Abstract
Background
Noroviruses (NoVs) are a leading cause of viral diarrhea in young children. Secretor status has been confirmed to be linked with Norwalk virus (NoV GI.1) infection but there is limited information about whether secretor genotypes are associated with pediatric NoV epidemic strains in vivo.
Methods
In this study, fecal specimens and serum samples were collected from 124 hospitalized children with acute diarrhea in Xi'an, China. TaqMan real-time RT-PCR was used to detect NoV in fecal samples and NoV positive samples were further verified using conventional RT-PCR and sequenced. DNA was extracted from sera and TaqMan single nucleotide polymorphism genotyping assay was applied to determine FUT2 A385T polymorphism.
Results
Only NoV GII.3 and GII.4 genotypes were found in NoV positive samples and NoV were detected in 25% (15/60), 40.5% (17/42), and 9.1% (2/22) of children with homozygous secretor genotype (Se385Se385), heterozygous secretor genotype (Se385se385), and homozygous weak secretor genotype (se385se385), respectively. Children with secretor genotypes (Se385Se385 and Se385se385) were significantly (P < 0.05) more susceptible to combined NoV GII.3 and GII.4 infections than children with weak secretor genotype (se385se385).
Conclusions
These findings indicate that secretor positive is significantly associated with GII.3 and GII.4 infections in Chinese pediatric diarrheal children and weak secretor is not a complete protection of children from GII.3 and GII.4 infections.
Keywords: Norovirus, diarrhea, secretor, genotype, FUT2
Introduction
Noroviruses (NoVs) have replaced rotavirus as the leading cause of acute gastroenteritis among young children in several countries since the introduction of rotavirus vaccines (1-3). This virus has a single stranded, plus-sense RNA, approximately 7.6 kb in length that contains three open-reading frames (ORFs). ORF2 and ORF3 encode a single major (VP1) and a minor (VP2) capsid protein, respectively (4). Expression of the major capsid protein VP1 in Sf9 insect cells results in the formation of virus-like particles (VLPs) that are morphologically and antigenically similar to native NoV (5). Serological studies using the VLPs have shown a high prevalence of NoV specific IgG both in children and adults (6-9). A recent Norwalk virus (NoV GI.1) vaccine trial indicates that vaccine recipients were significantly less likely to become ill or to be infected with homologous virus compared with placebo recipients (10), suggesting the serum antibodies can protect individuals from subsequent infection.
NoVs are classified into five genogroups (GI to GV), and each genogroup is subdivided into genotypes. Previous NoV genotyping studies have classified human NoVs into at least 26 genotypes (11) and NoV GII.3 and GII.4 are the most common genotypes responsible for the majority of sporadic diarrhea cases in children (12) and outbreaks in adults (13, 14).
The first report of a genetic factor likely involved in NoV infection was documented in a human challenge study in the 1970s (15). In 2003, human challenge studies (16) found that susceptibility to NV infection is associated with secretor status that is determined by the FUT2 gene. FUT2 encodes an alpha(1,2)fucosyltransferase which is responsible for the synthesis of H antigen and individuals with H antigen expressions are considered secretor positive. This gene has a significant polymorphism with ethnic specificity and several single nucleotide polymorphisms (SNP) in the FUT2 locus have been reported (17, 18). A missense mutation at nucleotide 385 (A>T) is found commonly in Asian populations (17) and homozygous carriers of this mutation are considered weak secretors, leading to low levels of ABH antigens. A secretor individual has at least 1 functional allele, either the Se385Se385or Se385se385 genotype, while a weak secretor is homozygous for the weak functional allele, the se385se385genotype.
Previous human challenge studies (16) indicates that secretor negative individuals do not become infected regardless of NV dose. However, Snow Mountain virus (SMV, GII.2) (19) and genotype GI.3 infections (22, 23) do not have any association with secretor status but genotypes GII.3 and GII.4 have been shown to be significantly associated with the secretor phenotype in previous challenge (20) and outbreak (21) studies. In contrast to Norwalk virus (16), secretor negative subjects are not completely protected from GII.4 infections (20). The objective of this study was to determine whether secretor genotypes are associated with GII.3 and GII.4 in a pediatric setting in Xian, China.
Materials and Methods
Study Population
Between March 2009 and March 2011, fecal specimens, serum samples, and clinical symptoms were collected from all hospitalized children aged <5 years, clinically diagnosed with acute gastroenteritis (defined as ≥3 loose or watery stools per day) in the Department of Digestive Diseases of Xi'an Children's Hospital, the largest children's healthcare center in Xi'an, China. Fecal and serum samples were collected only once within 48 hrs of admission when children were in the course of illness. Parents/guardians were asked to sign an informed consent form (approved by the IRB committee at Emory University) before their children's participation into this study.
