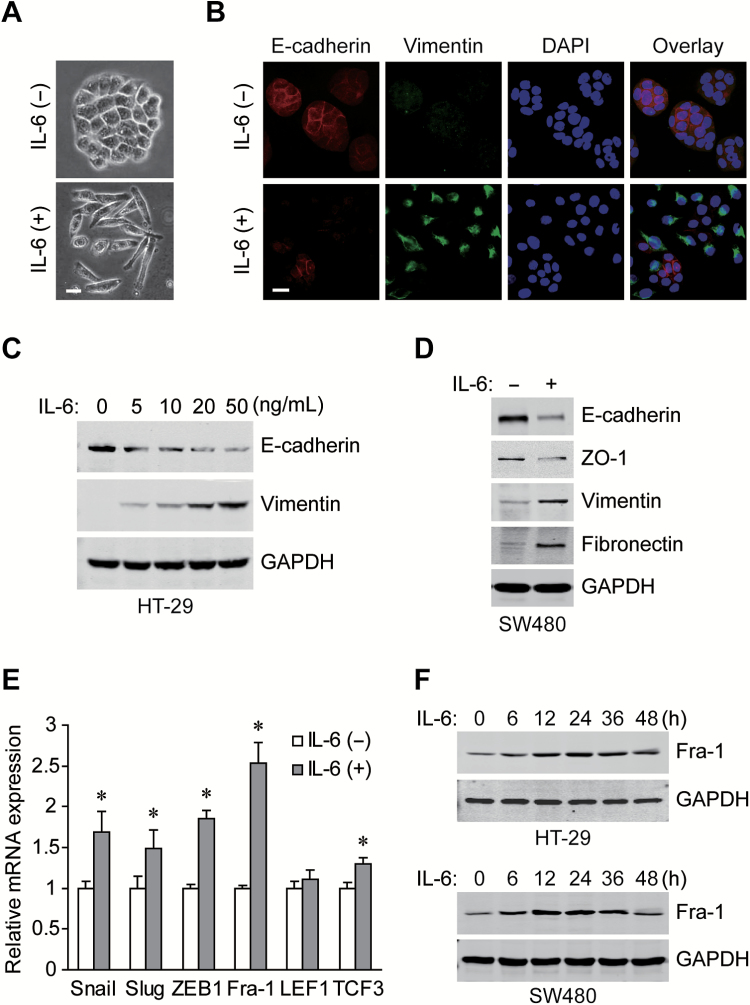
Clinical studies have shown that the levels of the pro-inflammatory cytokine IL-6 are frequently elevated in CRC and correlate with its aggressive behavior and metastatic potential (8,9), we set out to determine the potential role of IL-6 in EMT induction in CRC cells. After exogenous IL-6 treatment, the morphology of cultured HT-29 cells was found changing from adherent polygonal cells to scattered spindly cells (Figure 1A). Moreover, IL-6 exposure decreased the expression of membranous epithelial marker E-cadherin and increased the expression of cytoplasmic mesenchymal marker vimentin in a dose-dependent manner (Figure 1B and C). Similarly, the repression of E-cadherin and ZO-1 accompanied by the induction of vimentin and fibronectin were observed in IL-6 treated SW480 cells (Figure 1D), and the expression changes of EMT markers were further confirmed at the mRNA level (Supplementary Figure S1, available at Carcinogenesis Online). Mechanistically, key EMT regulators were screened by qRT-PCR after IL-6 treatment, among which, Fra-1 showed most significant upregulation (Figure 1E). The protein levels of Fra-1 were also increased upon IL-6 stimulation in a time dependent manner (Figure 1F). Together, it can be concluded that IL-6 exposure leads to EMT process and Fra-1 induction in CRC cells.
Figure 1.
IL-6 induces EMT changes and Fra-1 expresison in CRC cells. (A) Morphology of HT-29 cells treated with or without 50ng/ml IL-6 for 72h under phase contrast microscopy. Scale bar, 20 μm. (B) Immunofluorescent staining for E-cadherin and vimentin in HT-29 cells treated with IL-6 for 72h (nuclei stained with DAPI). Scale bar, 20 μm. (C) Western blots of E-cadherin and vimentin for HT-29 cells incubated with gradient concentrations of IL-6 for 72h. (D) Western blots of EMT markers with specific antibodies in SW480 cells exposed to IL-6 for 72h. (E) Relative mRNA levels of EMT regulators in HT-29 cells treated with IL-6 for 24h (normalized to GAPDH). *P < 0.05. (F) Western blots of Fra-1 and GAPDH from whole-cell lysates extracted from HT-29 and SW480 cells treated with 50ng/ml IL-6 for the indicated times.
