Abstract
Purpose of review
HIV genetic diversity poses major challenges for the prevention, control, and cure of infection. Characterizing the diversity and evolution of HIV populations within the host provides insights into the mechanisms of HIV persistence during antiretroviral therapy (ART). This review describes the HIV diversity within patients, how it is affected by suppressive ART, and makes a case for early treatment after HIV infection.
Recent findings
HIV evolution is effectively halted by ART. However, cells that were infected prior to initiating therapy can proliferate to very high numbers both before and during treatment. Such clonal expansions result in the persistence of integrated proviruses despite therapy. These expanding proviruses have been shown to be a source for residual viremia during ART, and they may be a source for viral rebound after interrupting ART.
Summary
Plasma HIV RNA shows no evidence for evolution during ART, suggesting that HIV persistence is not driven by low-level, ongoing replication. The emergence of identical viral sequences observed in both HIV RNA and DNA is likely due to proliferation of infected cells. Early treatment restricts the viral population and reduces the number of variants that must be targeted for future therapeutic strategies.
Keywords: antiretroviral therapy, HIV diversity, HIV early infection, HIV genetics, HIV replication
INTRODUCTION
An important feature of the HIV is its vast genetic diversity and rapid evolution that influence the pathogenesis, transmission, diagnosis, and clinical management of the infection [1]. Such a high genetic variation is due to the lack of a proofreading mechanism by reverse transcriptase, resulting in a mutation rate of 10–5 changes per replicative cycle. The virus also has a large population size and high capacity to recombine, adding to its genetic complexity [2,3]. After transmission, HIV evolves rapidly from the transmitted/founder virus within an infected individual. The immune response contributes to the intrahost diversification, selecting for both cell-mediated and antibody-mediated escape mutants [4,5]. Moreover, different cellular milieu or changes in the population of susceptible cells can drive diversification within a particular compartment, as well as change cell tropism and coreceptor usage [6]. Other host factors, such as apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC), play a role in HIV diversity by introducing G to A mutations. The interplay of all these factors explains why viral sequences within an individual can differ by 10% or more [7]. The major implications of viral diversity are the rapid selection for drug-resistant mutations by suboptimal antiretroviral treatment (ART) and the difficulties it imposes on the development of vaccines and curative strategies. In this review article, we will address how viral diversity and evolution are affected by ART and why early treatment may be a prerequisite to achieving a functional cure for HIV.
INTRAPATIENT HIV DIVERSITY
HIV populations in acute infection have been described as being mostly homogeneous, resulting from the productive transmission of a single viral variant or, less frequently, heterogeneous, resulting from the transmission of multiple variants [8,9]. After transmission, viral diversity typically accumulates in early infection, strongly influenced by pressures imposed by our immune systems, and then plateaus in chronic infection [8–17,18■]. Kearney et al. [9] showed that transmitted HIV variants carried cytotoxic T lymphocyte (CTL) escape mutations that were selected along the chain of transmission. These mutations persisted for years in the recipients, even at CTL epitopes that were not recognized by the new host’s major histocompatibility complexes. The persistence of these CTL mutations was concurrent with the selection of new escape mutations in epitopes that were recognized by the recipient’s own major histocompatibility complexes, contributing the accumulation of HIV diversity within patients and in the population as a whole [9]. Maldarelli et al. [18■] subsequently showed that CTL escape mutations continued to be selected during chronic infection, despite plateaus in the overall diversity of the virus populations. Rare HIV-infected individuals, termed elite controllers, do not have the typical pattern of HIV replication and diversification. Plasma HIV RNA levels in these patients are below the limit of detection by standard clinical assay seven without therapy. Mens et al. [19] analyzed the viral genetics of longitudinally sampled plasma from elite controllers and showed that HIV continued to evolve throughout the course of infection but that the immune systems of the elite controllers are likely more successful in limiting the number of cells that become infected. Such controllers are of special interest because they may reveal specific mechanisms of immune control without ART and, thus, be of great importance for the development of future vaccine and curative strategies.
