Abstract
Toll-like receptors (TLRs) are key innate immune receptors that recognize conserved features of biological molecules that are found in microbes. In particular, TLR2 has been reported to be activated by different kinds of microbial ligands. To advance our understanding of the interaction of TLR2 with its ligands, the recombinant human TLR2 ectodomain (hTLR2ED) was expressed using a baculovirus/insect cell expression system, and its biochemical as well as ligand binding properties were investigated. The hTLR2ED binds synthetic bacterial and mycoplasmal lipopeptides, lipoteichoic acid (LTA) from Staphylococcus aureus, and synthetic lipoarabinomannan precursors from Mycobacterium at extracellular physiological conditions, in the absence of its co-receptors TLR1 and TLR6. We also determined that lipopeptides and glycolipids cannot bind simultaneously to hTLR2ED and that the phosphatidyl inositol mannoside 2 (Pim2) is the minimal lipoarabinomannan structure for binding to hTLR2ED. Binding of hTLR2ED to Pim4, which contains a diacylglycerol group with one of its acyl chain containing 19 carbon atoms, indicates that hTLR2ED can bind ligands with acyl chains longer than 16 carbon atoms. In summary, our data indicate that diacylglycerol is the ligand moiety of microbial glycolipids and lipoproteins that bind to hTLR2ED and that both types of ligands bind to the same binding site of hTLR2ED. The design of novel inhibitors of TLR2, based on their ability to bind to TLR2 but not activate the TLR2 signaling pathway, may lead to the development of novel treatments for septic shock caused by Gram- positive bacteria.
Keywords: diacylglycerol ligands, lipoarabinomannan, lipoteichoic acid, FSL-1, TLR2
Toll-like receptors (TLRs) have emerged as one of the most important group of pattern-recognition receptors in innate immunity. TLRs are type I transmembrane receptors that are important for the initiation of immune responses against microbes. Ten different human TLRs are known, some of which are located in the plasma membrane (TLR1 TLR2, TLR4, TLR5, TLR6), while others are sequestered in internal compartments, including endosomes (TLR3, TLR7, TLR8 and TLR9) [1–2].
TLR2 has been reported to recognize different kinds of microbial ligands that include synthetic triacyl-lipopeptides [3], surfactant protein A [4], meningococcal porin b [5], mycoplasmal lipopeptides and mycobacterial lipoarabinomannan[6]. This promiscuity in the recognition of ligands has been partially explained by the association of TLR2 with other TLRs, such as TLR1 and TLR6[7], but in other cases, the activation of TLR2 was the result of the presence of contaminants in the ligand preparations [8]. The class B scavenger receptors, cluster of differentiation 36 (CD36) and cluster of differentiation 14 (CD14), also contribute to the ligand recognition process by TLR2[9–11]. The crystal structure of a chimera between the human TLR2/TLR1 ectodomain fused to LRR repeats of a variable lymphocyte receptor (VLR) in complex with the triacyl-lipopeptide Pam3CSK4 revealed that a hydrophobic pocket composed of the central leucine-rich repeats 9-12 (LRRs 9-12) of TLR2 is the binding site for Pam3CSK4 [12]. The crystal structures of a mouse TLR2 ectodomain (mTLR2ED)/VLR hybrid in complex with TLR6 and Pam2CSK4, as well as mTLR2ED/VLR in complex with PE-DTPA, a synthetic derivative of phosphatidylethanolamine, and LTA from Streptococcus pneumonia (S.pneumoniae) also uncovered similar findings [13]. While these are extremely important steps to determine the recognition of ligands by TLR2, our knowledge of TLR2 interactions with its different ligands is far from complete [14].
In order to investigate the binding of human TLR2 with the synthetic diacyl-lipopeptide FSL-1 (fibroblast stimulating lipopeptide-1) and LTA from S. aureus and lipoarabinomanan precursors, hTLR2ED was expressed employing a baculovirus/insect cell expression system. The biochemical properties of hTLR2ED and the interaction with its ligands were investigated.
Experimental procedures
Cell Culture and cell line
Sf9 (Invitrogen) and High Five (Invitrogen) insect cells were employed for baculovirus generation and protein expression, respectively. Both cell lines were cultured in suspension cultures in serum-free HyQ media (Hyclone) at 25°C under shaking conditions.
Generation of hTLR2ED Profold ER1 baculovirus
hTLR2ED (residues 19 to 590, NCBI accession number NP_003255) with a C-terminal His6-tag was amplified by PCR using pCDNA3.1 CMV TLR2 flag plasmid as template. The PCR product was cloned in TopoXL PCR (Invitrogen), and subcloned into the BamHI and NotIsites of the pAcGP67A transfer vector (BD Pharmigen). A baculovirus, which we call hTLR2ED Profold ER1, was generated by homologous recombination of hTLR2ED pAcGP67A with Profold ER1 linearized baculovirus vector (AB vector). After 5 rounds of viral amplification, a titer of 1×108 viruses/ml was achieved. Fluorescence from Green Fluorescent Protein (GFP) encoded in the Profold ER1 linearized baculovirus vector was monitored using an Axioplan Zeiss fluorescent microscope.
Purification of recombinant hTLR2ED protein
Six liter cultures of 1 × 106 High Five cells/ml were infected with hTLR2ED Profold ER1 at an MOI of 1. The infected cells were incubated at 27°C for 5 days under shaking conditions. The supernatant collected after centrifugation was buffer exchanged into phosphate-buffered saline (PBS) and concentrated to 100 ml with a Pall Filtron Centramate concentrator device (Millipore) using membranes with a 30,000 Da molecular weight cut-off (Amersham). Next, 5 ml Ni (II) Nitrilotriacetic acid (Ni-NTA) beads and 20 mM imidazole were added and incubated overnight at 4°C with agitation. Subsequently, Ni-NTA beads were separated from the supernatant by filtration and washed 3 times with 20 mM imidazole and 300 mM NaCl, pH 8. Bound proteins were eluted from the Ni-NTA beads with 100 mM imidazole and 300 mM NaCl. Anionic exchange chromatography using a Mono Q column (Pharmacia) followed by size exclusion chromatography using a Superdex 200 10/30 column (Pharmacia) were employed to purify hTLR2ED to homogeneity.