STAT3 binds directly to the Fra-1 gene promoter to activate its transcription in response to IL-6 stimulation
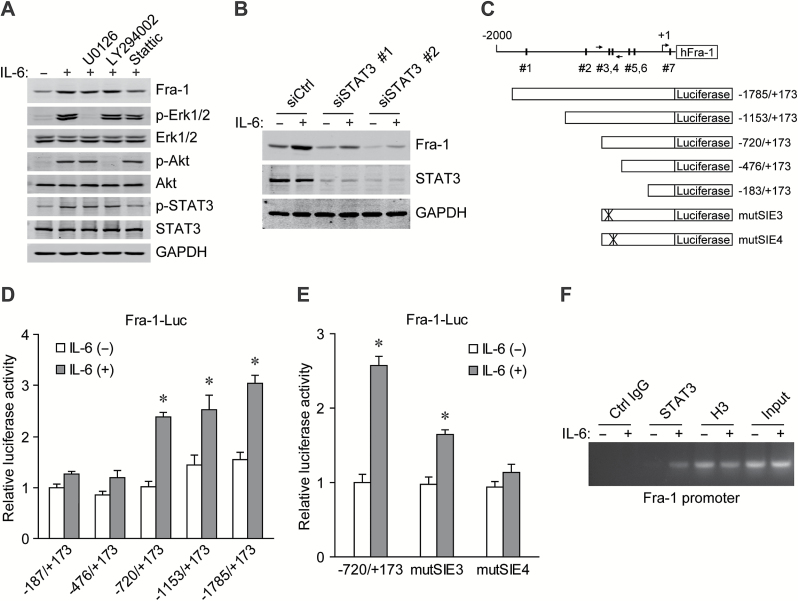
As IL-6 can activate several signaling pathways (7), specific inhibitors were used to identify possible upstream molecules responsible for the Fra-1 induction. In cells exposed to IL-6, the upregulation of Fra-1 was abrogated by pretreatment with stattic, an inhibitor of STAT3, but not with U0126 or LY294002, which are inhibitors of MEK and PI3K respectively (Figure 2A). In addition, siRNA-mediated STAT3 knockdown markedly attenuated the IL-6 induced Fra-1 expression in CRC cells (Figure 2B), indicating that STAT3 is critical in the upregulation of Fra-1. The promoter sequence of the Fra-1 gene was analyzed and in which seven potential STAT-inducible elements (SIEs) were predicted (Figure 2C). Serial deletion constructs of the Fra-1 gene promoter were examined by luciferase reporter assays to identify the transcriptional regulatory region responsive to IL-6/STAT3 signaling. It was shown that the Fra-1 promoter without the region between −720 and −476 lost the ability to respond to IL-6 stimulation (Figure 2D). This region contained two predicted SIEs (SIE3 and SIE4). Mutation in either the SIE3 or SIE4 site, especially the latter, markedly reduced the reporter activity induced by IL-6 (Figure 2E). ChIP-PCR analysis further demonstrated that STAT3 directly bound to the Fra-1 promoter region around −600bp upstream of the transcription start site upon IL-6 exposure (Figure 2F). The evidence suggests that IL-6-activated STAT3 regulates transcription of the Fra-1 gene by direct binding to the promoter.
Figure 2.
Fra-1 is transcriptionally regulated by STAT3 in response to IL-6 stimulation. (A) Western blots with the indicated antibodies from HT-29 cells pretreated for 1h with LY294002, U0126, or Stattic (specific inhibitors of PI3K, MEK and STAT3, respectively) and exposed to IL-6 for 12h. (B) Western blots of Fra-1 and STAT3 from HT-29 cells transfected with control or STAT3 siRNAs and incubated with IL-6 for 12h (GAPDH was used as a loading control). (C) Schematic representation of Fra-1 promoter with seven potential SIEs and the primer pair used in ChIP-PCR assays. The reporter construct Fra-1-Luc and its truncated and mutated derivatives are also shown. (D) Transcription activity in response to IL-6 treatmemt for 6h measured by luciferase assay in 293T cells with a series of deletion mutants of Fra-1-luc (internal control, pRL-TK). *P < 0.05. (E) Relative luciferase activity 6h after IL-6 incubation in 293T cells transfected with the wild-type or SIE mutated Fra-1 promoter reporter construct. *P < 0.05. (F) Chromatin prepared from HT-29 cells stimulated with IL-6 for 1h was immunoprecipitated with the indicated antibodies; PCR was performed on immunoprecipitated DNAs or soluble chromatin using specific primer pair for the Fra-1 promoter.
Both acetylated and phosphorylated modifications of STAT3 induced by IL-6 are needed for Fra-1 gene transactivation
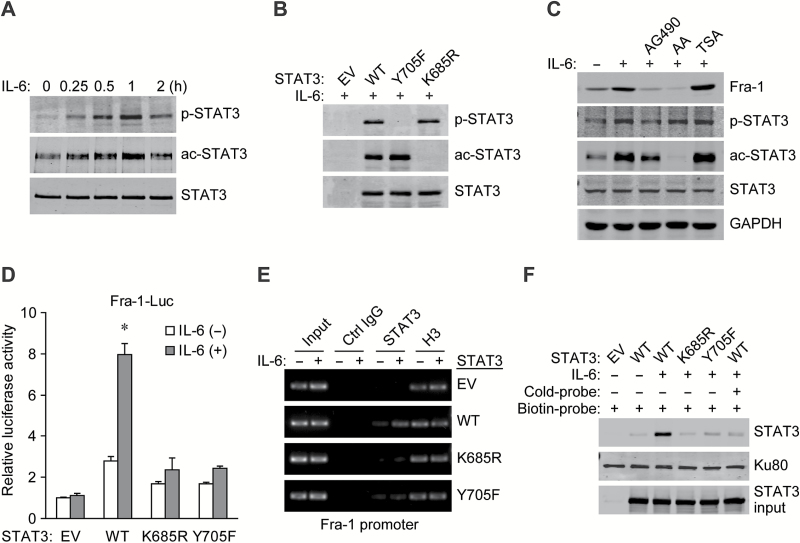
Forming a homodimer is a prerequisite for the activation of STAT3 and transcription of its target genes, and post-translational modifications (PTMs) are crucial for STAT3 dimerization (16,17). Our results showed that both Y705 phosphorylation and K685 acetylation of STAT3 were simultaneously increased in a time-dependent manner after IL-6 stimulation in CRC cells (Figure 3A). In STAT3-null PC3 cells, ectopic expression of the PTM-deficient Y705F or K685R mutant of STAT3 abolished the IL-6-induced site-specific phosphorylation or acetylation, respectively, and mutation in each site did not interfere with the modification of the other one (Figure 3B), indicating the independence of the two different PTM types of STAT3 in response to IL-6 stimulation.