The pool of HIV-susceptible cells in vivo is diverse and includes T cells, macrophages, dendritic cells, and other cell types. Long-lived, resting CD4+ T cells constitute what has been defined as the latent reservoir for HIV and area of significant target of eradication strategies. A molecular characterization of noninduced proviruses in resting CD4+ T cells revealed that most proviruses were rendered defective due to mutations introduced during reverse transcription or by APOBEC3G (A3G) [20■■,21,22]. A3G is a host enzyme with cytidine deaminase activity that results in the alteration of the nucleotide sequence from cytidine-to-uridine at the RNA level in the single-stranded negative strand and, therefore, become fixed in the viral DNA as guanosine-to-adenosine mutations in the plus strand. However, the complementary DNA integration can take place despite these mutations, resulting in the accumulation of hypermutated proviruses within the pool-infected cells. A recent analysis of the gag region from noninduced proviruses showed that about one out of three sequences contained A3G-mediated G-to-A changes [20■■].
HIV COMPARTMENTALIZATION
Understanding the distribution and persistence of infected cells across tissues is critical to characterizing the sources of viremia before, during, and after therapy. The term compartmentalization was introduced to indicate tissues or cell types wherein viral replication is ongoing, but viral gene flow (in and out) is restricted because of anatomical barriers [23]. Extensive compartmentalization in the central nervous system was reported and found to be strongly associated with the development of HIV-associated dementia [24]. However, studies of compartmentalization of HIV populations in gut-associated lymphoid tissues (GALT) have yielded contrasting results. Van Marle et al. [25] analyzed sequences from the esophagus, stomach, duodenum, and colorectum and reported compartmentalized viral replication in the gut, suggesting a role of the gut milieu in shaping HIV diversity, but Imamichi et al. [26■] characterized sequences from blood, ileum, and colon and found variants to be indistinguishable from those in the plasma. Such contrasting results may be explained by sampling bias, inadequate phylogenetic signal of the genomic regions analyzed, or varying levels of expansion of infected cells across tissues. Multiple reports have suggested HIV compartmentalization between the genital tract and the blood [27,28]. However, two studies from Bull et al. [29,30] revealed that the tissue-specific sequences detected had very low genetic diversity, suggesting that expanded infected cells could lead to local bursts of expression, which may cause a bias for statistical analyses of compartmentalization. Studies [31,32] have also investigated whether or not tissues are sanctuary sites for ongoing HIV replication during ART. Evering et al. [33■] analyzed viral diversity in longitudinal GALT samples from three patients treated during an early infection who were also categorized by having high, intermediate, or low levels of immune activation. Their results showed that the diversity did not increase after 1 year of therapy and there was no divergence from pretherapy sequences, suggesting the absence of de-novo rounds of replication in this compartment even in patients with higher levels of immune activation [33■]. These findings were confirmed by studies from Josefsson et al. [34■■] and (Simonetti et al., presented at Conference on Retroviruses and Opportunistic Infections; 2014) who found few genetic changes over time and no compartmentalization between peripheral blood mononuclear cells and GALT samples after prolonged treatment (Simonetti et al., Conference on Retroviruses and Opportunistic Infections; 2014) suggesting that GALT is not a sanctuary site for ongoing replication during ART.