Norovirus RNA Extraction and Detection Using TaqMan Real-time RT-PCR
A 20% (wt/vol.) stool suspension was prepared in RNAse- and DNAse-free water (Mediatech, Manassas, VA) and 500 μl of the suspension was mixed with an equal volume of Vertrel (Miller-Stephenson, Danbury, CT). After incubation at 4°C overnight, the samples were centrifuged at 12,000 × g for 15 min at 4°C. Subsequently, a 140-μl supernatant was removed and used for norovirus RNA extraction using the QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA.) in accordance with the manufacturer's instructions.
Separate NoV TaqMan real-time RT-PCR (RT-qPCR) assays were performed to detect GI and GII noroviruses using the Qiagen OneStep RT-PCR Kit (QIAGEN, Valencia, CA) and NoV GI and GII broadly-reactive primers and probes (Sigma, St. Louis, MO) that were previously described (24, 25). A total of 45 amplification cycles were carried out, each consisting of 95°C for 15 sec and 56°C for 1 min.
NoV Conventional RT-PCR and Phylogenetic Analysis
NoV positive samples by the TaqMan real-time RT-PCR were further confirmed using the conventional RT-PCR with a different set of primers spanning the 3′-end of NoV region B and 5′-end of region C (26). PCR amplicons from the conventional RT-PCR were sent to the Beijing Genomic Institute (Beijing, China) for determining NoV sequences. NoV sequences obtained in this study were cleaned with EditSeq program in the DNASTAR software package (Madison, WI). For GII.4 genotypes, two additional PCR and sequencing systems were performed so the full-length sequence of the capsid region was obtained, assembled, and submitted to the GenBank. The accession numbers (JX155737, JX155738, JX155739, JX155740, JX155741, JX155742, JX155743, JX155744, JX155745, JX155746, JX155747, JX155748, JX155749, JX155750, JX155751, JX155752, JX155753, JX155754) have been obtained and the NoV reference sequences downloaded from the GenBank database were aligned using the Clustal W program implemented in the MEGA version 5 (Tamura, Dudley). The phylogenetic tree was constructed using Maximum-likelihood method in the MEGA 5 software package with 1,000 pseudoreplicate data sets.
GII.3 and GII.4 VLPs Development
Because GII.3 and GII.4 were the only two NoV genotypes identified in this study, VLPs from these two strains were developed. The full length cDNA fragment (2.3 kb) of the VP1 region (NoV capsid protein) and the VP2 region of norovirus 2010 GII.3 and GII.4 2006b genotypes identified in this study was amplified by RT-PCR with the primers adding restriction enzyme sites (Table 1). The full-length cDNA fragment was cloned into a PCR®4-TOPO vector (Invitrogen, Carlsbad, CA) and then subcloned into a baculovirus pFastBac1 donor plasmid (Invitrogen, Carsbad, CA). The recombinant bacmids were transfected into Spodoptera frugiperda ovarian cell line Sf9 (Invitrogen, Grand Island, NY, USA) at a multiplicity infection (MOI) of 1.0 using the Bac-to-Bac®Baculovirus expression system (Invitrogen, Carlsbad, CA). After four days of incubation, infected Sf9 cells were clarified by centrifugation at 9,790 × g for 30 min at 4°C. The VLPs in the supernatant were concentrated by a Vivaflow 200 ultrafiltration system (Sartorius, Gottingen, Germany) at 4°C. The concentrated VLPs were applied to a discontinuous sucrose gradient (10 ∼ 50% w/v) and centrifugated at 115,255 × g for 1 hr. The fractionated VLP bands from the sucrose gradient were resuspended in 1× PBS buffer and pelleted by ultracentrifugation at 115,255 × g for 1 hr. The pellet was resuspended in 1× PBS buffer containing proteinase inhibitor cocktail (Pierce, Rockford, IL). The purified VLPs were analyzed by western blot, enzyme-linked immunosorbent assay (ELISA) and electron microscopy. Protein concentrations were determined by the Bradford method (BioRad, Hercules, CA, USA) using bovine serum albumin as a standard.
Table 1. Primers used in this study to construct the recombinant GII.3 and GII.4 baculoviruses.
