Abstract
At the core of every dynamic epithelium resides a population of carefully regulated stem cells ensuring its maintenance and balance. The complex mammalian epidermis is no exception to this rule. The last decade has delivered a wealth of knowledge regarding the biology of adult stem cells, but questions still remain regarding the intricate details of their function and maintenance. To help address these gaps, we turn to the small, single-stranded RNA molecules known as microRNAs. Since their discovery, microRNAs have provided us with novel insights and ground-breaking impulses to enhance our understanding of the biological sciences. Due to their unique role in posttranscriptional regulation, microRNAs are essential to cutaneous biology as well as the epidermal stem cell. By serving as buffers to balance between epithelial stemness, proliferation, and differentiation, microRNAs play essential roles in the maintenance of cutaneous stem cells and their transition out of the stem cell compartment. Following an updated overview of microRNA biology, we summarize the current knowledge of the role of microRNAs in cutaneous stem cells, focusing on three major players that have dominated the recent literature: miR-205, miR-203, and miR-125b. We then review clinical applications, discussing the potential of microRNAs as therapeutic targets in regenerative and oncological stem cell-based medicine.
Keywords: microRNAs, stem cells, epidermis, hair, regenerative medicine, carcinoma, squamous cell
Introduction
Recent years have brought a wealth of new data on the biology of adult stem cells. We have had glimpses on where they live, what they do, and how they are maintained. It has become clear that their regulation is of utmost importance to maintain an epithelium as dynamic as the epidermis; however, many of the details regarding how this works are still unclear. Decisions in the daily life of an epidermal stem cell have to be definitive and unambiguous, because mistakes may result in an imbalance between stemness, proliferation, and differentiation in the epithelium, eventually resulting in disease.
Fortunately, organisms harbor a post-transcriptional regulatory system that improves the precision of gene expression patterns. In animals, this system is based on the small single-stranded RNA molecules known as microRNAs. The literature on microRNAs has shown that this system is indispensable for cutaneous biology. Research has further demonstrated that microRNAs also play key roles in the maintenance of cutaneous stem cells, as well as their transition out of the stem cell compartment. In this review, we evaluate key findings regarding the role of microRNAs in cutaneous stem cell biology and explore their significance in the context of regenerative medicine and cancer therapy.
A brief overview of microRNAs
“You need to let the little things that would ordinarily bore you suddenly thrill you” – Andy Warhol
MicroRNAs and other small RNA molecules have been overlooked for some time. Only recently, with the discoveries of lin-4 in C. elegans in 1993[1] and let-7 in humans in 2000[2], has their true significance been realized. Since their unearthing, an avalanche of publications, hypotheses, and ideas has ensued. It is now known that thousands of functional microRNAs exist which are widely utilized in plants and animals.
The importance of microRNAs in the skin has been well-documented via knockout models for the microRNA-manufacturing machinery. The enzymes Dicer and Drosha, in particular, process original RNA transcripts to produce mature single-stranded microRNAs. Deletion of either enzyme results in severe defects in normal cutaneous functions and hair follicle programs in these models, even frequently leading to premature death[3]. In adult rodent skin, for example, the absence of these enzymes (and thus microRNAs) results in failure to maintain proliferation of the transit-amplifying (TA) hair follicle cells. Eventually, the stem cells of the hair follicle are lost as well[4].
Novel discoveries such as these continue to spark interest in the functions of these small single-stranded RNA molecules. Before we delve into their role in stem cells, however, we will review some basics regarding microRNA biology.
“A mighty flame followeth a tiny spark.” – Dante Alighieri
MicroRNAs are on average only a mere twenty nucleotides in length[5, 6]; yet, these “tiny sparks” can have profound effects due to their unique role in posttranscriptional gene expression control. While the features of microRNA function are still hotly debated, several axioms or dogmas have emerged as basic guides of how microRNAs work:
MicroRNAs direct the RNA-induced silencing complex (RISC) to silence target partially complementary mRNA transcripts
Once the microRNA-RISC complex is bound to the 3’UTR of the target mRNA, translation of the mRNA transcript will not occur
MicroRNAs are used by the cell to reduce transcriptional noise and to buffer and/or fine-tune gene expression
Each microRNA may regulate an entire set of genes simultaneously
In general, microRNAs modulate gene expression in a range of roughly 2- to 3-fold, though the true effect is far less in most cases
Redundancy exists within the microRNA system: several microRNAs can work in tandem to regulate a set of genes and complete a specific task
Many microRNAs probably work in regulatory networks such as feedback loops or bifan motifs
While several points are fairly dogmatic by now (though certainly not undisputed), some require particular emphasis. One of the most confusing aspects of microRNA biology is the notion that microRNAs are actually poor direct manipulators of gene expression. MicroRNAs do not serve as simple on-and-off switches for the expression of single genes—they are not master regulators in most cases. Instead, these small single-stranded RNA molecules serve to fine-tune gene expression overall, by modestly modulating the expression of entire sets of genes, thus essentially serving as gene expression buffers[7, 8].
This notion is exemplified by the fact that dramatic changes in microRNA levels are typically required to produce a phenotype[9, 10], and a phenotype is not guaranteed. Even the majority of microRNA knockout models demonstrate a lack of phenotype, except under stressful conditions[11]. This implies that while their loss is generally tolerated under homeostatic conditions, extreme conditions can result in a sufficient breakdown in gene expression control to produce phenotypes. Such findings support their role as gene expression buffers in lieu of direct effectors and highlight the challenges and controversy associated with the study of microRNAs.
Why microRNAs?