DYNAMICS OF VIRAL DECAY ON ANTIRETROVIRAL THERAPY
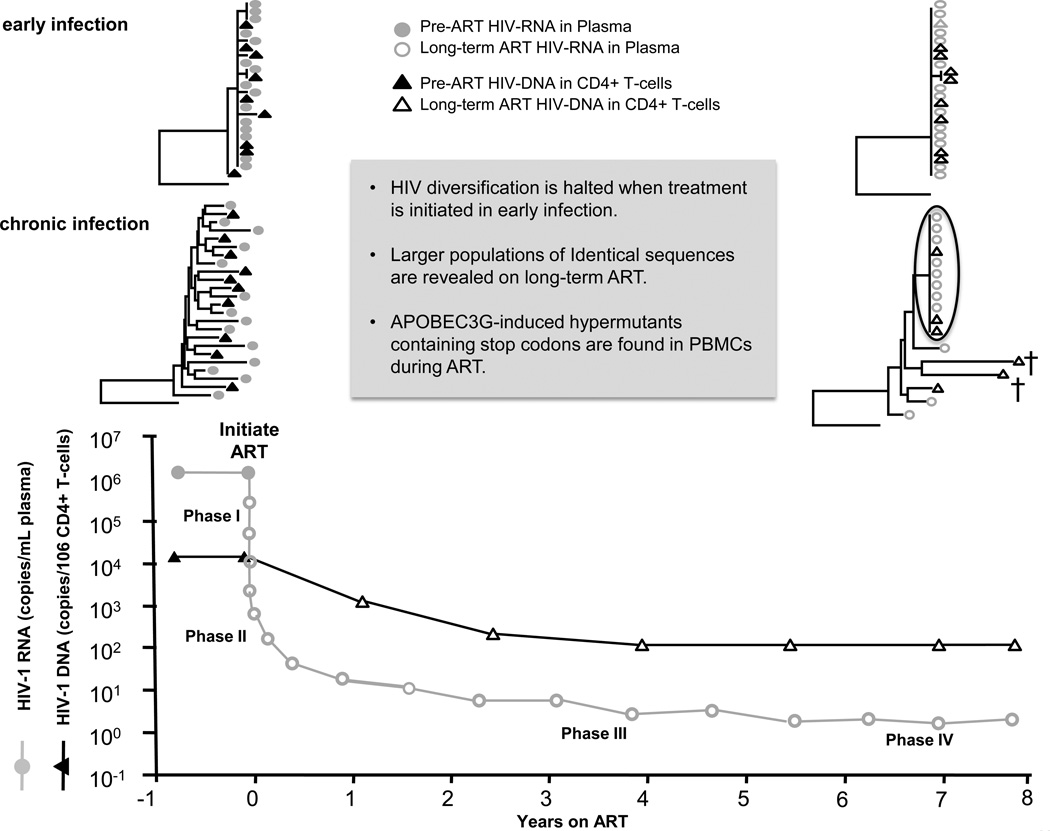
The dynamics of plasma HIV RNA decay after initiating ART can be divided into four phases [35–37]. The phases of decay are shown in Fig. 1 [34■■,35–37, 38■,39,40■■,41]. The first phase reflects the rapid clearance of about 90% of productively infected cells with half-lives of only 1–2 days. These are the short-lived, immune cells that are infected and killed daily en masse without treatment and, ultimately, result in the immune deficiency that is characteristic of HIV infection. The first phase is followed by a second phase, a more gradual decline resulting from the clearance of cell populations with half-lives of about 2–3 weeks, may be consisting of different subsets of immune cells, such as central, effector, or transitional memory CD4+ T cells. A study by Palmer et al. [35] in 2008 described a third phase of decay that consisted of longer lived cells, perhaps macrophages or monocytes, with half-lives of 6–44 months, and, finally, a fourth phase having a slope that was not significantly different from zero, likely resulting from the persistence of infected resting CD4+ T cells [35–37]. The plateau in the fourth phase suggests that ART effectively inhibits HIV infection of new cells and that the source of persistent viremia is either virus-expressing long-lived cells or virus expression from previously latently infected cells upon stochastic activation. A more recent study by Besson et al. [38■] investigated the decay of HIV DNA on ART. Their results show that HIV infected cells decline initially on ART but then achieve a steady state by maintaining a balance between infected long-lived cells, expanding cells, and dying cells.
FIGURE 1.
Change in HIV plasma RNA and peripheral blood mononuclear cell DNA over 8 years of treatment with ART. Levels of plasma HIV RNA decline in four phases; the last phase has a slope close to zero [35–37]. HIV DNA declines initially on ART, but then achieves a steady state at higher levels than HIV RNA [38■]. When ART is initiated early after infection, diversity is halted and only one or a few variants remain after a long-term treatment [34■■,39]. When ART is initiated early after infection, diversity is halted and only one or a few variants remain after long-term treatment [34■■,39,40■■,41]. ART, antiretroviral therapy; PBMC, peripheral blood mononuclear cells.