Identification of recombinant hTLR2ED by liquid chromatography-mass spectrometry (LC-MS) and Western blot analysis
hTLR2ED was concentrated to 5 mg/ml and its purity was analyzed by sodium dodecyl sulphate (SDS) PAGE. The band between 50 and 75 kDa was excised, reduced with dithiothreitol (DTT, 10 mM), and digested with trypsin overnight before being analyzed by nano LC-MS/MS (The Scripps Research Institute Center for Mass Spectrometry). Peptides were identified using MASCOT. Protein expression of hTLR2ED was also confirmed by a Western blot probing for the presence of the C-terminal His-tag of the hTLR2ED construct. After proteins were electroblotted from an SDS PAGE to a PVDF membrane, the PVDF membrane was blocked for 1 hour at room temperature with 3% bovine serum albumin (BSA), 0.5 % Tween 20. Subsequently, the membrane was incubated overnight with a 1/1000 dilution of a mouse IgG anti-penta His-tag antibody (Qiagen), washed 3 times with 0.5 % Tween 20 in PBS, and incubated for 1 hour at room temperature with a 1/5000 dilution of rabbit peroxidase-conjugated anti-mouse IgG antibody (Pierce). After three washes with 0.5% Tween 20 in PBS, the Western blot was developed using the Super Signal West Pico ECL substrate (Pierce).
Presence of disulfide bonds in hTLR2ED
Purified hTLR2ED was incubated for 10 minutes in a reducing buffer containing β-mercaptoethanol or in a non-reducing protein gel loading buffer. The samples were then run on SDS PAGE to determine any difference in electrophoretic mobility that would indicate the presence of disulfide bonds in hTLR2ED. The presence of disulfide bonds was also monitored in hTLR2ED deglycosylated with endoglycosidases Endo H and PNGase F. 10 μg of purified hTLR2ED were digested with 50 units of PNGase F (New England Biolab) or Endo H (New England Biolab) in native conditions overnight at 37°C. The samples were examined by SDS PAGE under reducing and non-reducing conditions.
Analysis of hTLR2ED glycosylation
The hTLR2ED band was excised from an SDS PAGE gel, cut into 1 mm cubes, and frozen overnight. The gel pieces were washed with acetonitrile to remove any reagents and then dried in a vacuum centrifuge. The dried gel pieces were rehydrated and incubated with PNGase F (Boehringer) for 16 hours at 37°C. The supernatant was collected. Distilled water was added to the gels, which were sonicated for 30 minutes, and the supernatant retained. This procedure was repeated twice with acetonitrile, followed by sbpd water, finishing with acetonitrile to maximize the recovery of glycans. Samples were then treated with an AG 50 × 12 (Biorad, HemelHemsptead UK) anion exchange resin to eliminate sodium. The solution was filtered through a filter syringe to eliminate the resin and then labeled with 2-aminobenzamide (2AB) using the Ludger Tag 2-AB labeling kit (Ludger limited, Oxford, UK). Excess label was removed by ascending chromatography on 3 mm Whatman paper with acetonitrile as the solvent and the glycans were eluted with sbpd water. Glycans were examined by normal-phase high performance liquid chromatography (NP-HPLC) as previously reported [15] and by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. The HPLC system consisted of a 2690 Alliance separation module (Waters, Milford, MA) with a 4.6 × 250 mm TSK Amide-80 column (Anachem, Luton, UK) and was equipped with a fluorescence detector set at 420 nm and was calibrated against a dextran standard. The retention time of each glycan was converted into glucose units (GU) by comparison with the elution times of a dextran ladder. The experimental GU values were then referred to Glycobase, a relational database (http://glycobase.ucd.ie/cgi-bin/public/glycobase.cgi), which stores information for glycan structure according to its GU values. Preliminary structure assignments were made for each peak. Exoglycosidase array digestions further confirmed these assignments. The 2-AB labeled glycans were digested with α-glucosidase II (EC 3.2.1.20) and jack bean α-mannosidase (EC 3.2.1.24) at 37°C for 18 hours. Digested samples were passed through a protein-binding filter (Micropure-EZ centrifugal devices, Millipore Corporation) to remove the enzymes from the glycans, which were washed thoroughly with sbpd water. Finally, the glycans were separated by NP-HPLC to determine the changes in GU values as a result of the enzyme digestions.
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry
Samples were cleaned with a Nafion membrane and MALDI-TOF mass spectra were recorded with a Waters TofSpec 2E mass spectrometer (Waters MS-Technologies, Manchester UK) in positive ion reflectron mode with delayed extraction. The acceleration voltage was 20.0 kV and the pulse voltage was 3.0 kV. Samples in 0.3 μl of water were mixed with 0.3 μl of matrix (a saturated solution of 2,5-dihydroxybenzoic acid in acetonitrile) on the MALDI target and allowed to dry under ambient conditions. The dry sample was then recrystallized from ethanol.
Electrospray ionization (ESI) mass spectrometry
Negative ion ESI MS/MS spectra were recorded with a Waters Q-TOF Ultima Global mass spectrometer. Samples (about 50 pmoles/μL), cleaned with a Nafion membrane as above, in 1:1 (v:v) methanol:water were infused with a Proxeon (ProxeonBiosystems, Odense, Denmark (now part of Thermo Scientific)) borosilicate capillary. The ion source was maintained at 120°C, the infusion needle potential was 1.1 kV, the cone was at 100 V and the RF-1 voltage was 180 V. Spectra (2 sec scans) were acquired with a digitization rate of 4 GHz. Collision-induced fragmentation (CID) data acquisition used a 4 m/z mass window for parent ion selection and argon at approx 0.5 mBar as the collision gas. The collision cell voltage was 80–120 V, depending on the parent ion mass and other voltages were as recommended by the manufacturer. The instrument was externally calibrated with N-glycans released from bovine fetuin and monoisotopic masses are cited to one decimal place. Instrument control, data acquisition and processing were performed with a MassLynx data system (Version 4.0).
Circular Dichroism (CD) analysis of purified hTLR2ED
CD measurements of hTLR2ED were performed in 50 mM NaCl, 10 mM Tris-HCl buffer at a concentration of 3.5 μM on an AVIV 202 spectropolarimeter equipped with a thermostat (Hellma, Mullheim, Baden, Germany). The purified hTLR2ED solution was placed in a 1 mm path length quartz cuvette and heated from 25°C–95°C at a rate of 1°C/minute. Spectra were recorded every 5°C after equilibration of 5 minutes at the corresponding temperature. The scans were performed in duplicate from 260 to 200 nm with 0.5 nm resolution. The spectra from duplicate scans were averaged and base-line corrected by subtracting the corresponding buffer spectrum obtained under identical conditions. Results were expressed as a mean residue ellipticity (Θ) at a given wavelength.