Detection of GII.3 and GII.4 Specific IgG in Sera by ELISA
Direct ELISAs were used to determine the IgG titers to GII.3 and GII.4 in serum samples collected at the acute-phase of diarrhea. Polystyrene plates were coated with 2 μg/ml of the GII.3 or GII.4 VLPs and the plates were blocked overnight at 4°C with 5% Blotto in phosphate buffered saline-Tween (PBS-T). After washes, the plates were incubated for 1 hr at room temperature with duplicate serum samples diluted 1:400 in blocking solution. The secondary antibody (alkaline phosphatase-labeled rabbit α-human IgG, Sigma-Aldrich Co., St. Louis, MO) was diluted 1:2500 and incubated for 30 min at room temperature. After final washes, 100 μl of p-nitrophenyl phosphate solution (Sigma-Aldrich Co., St. Louis, MO) was added to each well and the plate was incubated at room temperature in dark conditions for 10-30 min. The optical density at 405 nm was determined using an ELx800 plate reader (BioTek Instruments, Inc. Winooski, VT). Two-fold serial dilutions of a purified IgG standard (range of 1000 ng/ml to 15.6 ng/ml) were included in the ELISA and the standard curve was used to calculate the GII.3 and GII.4 specific IgG concentrations in serum samples.
FUT2 Genotyping
Genomic DNA was extracted from 200 μl of serum samples using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA). FUT2 A385T polymorphism was assayed using the TaqMan assay and fluorogenic probes as previously described (27). To determine the A385T polymorphism, a 285-bp fragment of the FUT2 gene was amplified using a conventional PCR incorporated with the forward primer 5′-CCCATCTTCAGAATCACCCTGCCGGTGCTG-3′ and the reverse primer 5′-ATGGACCCCTACAAAGGTGCCCGGCCGGCT-3′. The SNP of the A385T was determined using the Custom TaqMan SNP Genotyping Assay (Applied Biosystems) developed by Dr. Le Pendu with the fluorescent probes FAM-CGGGAAGTGGCGGT-BHQ1 and VIC- CGGGATGTGGCGGT-BHQ1 for A/T allele discrimination. The PCR program included a 40-cycle of amplification with 95°C for 10 min, 92°C for 15 sec, and 60°C for 1 min using the CFX96 Touch Deep Well™ Real-Time PCR Detection System (BioRad, Hercules, CA). The amplification results were analyzed using the Allelic Discrimination program in the BioRad PCR system.
Statistical Analysis
The optical density (OD) values obtained by ELISA were transformed to μg/ml according to the standard curve of the purified IgG and the dilution factor of serum samples. The normality and constant variance assumption were tested for the GII.3 and GII.4 IgG titers using the Shapira-Wilk test in R program. If such assumption was violated, the Mann-Whitney two-tailed test was used to compare serum IgG responses between the ranks in the three age groups and the three secretor genotyping groups. In addition, if IgG levels were not normally distributed, median and interquartile were reported in each group. NoV positive rates obtained by RT-PCR in fecal samples were compared with secretor genotypes by the Fisher exact test due to the fact of small sample size. Odds ratios and confidence intervals were calculated. Data were analyzed using SAS 9.2 (SAS, Cary, NC) and the tests were evaluated at a P-value <0.05.
Results
NoV Prevalence and Genotyping in Diarrheal Children
In this study, all hospitalized children with acute diarrhea between March 2009 and March 2011, under 5 years were targeted for sample collection. Finally, fecal and/or serum samples were collected from 385 diarrhea children but only a total of 124 children had matched fecal and serum samples and complete clinical information. When age distribution was analyzed, 122 (98.4%) of 124 eligible children were under 2 years old and two children (1.6%) were between 2 and 5 years old. The median age was 9.1 months (interquartile range, 6.3-12.8 month). NoV was detected in 27.4% (34/124) of children with acute gastroenteritis using the NoV real-time RT-PCR assays (25). All the 34 NoV positive samples were NoV GII infections; no NoV GI genotypes were detected. Phylogenetic analysis of these 34 NoV sequences clearly showed two distinct NoV genotypes (GII.3 and GII.4) and GII.4 belonged to 2006b Minerva variant (Table 2 and Figure 1).
Table 2. Association between NoV GII.3 and GII.4 Infections and Secretor Genotypes.
| Secretor genotype | No. | GII.3* | GII.4# | GII.3 +GII.4† | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| (+) | % | P | (+) | % | P | Pos (%) | P | Odds Ratio (95% CI) | ||
| SeSe(AA) | 60 | 6 | 10.0 | 0.668 | 9 | 15.0 | 0.275 | 15(25.0) | 0.1 | 3.3 (0.7- 16.0) |
| Sese(AT) | 42 | 9 | 21.4 | 0.144 | 8 | 19.0 | 0.147 | 17(40.5) | 0.01 | 6.8(1.4-33.0) |
| sese(TT) | 22 | 1 | 4.6 | Ref. | 1 | 4.6 | Ref. | 2(9.1) | Ref. | 1.0 |
|
| ||||||||||
| Total | 124 | 16 | 12.9 | 18 | 14.5 | 34(27.4) | ||||
When compared SeSe + Sese with sese, the two-tailed Fisher exact test was used. Unadjusted crude odds ratio was estimated.