EFFECT OF ANTIRETROVIRAL THERAPY ON HIV DIVERSITY
Recent studies have assessed the effect of ART on HIV genetic diversity and evolution when treatment is initiated in early or chronic infection. Their findings are summarized in Fig. 1. Josefsson et al. [34■■] and Kearney et al. [40■■] revealed that patients who were treated early had low viral diversity in both pretherapy plasma and plasma or cells isolated after 4–12 years on therapy. Additionally, patients treated in the earliest phases of infection achieved a reduced burden of infected cells during ART, indicating a smaller reservoir for HIV [42■–44■]. Studies by Josefsson et al. [34■■], Bailey et al. [39], Kearney et al. [40■■], and Wagner et al. [41] also investigated the evolution of HIV during ART in patients who initiated treatment during chronic infection. Each of these studies [34■■,39,40■■,41] showed that HIV evolution is effectively halted during ART and that long-term therapy reveals an underlying population of cells carrying, and sometimes expressing, identical viral variants. These findings suggest that persistent viremia during ART is not derived from ongoing, full cycles of replication but rather from the expression of proliferating cells that were infected prior to initiating treatment. This conclusion is consistent with studies by Dinoso et al. [45], Gandhi et al. [46], and McMahon et al. [47] that failed to show a decrease in the level of persistent viremia when additional antiretroviral drugs were added to suppressive regimens. Studies by Kearney et al. [40■■] and Joos et al. [48] also showed that rebound viremia after long-term cART consisted of homogeneous populations, suggesting they may originate from the expression of the identical sequences that are present during suppression. Determining the sources for viral persistence during cART and during rebound is essential for designing strategies to eradicate HIV infection.
PROLIFERATION OF INFECTED CELLS REVEALED BY ANTIRETROVIRAL THERAPY
Homeostatic proliferation and long-term survival of infected cells have been proposed to have a leading role in HIV persistence during ART [49]. Identification of clonal sequences from clearly defective or A3G-hypermutated proviruses strongly suggested that the only way such variants could dominate and persist would be through expansion of a progenitor cell seeded by a single infection event [34■■] (Simonetti et al., Conference on Retroviruses and Opportunistic Infections; 2014). Two very recent studies using HIV integration site analysis on patients during ART described unique methods to identify the expansion of infected cells. Both Maldarelli et al. [50■■] and Wagner et al. [51■■] showed clonal expansion by identifying enriched integration sites, especially those near or within genes involved in cell cycle regulation (e.g., BACH2 and MKL2), suggesting that the integration site in certain loci provides the infected cell with a phenotype more prone to survival and expansion [50■■,51■■]. In one patient, Maldarelli et al. [50■■] found a highly expanded cell clone responsible for an HIV variant that dominated the residual viremia during ART. This is the strongest evidence that persistent viremia during ART is due to the expression of HIV from long-lived infected cell populations and not from ongoing, low-level viral replication. Further investigations are needed to assess whether clonally expanded variants in plasma are replication-competent and, thus, capable of resulting in rebound viremia, and representing a true reservoir for HIV that must be targeted for eradication. To investigate the extent of clonal expansion of infected cells during long-term ART, Kearney et al. (Conference on Retroviruses and Opportunistic Infections; 2014) estimated the number of CD4+ T cells in infected individuals that carried clonal proviruses by multiplying the fraction of identical sequences (normalized for HIV DNA copy number) by the total number of CD4+ T cells. The fraction of identical sequences in peripheral blood mononuclear cells during long-term ART ranged from 7 to 55%. Estimates for the total number of CD4+ T cells in an individual containing clonal HIV sequences ranged from 1×109 to 7×1010. These data indicate that some infected CD4+ T cells can massively expand to constitute billions of infected cells during ART.
