Abstract
Gene duplications allow evolution of genes with new functions. Here, we describe the innovation-amplification-divergence (IAD) model in which the new function appears before duplication and functionally distinct new genes evolve under continuous selection. One example fitting this model is a preexisting parental gene in Salmonella enterica that has low levels of two distinct activities. This gene is amplified to a high copy number, and the amplified gene copies accumulate mutations that provide enzymatic specialization of different copies and faster growth. Selection maintains the initial amplification and beneficial mutant alleles but is relaxed for other less improved gene copies, allowing their loss. This rapid process, completed in fewer than 3000 generations, shows the efficacy of the IAD model and allows the study of gene evolution in real time.
The origin of new genetic functions poses a fundamental biological question. In many bacteria, most new genes are acquired by horizontal gene transfer (HGT) from related organisms (1), but new genes with novel functions can also evolve from extra copies of duplicated genes in both bacteria and eukaryotes (2). New genes can evolve from a redundant copy of a duplicated parental gene, by removing one copy from selection. This extra copy is then able to acquire one or more mutations providing a new beneficial function. At this point, natural selection would maintain the duplication and drive further evolution. However, this model requires that the extra gene be maintained without selection long enough for rare improvements to occur; because tandem duplications are generally very unstable and short-lived, they are unlikely to remain long enough to acquire mutations (3, 4). Furthermore, duplications typically have fitness costs (3, 4), and deleterious mutations outnumber beneficial mutations, making inactivation of the gene (pseudo-gene formation) the most likely fate for any redundant gene copies.
We propose the innovation-amplification-divergence (IAD) model (Fig. 1A), which allows the evolution of new genes to be completed under continuous selection that favors maintenance of the functional duplicate copies and divergence of the extra copy from the parental allele (5). The IAD model proposes that the ancestral gene has a weak secondary activity (innovation) (6, 7), and when a change in conditions makes this activity useful, selection favors increased gene dosage (amplification), resulting in two or more copies of the parent allele. The increased copy number provides multiple targets for beneficial mutations and buffers any negative effects a new mutation may have on the original activity. During continuous growth under conditions that select for both the original activity and the new activity, beneficial mutations will accumulate (divergence) in the copies. Any improved copy can be further amplified, whereas less functional copies, including the parental gene, can be lost. Ultimately, this results in a gene duplication in which one gene copy encodes the parent activity and another copy provides an improved, new activity.
Figure 1.
(A) The IAD model. Innovation occurs when the ancestral gene (green) encodes a protein with the main function “A” and a minor activity “b.” Amplification occurs when an environmental change makes the b activity beneficial and selection favors variants with increased b activity. Divergence may occur in any one of the amplified gene copies that acquires a beneficial mutation that increases “B” activity (blue gene copy). After a B mutation, selection for the amplified array is relaxed, and segregation occurs to leave alleles with original A activity and the evolved B activity. (B) The isomerization reaction catalyzed by HisA and TrpF. The respective substrates and products differ in which chemical group (R) is attached to the 1′-amino group of the phosphoribosylamine. (C) Structure of the T-his element (linear insert) and its location on F′128 (circle) with the relative genetic elements on the F′ as shown (transposons; IS elements; replication origins; and the tra, lac, and mhp operons).
To experimentally test the IAD model, we examined a histidine biosynthetic enzyme (HisA), and through continuous selection we created, by duplication and divergence, a new gene that catalyzes a step in tryptophan synthesis. The original HisA and TrpF enzymes both catalyze isomerization of a phosphoribosyl compound, but each acts on different substrates in the biosynthesis of the amino acids histidine and tryptophan (Fig. 1B). HisA and TrpF enzyme activities are selectable by growth in minimal media lacking histidine and tryptophan. In addition, the enzymes are structurally related and evolved from a common ancestor (8). Furthermore, Streptomyces and Mycobacteria lack TrpF but instead have one enzyme, PriA, that is a HisA ortholog and catalyzes both reactions (9).
In a strain lacking trpF, we selected a spontaneous hisA mutant of Salmonella enterica that maintained its original function (HisA) but acquired a low level of TrpF activity sufficient to support slow growth on a medium lacking histidine and tryptophan, representing the innovation of the IAD model (see table S1 for strains). Two mutations were required for this innovation: First, an internal duplication of codons 13 to 15 (dup13-15) gave a weak TrpF activity but led to a complete loss of HisA activity. A subsequent amino acid substitution [Asp10→Gly10 (D10G)] restored some of the original HisA activity (10). We also isolated two other bifunctional derivatives of hisA that had acquired TrpF activity, but we will not discuss these mutants in this paper (fig. S1, A to C) (10).