P = 0.3 for GII.3
P = 0.193 for GII.4
P = 0.03 for GII.3 + GII.4
Figure 1.
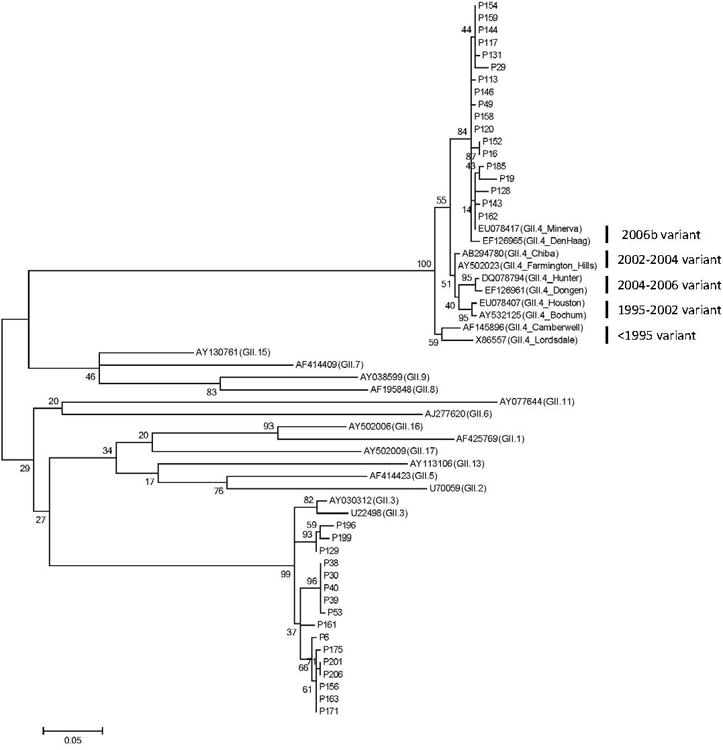
Phylogenetic tree of partial capsid nucleotide sequences [(288 bp, nt 5085-5372 of GII.4 Lordsdale (X86557)] spanning the 3′-end of region B and 5′-end of region C from 34 fecal samples in children with NoV infection and NoV GII reference sequences. Dendrograms were generated using the Maximum-likelihood method, and the statistical analysis of the phylogenies was performed by bootstrap test with 1,000 replicates. The numbers in the branches indicate the bootstrap values. Sequence names beginning with p represent sequences from diarrheal patients.
Serum IgG Antibodies against GII.3 and GII.4
GII.3 and GII.4 VLPs were used to detect GII.3 and GII.4 specific IgG in serum samples by ELISAs separately. Serial two-fold dilutions (2000 ng/ml to 15.6 ng/ml) of a purified IgG standard with known concentration were used as the standard to quantify NoV specific IgG levels. Of the 122 available serum specimens, the median of anti-GII.4 serum IgG was 112 μg/ml in the age group <6 months, 144 μg/ml in the age group 6-12 months, and 168 μg/ml in the age group >12 months. There was no significant difference (P > 0.05) between the ranks in the three age groups using the Mann-Whitney two-tailed test. The median of anti-GII.3 GMTs serum IgG was 100 μg/ml in the age group <6 months, 114 μg/ml in the age group 6-12 months, and 109 μg/ml in the age group >12 months, respectively (Data not shown).
The median of NoV GII.4 specific IgG were 151 μg/ml and 152 μg/ml among homozygous and heterozygous secretor children, respectively, which were higher than the mean titer of 121 μg/ml measured among homozygous weak secretor children, but there was no significant difference (P > 0.05). Comparison of GII.3 specific IgG among homozygous secretor, heterozygous secretor, and homozygous weak secretor children showed a similar trend (Figure 2)
Figure 2.