Native PAGE experiments
The following synthetic lipopeptides were employed for native PAGE experiments: Cys-Ser-Lys4 (CSK4), N-palmitoyl Cys-Ser-Lys4 (PamCSK4), S-(2,3-bis-palmitoyloxypropyl)-Cys-Ser-Lys4 (Pam2CSK4), Pam2CSK4 fluorescein, N-palmitoyl-S-(2,3-bispalmitoyloxypropyl)-Cys-Ser-Lys4 (Pam3CSK4), N-palmitoyl-S-(2,3-bis-palmitoyloxypropyl)-Cys-Ser-Lys4, and S-(2,3-bis-palmitoyloxypropyl)-Cys-Gly-Asp-Pro-Lys-His-Pro-Lys-Ser-Phe-K-Aca-Aca-Fluorescein (FSL-1-fluorescein) activator. All of these synthetic lipopeptides were obtained from EMC Microcollection (Tubingen, Germany). Lipoteichoic acid (LTA) and the human TLR9 ligand CpG-ODN2006 was obtained from Invivogen (San Diego, CA, USA). Synthetic phosphatidylinositol mannosides (Pim2 and Pim4) were synthesized as described previously[16–17]. The gp120 from HIV-1 clade B was a gift from Dr. Robert Pejchal in our laboratory. 1 μg of each of the mentioned compounds was separately incubated with 10 μg of hTLR2ED overnight at 37°C, pH 7.4. In case of the simultaneous incubation with LTA and Pam2CSK4-fluorescein, 1 μg of each compound was mixed and incubated with 10 μg of hTLR2ED. For the lipid ligands, an excess of ligands was added with respect to hTLR2ED (ratio ligand/hTLR2ED: 11.3 (CSK4), 8 (PamCSK4), 5.41 (Pam2CSK4), 4.55 (Pam3CSK4), 6.53 (FSL-1), 3.7 (Pim2), 4.55 (Pim4), 2.9 (LTA)) because the lipid alone forms aggregates due to hydrophobic interactions and, thus, it is difficult to calculate an accurate ratio of monomeric ligand to hTLR2ED. The molar ratio of hTLR2ED to gp120 was 1.7:1. All samples were run for 3 hours at 100 volts in a 4–20% native PAGE. Fluorescence was determined with a Versadoc imaging system equipped with an emission filter with a 60 nm bandwidth (from 500 nm to 560 nm). Subsequently, the gel was stained with Coomassie Blue and destained with a solution containing 20% methanol, 10% acetic acid and 1% glycerol. Gel images were taken with a Versadoc gel imaging system.
Modeling of hTLR2ED-FSL-1, hTLR2ED-LTA, hTLR2ED-LTA C19 hTLR2ED-Pim-2 and hTLR2ED-Pim-4 complexes
Molecular modeling was performed on a Silicon Graphics Indigo 2 workstation using Insight II and Discover software (Accelrys). Figures were produced using the program Molscript [18]. The models of hTLR2ED in complex with various ligands were based on the crystal structures of the hTLR2ED/VLR hybrid in complex with Pam3CSK4 (Protein Data Bank (PDB) 2Z7X) [12]and the Nogo receptor (PDB 1OZN) [19]. Residues 27-508 were taken from 2Z7X, while residues 509-585 are based on residues 236-309 of 1OZN. The model was created by overlaying the backbone atoms of residues 500-509 of 2Z7X with residues 227-236 of 1OZN. Five disulfide bonds were added to the model, one in the N-terminal cap, two in the LRR domain (LRR12-13 and LRR15-16) as present in 2Z7X and two C-terminal disulfide bonds, which are conserved in Nogo receptor and hTLR2ED (Figure 1). High-mannose glycans were attached to each of the four glycosylation sites (Asn-114, Asn-199, Asn-414, and Asn-442) (Figure 1). All four potential glycosylation sites were solvent accessible. N-glycan structures were generated using the database of glycosidic linkage conformations [20]and in vacuo energy minimization to relieve unfavorable steric interactions. The Asn-GlcNAc linkage conformations were based on the observed range of crystallographic values [20], the torsion angles around the Asn Cα-Cβ and Cβ-Cγ bonds then being adjusted to eliminate unfavorable steric interactions between the glycans and the protein surface. The complexes involving ligands with C16 acyl chains were modeled simply by replacing the ligand head group of the crystallographic Pam3CSK4 as appropriate. All of the modeled head groups could be accommodated with no need to adjust any of the rest of the structure. The complexes of hTLR2ED with a C19 ligand (LTA C19) and Pim4 were modeled by keeping the head group position constant and allowing the protein structure to alter to accommodate the increased acyl chain length. This process was carried out by increasing the chain length one carbon at a time and using molecular dynamic simulations to allow relaxation of the local protein structure, while keeping the ligand head group and the regions of the protein that do not interact with the acyl chains fixed. This resulted in three of the outer-face loops changing their conformation slightly.
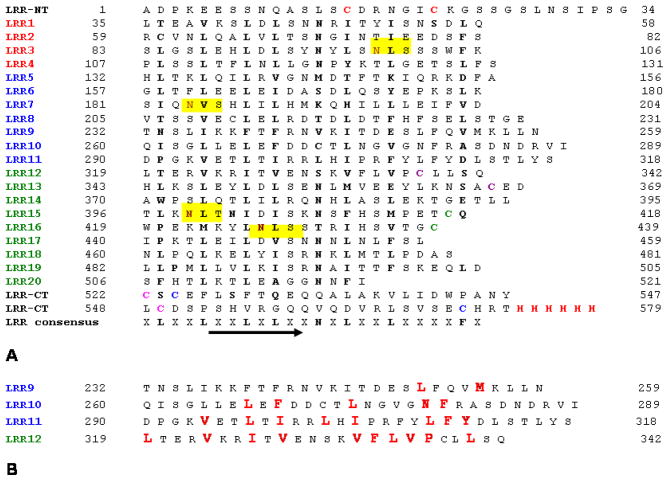
Figure 1. Protein sequence of hTLR2ED employed for modelling with its ligands.
hTLR2ED has 20 predicted LRR repeats which are capped at its N-terminal (LRR-NT) and a C-terminal (LRR-CT). The individual LRRs of hTLR2ED were aligned based on the hTLR2ED/VLR crystal structure solved by Jin et.al[12]. The N-terminal, Central and C-terminal LRRs are indicated in red, blue and green, respectively. Cysteines that are shown to form disulfide bonds in the crystal structure solved by Jin et.al[12] and cysteines from the C- terminal cap that are predicted to form disulfide bonds are color matched. Asparagines that are the potential glycosylation sites are highlighted in dark brown and the N-linked glycosylation consensus sequences are highlighted in yellow. The LRR consensus sequence are depicted and the position of the β strand is indicated by an arrow. The binding site in hTLR2ED is formed by LRR9,LRR 10,LRR 11 and LRR 12[12]. Mutations to Isoleucine 319 in the LRR11 (depicted in orange) have been reported [74] to abrogate the binding of TLR2 to Pam3CSK4.