Figure 3.
Acetylation and phosphorylation are both required for STAT3 activation to transactivate the Fra-1 gene. (A) Western blots of STAT3 and its PTM status with site-specific antibodies in serum-starved HT-29 cells treated with 50ng/ml IL-6 for the indicated times. (B) Western blots for the STAT3-null PC3 cells transfected with empty vector (EV), wild-type (WT) or mutated STAT3 and stimulated with IL-6 for 1h. (C) Western blots with specific antibodies from whole-cell extracts of HT-29 cells pretreated with AG490, anacardic acid or trichostatin A (inhibitors of JAK2, histone acetyltransferases and histone deacetylases, respectively) and further stimulated with IL-6 for 12h. (D) Relative luciferase activity in PC3 cells co-transfected with wild-type or mutated STAT3, Fra-1 promoter reporter (−720/+173) and an internal control reporter pRL-TK and administered with IL-6 24h later. *P < 0.05. (E) Upon IL-6 stimulation for 1h, ChIP assays were performed in PC3 cells reintroduced with wild-type STAT3 or derived mutants using the indicated antibodies. The Fra-1 promoter region containing STAT3 binding sites was amplified by PCR. (F) PC3 cells transfected with STAT3-WT or its mutant were treated with IL-6 for 1h, the binding ability of STAT3 to the biotin-labeled Fra-1 promoter probe (−760/−524) was analyzed by DNA pull-down assay. Unlabeled Fra-1 promoter probe (cold-probe) was used for competitive inhibition. Ku80 served as a control.
It is known that JAK2 is a critical kinase of STAT3, and the acetylation of STAT3 is reversibly regulated by histone acetyltransferases and histone deacetylases (16). Western blot analyses showed that the JAK2 inhibitor AG490 and the histone acetyltransferases inhibitor anacardic acid abrogated their corresponding PTMs in STAT3 and impeded Fra-1 induction, respectively, while the histone deacetylases inhibitor trichostatin A further elevated STAT3 acetylation and hence Fra-1 expression in IL-6-treated cells (Figure 3C). Reintroduction of wild-type STAT3 but not the K685R or Y705F mutant into PC3 cells restored the IL-6 induced Fra-1-Luc reporter gene activity (Figure 3D). ChIP-PCR and DNA pull-down analyses confirmed the inability of two STAT3 mutants to bind to the Fra-1 promoter compared with the wild-type in IL-6-treated PC3 cells (Figure 3E and F). Therefore, both K685 acetylation and Y705 phosphorylation of STAT3 induced by IL-6 contribute to its activation in the regulation of Fra-1 gene expression.