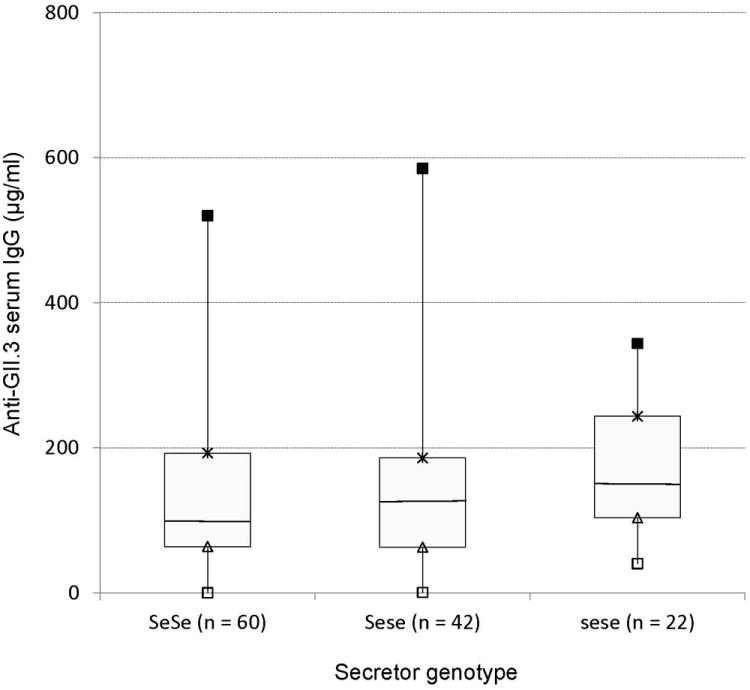
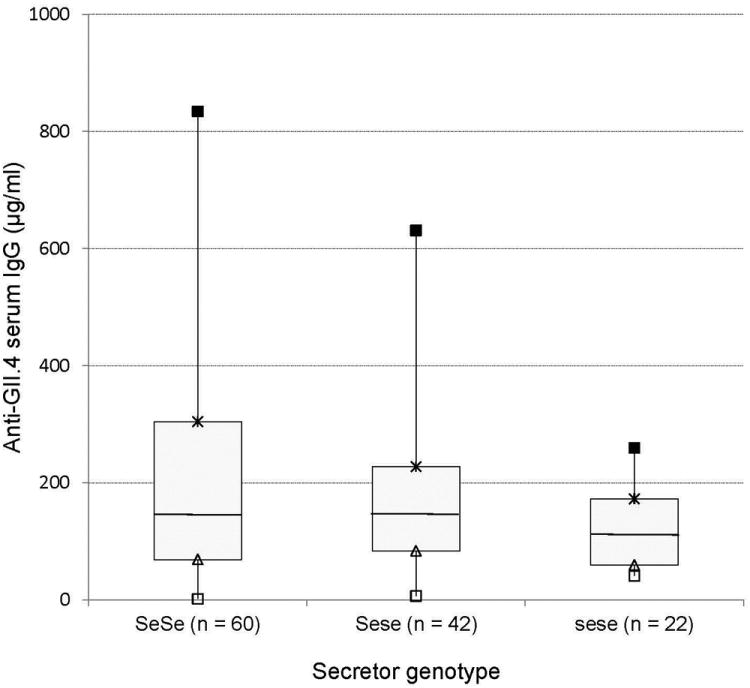
GII.3 and GII.4 specific serum IgG levels (μg/ml) in children with homozygous secretor, heterozygous secretor and weak secretor. Box plots interpreted as follows: ■=maximal IgG; × = 75th percentile; the horizontal lines in the boxes represent the medians; ▲ = 25th percentile; □ = minimal IgG.
Association between Secretor Genotypes and NoV GII.3/GII.4 Infections
Of the 124 eligible children, 60 (48.4%) were typed as homozygous secretor genotype (Se385Se385), 42 (33.9%) were heterozygous secretor genotype (Se385se385), and 22 (17.7%) were grouped as weak secretor genotype with homozygous carriers of the A385T missense mutation (se385se385) (Table 2). When GII.3 and GII.4 positive rates among homozygous and heterozygous secretor children were compared with weak secretor children separately, there were no statistically significant differences when types were analyzed individually or combined (Table 2). However, when GII.3 and GII.4 positive rates were combined, heterozygous secretor children were significantly more susceptible (P = 0.01) to NoV GII.3 and GII.4 infections than homozygous weak secretor children while comparison of homozygous secretor children to weak secretor children was not significant (P = 0.1). When homozygous and heterozygous secretor children were combined (equivalent to secretor positive phenotype), secretor positive children were at higher risk of GII.3 and GII.4 infections than weak secretor children (P = 0.03) (Table 2). Therefore, the significance was obtained only when combining GII.3 and GII.4 infectious together.