We placed this bifunctional parental gene (dup13-15, D10G) under the control of a constitutive promoter that cotranscribed a yellow fluorescent protein (yfp) gene. We also placed the T-his operon in a transposition-inactive transposable element Tn10dTet close to the lac operon on the low–copy number (about two copies per chromosome) (11) F′128 plasmid (Fig. 1C). Duplications and amplifications of this region are frequent and have low fitness cost (3), allowing experimental study of the process within a reasonable time frame. An F′ plasmid with the bifunctional gene inside T-his was introduced into a S. enterica strain with deleted hisA and trpF genes, dependent on the bifunctional gene for synthesis of both histidine and tryptophan. In the absence of both amino acids, the bifunctional gene supported a generation time of ~5.1 hours in minimal medium with doubling times of ~2.8 hours in the presence of tryptophan alone, ~2.6 hours in the presence of histidine alone, and ~1.5 hours in presence of both amino acids.
Several independent lineages of this strain evolved under continuous selection for improved growth and increased HisA and TrpF activities by serial passages in minimal glycerol medium lacking both amino acids (10).Within a few hundred generations, the growth rate increased from 5 hours per division to 1.9 to 2.5 hours, depending on the lineage. Associated with the increased growth rate, expression of the parental bifunctional gene (fig. S2 and table S2) increased stepwise (up to 20-fold) in most cultures due to amplifications of a region of the plasmid that includes the bifunctional parental gene and yfp (see fig. S3 and table S3 for structures of amplified units).
After evolution for up to 3000 generations, all lineages acquired mutations resulting in faster growth. In many of the lineages, different gene copies in the amplified array diverged by mutations that allowed enzymatic specialization (Fig. 2 and fig. S1, A to C). As predicted from the IAD model, we observed the appearance of a diverged gene copy with improved activity, relaxed selection for maintenance of the unimproved copies in the amplified array, loss of the unimproved copies, and, in some cases, reduction in the total gene copy number. (fig. S2 and table S2).
Figure 2.
Trajectory for 3000 generations of evolution of the bifunctional parental gene (dup13-15, D10G) during selection for improved TrpF and HisA activities from one main parental lineage to the numerous variants found in daughter lineages. Mutations verified by sequencing are shown below the gene symbols. Red text indicates the identification of a new mutation for that lineage after the indicated number of generations. Additional lineages are shown in fig. S1, A to C. Asterisks next to a mutation indicate the presence of more than one subpopulation, differing in which of the indicated mutations they contain. Two asterisks indicate that only a subpopulation of the cells in the culture contained the indicated gene copy. A, Ala; Q, Gln; L, Leu; S, Ser; N, Asn; V, Val; M, Met; E, Glu; I, Ile.
To test the HisA and TrpF activities of the evolved enzymes, 22 different genes from the evolved strain were individually cloned into the chromosomal cobA gene of a strain (lacking both the hisA and trpF genes) that had never been subjected to a histidine-tryptophan selection (10). This assured that the strain had only one copy of the gene to be tested, and no outside mutations contributed to activity. Strains with these single-copy evolved genes were tested for their ability to grow on a minimal medium with single amino acids (Trp or His), both, or neither. In every case, the evolved mutated gene increased the growth rate in the absence of either histidine or tryptophan or when both amino acids were absent. The evolved genes fell into three classes: (i) specialized genes with strongly improved HisA activity and loss of TrpF activity, (ii) specialized genes with strongly improved TrpF activity and loss of HisA activity, and (iii) generalist genes whose encoded enzyme showed a moderate increase in both activities (Fig. 3; Fig. 4, A to D; and table S4). In several cases, specialized mutant genes of both types (i) and (ii) were found in single bacterial clones, demonstrating that gene copies within a single amplified array had diverged to become specialized to perform either the HisA- or TrpF-specific reactions (Fig. 4, A to C). In other cases, the ancestral gene evolved into an individual gene with an improved level of both HisA and TrpF activities (Fig. 4D). In some strains an improved generalist enzyme evolved first and then duplicated with copies, subsequently diverging and becoming specialized (Fig. 4, B and C). Figure S4 shows the locations of the identified mutations on the HisA structure from Thermotoga maritima.