Results
hTLR2ED is expressed a monomer in insect cells with internal disulfide bonds and high-mannose sugars
The hTLR2ED, consisting of an N-terminal cap, an LRR domain, a C-terminal cap, and a C-terminal His-tag, was successfully expressed using a baculovirus/insect cell expression system. Baculovirus for expression of hTLR2ED was generated by homologous recombination of a plasmid encoding hTLR2ED and linearized baculovirus DNA. Baculovirus amplification was monitored by fluorescent microscopy to detect expression of GFP encoded by the baculovirus DNA. After three steps of purification, highly purified hTLR2ED was obtained (Figure 2A). Moreover, hTLR2ED ran as an apparent monomer on size exclusion chromatography by comparison with molecular weight standards (Figure 2B). Digestion of hTLR2ED with PNGase F under native condition indicated the presence of N-linked glycosylation (Figure 2C). The decreased electrophoretic mobility of native and deglycosylated hTLR2ED under reducing conditions in the presence of β-mercaptoethanol was indicative of the presence of disulfide bonds (Figures 2D, E). This finding is consistent with the reported crystal structure of hTLR2ED, which reveals disulfide bonds in the N-terminal cap and in the LRR domain [12]. The identity of hTLR2ED was confirmed by anti-His-tag Western blot and by LC-MS peptide fingerprinting (Figures 3A–B). The theoretical molecular weight of the hTLR2ED protein is 65,734 Da (Protparam, SwissProt), while the molecular weight of the hTLR2ED determined by MALDI TOF MS is 69,903 Da (Figure 3C). Digestion of hTLR2ED with PNGase F under native condition indicated that this difference in mass is due to the presence of N-linked glycosylation (Figure 2C). The hTLR2ED has four N-linked glycosylation sites, which are necessary for its secretion [21], highlighting the importance of N-linked glycosylation in correct folding and stability [22–24]. Furthermore, we determined the type of glycosylation by releasing the N-linked glycans from recombinant hTLR2ED using in-gel digestion with PNGase F. We labeled the glycans with a fluorescent tag, digested with glucosidase II and α-jack bean mannosidase and analyzed their composition by HPLC with fluorescence detection (Figure 4A). The N-linked glycans were also identified by MALDI-TOF (Figure 4B) and negative ion CID mass spectrometry. These data indicate that hTLR2ED glycosylation is predominantly of the high-mannose type, which is consistent with the absence of secondary glycan processing in insect cells [25–26]. This result was further confirmed by glucosidase II and α-jack bean mannosidase digestions. Thus, the main glycan structures found in hTLR2ED were Fuc1Man3GlcNAc2, Man3GlcNAc2, Man9GlcNAc2 and Man9Glc1GlcNAc2 (Table 1).
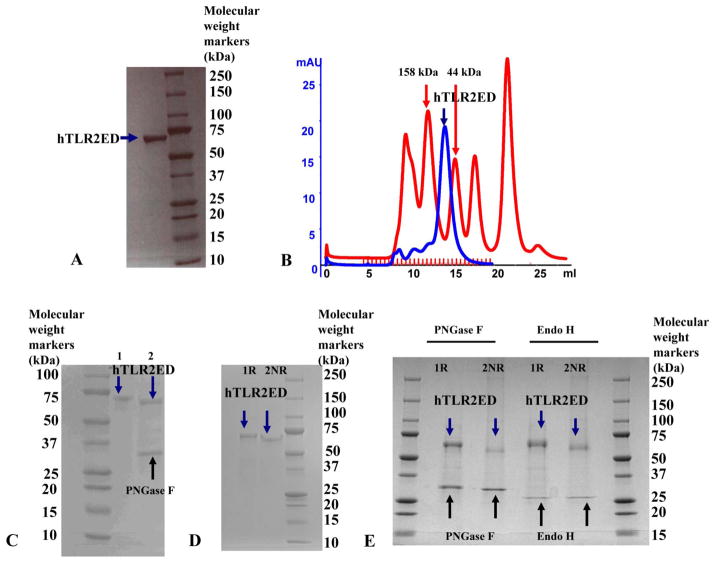
Figure 2. hTLR2ED expression and purification from insect cell culture.
A) Culture supernatants of insect cells infected with hTLR2ED Profold ER1 were concentrated and purified to homogeneity by Ni-NTA affinity, anion exchange and gel filtration chromatography. Samples were analyzed on 4–20% SDS-PAGE and stained with Coomassie Blue. B)hTLR2ED is a monomer in solution. Purified hTLR2ED and molecular weight standards were run on a Superdex 200 10/30 gel filtration column. Purified hTLR2 ED elutes at 14.2 ml, which corresponds to the elution volume of a protein of 70,000 Da, which is approximately the molecular mass of the hTLR2ED monomer(n.b. the molecular weight of the hTLR2ED monomer as determined by MALDI TOF mass spectrometry is 69,903 Da). C)hTLR2ED has N-linked glycosylation. hTLR2 undigested (1) and digested overnight with PNGase F in native conditions (2). D–E) hTLR2ED contains intramolecular disulfide bonds. D) Purified hTLR2ED was run under reducing (1R) and non-reducing (2NR) conditions in an SDS PAGE gel. The difference in electrophoretic mobility indicates the presence of disulfide bridges. E)hTLR2ED was digested overnight at 37°C with PNGase F and Endo H and then run in SDS PAGE under reducing (1R) and non-reducing (2NR) conditions (E). Molecular weight markers are shown on each gel in kDa.
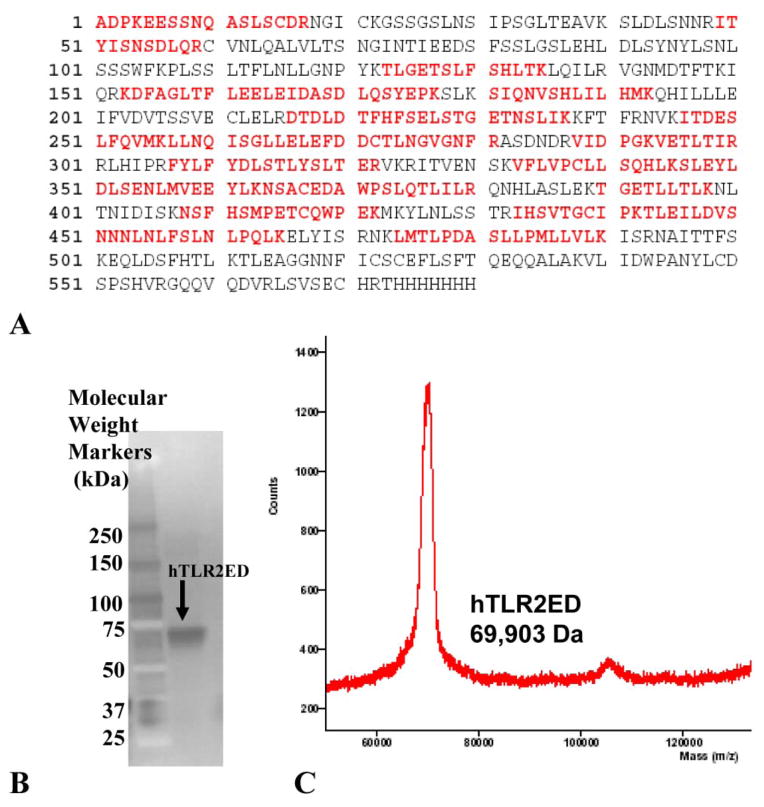
Figure 3. hTLR2ED expression using a baculovirus/insect cell expression system.