A CASE FOR EARLY ANTIRETROVIRAL THERAPY
Early initiation of ART has been associated with lower levels of HIV proviruses persisting during therapy [34■■,39,40■■,41,52,53]. Fewer persisting infected cells may limit the viral reservoir that can rebound after ART is interrupted. Additionally, early ART results in freezing of the virus population diversity at an early stage when it is still relatively homogeneous and prior to the emergence of new immune-escape variants [40■■]. Early ART may also prevent the loss of HIV-specific immune responses to the existing variants [53,54]. Together, the reduced size and diversity of the HIV reservoir that occurs when ART is initiated early after infection may favor immune-mediated control, as is suggested by posttherapy control of virus replication in patients from the Visconti cohort [55■]. The recent finding that specific HIV integration sites can lead to clonal cell expansion [50■■] as a mechanism of infected cell persistence suggests that there may be a time-limited opportunity, through early initiation of ART, to prevent HIV integrations in key locations in the human genome that influence cell growth and survival.
CONCLUSION
Killing or permanently inactivating infected cells that persist during ART and carry proviruses that are capable of producing replication competent virions is necessary to achieve long-term remission off ART. Unfortunately, it is now clear that the reservoir for HIV is established extremely early after transmission [42■] and that even an extensive reduction of the reservoir to below the limit of detection of our current assays is not sufficient to achieve a long-term remission [56■]. However, treating patients during primary HIV infection does reduce the size and diversity of the reservoir [44■] and may minimize the number of cells that are capable of proliferation and harbor intact proviruses. Future studies will address the distribution of integration sites in acute infection and whether clonal expansion is affected by early treatment. There is sufficient data showing that treating early halts the diversification of HIV resulting in more monomorphic populations. This restricted pool of variants will allow more focused targeting by therapeutic vaccines and other immune approaches, such as those inducing CTL responses to eliminate infected cells after reactivation with latency reversing agents (Kai Deng et al., Conference on Retroviruses and Opportunistic Infections; 2014).
KEY POINTS.
Rapid HIV evolution results in diverse viral populations without treatment.
Optimal ART prevents further HIV evolution, suggesting that replication is effectively halted during treatment.
Some long-lived, infected cells can proliferate and express virions, making them a source of persistent viremia during ART.
Persistent cells harboring replication-competent proviruses are the main obstacles for HIV eradication.
Initiating treatment during early infection restricts HIV diversity and limits the size of the reservoir, reducing the pool of variants that must be targeted for eradication.
Acknowledgements
We thank Frank Maldarelli, Guillaume Besson, and John Mellors for sharing data used to generate the figure and we thank Connie Kinna, Sue Toms, and Valerie Turnquist for administrative support.
Financial support and sponsorship
The laboratories of F.R.S. and M.F.K. are funded by intramural NIH funding and NIH Bench-to-Bedside grants.
Footnotes
Conflicts of interest
F.R.S. and M.F.K. have no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Hemelaar J. Implications of HIV diversity for the HIV-1 pandemic. J Infect. 2013;66:391–400. doi: 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2:a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin J, Swanstrom R. HIV pathogenesis: dynamics and genetics of viral populations and infected cells. Cold Spring Harb Perspect Med. 2013;3:a012526. doi: 10.1101/cshperspect.a012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips RE, Rowland-Jones S, Nixon DF, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 5.Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 6.Swanstrom R, Coffin J. HIV-1 pathogenesis: the virus. Cold Spring Harb Perspect Med. 2012;2:a007443. doi: 10.1101/cshperspect.a007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korber B, Gaschen B, Yusim K, et al. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 8.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearney M, Maldarelli F, Shao W, et al. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol. 2009;83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart EL, Sheppard HW, Walker BD, et al. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuta Y, Bergstrom T, Norkrans G, Horal P. HIV type 1 V3 sequence diversity in contact-traced Swedish couples at the time of sexual transmission. AIDS Res Hum Retroviruses. 1994;10:1187–1189. doi: 10.1089/aid.1994.10.1187. [DOI] [PubMed] [Google Scholar]
- 12.McNearney T, Hornickova Z, Kloster B, et al. Evolution of sequence divergence among human immunodeficiency virus type 1 isolates derived from a blood donor and a recipient. Pediatr Res. 1993;33:36–42. doi: 10.1203/00006450-199301000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfs TF, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 15.Ritola K, Pilcher CD, Fiscus SA, et al. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J Virol. 2004;78:11208–11218. doi: 10.1128/JVI.78.20.11208-11218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poss M, Rodrigo AG, Gosink JJ, et al. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol. 1998;72:8240–8251. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salazar-Gonzalez JF, Bailes E, Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maldarelli F, Kearney M, Palmer S, et al. HIV populations are large and accumulate high genetic diversity in nonlinear fashion. J Virol. 2013;87:10313–10323. doi: 10.1128/JVI.01225-12. The authors showed that CTL escape mutations continued to be selected over years of chronic infection with HIV despite plateaus in the overall diversity of the virus populations.