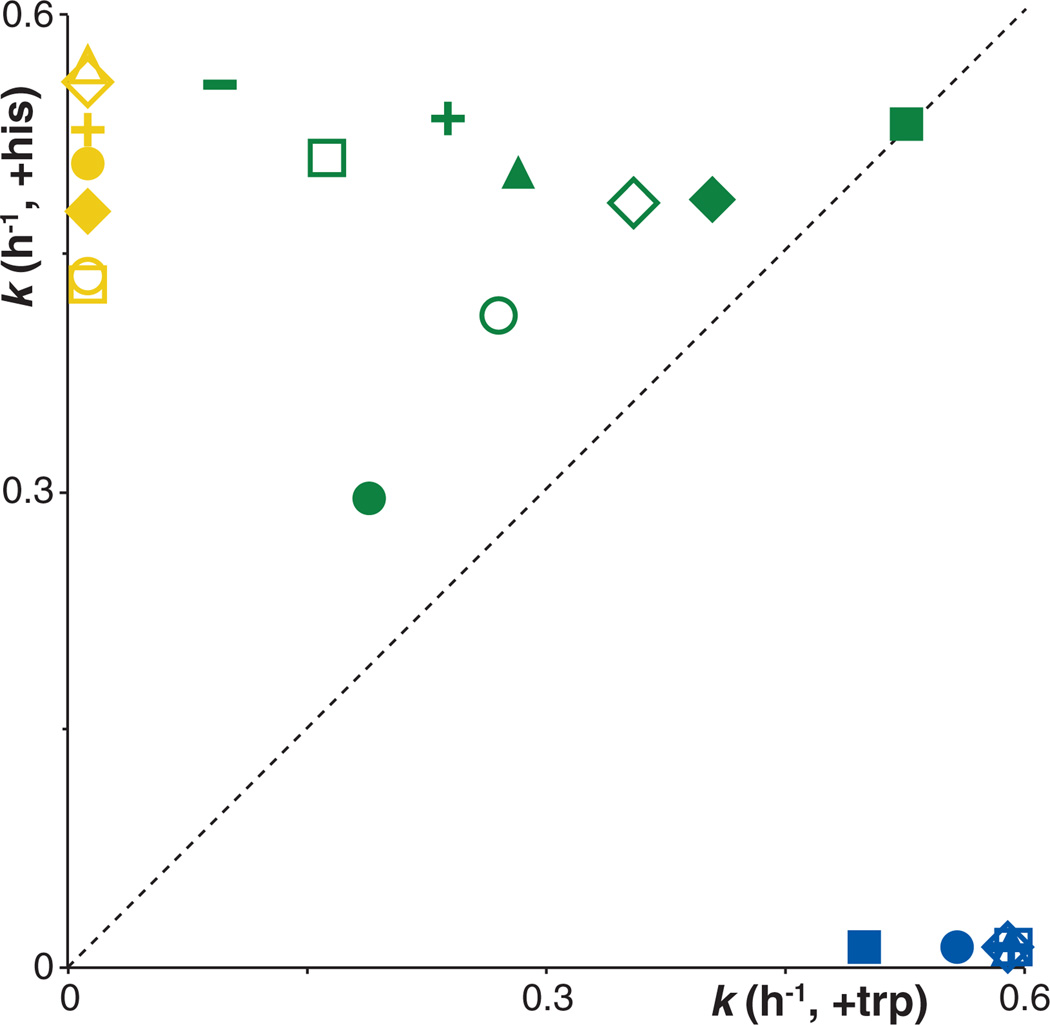
Figure 3.
Characteristics of 22 different evolved mutant gene variants. Each point represents the fitness of one specific mutant gene for its HisA activity on the x axis [assayed as growth rate (k) in minimal glycerol medium with added tryptophan] and TrpF activity on the y axis (assayed as growth rate in minimal glycerol medium with added histidine). Mutant genes fall into three main classes as indicated by the colors: Blue, HisA specialists [open diamond, hisA(wt); open triangle, D10G G102A; dash, D10G G102A V106M; cross, D10G; open square, D10G G102A S143N; solid circle, D10G R83C; solid square, D10G G102A V106M V88I]. Yellow, TrpF specialists (open triangle, dup13-15 D10G G102A Q24L V106L; open diamond, dup13-15 D10G G102A V106M V88I; cross, dup13-15 D10G G102A V106M Q24L; solid circle, dup13-15 D10G G102A Q24L V14:2M; solid diamond, dup13-15 D10G G102A V106M; open circle, dup13-15 D10G R83C; open square, dup13-15 D10G G81D). Green, generalist enzymes [solid circle, dup13-15 D10G (ancestral bifunctional gene); dash, dup13-15 D10G G102A V106M V45M; cross, dup13-15 D10G G102A Q24L G44E; solid square, dup13-15 D10G G102A G11D G44E; open square, dup13-15 D10G G102A Q24L; solid triangle, dup13-15 D10G G102A V88I; open diamond, dup13-15 D10G G102A S143N; solid diamond, dup13-15 D10G G102A G11D; open circle, dup13-15 D10G G102A].