A) The identity of the recombinant hTLR2ED was confirmed by LC-MS/MS. 19 peptides which cover 49% of hTLR2ED protein sequence were identified using MASCOT and depicted in red. B) The identity of the hTLR2ED was also confirmed by Western blot against its C-terminal His tag. C) The molecular weight was determined by mass spectrometry (TSRI mass spectrometry core).
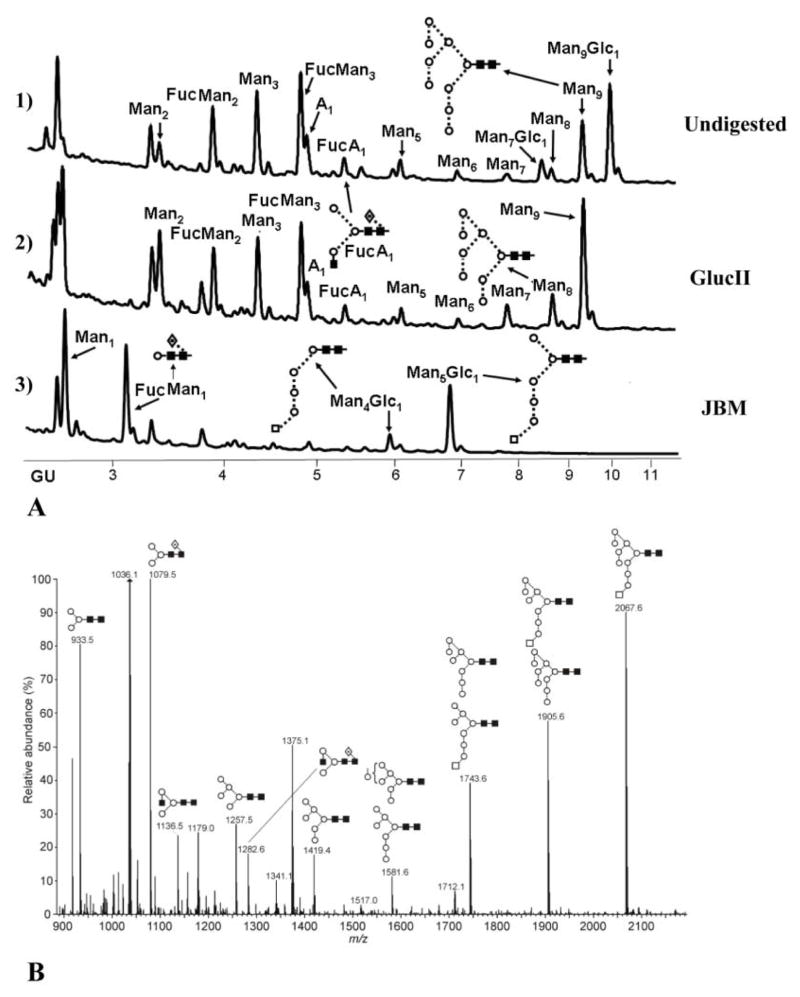
Figure 4. hTLR2ED expressed in insect cells is glycosylated with high mannose N-linked sugars.
A) HPLC chromatogram showing the profile of hTLR2ED glycans. The glycans were labeled with 2-AB and digested for 18 hours at 37°C with α-glucosidase II (Gluc II) and jack bean α-mannosidase (JBM) and run on a normal phase HPLC. An alternative nomenclature for naming the glycan structures was employed which does not mention the two core N-acetylglucosamine (GlcNAc2)which are present in all the glycan structures(for example: Man2 corresponds to Man2GlcNAc2). B) MALDI-TOF mass spectrum of N-linked glycans released from hTLR2ED by PNGaseF treatment. The spectrum has been processed by the MaxEnt 2 algorithm from MassLynx (Waters). Ions at m/z 1036, 1179, 1341, 1375 and 1712 are contaminants. Symbols and abbreviations used for the structures: Man: mannose (open circle), Glc: glucose (open square), Fuc: core fucose (diamond with central dot), N-acetylglucosamine (filled square), and A1: mono-antennary.
Table1. Masses and structures of TLR2 glycans.
Percentages and Glucose unit values (GU) of the N-linked glycans of hTLR2ED expressed in insect cells.