“Have no fear of perfection – you'll never reach it.”— Salvador Dalí
It may seem strange that plants and animals have evolved such an elaborate system to prevent the use of certain mRNAs. The notion that perfectly good nucleic acids are degraded after they are transcribed seems even wasteful at times. MicroRNAs were likely implemented throughout evolution due to inherent deficiencies in gene expression regulation. Such inadequacies could have otherwise hindered the emergence of complex organisms [7]. referring to the evolution of vertebrates and mammals in particular [12, 13].
Furthermore, the complex nature of microRNAs implies that they are capable of achieving amazing regulation of cellular function. For example, microRNAs have even been demonstrated to regulate vertebrate embryonic development through managing gradient expression[14].
We refer back to the rodent hair follicle to put this notion into the context of squamous epithelia. The hair follicle is one of the most complex structures in the skin, consisting of many cell types undergoing various stages of differentiation, and is thus highly sensitive to aberrant microRNA function. As demonstrated in knockout models of microRNA-processing enzymes, improper decisions in a large set of cells quickly result in structural defects in the growing hair follicle, eventually leading to its demise[3, 4].
Similarly, it is easy to imagine how these “tiny sparks” may contribute to stem cell biology. Stem cells sit at the core of epithelia to provide the constant flow of mature cells that fulfill the function of the tissue. As in the hair follicle or with vertebrate embryogenesis, this is a complex system, and not surprisingly, microRNAs appear to play critical roles in the regulation of the stemness program, by smoothly facilitating the transition from stem cell to TA cell.
With this framework of ideas set, we will now explore the role of three specific microRNAs that have dominated the literature on microRNAs in cutaneous stem cell biology.
MiR-205: activator of the reserves
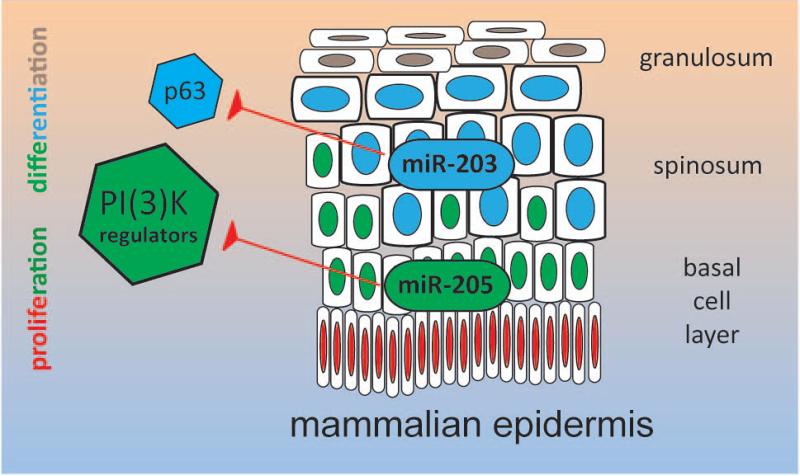
There are currently over 2,500 human mature microRNAs documented in miRbase[15]; yet, not all of these are significant to every tissue. Analysis of the microRNA expression levels serves as a simple method to help identify those that are important[9]. Different organs within the human body exhibit diverse miRNomes composed of sets of microRNAs which are more-or-less tissue-specific. For example, some of the most abundant microRNAs in the skin are also relatively specific to the cutaneous epithelium[7]. The microRNAs miR-203 and miR-205, in particular, have been intensively studied and connected to important decisions in the life of a cutaneous stem cell (Figure 1).
Figure 1.
MicroRNAs crucial to cutaneous stem cell biology: miR-205 and miR-203. MiR-205 is enriched in stem cells and modulates their activation via regulation of PI(3)-kinase signaling, whereas miR-203 occupies the differentiated compartment of the epithelium to promote and maintain differentiation while inhibiting proliferation via control of p63.
MiR-205 is the better studied of the two, with a well-documented knockout model in existence[16]. In vivo, miR-205 is particularly enriched in stem cells and is highly associated with undifferentiated cell populations in the epidermis and the hair follicle. A lack of miR-205 results in defects in the epidermis and hair follicle, related to impaired proliferation[17]. The complete absence of miR-205 even results in death around postnatal day 10.
Regarding its specific function, miR-205 does not seem to affect the generation and maintenance of stem cells as much as it modulates their activation. The major mechanism underlying this activation is thought to be the regulation of the PI(3)-kinase signaling. In this pathway, miR-205 lowers the threshold for phosphorylation and activation of Akt1, thus stimulating proliferation[17]. Evidence by Lavker et al. implies that the effect on Akt signaling occurs through regulation of the lipid phosphatase, SHIP2, and that another microRNA, miR-184, may interfere with this mechanism[18, 19].
The group showed that through its pro-migratory functions, miR-205 may play an essential role in wound healing[18, 19]. While this specific aspect of miR-205 function has not been formally tested in knockout mice, one can speculate that miR-205 may have functions in stress and emergency conditions (such as wounding), requiring the keratinocytes to adopt specific behaviors beneficial to closing the wound (e.g. migration, proliferation, cytokine production, etc.) The correct balance of miR-205 is likely required in making these decisions.
However, the PI(3)-kinase model is not as clear as often presented. For example, it is uncertain as to why miR-205 is most highly expressed in the quiescent stem cell compartment, and how its broad expression in other squamous epithelia fits into its role in the control of progenitor proliferation[17]. In the hair follicle, non-stem cells of the outer root sheath suffer from miR-205 loss in the same fashion as stem cells, despite the fact that their expression levels differ by nearly 4-fold. Perhaps the absolute level of miR-205 is not so essential that expression differences of 4-fold cannot be tolerated, and miR-205 still functions as a PI3K signaling regulator in these settings; yet, such an explanation does not hold for the highly proliferative matrix cells, whose fast cell cycle progression remains unperturbed by changes in miR-205, implying independence from PI(3)-kinase signaling.