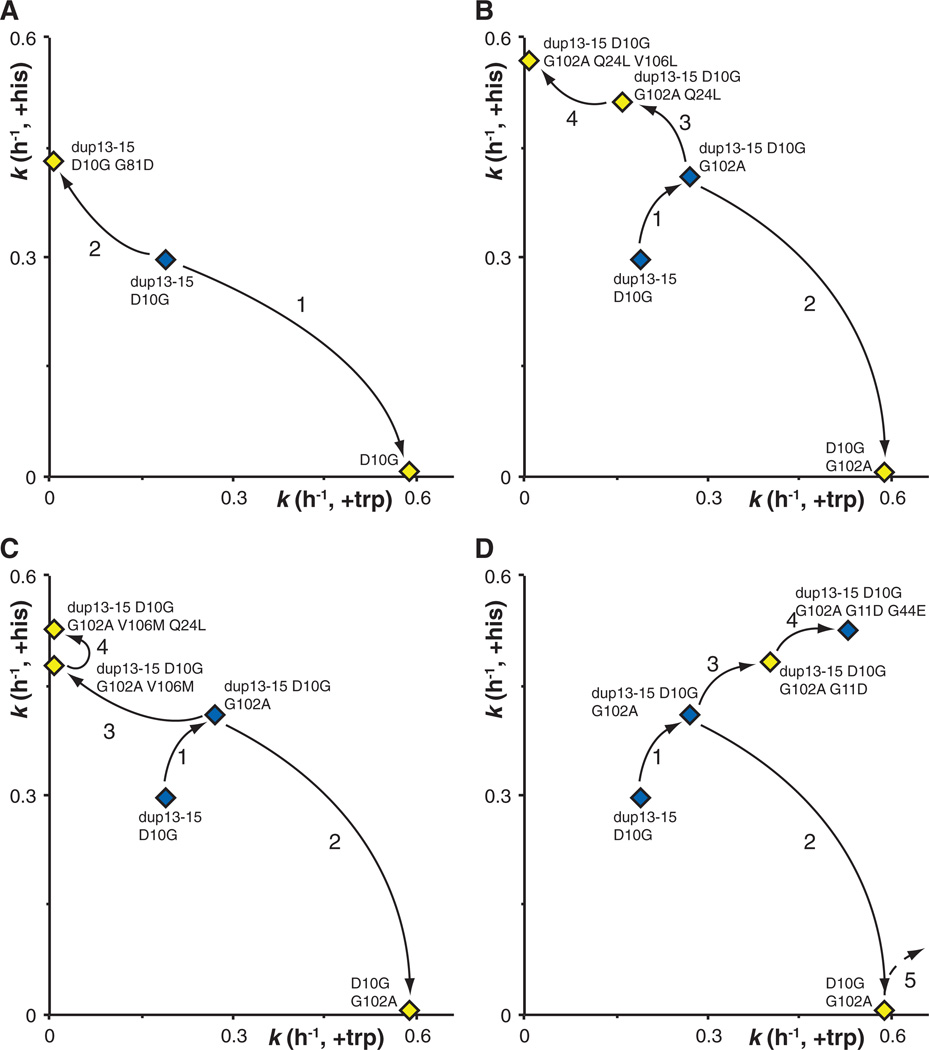
Figure 4.
Multiple evolutionary trajectories recovered through IAD. The x axis indicates the HisA activity (assayed as growth rate in minimal glycerol medium with added tryptophan); the y axis indicates the TrpF activity (assayed as growth rate in minimal glycerol medium with added histidine). (A) Evolution of specialist enzymes in which one activity is improved at the expense of the other. (B and C) Evolution of specialist enzymes after initial evolution of a generalist enzyme. (D) Evolution of a generalist enzyme with improvement of both activities. Arrows and numbers indicate the sequential order of appearance of the various mutations in the population. Yellow symbols denote gene variants that were always accompanied by another gene variant (generalist or with the complementary activity) in the same amplified array. Blue symbols denote gene variants that, at some point during the evolution, were the only variants found in the population.