| Glycan assignment | GU(1) | m/z ( [M+Na]+) | m/z ([M+H2 PO4]−) | Composition | Structure | % Total (6) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Found | Calc. | Found | Calc. | Hex | HexNAc | Fuc | ||||
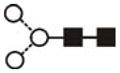
| Man2GlcNAc2 (Man2) | 3.40 | - | 771.3 | - | 845.2 | 2 | 2 | 0 |
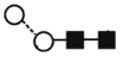 (2) (2) |
6.5 |
| FucMan2GlcNAc2 (FucMan2) | 3.94 | 917.5 | 917.3 | - | 991.3 | 2 | 2 | 1 |
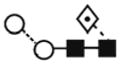 (3) (3) |
10.3 |
| Man3GlcNAc2 (Man3) | 4.37 | 933.5 | 933.3 | 1007.3 | 1007.3 | 3 | 2 | 0 |
|
12.6 |
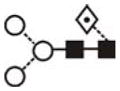
| FucMan3GlcNAc2 (FucMan3) | 4.84 | 1079.6 | 1079.4 | 1153.4 | 1153.4 | 3 | 2 | 1 |
|
17.5 |
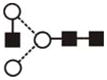
| GlcNAc Man3GlcNAc2 (A1) | 4.91 | 1136.5 | 1136.4 | 1210.4 | 1210.4 | 3 | 3 | 0 |
|
5.7 |
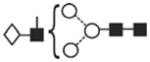
| GalGlcNAc Man3GlcNAc2 (A1G1) | (4) | - | 1298.4 | 1372.4 | 1372.4 | 4 | 3 | 0 |
 (5) (5) |
(4) |
| FucGalGlcNAc Man3GlcNAc2 (FucA1G1) | (4) | - | 1444.5 | - | 1518.5 | 4 | 3 | 1 |
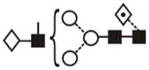 (3) (3) |
(4) |
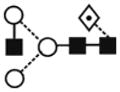
| Man5GlcNAc2 (Man5) | 6.14 | 1257.5 | 1257.4 | 1331.4 | 1331.4 | 5 | 2 | 0 |
|
2.9 |
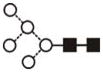
| FucGlcNAcMan3GlcNAc2 (FucA1) | 5.36 | 1282.6 | 1282.5 | 1356.5 | 1356.4 | 3 | 3 | 1 |
|
3.1 |
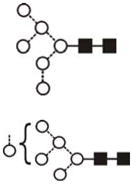
| Man6GlcNAc2 (Man6) | 7.06 | 1419.6 | 1419.5 | 1493.5 | 1493.5 | 6 | 2 | 0 |
 (Trace) (Trace) |
1.8 |
| Man7GlcNAc2 (Man7) | 7.96 | 1581.6 | 1581.5 | 1655.5 | 1655.5 | 7 | 2 | 0 |
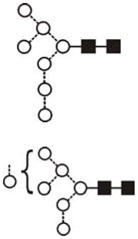
|
1.5 |
| Man8GlcNAc2 (Man8) | 8.85 | 1743.5 | 1743.6 | 1817.6 | 1817.6 | 8 | 2 | 0 |
|
2.3 |
| Glc1Man7GlcNAc2 (Man7Glc1) | 8.65 |
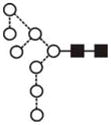
|
4.2 | |||||||
| Man9GlcNAc2 (Man9) | 9.53 | 1905.5 | 1905.6 | 1979.6 | 1979.6 | 9 | 2 | 0 |
|
10.7 |
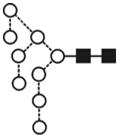
| Glc1Man8GlcNAc2 (Man8Glc1) | (4) |
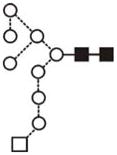
|
(4) | |||||||
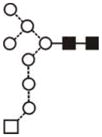
| Glc1Man9GlcNAc2 (Man9 Glc1) | 10.16 | 2067.7 | 2067.7 | 2141.7 | 2141.7 | 10 | 2 | 0 |
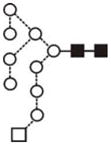
|
17.5 |
| Unknown | 9.73 10.36 |
- | - | - | - | - | - | - | - | 3.4 |
The most abundant glycan structures are Man9Glc1GlcNAc2, Man9GlcNAc2, fucosylated-Man3GlcNAc2, Man3GlcNAc2. The alternative nomenclature of glycan structures, depicted in the HPLC Chromatogram, is shown in parentheses. References: GU glucose units: Man: mannose (open circle), Glc: glucose (open square), Gal: galactose (diamond), Fuc: fucose (diamond with central dotGlcNAc:N-acetylglucosamine (filled square), A1: mono-antennary. (1) Listed in order of GU value. (2) Not detected by mass spectrometry. (3) Not detected by ESI mass spectrometry; structure not confirmed by fragmentation.(4) Not detected by HPLC.(5) Trace amount by mass spectrometry; structure not confirmed by fragmentation.(6) As measured by HPLC.
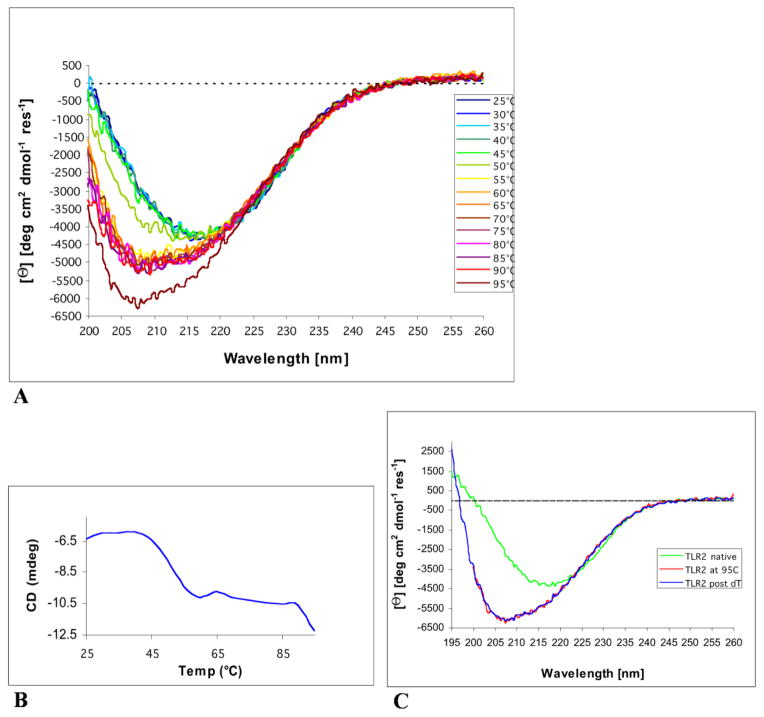
Thermal denaturation of hTLR2ED is an irreversible two-stage process
The CD spectrum of hTLR2ED at 25°C revealed a minimum between 215 and 220 nm (Figure 5A), which is indicative of a protein fold dominated by β-sheet secondary structure[27]. Furthermore, thermal denaturation of hTLR2ED, as monitored by CD spectroscopy, uncovered a two stage process (Figures 5A, B). The first stage occurred between 45°C and 55°C and another transition occurs between 90°C and 95°C (Figure 5A, B). After denaturation at 95°C, hTLR2ED was slowly cooled back down to 25°C and another CD spectrum was recorded. This spectrum showed no difference from the spectrum recorded at 95°C demonstrating that thermal denaturation of hTLR2ED appears to be an irreversible process (Figure 5C).
Figure 5. Circular Dichroism analysis of hTLR2ED.
A) CD spectra of hTLR2ED recorded at various temperatures. The CD spectrum of hTLR2ED at 25°C reveals a minimum around 215 nm that is indicative of a folded protein with a significant portion of β-sheet secondary structure. At temperatures greater than 50°C, the β-sheet character of the hTLR2ED CD spectra is lost and the shape of the spectra indicates that the hTLR2ED is now mostly unfolded. B) Thermal denaturation of hTLR2ED is a two-stage process. The CD signal at 208 nm was plotted against the corresponding temperatures. hTLR2ED undergoes two unfolding transitions: at the first Tm of 45–55°C, and a second Tm of 90–95°C, the protein becomes mostly unfolded. C) The thermal denaturation of hTLR2ED is irreversible. After the CD spectra of hTLR2ED were recorded at 95°C, hTLR2ED was slowly cooled back down to 25°C and new CD spectra were recorded (TLR2 post dT). There were no differences between the spectra at 95°C and after re-cooling, indicating that no secondary structure is regained and that the thermal denaturation of hTLR2ED is irreversible.