- 19.Mens H, Kearney M, Wiegand A, et al. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J Virol. 2010;84:12971–12981. doi: 10.1128/JVI.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. The authors showed that about 88% of noninduced HIV proviruses are defective.
- 21.Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu X, Yu Y, Liu B, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 23.Nickle DC, Jensen MA, Shriner D, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritola K, Robertson K, Fiscus SA, et al. Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J Virol. 2005;79:10830–10834. doi: 10.1128/JVI.79.16.10830-10834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Marle G, Gill MJ, Kolodka D, et al. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology. 2007;4:87. doi: 10.1186/1742-4690-4-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Imamichi H, Degray G, Dewar RL, et al. Lack of compartmentalization of HIV-1 quasispecies between the gut and peripheral blood compartments. J Infect Dis. 2011;204:309–314. doi: 10.1093/infdis/jir259. The authors characterized HIV sequences from blood, ileum, and colon and found variants to be indistinguishable from those in the plasma.
- 27.Chomont N, Hocini H, Gresenguet G, et al. Early archives of genetically-restricted proviral DNA in the female genital tract after heterosexual transmission of HIV-1. AIDS. 2007;21:153–162. doi: 10.1097/QAD.0b013e328011f94b. [DOI] [PubMed] [Google Scholar]
- 28.Chaudhary S, Noel RJ, Rodriguez N, et al. Correlation between CD4 T cell counts and virus compartmentalization in genital and systemic compartments of HIV-infected females. Virology. 2011;417:320–326. doi: 10.1016/j.virol.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bull M, Learn G, Genowati I, et al. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diversity variants not distinct viral populations. PLoS One. 2009;4:e7122. doi: 10.1371/journal.pone.0007122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bull ME, Learn GH, McElhone S, et al. Monotypic human immunodeficiency virus type 1 genotypes across the uterine cervix and in blood suggest proliferation of cells with provirus. J Virol. 2009;83:6020–6028. doi: 10.1128/JVI.02664-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yukl SA, Gianella S, Sinclair E, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yukl SA, Shergill AK, McQuaid K, et al. Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. AIDS. 2010;24:2451–2460. doi: 10.1097/QAD.0b013e32833ef7bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evering TH, Mehandru S, Racz P, et al. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 2012;8:e1002506. doi: 10.1371/journal.ppat.1002506. The authors showed that HIV diversity in the gut did not increase after 1 year of therapy and there was no divergence from pretherapy sequences, suggesting the absence of de-novo rounds of replication even in patients with higher levels of immune activation.
- 34. Josefsson L, von Stockenstrom S, Faria NR, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A. 2013;110:E4987–E4996. doi: 10.1073/pnas.1308313110. The authors and [40■■] revealed that patients who were treated early had low viral diversity in both pretherapy plasma and plasma or cells isolated after 4–12 years on therapy, indicating a lack of HIV replication during therapy.
- 35.Palmer S, Maldarelli F, Wiegand A, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldarelli F, Palmer S, King MS, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perelson AS, Essunger P, Cao Y, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 38. Besson GJ, Lalama CM, Bosch RJ, et al. Hiv-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis. 2014 doi: 10.1093/cid/ciu585. [Epub ahead of print] The authors investigated the decay of HIV DNA on ART and found a steady state was achieved after 4 years of therapy, suggesting a very stable reservoir for HIV.
- 39.Bailey JR, Sedaghat AR, Kieffer T, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kearney MF, Spindler J, Shao W, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10:e1004010. doi: 10.1371/journal.ppat.1004010. The authors and [34■■] revealed that patients who were treated early had low viral diversity in both pretherapy plasma and plasma or cells isolated after 4–12 years on therapy, indicating a lack of HIV replication during therapy.