Fra-1 is a major player in the IL-6 induced EMT and mobility of CRC cells
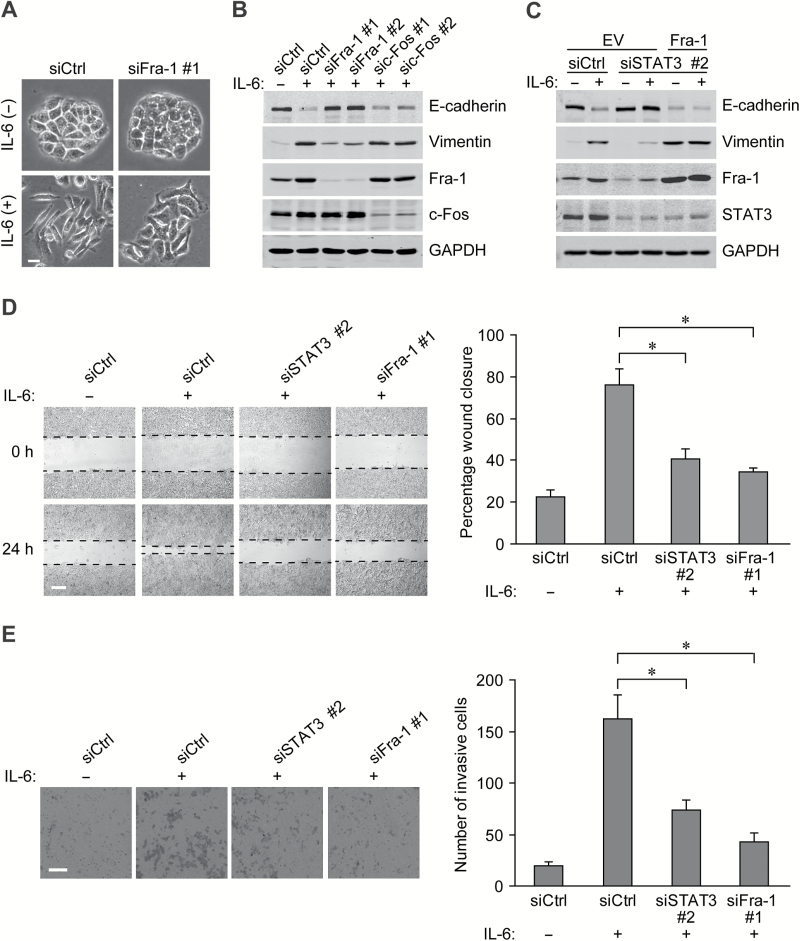
As the activation of IL-6/STAT3 signaling resulted in a robust increase in Fra-1 expression, we next investigated the functional role of Fra-1 in the IL-6 induced EMT process. When Fra-1-siRNA was transfered into HT-29 cells, the mesenchymal-like morphological conversion stimulated by IL-6 was substantially prevented (Figure 4A). The expression changes in EMT markers (decreased E-cadherin and induction of vimentin) were also blunted in the Fra-1-silenced CRC cells, while knockdown of c-Fos, another Fos family member, had no evident impact on the IL-6-induced EMT changes (Figure 4B). Moreover, STAT3 silencing had a destructive effect similar to that of Fra-1 knockdown on the EMT process, while ectopic expression of Fra-1 largely restored the molecular changes in CRC cells (Figure 4C).
Figure 4.
Fra-1 plays a critical role in IL-6 induced EMT and cell mobility. (A) Morphology of HT-29 cells with siRNAs against Fra-1 or scrambled control and stimulated with 50ng/mL of IL-6 for 72h. Scale bar, 20 μm. (B) Western blots of 72h IL-6 treated HT-29 cells receiving the indicated siRNAs using specific antibodies. (C) Western blots of HT-29 cells transfected with Fra-1 expression vector or empty vector and indicated siRNAs and incubated with IL-6 for 72h afterwards. (D) Left panels: images from scratch assays with HT-29 cells transfected with indicated siRNAs. Scale bar, 200 μm. Right panel: percentage wound closure 24h after stimulation with IL-6. *P < 0.05. (E) Left panels: representative images of HT-29 tumor cells penetrating the Matrigel in invasion assays. Scale bar, 100 μm. Right panel: numbers of invasive cells transfected with the indicated siRNAs and exposed to IL-6 for 48h. *P < 0.05.
EMT-related functional assays were carried out to determine the roles of STAT3 and Fra-1 in the IL-6-regulated migration and invasion of CRC cells. Scratch assays showed that IL-6 increased migration in HT-29 cells, and siRNA-mediated knockdown of either STAT3 or Fra-1 retarded the cell motility and wound healing (Figure 4D). Consistently, the acquisition of invasive capacity in the CRC cells exposed to IL-6 was also attenuated by STAT3 or Fra-1 silencing (Figure 4E). The above results indicate that Fra-1 transactivation by STAT3 signaling is essential for IL-6-induced EMT program in CRC cells.
Fra-1 enhances migration and invasion of CRC cells by regulating the expression of EMT-promoting factors
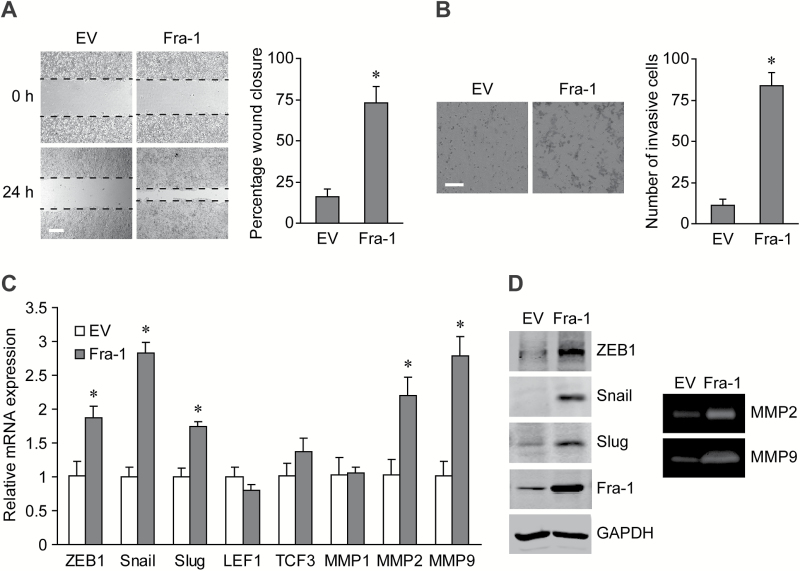
To further characterize the function of Fra-1 in cell motility, stably overexpression of Fra-1 facilitated the wound healing and invading abilities of CRC cells (Figure 5A and B). Then, the expression of EMT-inducing transcription factors (EMT-TFs) and matrix metalloproteinases (MMPs) in these cells were analyzed to explore the possible molecular mechanism. Fra-1 increased the mRNA levels of ZEB1, Snail, Slug, MMP2 and MMP9, while the LEF1, TCF3 and MMP1 levels showed no significant changes (Figure 5C), and the changes of protein levels and enzyme activities were also verified (Figure 5D). Thus, Fra-1 plays an important role in promoting aggressive behavior in CRC cells through the EMT-TFs and MMPs.