Discussion
The present study demonstrates that weak secretor Chinese children with homozygous carriers of the A385T missense mutation can be infected with GII.3 or GII.4 norovirus in pediatric clinical setting in agreement with previous studies (20,21), but the probability of infection appears much smaller than in secretor-positive children with at least one functional allele. A strong association between secretor genotypes and GII.3 and GII.4 infections was observed in this study. Children with homozygous and heterozygous secretor were significantly more susceptible to norovirus infections than individuals with weak secretor when combining GII.3 and GII.4 for analysis.
NoV seroprevalence studies using baculovirus-expressed virus-like particles have been performed in several countries (6-9, 28-30). Those early studies all used Norwalk virus (GI.1) and Mexico virus (GII.2) capsid antigens to detect NoV serological antibodies, suggesting that the prevalence of NoV antibodies followed an age-related pattern. Children appear to acquire antibodies against NoV at an early age and seroprevalence remains high at older ages; however, Norwalk virus and Mexico virus were not epidemic genotypes when the studies were performed; therefore, presence of cross-reactive antibodies against NoV epidemic strains may result in biased results. In this study, we used NoV GII.3 and GII.4 VLPs to detect homologous IgG immune response and observed an age-related increase of GII.3 and GII.4 antibodies in the 6-12 months age group compared with children < 6 months, and NoV antibodies remain at high levels in older children (>12 months) thereafter. Since all serum samples in this study were collected in the course of illness, it is likely that the antibodies against the contemporary GII.3 and GII.4 infections have not yet been mounted because NoV specific antibodies are usually detected after 5 days of infection when clinical symptoms disappear (31, 32).
Early human challenge studies (16) with Norwalk virus showed that the risk of NV infection was exclusively associated with secretor status as determined by the FUT2 gene, and nonsecretors were naturally resistant to NV infection. However, it is not well documented whether other NoV genotypes are associated with secretor status. Recent in vitro studies have found that different NoV genotypes showed different binding patterns and the worldwide dominating NoV GII.4 demonstrates the broadest histo-blood group-binding patterns (33). Furthermore, Lindesmith et al., have shown that the 2002 NoV GII.4 variant bound not only secretor-positive but also to secretor-negative saliva (34). In this study, we identified NoV GII.3 and GII.4 in both secretor-positive and weak secretor children, but with significantly higher susceptibility among secretor-positive children (31.4%, 32/102) compared to weak secreor children (9.1%, 2/22). The findings from clinical setting are agreement withprevious GII.4 human challenge study (20) and outbreak investigation (21-23, 35).
Although several single nucleotide polymorphisms including Se357, Se385, Se428, Se480, and Se739 in the FUT2 locus have been identified in different ethnic populations, Se428 is mainly reported in Europeans and Caucasians and this is a nonsense mutation that causes an early stop codon and the homozygous carriers of this mutation in the FUT2 gene are called non-secretor. In contrast to Se428, Se385 is a missense mutation at position 385(A>T) and the homozygous carriers of this mutation is called “weak secretor” and this mutation is commonly found in South East and East Asians. In this study, we investigated the association between A385T mutation and NoV infections in Chinese children in clinical setting. While NoV infection was highly likely to be detected in individuals with homozygous and heterozygous secretor genotypes, NoV GII.3 and GII.4 were also identified in homozygous carriers of the A385T mutation, respectively, suggesting that the weak secretor phenotype does not completely protect susceptible children from GII.3 and GII.4 infections but children with this mutation tend to have low risk of GII.3 and GII.4 infections compared with homozygous and heterozygous secretor genotypes.
Secretor status can be determined by either phenotyping or genotyping methods. In Chinese population, 12.0% and 88% of individuals showed secretor negative and positive using phenotyping method, respectively (35). Typed by genotyping method, homozygous Se385Se385, heterozygous Se385se385 and homozygous se385se385 were detected in 28.2%, 54.0% and 17.8% of the Chinese populations (17). The majority of previous NoV human challenge (16, 20), outbreak studies (21, 35), and pediatric research (36) determined secretor status by phenotypes (only two types: either positive or negative) from saliva and the genotyping method has been rarely used. Secretor genotyping is an important alternative method when saliva is not available and only serum is collected. Prior to this study, the genotyping method was validated using our archived serum and saliva samples collected from a previous human challenge study (16) and complete agreement was founded between the secretor phenotyping and the genotyping results of all test samples (data not shown).
In summary, one important finding of this pediatric NoV study indicates that secretor genotypes determine susceptibility to GII.4 and GII.3 infections but individuals with weak secretor are not fully protected from NoV GII.3 and GII.4 infections.