- 41.Wagner TA, McKernan JL, Tobin NH, et al. An increasing proportion of monotypic hiv-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol. 2013;87:1770–1778. doi: 10.1128/JVI.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whitney JB, Hill AL, Sanisetty S, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–77. doi: 10.1038/nature13594. This study shows that treatment in the earliest phases of infection achieved a reduced burden of infected cells during ART indicating a smaller reservoir for HIV.
- 43. Hocqueloux L, Avettand-Fenoel V, Jacquot S, et al. Long-term antiretroviral therapy initiated during primary HIV-1 infection is key to achieving both low HIV reservoirs and normal T cell counts. J Antimicrob Chemother. 2013;68:1169–1178. doi: 10.1093/jac/dks533. This study shows that patients treated in the earliest phases of infection achieved a reduced burden of infected cells during ART indicating a smaller reservoir for HIV.
- 44. Buzon MJ, Martin-Gayo E, Pereyra F, et al. Long-term antiretroviral treatment initiated in primary HIV-1 infection affects the size, composition and decay kinetics of the reservoir of HIV-1 infected CD4 T cells. J Virol. 2014;88:10056–10065. doi: 10.1128/JVI.01046-14. This study shows that patients treated in the earliest phases of infection achieved a reduced burden of infected cells during ART indicating a smaller reservoir for HIV.
- 45.Dinoso JB, Kim SY, Wiegand AM, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 2009;106:9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandhi RT, Zheng L, Bosch RJ, et al. AIDS Clinical Trials Group A5244 team. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 2010;7:e1000321. doi: 10.1371/journal.pmed.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMahon D, Jones J, Wiegand A, et al. Short-course raltegravir intensification does not reduce persistent low-level viremia in patients with HIV-1 suppression during receipt of combination antiretroviral therapy. Clin Infect Dis. 2010;50:912–919. doi: 10.1086/650749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joos B, Fischer M, Kuster H, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A. 2008;105:16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maldarelli F, Wu X, Su L, et al. HIV latency specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. The authors and [51■■] showed clonal expansion of HIV infected cells by identifying enriched integration sites, especially those near or within genes involved in cell cycle regulation (e.g.,BACH2 and MKL2), suggesting that the integration site in certain loci provides the infected cell with a phenotype more prone towards survival and expansion.
- 51. Wagner TA, McLaughlin S, Garg K, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. The authors and [50■■], showed a clonal expansion of HIV-infected cells by identifying enriched integration sites, especially those near or within genes involved in cell cycle regulation (e.g.,BACH2 and MKL2), suggesting that the integration site in certain loci provides the infected cell with a phenotype more prone toward survival and expansion.
- 52.Ananworanich J, Puthanakit T, Suntarattiwong P, et al. HIV-NAT 194 Study Group. Reduced markers of HIV persistence and restricted HIV-specific immune responses after early antiretroviral therapy in children. AIDS. 2014;28:1015–1020. doi: 10.1097/QAD.0000000000000178. [DOI] [PubMed] [Google Scholar]
- 53.Ananworanich J, Schuetz A, Vandergeeten C, et al. Impact of multitargeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One. 2012;7:e33948. doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cellerai C, Harari A, Stauss H, et al. Early and prolonged antiretroviral therapy is associated with an HIV-1-specific T-cell profile comparable to that of long-term nonprogressors. PLoS One. 2011;6:e18164. doi: 10.1371/journal.pone.0018164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saez-Cirion A, Bacchus C, Hocqueloux L, et al. ANRS VISCONTI Study Group. Posttreatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI study. PLoS Pathog. 2013;9:e1003211. doi: 10.1371/journal.ppat.1003211. The authors show that immune-mediated control after ART is achieved by the posttherapy controllers from the VISCONTI cohort.
- 56. Henrich TJ, Hanhauser E, Marty FM, et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161:319–327. doi: 10.7326/M14-1027. The authors showed that even an extensive reduction of the reservoir to below the limit of detection of our current assays is not sufficient to achieve a long-term remission.