Figure 5.
Fra-1 facilitates cell migration and invasion via EMT-promoting factors. (A) Left panels: images from scratch assays with Fra-1 or EV stably transfected HT-29 cells. Scale bar, 200 μm. Right panel: percentage wound closure 24h after scratch. *P < 0.05. (B) Left panels: representative images of stably transfected HT-29 cells invading the Matrigel. Scale bar, 100 μm. Right panel: numbers of invasive cells stably transfected with Fra-1 or EV. *P < 0.05. (C) The mRNA levels of EMT-TFs and MMPs in Fra-1 stably transfected HT-29 cells. *P < 0.05. (D) Left panels: western blots of total proteins from stably transfected HT-29 cells (equal protein loading confirmed by GAPDH expression). Right panels: gelatin zymography for MMPs activity in conditioned medium of stably transfected HT-29 cells.
Increased Fra-1 expression is associated with cancer aggressiveness and activated IL-6/STAT3 signaling in CRC samples
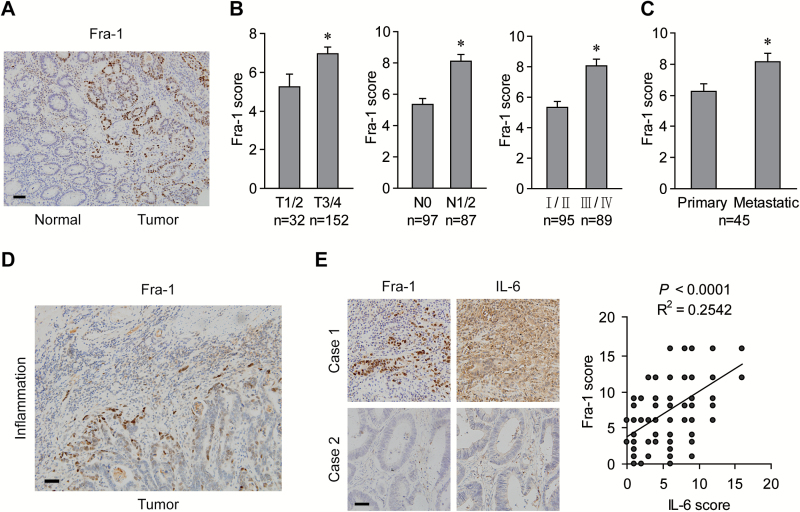
Immunohistochemical staining was performed to evaluate the clinical relevance of Fra-1 expression in human CRC specimens. Fra-1 immunoreactivity was observed in the nuclei of cancer cells but not in the adjacent normal epithelial cells (Figure 6A). Fra-1 expression was positively associated with local invasion depth as well as lymph node and distant metastasis (Figure 6B). In liver metastatic lesions, Fra-1 levels were significantly higher than in the primary tumor tissues (Figure 6C). Notably, cancer cells at the marginal area adjacent to the inflammatory region displayed much stronger Fra-1 immunoreactivity than those in the central cancer region, implying a paracrine effect of inflammatory cells on Fra-1 expression in CRC cells (Figure 6D). Furthermore, immunostaining using consecutive sections positively correlated the expression levels of Fra-1 with the degree of IL-6 secretion, and both were increased in CRC tissues with inflammatory infiltration (Figure 6E). Besides, the significant correlation of Fra-1 expression with acetylation or phosphorylation of STAT3 was also manifested in the consecutive sections (Supplementary Figure S2, available at Carcinogenesis Online). These data suggest that the pro-inflammatory cytokine IL-6 is positively correlated with Fra-1 expression, which is involved in CRC progression and metastasis.
Figure 6.
Expression of Fra-1 is upregulated coincident with IL-6 secretion in advanced CRC. (A) Representative images of Fra-1 immunostaining in CRC tissues and adjacent non-cancerous tissues. Scale bar, 50 μm. (B) Expression of Fra-1 protein in primary tumors versus pathological parameters. *P < 0.05. (C) Fra-1 levels in the primary colorectal and liver metastatic tumors. *P < 0.05 (D) Representative images of Fra-1 immunostaining in tumor cells at the marginal area neighboring inflammatory infiltrations. Scale bar, 50 μm. (E) Left panels: representative images of concurrent expression of Fra-1 and IL-6 in consecutive sections of CRC tissues. Scale bar, 50 μm. Right panel: linear regression between immunostaining intensity of Fra-1 and IL-6.