Furthermore, while studies of miR-205 in the cutaneous epithelium are still limited, the microRNA has been demonstrated to have roles unrelated to PI(3)-kinase in other epithelia as well. Chao et al. showed that miR-205 alters Notch-signaling via Notch2 suppression in the mouse mammary epithelium[20]. This model is not without its flaws, as a miR-205 inhibitor is utilized in lieu of a classic knockdown approach; and the publication remains elusive about the normal miR-205 expression and function in breast epithelium (myoepithelial or luminal) as well as the significance of breast cancer tumor type. However, this paper does confirm the notion that miR-205 is a true epithelial marker and that its loss may be accompanied by epithelial-tomesenchymal transition, as previously indicated by Gregory et al[21].
Perhaps miR-205 serves as more than a mere PI3K pathway regulator. In vitro, miR-205 is actually upregulated during differentiation, and downregulated after treatment with immortalizing factor E6/E7 of HPV[22]. Review of the most recent collection of published miR-205 target genes is confusing due to lack of consistency: PTEN[23]; ZEB1 and ZEB2[24]; CGNL1, TJP1, and CDC42[25]; and NOTCH2[20], among others. Such publications have little uniformity with one another and rarely demonstrate a relationship with PI(3)K signaling.
With such contrasting findings in the literature, the role of miR-205 remains uncertain. Such disconnect may be due to common flaws in methodology which plague microRNA studies, including: the inadequacy of in vitro studies to accurately replicate microRNA biology in vivo; investigations in cell types with negligible levels of the microRNA in question; biased approaches in identifying target genes (e.g. focus on a single target gene); or highly rigorous cutoffs for regulation (>2-fold change in target genes).
Regardless, the best data thus far on miR-205 in the cutaneous epithelia indicate that its key functions entail the regulation of the PI(3)K-signaling pathway to modulate the migration of keratinocytes.
MiR-203: gatekeeper of the differentiated state
In striking contrast to miR-205, miR-203—the other hallmark microRNA of squamous epithelia— occupies the differentiated compartment of the epithelium. Here, this microRNA promotes and maintains differentiation, while actually suppressing proliferation[26, 27]. As one can imagine, these roles are pivotal in maintaining the overall function of the epidermis as a barrier to infection and water loss[28].
The mechanism underlying this control involves p63, which may be one of the primary targets of miR-203 in keratinocytes and the master regulator of squamous cell fate. MiR-203 is induced in differentiating cells and subsequently downregulates p63 during epidermal stratification to suppress proliferation and facilitate the induction of differentiation[26, 29]. Thus, miR-203 serves as an excellent example of a microRNA sharpening the transition from an undifferentiated, proliferative state to a differentiated, post-mitotic compartment within the squamous epithelium.
Due to these significant roles, several groups even refer to miR-203 as a stemness repressor[26, 29]; however, it is difficult to determine the actual effect miR-203 may or may not have on stem cells. For one, miR-203 does not appear to be induced during the stem cell-TA transition, as would be expected from a stemness repressor. Thus, it is perhaps more appropriate to refer to miR-203 as more of a “blockade” to proliferation, which is put up only after initiation of differentiation.
It should also be noted that, while miR-203 does serve as a gatekeeper of the differentiated state, several other microRNAs (e.g. let-7, miR-23, miR-24) exist which hold expression patterns reminiscent of miR-203 and assist miR-203 in promoting and maintain differentiation while inhibiting proliferation[30]. Thus, while miR-203 is indeed a significant roadblock to proliferation in itself, there are other microRNAs functioning as traffic spikes to aid miR-203 in its task[7].
MiR-125b: regulator of proliferation
“We must always change, renew, rejuvenate ourselves; otherwise, we harden.” – Johann Wolfgang von Goethe
Stem cells successfully bestow the impression of a constant cutaneous epithelium on gross appearance. In reality, the cutaneous epithelia are undergoing constant change to maintain this steady appearance. This self-renewal process is once again regulated by microRNAs, such as miR-125b, which mediate the flow of cells from the stem cell compartment to the most differentiated cell layers.
MiR-125b is similar to miR-205, inasmuch as it is preferentially expressed in progenitor cells in the epidermis and hair follicles. This microRNA has been associated with differentiation in human skin and inhibition of proliferation in keratinocytes via targeting FGFR2[31]. Overexpression of miR-125b in the skin results in impaired hair growth due to reduced proliferation and differentiation in the matrix of the hair follicle[32]. MiR-125b appears to maintain a healthy stem cell pool, with inappropriate levels of miR-125b leading to dysregulated expansion of stem cells and reduced TA cell activity.
An unbiased screen for miR-125 target genes has identified VDR as a critical for the function of this microRNA, both in vivo and in vitro[32]. VDR explains many of the differentiation issues witnessed in the hair follicle following miR-125b overexpression; however, additional target genes have also been identified that may hold even more insight into the microRNA's role in the regulation of stem cell behavior. Interestingly, miR-125b has similar effects on stem cell expansion in competitive repopulation assays of hematopoietic stem cells in vivo[33], though the expansion in these cells was attributed to the inhibition of TGF-β and the activation of Wnt signaling, rather than VDR. The targets identified for miR-125b are not conserved across different species, but the pathways that are affected, such as the p53 network, are conserved[34]. Many of these targets support the idea that miR-125 serves as a general regulator of proliferation and apoptosis, in addition to functioning as a controller of stem cell proliferation[31].
We will further elaborate on one of these key targets, Lin28, due to its particular significance and recent updates in the literature. Thus begins our discussion of the clinical implications of microRNAs in epithelial stem cell biology.