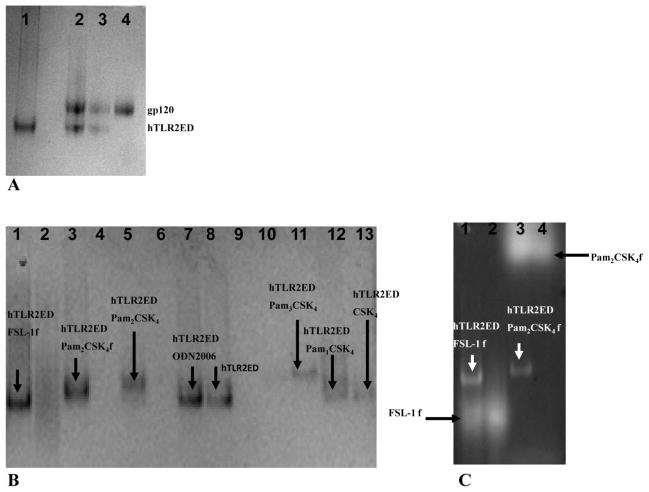
hTLR2ED does not bind gp120 from HIV-1
The hTLR2ED was employed for binding to gp120 from HIV-1, different lipopeptides, and glycolipids (Figure 6). Because levels of soluble TLR2 have been shown to correlate with HIV progression and increase after anti-retroviral treatment [28], we tested whether hTLR2ED ectodomain is able to recognize the surface viral glycoprotein gp120 from HIV-1. We determined that hTLR2ED does not bind gp120 as no complex was observed in native PAGE (Figure 7A).
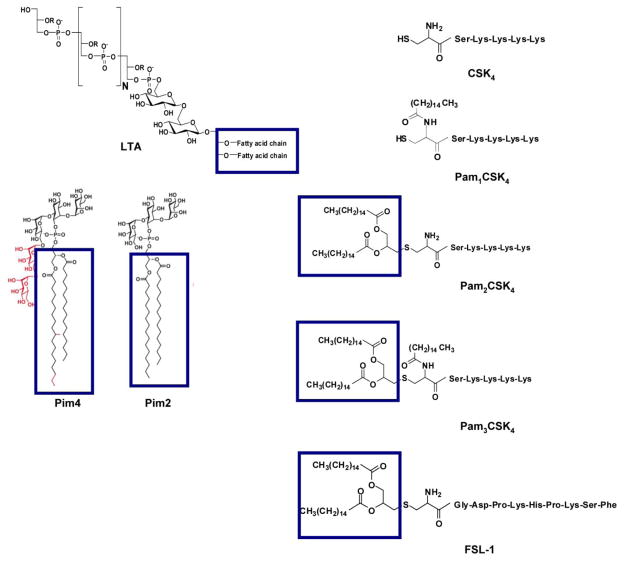
Figure 6. The chemical structures oflipopeptides and glycolipids employed for binding studies with hTLR2ED.
The diacylglycerol moieties that are involved in the interaction with hTLR2ED are highlighted by boxes.
Figure 7. hTLR2ED binds diacyl and triacyllipopeptides, but not HIV-1 gp120.
Samples were incubated overnight at 37°C and run for 4 hours at 100 volts on a native 4–20% native PAGE. Native gels stained with Coomassie Blue. A) hTLR2EDincubated with a clade B HIV-1gp120. Lane 1 1- 10 μg of hTLR2ED; lane 2- 10 μg hTLR2ED with 10 μg gp120; lane 3- 1 μg hTLR2ED with 1μg gp120; lane 4- 10 μg gp120. B–C) hTLR2ED incubated with lipopeptides. Native gel stained with Coomassie Blue (B) and corresponding fluorescence determined using a Versadoc imaging system (C). Lane 1-10 μg hTLR2ED with 1μg FSL-1 fluorescein (FSL-1f); Lane 2- 1 μg FSL-1f; Lane 3-10 μg hTLR2ED with 1μg Pam2CSK4 fluorescein (Pam2CSK4f); Lane 4-1μg Pam2CSK4f; Lane 5-10 μg hTLR2ED with1μg Pam2CSK4: Lane 6-1μg of Pam3CSK4; ); Lane 7- 10 μghTLR2ED; Lane 8-10 μg hTLR2ED with1μg ODN2006 (human TLR9 activator; Lane 9- 1 μg ODN2006; Lane 10-1μg Pam2CSK4; Lane 11- 10 μg of hTLR2ED with1μg Pam3CSK4; Lane 12-10 μg hTLR2ED with1μg Pam1CSK4: Lane 13-10 μg hTLR2ED with 1μg CSK4. Fluorescence of Pam2CSK4f-hTLR2ED and FSL1f-hTLR2EDis indicated by arrows.
hTLR2ED binds synthetic diacylglycerol lipopeptides but not monoacylated lipopeptides
Native polyacrylamide gel electrophoresis (native PAGE) experiments showed that hTLR2ED binds synthetic lipopeptides (Figures 7B, C). A gel shift in the presence of Pam2CSK4 and Pam2CSK4-fluorescein (Pam2CSK4f), but not CSK4 and Pam1CSK4, indicated that hTLR2ED binds the two fatty acid chains linked to cysteinyl S-glycerol (Figures 7B, C) in accord with the crystal structure of hTLR2ED-Pam3CSK4[12]. Likewise, the gel shift of hTLR2ED incubated with Fibroblast Stimulating Factor-1 fluorescein (FSL-1f) also reflects recognition of the palmitic acid chains connected to the cysteinyl S-glycerol of FSL-1(Figure 7B). Binding of FSL-1f to hTLR2ED was confirmed by co-localization of hTLR2ED with the fluorescence as with Pam2CSK4f (Figure 6C). As expected, hTLR2ED does not bind synthetic human TLR9 ligand CpG-ODN2006 [29–31] or the synthetic lipopeptide Pam3CSK4 (Figure 7B).