Discussion
In the early stage of CRC, tumor cells are generally arranged in tubular structures with an epithelial phenotype. During progression to advanced stages involving invasion into surrounding tissues and further metastasis to regional lymph nodes and distant organs, the CRC cells localized at the tumor-host tissue interface often exhibit the hallmarks of EMT, that is loss of epithelial cell markers like E-cadherin and gain of mesenchymal cell markers like vimentin, resulting in the dissolution of adhesion junctions and the acquisition of aggressive behaviors (18). As a complex cellular program, EMT mediates switches of molecular signatures, morphological characteristics and motile attributes (19). That the EMT process mainly occurs at the marginal areas of metastatic tumors suggests its enhancement by substances from the microenvironment, including various chemokines, cytokines and growth factors. Among these, TGF-β, VEGF and TNFα are known to promote the malignant traits of CRC cells by inducing EMT (20). The clinical relevance of pro-inflammatory cytokine IL-6 to cancer development is evident in both colitis-associated and sporadic CRC patients (8,9). Here, we demonstrate a previously unrevealed IL-6/STAT3/Fra-1 signaling axis which promotes EMT program and metastasis in CRC.
As a major mediator of IL-6 signaling, STAT3 is often constitutively activated in many solid and hematological tumors to transactivate the expression of genes responsible for cell growth, survival, angiogenesis and invasion (17). In this study, we showed that aberrant activation of STAT3, but not the Akt or ERK, mediated IL-6-induced Fra-1 upregulation, which was required for the EMT and aggressiveness of CRC cells. The correlation between STAT3 activation and Fra-1 expression in clinical CRC specimens supported the role of IL-6/STAT3 signaling in Fra-1 transactivation. However, in addition to IL-6, STAT3 activation is also associated with the EMT programs induced by other cytokines such as TGF-β, EGF and OSM (21–23). The possible pathway crosstalks among these signals in EMT regulation are interesting and need further investigations in the future.
Recently, the acetylation of proteins other than histones, especially that of transcription factors, has attracted increasing attention to its roles in cancer (24,25). Besides the well-known tyrosine (Y705) phosphorylation in the functional regulation of STAT3, the lysine (K685) acetylation also facilitates STAT3 dimerization, nuclear translocation and sequence-specific DNA binding, thereby promoting the transcription of target genes such as IDO and cyclin D1 (26,27). Elevated STAT3 acetylation in tumor tissues can combine with DNMT1 to silence tumor-suppressor genes via methylation of promoter CpG islands (28,29). Moreover, nuclear CD44 exploits K685-acetylated STAT3 to confer stem cell properties on cancer cells (30,31). Here, we revealed a correlation between STAT3 acetylation and Fra-1 transactivation in IL-6 stimulated CRC cells and clinical CRC tissues, indicating the importance of this modification in the EMT process during cancer progression. Although the interdependence between phosphorylation and acetylation in STAT3 modulation has been reported (32,33), we found that the two PTMs occurred simultaneously after IL-6 treatment in an independent manner, and both of them were needed to control STAT3 activation. This discrepancy could be explained by the strong dependency of the regulation of PTMs on cell type and gene promoter context, as well as the stimulus applied (34). Considering the autonomous function of STAT3 acetylation even in the Y705F mutant (30,31), the development of chemicals targeting the acetylation in addition to quenching the phosphorylation would be attractive for the full inhibition of STAT3 activity in human malignancies (35).
We observed that exogenous expression of Fra-1 restored EMT changes in STAT3-depleted cells and promoted cell migration and invasion, indicating a driving role of Fra-1 in the EMT and aggressiveness of CRC. The positive role of Fra-1 in the progression of various tumors has been noted in recent years. It is reported that Fra-1 is negatively correlated with E-cadherin levels (11) and positively regulates the expression of vimentin (36) and pro-EMT microRNAs (37). Here, we showed that Fra-1 overexpression led to the induction of EMT-TFs Snail, Slug and ZEB1, as well as the secretion of extracellular matrix-degrading enzymes MMP2 and MMP9. However, the expression of Twist, another important repressor of E-cadherin in breast cancer cells (38), was not detectable in our experiments, which is possibly related to the tissue-dependent expression patterns of EMT-TFs (39,40).
Several previous studies have implied a correlation between STAT3 and Fra-1 using cell models. Both STAT3 activation and Fra-1 elevation are found in the EMT process induced by annexin A1 knockdown (41), and microarray analyses have revealed the possible upregulation of Fra-1 mRNAs in cells treated with IL-6 or STAT3 overexpression (42,43). However, the function and regulatory mechanism by which IL-6/STAT3 induces Fra-1 expression had not yet been determined. Here, we showed that IL-6 stimulated the phosphorylation and acetylation of STAT3, which directly bound to the promoter of the Fra-1 gene to increase its transcription. This was verified by the positive association between Fra-1 levels and IL-6 secretion as well as STAT3 modifications in the CRC cases. In addition, Fra-1 expression is reported to be regulated by multiple means, including transcriptional activation, microRNA regulation, phosphorylation and protein stabilization (44–46). It needs further investigation whether these mechanisms are involved in the functional regulation of Fra-1 in tumor-promoting inflammatory microenvironment.