Regenerative medicine: lin28
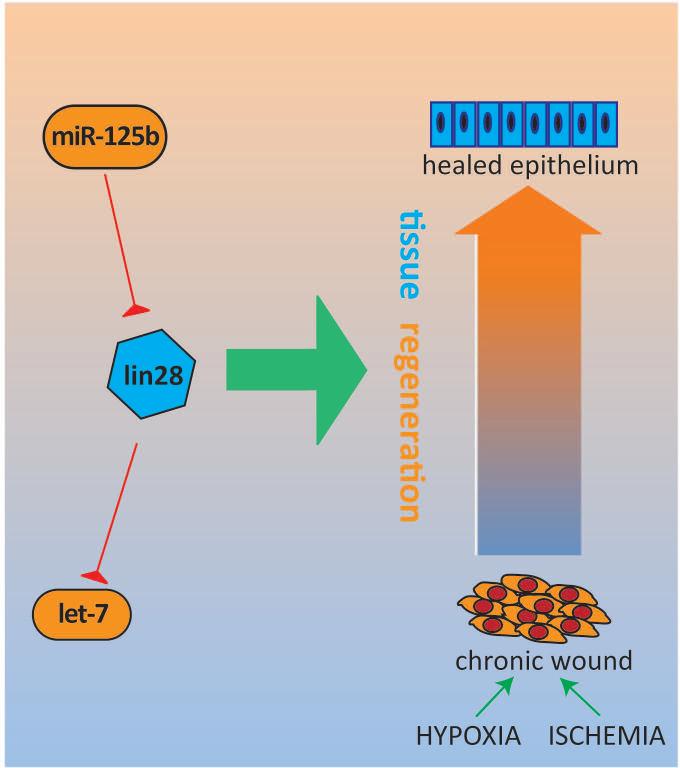
One of the most exciting aspects of cutaneous biology is the skin's ability to serve as a canvas for studying tissue regeneration. Non-mammalian vertebrates dominate the classical models of regeneration and wound healing[35]; therefore, most of the limited information we have on microRNAs in this context stem from these models, which include the regeneration of zebrafish fins[36] and salamander tails[37]. However, recent research on the miR-125b target gene, lin28, has advanced our understanding of the role microRNAs play in mammalian tissue regeneration and stem cell biology.
Lin28 is a highly conserved RNA-binding protein expressed in human stem cells and thought to regulate their growth, metabolism, and self-renewal[38, 39]. Given what we know about miR-125b, it makes sense that Lin28 serves as one of its targets. This protein holds obvious implications for regenerative medicine, as stem cells are necessary for regeneration and repair across all tissues.
As expected, Lin28 is almost completely absent in mature adult tissues and has low-to-absent baseline expression across non-stem cell lines. However, recent findings indicate that the re- introduction of Lin28 in adult tissues results in improved wound healing and hair growth by enhancing the metabolic activity of the tissue. For example, mice genetically modified to produce Lin28 demonstrate improvements in tissue regeneration following various wound insults even in later life stages[40]. These mice could even regenerate limbs, though they were not able to repair cardiac tissue.
Adding complexity to the network, the major targets of Lin28 are also microRNAs, particularly members of the let-7 family. This microRNA family has a range of highly conserved target genes which carry out various cellular functions, including stemness, proliferation, and differentiation. In the skin, let-7 members are mainly expressed in the suprabasal cell layers to facilitate differentiation[41], similar to miR-203, in essence functioning as anti-stemness factors. Lin28 and let-7 mediate their contrasting effects on growth through the opposing manipulation of key metabolic enzymes[40, 42, 43].
In the context of the relatively quiescent and slow cycling stem cell, re-expression of Lin28 with suppression of let-7 may promote speedier self-renewal in these populations, which may one day translate into clinical applications, such as adjunctive therapy for wound healing (e.g. diabetic ulcers). As microRNAs are so well-intertwined with the Lin28 system (e.g. miR-125b, let-7), these small, single-stranded RNA molecules may potentially serve as components of a novel Lin28-based therapeutic approach.
It should be mentioned that wound healing is a coordinated process often involving other tissues in addition to the skin. MicroRNAs have been implicated in the self-renewal and differentiation of mesenchymal stem cells[44] and angiogenic cells[45] as well; and have been implicated in diabetic wound healing in murine models[46, 47] in addition to poorly healing, ischemic wounds[48].
Finally, while research on the Lin28/let-7 axis continues, we should keep in mind the potential risks of Lin28 activation. In vitro, re-expression of Lin28 in cells results in increased oxidative stress and subsequently reduced life spans for the cell lines[40]. The in vivo costs for enhanced tissue repair are unclear, but potential adverse effects of Lin28 overactivation include tumorigenesis[49, 50] as well as stem cell exhaustion[40, 43].
Cancer stem cells and microRNAs
Interestingly, the vast majority of candidate microRNAs (e.g. let-7, miR-34, miR-200) that have been implicated in the regulation of cancer stem cells (CSCs) appear to do so via inhibition of cancer stem cell status[51–53]. In general, this is consistent with the recurring trend of microRNA function: they control proliferation and inhibit stemness, preventing cancer in the process. However, as this is a field that basically incorporates two controversial topics, it is often riddled with contradictions. A case in point is miR-125: while most reports show a positive association of miR-125 with the CSC phenotype[54–56], others suggest that the microRNA is of an inhibitory nature[57–59]. MicroRNAs are frequently depicted as potential therapeutic targets in the treatment of numerous carcinomas, but admittedly, little is known about the involvement of microRNAs in squamous epithelial cancer stem cell biology. We will specifically review the role of five microRNAs, referring back to some previously mentioned and introducing some new ones.