Acknowledgments
We are grateful to Dr. Robert Atmar at Baylor College of Medicine and Dr. Jacques LePendu at the Institute of Biology in Nantes, France for assistance with the secretor genotyping. We also thank Dr. Jan Vinje, Dr. Everardo Vega, and Dr. Veronica Costantini at the Centers for Disease Control for help in the production of GII.3 and GII.4 virus-like particles.
Source of Funding. This study was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Partial funding was provided by the Food Virology Collaborative grant (# 1111-2011-0494) of the National Institute of Food and Agriculture (NIFA) at the US Department of Agriculture (USDA).
Footnotes
Conflicts of Interest. The authors have no conflicts of interest or funding to disclose.
References
- 1.Payne DC, Vinje J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med. 2013;368:1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemming M, Rasanen S, Huhti L, Paloniemi M, Salminen M, Vesikari T. Major reduction of rotavirus, but not norovirus, gastroenteritis in children seen in hospital after the introduction of RotaTeq vaccine into the National Immunization Programme in Finland. European Journal of Pediatrics. 2013;172:739–746. doi: 10.1007/s00431-013-1945-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucardo F, Lindgren PE, Svensson L, Nordgren J. Low prevalence of rotavirus and high prevalence of norovirus in hospital and community wastewater after introduction of rotavirus vaccine in Nicaragua. PLoS One. 2011;6:e25962. doi: 10.1371/journal.pone.0025962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang X, Graham D, Wang K, Estes M. Norwalk virus genome cloning and characterization. Science. 1990;250:1580–1583. doi: 10.1126/science.2177224. [DOI] [PubMed] [Google Scholar]
- 5.Jiang X, Wang M, Graham DY, Estes MK. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker SP, Cubitt WD, Jiang X. Enzyme immunoassay using baculovirus-expressed human calicivirus (Mexico) for the measurement of IgG responses and determining its seroprevalence in London, UK. J Med Virol. 1995;46:194–200. doi: 10.1002/jmv.1890460305. [DOI] [PubMed] [Google Scholar]
- 7.Smit TK, Bos P, Peenze I, Jiang X, Estes MK, Steele AD. Seroepidemiological study of genogroup I and II calicivirus infections in South and southern Africa. J Med Virol. 1999;59:227–231. doi: 10.1002/(sici)1096-9071(199910)59:2<227::aid-jmv17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Dimitrov DH, Dashti SA, Ball JM, et al. Prevalence of antibodies to human caliciviruses (HuCVs) in Kuwait established by ELISA using baculovirus-expressed capsid antigens representing two genogroups of HuCVs. J Med Virol. 1997;51:115–118. [PubMed] [Google Scholar]
- 9.Nurminen K, Blazevic V, Huhti L, et al. Prevalence of norovirus GII-4 antibodies in Finnish children. J Med Virol. 2011;83:525–531. doi: 10.1002/jmv.21990. [DOI] [PubMed] [Google Scholar]
- 10.Atmar RL, Bernstein DI, Harro CD, et al. Norovirus vaccine against experimental human Norwalk Virus illness. N Engl J Med. 2011;365:2178–2187. doi: 10.1056/NEJMoa1101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Jin M, Xie HP, Duan ZJ, et al. Emergence of the GII4/2006b variant and recombinant noroviruses in China. J Med Virol. 2008;80:1997–2004. doi: 10.1002/jmv.21308. [DOI] [PubMed] [Google Scholar]
- 13.Iritani N, Kaida A, Abe N, et al. Increase of GII.2 norovirus infections during the 2009-2010 season in Osaka City, Japan. J Med Virol. 2012;84:517–525. doi: 10.1002/jmv.23211. [DOI] [PubMed] [Google Scholar]
- 14.Bruggink LD, Marshall JA. Molecular and epidemiological features of GIIb norovirus outbreaks in Victoria, Australia, 2002-2005. J Med Virol. 2009;81:1652–1660. doi: 10.1002/jmv.21582. [DOI] [PubMed] [Google Scholar]
- 15.Parrino TA, Schreiber DS, Trier JS, Kapikian AZ, Blacklow NR. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N Engl J Med. 1977;297:86–89. doi: 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- 16.Lindesmith L, Moe C, Marionneau S, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 17.Chang JG, Yang TY, Liu TC, et al. Molecular analysis of secretor type alpha(1,2)-fucosyltransferase gene mutations in the Chinese and Thai populations. Transfusion. 1999;39:1013–1017. doi: 10.1046/j.1537-2995.1999.39091013.x. [DOI] [PubMed] [Google Scholar]