Another Fos family member, c-Fos, is known to be a STAT3 downstream target gene as well. However, the STAT3 binding regions in the promoters of Fra-1 and c-Fos are not conserved. Although Fra-1 can substitute for c-Fos functionally in osteoclast differentiation and bone development, their underlying mechanisms and downstream genes are markedly different (47,48). In addition, c-Fos and Fra-1 show reciprocal expression patterns in tumor cell lines at different stages of progression from epithelial to mesenchymal phenotypes (11). Our results showed that Fra-1 silencing largely prevented the IL-6-induced expressional changes of epithelial and mesenchymal molecules, whereas c-Fos depletion had hardly any effects. As their profiles and functions are disparate in many cellular processes like the EMT and its reversal, the MET, the alternation of Fra-1 and c-Fos expression may determine reversible switches between proliferative and invasive phenotypes and manifest the plasticity of tumor cells in aggressive carcinomas.
In summary, our finding demonstrated that the EMT program in CRC cells can be initiated by the pro-inflammatory cytokine IL-6 stimulation. Mechanistic studies revealed that transactivation of Fra-1 was induced by IL-6 through activating the phosphorylation and acetylation of STAT3 simultaneously. Clinical sample analyses supported the critical role of Fra-1 aberrantly induced by IL-6/STAT3 in mediating the EMT and aggressiveness of CRC. In addition to metastatic dissemination, EMT confers a cancer stem cell phenotype and drug resistance on carcinoma cells (49). It would be interesting to further investigate whether Fra-1 is involved in these malignant properties. The observed correlation of Fra-1 level with tumor invasion and metastasis raises the possibility of developing it as a new biomarker and therapeutic target for human colorectal malignancy. Furthermore, since EMT and dissemination induced by inflammation could occur in the early stage of tumorigenesis (50), the present study may also shed light on prophylactic treatment for the primary CRC as well as the inflammatory bowel diseases.
Supplementary material
Supplementary Tables S1 and S2 and Figures S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
National Natural Science Foundation of China (81372138, 30873094 and 81090421), the 863 National High Technology Research and Development Program of China (2012AA020206), the Specialized Research Fund for the Doctoral Program of Higher Education of China (J20100041) and the Zhejiang Provincial Natural Science Foundation of China (LY13H160001).
Supplementary Material
Acknowledgments
The authors thank Drs. Chenfang Dong and Yingjie Wang for critically reading the manuscript and valuable suggestions.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- CRC
colorectal cancer
- EMT
epithelial–mesenchymal transition
- EMT-TF
EMT inducing transcription factor
- Fra-1
Fos-related antigen-1
- IL-6
interleukin-6
- MMP
matrix metalloproteinase
- PTM
post-translational modification
- SIE
STAT-inducible element
- STAT3
signal transducer and activator of transcription 3
References
- 1. Center M.M., et al. (2009). International trends in colorectal cancer incidence rates. Cancer Epidemiol. Biomarkers Prev., 18, 1688–1694. [DOI] [PubMed] [Google Scholar]
- 2. Andre N., et al. (2005). Chemoradiotherapy for colorectal cancer. Gut, 54, 1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ullman T.A., et al. (2011). Intestinal inflammation and cancer. Gastroenterology, 140, 1807–1816. [DOI] [PubMed] [Google Scholar]
- 4. Grivennikov S., et al. (2009). IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell, 15, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bollrath J., et al. (2009). gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell, 15, 91–102. [DOI] [PubMed] [Google Scholar]
- 6. Rokavec M., et al. (2014). IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest., 124, 1853–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ataie-Kachoie P., et al. (2013). Inhibition of the IL-6 signaling pathway: a strategy to combat chronic inflammatory diseases and cancer. Cytokine Growth Factor Rev., 24, 163–173. [DOI] [PubMed] [Google Scholar]
- 8. Komoda H., et al. (1998). Interleukin-6 levels in colorectal cancer tissues. World J. Surg., 22, 895–898. [DOI] [PubMed] [Google Scholar]
- 9. Knüpfer H., et al. (2010). Serum interleukin-6 levels in colorectal cancer patients—a summary of published results. Int. J. Colorectal Dis., 25, 135–140. [DOI] [PubMed] [Google Scholar]
- 10. Young M.R., et al. (2006). Fra-1 a target for cancer prevention or intervention. Gene, 379, 1–11. [DOI] [PubMed] [Google Scholar]
- 11. Kustikova O., et al. (1998). Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol. Cell. Biol., 18, 7095–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lombaerts M., et al. (2006). E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br. J. Cancer, 94, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tam W.L., et al. (2013). Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell, 24, 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caramel J., et al. (2013). A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell, 24, 466–480. [DOI] [PubMed] [Google Scholar]
- 15. Diesch J., et al. (2014). Widespread FRA1-dependent control of mesenchymal transdifferentiation programs in colorectal cancer cells. PLoS One, 9, e88950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan Z.L., et al. (2005). Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science, 307, 269–273. [DOI] [PubMed] [Google Scholar]
- 17. Al Zaid Siddiquee K., et al. (2008). STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res., 18, 254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brabletz T., et al. (2001). Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. U. S. A., 98, 10356–10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang J., et al. (2008). Epithelial–mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell, 14, 818–829. [DOI] [PubMed] [Google Scholar]