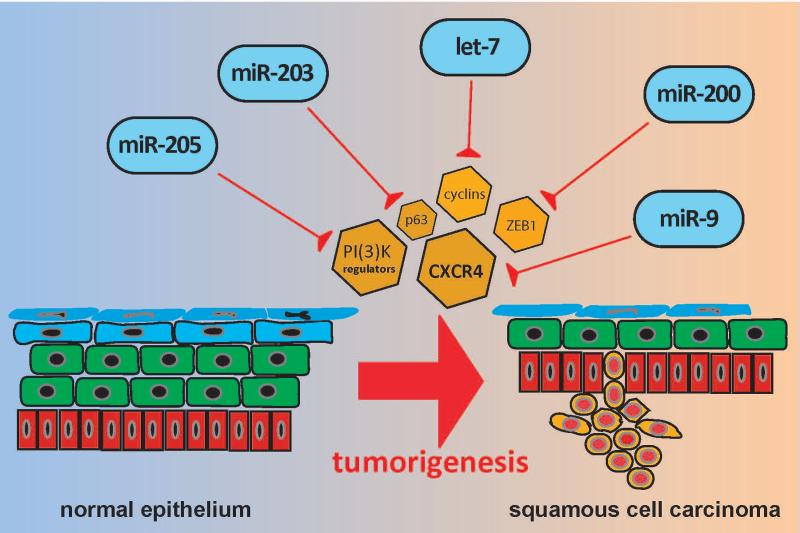
MiR-205 & miR-200 family
The miR-200 family and miR-205 are consistently shown to be downregulated in cancer stem cells from other non-epidermal tissue surfaces, such as the oral epithelium[60]. This regulation is thought to occur via a reciprocal relationship with the Zinc finger E-box-binding protein (ZEB1), a transcription factor essential in the activation of TGF-β-mediated EMT and cancer stem cell functions[61, 62].
It is thus easy to imagine how the repression of these microRNA families may contribute to driving tumor growth. In numerous epithelial carcinomas, the miR-200 family inhibits tumor growth and metastasis by upregulating E-cadherin and downregulating ZEB1, Snail, and N-cadherin[20, 60, 61, 63–66], ultimately inhibiting EMT in malignant SCC cells. Restoration of miR-200 levels in cancer stem cells or other utilization of the ZEB-1/miR-200 feedback loop to our advantage could potentially serve as the foundation for a novel treatment approach for multiple cancers.
MiR-203
As mentioned previously, miR-203 can promote differentiation in epidermal stem cells via inhibition of p63. Interestingly, the epigenetic silencing of miR-203 is required for EMT and cancer stem cell properties[67, 68], likely due to the effects on p63[29].
Referring to another epithelial example, miR-203 was shown to induce differentiation of esophageal SCC cells to restore epithelial tissue architecture and inhibit tumor growth in murine xenograft models[69]. Another laboratory demonstrated a similar inhibitory effect of miR-203 in vitro on esophageal cancer stem-like cells[70]. Similarly, miR-203 is also downregulated in laryngeal SCC, with the potential to inhibit growth via ectopic expression in murine xenograft models[71].
Taken together, these results imply that miR-203 too has potential as a novel therapeutic target in multiple cancers. As miR-203 is so important to epidermal differentiation, this microRNA could plausibly play a role in the differentiation therapy of skin cancer[28].
Let-7
Let-7 family members modulate the expression of stemness genes. These microRNAs have been demonstrated to serve as tumor suppressors via regulation of multiple oncogenic signaling networks. The ancient lineage of let-7 family members and widespread targets certainly make it a suitable candidate for further investigation.
Referring to examples of head and neck cancer, let-7a has been shown to repress chemoresistance and tumorigenesis through repression of multiple stem-like properties[72]. Another link has also been established between let-7 and lin-28[73]. The mRNA AU-rich element binding factor ZFP36 mediates the degradation of lin-28, subsequently elevating the expression of let-7. ZFP36 is thought to be a tumor suppressor in both SCC and in melanoma[74, 75].
And in human keratinocytes, levels of let-7a, let-7b, and let-7c are epigenetically repressed during neoplastic transformation induced by arsenite. This process entails methylation following activation of the Ras/NF-κB pathway, for which let-7c is an up-stream regulator. Consistent with these findings, overexpression of let-7c inhibits the development of cancer stem cell-like attributes and malignant potential of these transformed keratinocytes[76].
MiR-9
Expression levels of miR-9 are reduced in various human carcinomas. In nasopharyngeal carcinoma, this occurs via CpG island hypermethylation, with expression levels inversely correlated with clinical stages, marking the progression from locoregional to metastatic spread[77]. The methylation of miR-9 is also advertised as a biomarker for oral and oropharyngeal SCCs[78].
In oral SCC, lentivirus-mediated miR-9 overexpression inhibits the proliferation of tumor cell lines both in vitro and in vivo[79]. Such ectopic expression leads to similar effects in nasopharyngeal carcinoma[77]. The CXC chemokine receptor 4 (CXCR4) is implicated as a direct target in both cancers[77, 79]. In addition, miR-9 regulates lin28, which as mentioned previously can function as an oncogene in cancer cells[50]. Taken together, such results are evidence demonstrating the promise of miR-9 as a potential therapeutic target in SCCs.
Conclusions
“We shouldn't be looking for heroes, we should be looking for good ideas” – Noam Chomsky
Naturally, microRNAs are often associated with novel therapeutic ideas. Several budding examples exist, wherein they have already been successfully implemented to break barriers in treatment[80]. As outlined, there is much potential for their clinical applications in the context of stem cells as well. Despite significant progress, however, we must not be too quick to depict these small, single-stranded RNA molecules as the definitive targets and tools for the cure of disease.