Thus, under suitable selective conditions, the IAD process rapidly generates genes with distinct enzymatic activities. In Salmonella, duplications of any particular gene form at a rate of roughly 10−5 per cell per division and reach a high steady-state frequency in the population, providing a reservoir of standing copy number variation upon which selection can act (12). Amplification to higher copy numbers occurs at 10−2 per cell per division (3), several orders of magnitude more frequent than point mutations. Thus, whenever a limiting gene product restricts cell growth, initial escape from this restriction may initially occur by duplication events and higher amplification, rather than rare point mutations that alter catalytic activity (13). The delayed appearance of point mutations suggests that the accumulation of a point mutation is the rate-limiting step in the IAD process.
Other sequence-based evidence supports the predictions of the IAD model, particularly in eukaryotes where new genes often evolve by amplification-divergence processes. For example, the evolution of a new gene may be accompanied by the appearance of paralogs and pseudogenes in the genome (14, 15), new genes may show evidence of continuous selection (16, 17), and new genes and pseudogenes may be tandemly clustered with the parent gene (18, 19). On the contrary, in bacteria duplicate genes most commonly arise via HGT (20), but the IAD process could still generate new genes that can be distributed to other organisms by HGT. Conversely, horizontally acquired genes have a higher likelihood of possessing a new side-activity upon which selection and the IAD process could act, suggesting a potential coupling between the IAD process and HGT (21).
Supplementary Material
Acknowledgments
This work was supported by a grant from the Swedish Research Council to D.I.A. and from the NIH (grant GM27068) to J.R.R. The raw Illumina sequencing data sets have been deposited in the National Center for Biotechnology Information Sequence Read Archive (www.ncbi.nlm.nih.gov/Traces/sra/) with accession numbers SRX180378, SRX180382, SRX180383, SRX180384, SRX180385, SRX180387, SRX180388, SRX180390, SRX180391, and SRX180392.
Footnotes
Supplementary Materials
www.sciencemag.org/cgi/content/full/338/6105/384/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S4
Tables S1 to S4
References (22–34)
References and Notes
- 1.Ochman H, Lawrence JG, Groisman EA. Nature. 2000;405:299. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 3.Reams AB, Kofoid E, Savageau M, Roth JR. Genetics. 2010;184:1077. doi: 10.1534/genetics.109.111963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettersson ME, Sun S, Andersson DI, Berg OG. Genetica. 2009;135:309. doi: 10.1007/s10709-008-9289-z. [DOI] [PubMed] [Google Scholar]
- 5.Bergthorsson U, Andersson DI, Roth JR. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17004. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copley SD. Curr. Opin. Chem. Biol. 2003;7:265. doi: 10.1016/s1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 7.Khersonsky O, Tawfik DS. Annu. Rev. Biochem. 2010;79:471. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 8.Henn-Sax M, Höcker B, Wilmanns M, Sterner R. Biol. Chem. 2001;382:1315. doi: 10.1515/BC.2001.163. [DOI] [PubMed] [Google Scholar]
- 9.Barona-Gómez F, Hodgson DA. EMBO Rep. 2003;4:296. doi: 10.1038/sj.embor.embor771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supplementary materials on Science Online.
- 11.Frame R, Bishop JO. Biochem. J. 1971;121:93. doi: 10.1042/bj1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P, Roth J. Proc. Natl. Acad. Sci. U.S.A. 1981;78:3113. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson DI, Hughes D. Annu. Rev. Genet. 2009;43:167. doi: 10.1146/annurev-genet-102108-134805. [DOI] [PubMed] [Google Scholar]
- 14.Bergelson J, Kreitman M, Stahl EA, Tian D. Science. 2001;292:2281. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- 15.Michelmore RW, Meyers BC. Genome Res. 1998;8:1113. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 16.Lynch M, Conery JS. Science. 2000;290:1151. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 17.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Genome Biol. 2002;3:research0008. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner GP, Amemiya C, Ruddle F. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14603. doi: 10.1073/pnas.2536656100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M, et al. Proc. Biol. Sci. 2007;274:33. doi: 10.1098/rspb.2006.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treangen TJ, Rocha EP. PLoS Genet. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper SD, Berg OG. Genome Biol. 2003;4:R48. doi: 10.1186/gb-2003-4-8-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.