- 18.Yip SP, Lai SK, Wong ML. Systematic sequence analysis of the human fucosyltransferase 2 (FUT2) gene identifies novel sequence variations and alleles. Transfusion. 2007;47:1369–1380. doi: 10.1111/j.1537-2995.2007.01280.x. [DOI] [PubMed] [Google Scholar]
- 19.Lindesmith L, Moe C, Lependu J, Frelinger JA, Treanor J, Baric RS. Cellular and humoral immunity following Snow Mountain virus challenge. J Virol. 2005;79:2900–2909. doi: 10.1128/JVI.79.5.2900-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frenck R, Bernstein DI, Xia M, et al. Predicting susceptibility to norovirus GII.4 by use of a challenge model involving humans. J Infect Dis. 2012;206:1386–1393. doi: 10.1093/infdis/jis514. [DOI] [PubMed] [Google Scholar]
- 21.Jin M, He Y, Li H, et al. Two gastroenteritis outbreaks caused by GII Noroviruses: host susceptibility and HBGA phenotypes. PLoS One. 2013;8:e58605. doi: 10.1371/journal.pone.0058605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockx BH, Vennema H, Hoebe CJ, Duizer E, Koopmans MP. Association of histo-blood group antigens and susceptibility to norovirus infections. J Infect Dis. 2005;191:749–754. doi: 10.1086/427779. [DOI] [PubMed] [Google Scholar]
- 23.Nordgren J, Kindberg E, Lindgren PE, Matussek A, Svensson L. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerg Infect Dis. 2010;16:81–87. doi: 10.3201/eid1601.090633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P, Hsiao HM, Jaykus LA, Moe C. Quantification of Norwalk virus inocula: Comparison of endpoint titration and real-time reverse transcription-PCR methods. J Med Virol. 2010;82:1612–1616. doi: 10.1002/jmv.21851. [DOI] [PubMed] [Google Scholar]
- 25.Kageyama T, Kojima S, Shinohara M, et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dey SK, Nguyen TA, Phan TG, et al. Molecular and epidemiological trend of norovirus associated gastroenteritis in Dhaka City, Bangladesh. J Clin Virol. 2007;40:218–223. doi: 10.1016/j.jcv.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Hutson AM, Airaud F, LePendu J, Estes MK, Atmar RL. Norwalk virus infection associates with secretor status genotyped from sera. J Med Virol. 2005;77:116–120. doi: 10.1002/jmv.20423. [DOI] [PubMed] [Google Scholar]
- 28.Smit T, Steele A, Peenze I, Jiang X, Estes M. Study of Norwalk virus and Mexico virus infections at Ga-Rankuwa Hospital, Ga-Rankuwa, South Africa. J Clin Microbiol. 1997;35:2381–2385. doi: 10.1128/jcm.35.9.2381-2385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Ryan ML, Vial PA, Mamani N, et al. Seroprevalence of Norwalk virus and Mexico virus in Chilean individuals: assessment of independent risk factors for antibody acquisition. Clin Infect Dis. 1998;27:789–795. doi: 10.1086/514949. [DOI] [PubMed] [Google Scholar]
- 30.Jing Y, Qian Y, Huo Y, Wang LP, Jiang X. Seroprevalence against Norwalk-like human caliciviruses in beijing, China. J Med Virol. 2000;60:97–101. doi: 10.1002/(sici)1096-9071(200001)60:1<97::aid-jmv16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 31.Iritani N, Seto T, Hattori H, et al. Humoral immune responses against norovirus infections of children. J Med Virol. 2007;79:1187–1193. doi: 10.1002/jmv.20897. [DOI] [PubMed] [Google Scholar]
- 32.Erdman DD, Gary GW, Anderson LJ. Serum immunoglobulin A response to Norwalk virus infection. J Clin Microbiol. 1989;27:1417–1418. doi: 10.1128/jcm.27.6.1417-1418.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang P, Farkas T, Zhong W, et al. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classfication of two major binding groups among multiple binding patterns. J Virol. 2005;79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindesmith LC, Donaldson EF, Lobue AD, et al. Mechanism of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5:269–290. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan M, Jin M, Xie H, Duan Z, Jiang X, Fang Z. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J Med Virol. 2008;80:1296–1301. doi: 10.1002/jmv.21200. [DOI] [PubMed] [Google Scholar]
- 36.Nordgren J, Nitiema LW, Ouermi D, Simpore J, Svensson L. Host genetic factors affect susceptibility to norovirus infections in Burkina Faso. PLoS One. 2013;8:e69557. doi: 10.1371/journal.pone.0069557. [DOI] [PMC free article] [PubMed] [Google Scholar]