In the example of SCC, several cancer stem cell-targeting therapies have been tested thus far, such as RNA interference, microRNA precursors, and lentiviral microRNA vectors[81]. While some of these compounds demonstrate considerable promise, there is limited evidence demonstrating that such treatments are specific to cancer stem cells and more effective than standard therapy (e.g. radiation, cisplatin, etc.).
Indeed, many obstacles still remain. Given the complexities associated with this system—for instance: the fact that microRNAs only modulate gene expression in a limited range, as well as the inherent redundancy—microRNAs are unlikely to provide us with instant salvation from our ailments. Nevertheless, their strong associations underlying diseases imply that we will come closer to their utilization as we continue to study their nature. One day, microRNAs may serve as adjunctive therapies, rather than definitive treatments, for regenerative, oncological, or other stem cell-based medicine.
One thing is certain though: microRNAs have provided us with novel insights and ground-breaking impulses to advance our understanding of the biological sciences. Their discovery has reopened our eyes and exposed a new level of regulation neatly intertwined between the known transcriptional and posttranscriptional regulatory systems. MicroRNAs will continue to vastly improve our current concepts and notions on how biological systems are maintained in our cells, tissues, and organs. In the process, they will continue to provide us with novel clues in the study of cutaneous tissues and their stem cells.
Figure 2.
MicroRNAs in regenerative medicine: miR-125b and let-7. Lin28 is highly conserved in human stem cells and has the ability promote wound healing in adult tissues via enhancement of metabolic activity. This regenerative axis is intimately involved with microRNAs: lin28 is regulated by miR-125b, and in turn, the major targets of lin28 include members of the let-7 family of anti-stemness microRNAs.
Figure 3.
MicroRNAs in squamous cell carcinoma: miR-205, miR-203, let-7, miR-200, and miR-9. Each of these microRNAs has been implicated as a tumor suppressor in squamous cell carcinoma through regulation of stemness and/or EMT via inhibition of their respective targets.
Acknowledgements
Thomas Andl received support from a Dermatology Foundation Career Development Award. Support of this work by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (Grant number: 1R01AR061474-01) is also gratefully acknowledged.
Funding: None to declare
Footnotes
Author contributions:
Matthew S. Ning: conception and design, manuscript writing Thomas Andl: conception and design, manuscript writing, final approval of manuscript
Disclaimers: None to declare
Conflicts of interest: None to declare
References
- 1.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. CELL. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 2.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. NATURE. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 3.Andl T, Murchison EP, Liu F, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. CURR. BIOL. CB. 2006;16(10):1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teta M, Choi YS, Okegbe T, et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. DEV. CAMB. ENGL. 2012;139(8):1405–1416. doi: 10.1242/dev.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. SCIENCE. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 6.Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. SCIENCE. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 7.Ning MS, Andl T. Control by a hair's breadth: the role of microRNAs in the skin. CELL. MOL. LIFE SCI. CMLS. 2013;70(7):1149–1169. doi: 10.1007/s00018-012-1117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. GENES DEV. 2010;24(13):1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broderick JA, Zamore PD. Competitive endogenous RNAs cannot alter microRNA function in vivo. MOL. CELL. 2014;54(5):711–713. doi: 10.1016/j.molcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Denzler R, Agarwal V, Stefano J, et al. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. MOL. CELL. 2014;54(5):766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung AKL, Sharp PA. MicroRNA functions in stress responses. MOL. CELL. 2010;40(2):205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hertel J, Lindemeyer M, Missal K, et al. The expansion of the metazoan microRNA repertoire. BMC GENOMICS. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler BM, Heimberg AM, Moy VN, et al. The deep evolution of metazoan microRNAs. EVOL. DEV. 2009;11(1):50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 14.Martello G, Zacchigna L, Inui M, et al. MicroRNA control of Nodal signalling. NATURE. 2007;449(7159):183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 15.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. NUCLEIC ACIDS RES. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park CY, Jeker LT, Carver-Moore K, et al. A resource for the conditional ablation of microRNAs in the mouse. CELL REP. 2012;1(4):385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Zhang Z, O'Loughlin E, et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. NAT. CELL BIOL. 2013;15(10):1153–1163. doi: 10.1038/ncb2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Peng H, Ruan Q, et al. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. OFF. PUBL. FED. AM. SOC. EXP. BIOL. 2010;24(10):3950–3959. doi: 10.1096/fj.10-157404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Ryan DG, Getsios S, et al. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. PROC. NATL. ACAD. SCI. U. S. A. 2008;105(49):19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao C-H, Chang C-C, Wu M-J, et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J. CLIN. INVEST. 2014 doi: 10.1172/JCI73351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. NAT. CELL BIOL. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 22.McKenna DJ, Patel D, McCance DJ. miR-24 and miR-205 expression is dependent on HPV onco-protein expression in keratinocytes. VIROLOGY. 2014;448:210–216. doi: 10.1016/j.virol.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G, Hou X, Li Y, et al. MiR-205 inhibits cell apoptosis by targeting phosphatase and tensin homolog deleted on chromosome ten in endometrial cancer Ishikawa cells. BMC CANCER. 2014;14:440. doi: 10.1186/1471-2407-14-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J-Y, Park MK, Park J-H, et al. Loss of the polycomb protein Mel-18 enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. ONCOGENE. 2014;33(10):1325–1335. doi: 10.1038/onc.2013.53. [DOI] [PubMed] [Google Scholar]