- 20. Bates R.C., et al. (2005). The epithelial–mesenchymal transition (EMT) and colorectal cancer progression. Cancer Biol. Ther., 4, 365–370. [DOI] [PubMed] [Google Scholar]
- 21. Yang Y., et al. (2006). Regulation of transforming growth factor-beta 1-induced apoptosis and epithelial-to-mesenchymal transition by protein kinase A and signal transducers and activators of transcription 3. Cancer Res., 66, 8617–8624. [DOI] [PubMed] [Google Scholar]
- 22. Lo H.W., et al. (2007). Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial–mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res., 67, 9066–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo L., et al. (2013). Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial–mesenchymal transition. Oncogene, 32, 5272–5282. [DOI] [PubMed] [Google Scholar]
- 24. Singh B.N., et al. (2010). Nonhistone protein acetylation as cancer therapy targets. Expert Rev. Anticancer Ther., 10, 935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qi H., et al. (2012). Interferon regulatory factor 1 transactivates expression of human DNA polymerase η in response to carcinogen N-methyl-Nʹ-nitro-N-nitrosoguanidine. J. Biol. Chem., 287, 12622–12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun Y., et al. (2009). Cutting edge: negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J. Immunol., 182, 5899–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kundumani-Sridharan V., et al. (2012). Novel interactions between NFATc1 (nuclear factor of activated T cells c1) and STAT-3 (signal transducer and activator of transcription-3) mediate G protein-coupled receptor agonist, thrombin-induced biphasic expression of cyclin D1, with first phase influencing cell migration and second phase directing cell proliferation. J. Biol. Chem., 287, 22463–22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee H., et al. (2012). Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc. Natl. Acad. Sci. U. S. A., 109, 7765–7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J., et al. (2013). STAT3 acetylation-induced promoter methylation is associated with downregulation of the ARHI tumor-suppressor gene in ovarian cancer. Oncol. Rep., 30, 165–170. [DOI] [PubMed] [Google Scholar]
- 30. Lee J.L., et al. (2009). Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J. Cell Biol., 185, 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Su Y.J., et al. (2011). Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J., 30, 3186–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohbayashi N., et al. (2007). LIF- and IL-6-induced acetylation of STAT3 at Lys-685 through PI3K/Akt activation. Biol. Pharm. Bull., 30, 1860–1864. [DOI] [PubMed] [Google Scholar]
- 33. Nie Y., et al. (2009). STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell Biol., 11, 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Icardi L., et al. (2012). The HAT/HDAC interplay: multilevel control of STAT signaling. Cytokine Growth Factor Rev., 23, 283–291. [DOI] [PubMed] [Google Scholar]
- 35. Yue P., et al. (2009). Targeting STAT3 in cancer: how successful are we? Expert Opin. Investig. Drugs, 18, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andreolas C., et al. (2008). Fra-1 regulates vimentin during Ha-RAS-induced epithelial–mesenchymal transition in human colon carcinoma cells. Int. J. Cancer, 122, 1745–1756. [DOI] [PubMed] [Google Scholar]
- 37. Stinson S., et al. (2011). TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal, 4, ra41. [DOI] [PubMed] [Google Scholar]
- 38. Sullivan N.J., et al. (2009). Interleukin-6 induces an epithelial–mesenchymal transition phenotype in human breast cancer cells. Oncogene, 28, 2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peinado H., et al. (2007). Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer, 7, 415–428. [DOI] [PubMed] [Google Scholar]
- 40. Huang C., et al. (2011). The effects and mechanisms of blockage of STAT3 signaling pathway on IL-6 inducing EMT in human pancreatic cancer cells in vitro . Neoplasma, 58, 396–405. [DOI] [PubMed] [Google Scholar]
- 41. Maschler S., et al. (2010). Annexin A1 attenuates EMT and metastatic potential in breast cancer. EMBO Mol. Med., 2, 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dauer D.J., et al. (2005). Stat3 regulates genes common to both wound healing and cancer. Oncogene, 24, 3397–3408. [DOI] [PubMed] [Google Scholar]
- 43. Rojas A., et al. (2011). IL-6 promotes prostate tumorigenesis and progression through autocrine cross-activation of IGF-IR. Oncogene, 30, 2345–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adiseshaiah P., et al. (2005). Mitogen regulated induction of FRA-1 proto-oncogene is controlled by the transcription factors binding to both serum and TPA response elements. Oncogene, 24, 4193–4205. [DOI] [PubMed] [Google Scholar]
- 45. Yang S., et al. (2013). MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene, 32, 4294–4303. [DOI] [PubMed] [Google Scholar]
- 46. Belguise K., et al. (2012). The PKCθ pathway participates in the aberrant accumulation of Fra-1 protein in invasive ER-negative breast cancer cells. Oncogene, 31, 4889–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fleischmann A., et al. (2000). Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev., 14, 2695–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bakiri L., et al. (2007). Role of heterodimerization of c-Fos and Fra1 proteins in osteoclast differentiation. Bone, 40, 867–875. [DOI] [PubMed] [Google Scholar]
- 49. Singh A., et al. (2010). EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene, 29, 4741–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rhim A.D., et al. (2012). EMT and dissemination precede pancreatic tumor formation. Cell, 148, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.