- 25.Chung P-JK, Chi L-M, Chen C-L, et al. MicroRNA-205 Targets Tight Junction-related Proteins during Urothelial Cellular Differentiation. MOL. CELL. PROTEOMICS MCP. 2014;13(9):2321–2336. doi: 10.1074/mcp.M113.033563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi R, Poy MN, Stoffel M, et al. A skin microRNA promotes differentiation by repressing “stemness.”. NATURE. 2008;452(7184):225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonkoly E, Wei T, Janson PCJ, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLOS ONE. 2007;2(7):e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson SJ, Zhang Z, Feng D, et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. DEV. CAMB. ENGL. 2013;140(9):1882–1891. doi: 10.1242/dev.089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, et al. miR-203 represses “stemness” by repressing DeltaNp63. CELL DEATH DIFFER. 2008;15(7):1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 30.Büssing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. TRENDS MOL. MED. 2008;14(9):400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Xu N, Brodin P, Wei T, et al. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J. INVEST. DERMATOL. 2011;131(7):1521–1529. doi: 10.1038/jid.2011.55. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Stokes N, Polak L, et al. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. CELL STEM CELL. 2011;8(3):294–308. doi: 10.1016/j.stem.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emmrich S, Rasche M, Schöning J, et al. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. GENES DEV. 2014;28(8):858–874. doi: 10.1101/gad.233791.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le MTN, Shyh-Chang N, Khaw SL, et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLOS GENET. 2011;7(9):e1002242. doi: 10.1371/journal.pgen.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thatcher EJ, Patton JG. Small RNAs have a big impact on regeneration. RNA BIOL. 2010;7(3):333–338. doi: 10.4161/rna.7.3.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin VP, Thomson JM, Thummel R, et al. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. GENES DEV. 2008;22(6):728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sehm T, Sachse C, Frenzel C, et al. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. DEV. BIOL. 2009;334(2):468–480. doi: 10.1016/j.ydbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Richards M, Tan S-P, Tan J-H, et al. The transcriptome profile of human embryonic stem cells as defined by SAGE. STEM CELLS DAYT. OHIO. 2004;22(1):51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 39.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. CELL STEM CELL. 2013;12(4):395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shyh-Chang N, Zhu H, Yvanka de Soysa T, et al. Lin28 enhances tissue repair by reprogramming cellular metabolism. CELL. 2013;155(4):778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rybak A, Fuchs H, Hadian K, et al. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. NAT. CELL BIOL. 2009;11(12):1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 42.Peng S, Chen L-L, Lei X-X, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. STEM CELLS DAYT. OHIO. 2011;29(3):496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 43.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. NAT. REV. MOL. CELL BIOL. 2014;15(4):243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo L, Zhao RCH, Wu Y. The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. EXP. HEMATOL. 2011;39(6):608–616. doi: 10.1016/j.exphem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Wang J-M, Tao J, Chen D-D, et al. MicroRNA miR-27b rescues bone marrow-derived angiogenic cell function and accelerates wound healing in type 2 diabetes mellitus. ARTERIOSCLER. THROMB. VASC. BIOL. 2014;34(1):99–109. doi: 10.1161/ATVBAHA.113.302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lü M-H, Hu C-J, Chen L, et al. miR-27b represses migration of mouse MSCs to burned margins and prolongs wound repair through silencing SDF-1a. PLOS ONE. 2013;8(7):e68972. doi: 10.1371/journal.pone.0068972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J, Wu W, Zhang L, et al. The role of microRNA-146a in the pathogenesis of the diabetic wound-healing impairment: correction with mesenchymal stem cell treatment. DIABETES. 2012;61(11):2906–2912. doi: 10.2337/db12-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biswas S, Roy S, Banerjee J, et al. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. PROC. NATL. ACAD. SCI. U. S. A. 2010;107(15):6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan Y, Sun G, Wang Z, et al. miR-125b promotes cell proliferation by directly targeting Lin28 in glioblastoma stem cells with low expression levels of miR-125b. NEUROREPORT. 2014;25(5):289–296. doi: 10.1097/WNR.0000000000000085. [DOI] [PubMed] [Google Scholar]
- 50.Zhong X, Li N, Liang S, et al. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. BIOL. CHEM. 2010;285(53):41961–41971. doi: 10.1074/jbc.M110.169607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi R-U, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. FRONT. GENET. 2014;4:295. doi: 10.3389/fgene.2013.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu H-F, Lin S-C, Chang K-W. MicroRNA aberrances in head and neck cancer: pathogenetic and clinical significance. CURR. OPIN. OTOLARYNGOL. HEAD NECK SURG. 2013;21(2):104–111. doi: 10.1097/MOO.0b013e32835e1d6e. [DOI] [PubMed] [Google Scholar]
- 53.Chen C, Zimmermann M, Tinhofer I, et al. Epithelial-to-mesenchymal transition and cancer stem(-like) cells in head and neck squamous cell carcinoma. CANCER LETT. 2013;338(1):47–56. doi: 10.1016/j.canlet.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Wang H-J, Guo Y-Q, Tan G, et al. miR-125b regulates side population in breast cancer and confers a chemoresistant phenotype. J. CELL. BIOCHEM. 2013;114(10):2248–2257. doi: 10.1002/jcb.24574. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Liu H, Desai S, et al. miR-125b functions as a key mediator for snail-induced stem cell propagation and chemoresistance. J. BIOL. CHEM. 2013;288(6):4334–4345. doi: 10.1074/jbc.M112.419168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi L, Wan Y, Sun G, et al. miR-125b inhibitor may enhance the invasion-prevention activity of temozolomide in glioblastoma stem cells by targeting PIAS3. BIODRUGS CLIN. IMMUNOTHER. BIOPHARM. GENE THER. 2014;28(1):41–54. doi: 10.1007/s40259-013-0053-2. [DOI] [PubMed] [Google Scholar]
- 57.Luo G, Long J, Cui X, et al. Highly lymphatic metastatic pancreatic cancer cells possess stem cell-like properties. INT. J. ONCOL. 2013;42(3):979–984. doi: 10.3892/ijo.2013.1780. [DOI] [PubMed] [Google Scholar]
- 58.Chen J, Chen Y, Chen Z. CANCER INVEST. 2013;MiR-125a/b regulates the activation of cancer stem cells in paclitaxel-resistant colon cancer.31(1):17–23. doi: 10.3109/07357907.2012.743557. [DOI] [PubMed] [Google Scholar]
- 59.Wu N, Xiao L, Zhao X, et al. miR-125b regulates the proliferation of glioblastoma stem cells by targeting E2F2. FEBS LETT. 2012;586(21):3831–3839. doi: 10.1016/j.febslet.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 60.Wiklund ED, Gao S, Hulf T, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLOS ONE. 2011;6(11):e27840. doi: 10.1371/journal.pone.0027840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. NAT. CELL BIOL. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 62.Ohashi S, Natsuizaka M, Naganuma S, et al. A NOTCH3-mediated squamous cell differentiation program limits expansion of EMT-competent cells that express the ZEB transcription factors. CANCER RES. 2011;71(21):6836–6847. doi: 10.1158/0008-5472.CAN-11-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lo W-L, Yu C-C, Chiou G-Y, et al. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J. PATHOL. 2011;223(4):482–495. doi: 10.1002/path.2826. [DOI] [PubMed] [Google Scholar]
- 64.Tellez CS, Juri DE, Do K, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. CANCER RES. 2011;71(8):3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puhr M, Hoefer J, Schäfer G, et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. AM. J. PATHOL. 2012;181(6):2188–2201. doi: 10.1016/j.ajpath.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, Liao H, Deng Z, et al. miRNA-205 affects infiltration and metastasis of breast cancer. BIOCHEM. BIOPHYS. RES. COMMUN. 2013;441(1):139–143. doi: 10.1016/j.bbrc.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 67.Taube JH, Malouf GG, Lu E, et al. Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. SCI. REP. 2013;3:2687. doi: 10.1038/srep02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ju S-Y, Chiou S-H, Su Y. Maintenance of the stemness in CD44(+) HCT-15 and HCT-116 human colon cancer cells requires miR-203 suppression. STEM CELL RES. 2014;12(1):86–100. doi: 10.1016/j.scr.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 69.Okumura T, Shimada Y, Moriyama M, et al. MicroRNA-203 inhibits the progression of esophageal squamous cell carcinoma with restored epithelial tissue architecture in vivo. INT. J. ONCOL. 2014;44(6):1923–1932. doi: 10.3892/ijo.2014.2365. [DOI] [PubMed] [Google Scholar]
- 70.Yu X, Jiang X, Li H, et al. miR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. STEM CELLS DEV. 2014;23(6):576–585. doi: 10.1089/scd.2013.0308. [DOI] [PubMed] [Google Scholar]
- 71.Tian L, Li M, Ge J, et al. MiR-203 is downregulated in laryngeal squamous cell carcinoma and can suppress proliferation and induce apoptosis of tumours. TUMOUR BIOL. J. INT. SOC. ONCODEVELOPMENTAL BIOL. MED. 2014;35(6):5953–5963. doi: 10.1007/s13277-014-1790-7. [DOI] [PubMed] [Google Scholar]
- 72.Yu C-C, Chen Y-W, Chiou G-Y, et al. MicroRNA let-7a represses chemoresistance and tumourigenicity in head and neck cancer via stem-like properties ablation. ORAL ONCOL. 2011;47(3):202–210. doi: 10.1016/j.oraloncology.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 73.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. SCIENCE. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bourcier C, Griseri P, Grépin R, et al. Constitutive ERK activity induces downregulation of tristetraprolin, a major protein controlling interleukin8/CXCL8 mRNA stability in melanoma cells. AM. J. PHYSIOL. CELL PHYSIOL. 2011;301(3):C609–618. doi: 10.1152/ajpcell.00506.2010. [DOI] [PubMed] [Google Scholar]
- 75.Tubergen E Van, Vander Broek R, Lee J, et al. Tristetraprolin regulates interleukin-6, which is correlated with tumor progression in patients with head and neck squamous cell carcinoma. CANCER. 2011;117(12):2677–2689. doi: 10.1002/cncr.25859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang R, Li Y, Zhang A, et al. The acquisition of cancer stem cell-like properties and neoplastic transformation of human keratinocytes induced by arsenite involves epigenetic silencing of let-7c via Ras/NF-κB. TOXICOL. LETT. 2014;227(2):91–98. doi: 10.1016/j.toxlet.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 77.Lu J, Luo H, Liu X, et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. CARCINOGENESIS. 2014;35(3):554–563. doi: 10.1093/carcin/bgt354. [DOI] [PubMed] [Google Scholar]
- 78.Minor J, Wang X, Zhang F, et al. Methylation of microRNA-9 is a specific and sensitive biomarker for oral and oropharyngeal squamous cell carcinomas. ORAL ONCOL. 2012;48(1):73–78. doi: 10.1016/j.oraloncology.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu T, Liu K, Wu Y, et al. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. ONCOGENE. 2013 doi: 10.1038/onc.2013.448. [DOI] [PubMed] [Google Scholar]
- 80.Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. MOL. CANCER THER. 2008;7(12):3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 81.Kiang A, Yu MA, Ongkeko WM. Progress and pitfalls in the identification of cancer stem cell-targeting therapies in head and neck squamous cell carcinoma. CURR. MED. CHEM. 2012;19(35):6056–6064. [PubMed] [Google Scholar]