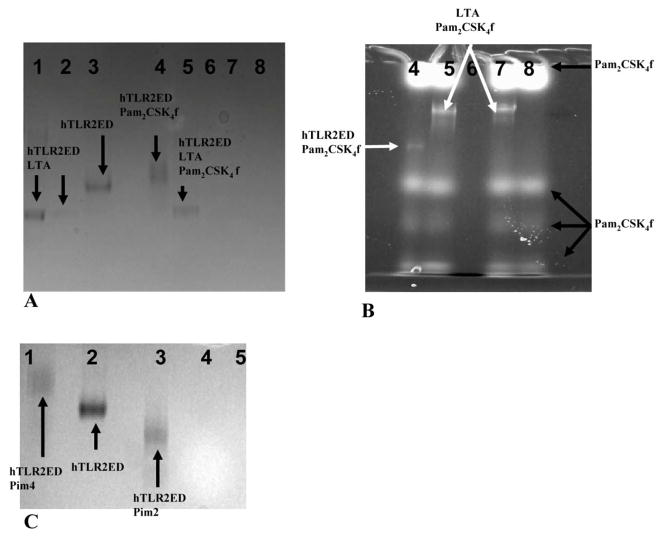
hTLR2ED binds LTA from S. aureus but cannot bind Pam2CSK4 and LTA simultaneously
A considerable increase in the electrophoretic mobility was observed for the hTLR2ED-LTA complex (Figure 8A), consistent with the presence of the polyglycerophosphate moiety in LTA Interestingly, native PAGE of hTLR2ED co-incubated with both LTA from S. aureus and synthetic Pam2CSK4f showed that hTLR2ED preferentially binds to LTA rather than to Pam2CSK4f (Figures 8A, 8B).
Figure 8. hTLR2ED binds LTA from S. aureus and mycobacterial Pims.
A–B) hTLR2ED incubated with LTA and/or Pam2CSK4f. A) Native gel stained with Coomassie Blue and B) fluorescence determined using a Versadoc imaging system. Lane 1-10 μg hTLR2EDwith 1μg LTA; Lane 2- 1 μg hTLR2ED with 1μg of LTA; Lane 3-10 μg hTLR2ED; Lane 4- 10 μg hTLR2ED with 1μg Pam2CSK4 fluorescein (Pam2CSK4f); Lane 5-10 μg hTLR2ED with 1μg Pam2CSK4f and 1 μg LTA; Lane 6- 1 μg LTA; Lane 7- 1 μg Pam2CSK4f plus 1 μg LTA; Lane 8- 1 μg Pam2CSK4f. C)hTLR2ED incubated with Pims. Native gel stained with Coomassie Blue. Lane 1-10 μg hTLR2ED with 1μg Pim4; Lane 2- 10 μg hTLR2ED; Lane 3-10 μg hTLR2ED with 1μg of Pim2; Lane 4-1μg Pim2: Lane 5-1μg Pim4. The chemical structures of Pim4 and Pim2 are shown in Figure 3, with the two extra mannoses and extra three carbon atoms of Pim4 highlighted in red.
hTLR2ED binds synthetic phopshatidylinositol mannosides (Pim2 and Pim4)
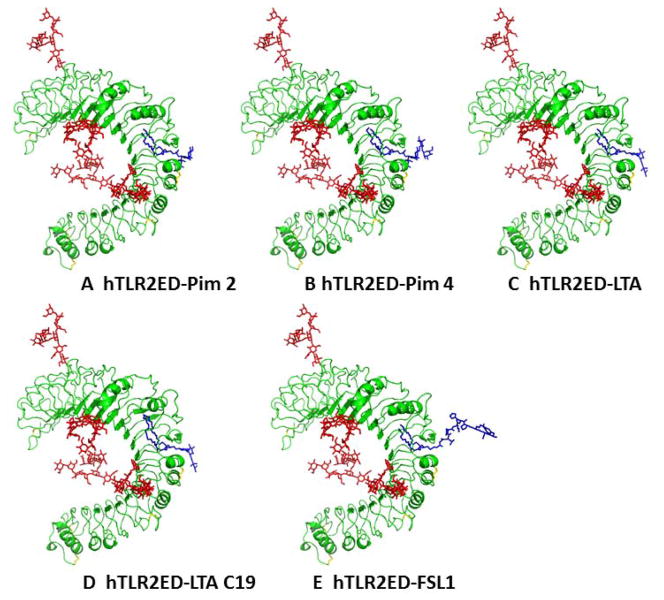
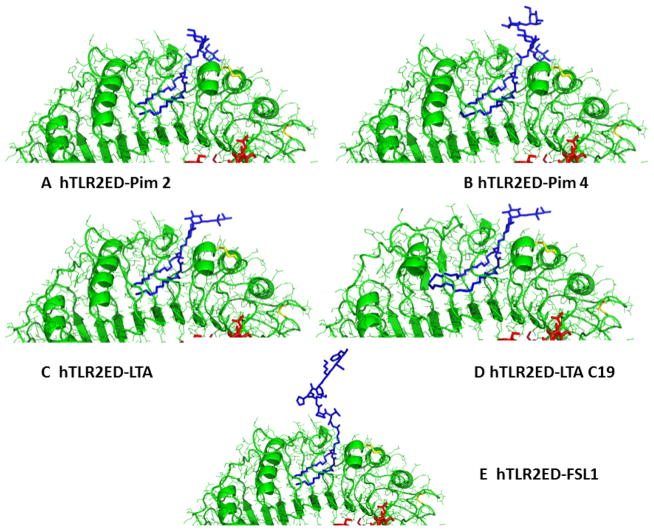
We also determined whether hTLR2ED binds other important glycolipids, such as mycobacterial phosphatidylinositol mannosides, which were previously shown to be ligands for the TLR2/TLR1 heterocomplex [32]. For these binding assays, we employed synthetic phosphatidyl inositol mannosides, which we had previously shown to bind CD36, but not induce TNF-α secretion by macrophages [10]. We also found that hTLR2ED binds to lipoarabinomannan precursors (Pim2 and Pim4) (Figure 8C). The negative charge of Pim2 increased the mobility in the native gel of the complex hTLR2ED-Pim2. In contrast, the extra two mannose and the extra three carbons in one of the acyl chains of Pim4 (Figure 6) increased the mobility of hTLR2ED-Pim4 (Figure 8C). Previous published results showed that human TLR2 has an impaired recognition of lipopeptides with acyl chains shorter than 16 carbons atoms (C16) [33] and can recognize longer acyl chains than C16 [34]. We report here that human TLR2 can bind glycolipids with an acyl chain of 19 carbons atoms (C19) demonstrating that human TLR2 can indeed accommodate acyl chains longer than C16 in its hydrophobic binding pocket. Taken together, these binding assays demonstrate that the presence of two acyl chains linked to glycerol is the essential ligand moiety recognized by TLR2 ligands, and that both glycolipids and lipopeptides bind to the same binding site in hTLR2ED. Based on the crystal structure of hTLR2ED-Pam3CSK4 [12], we modeled the hTLR2ED- FSL-1, hTLR2ED-LTA, hTLR2-LTA (C19), hTLR2ED-Pim2, hTLR2-Pim4 complexes (Figure 9, 10). No clashes were found between any of the ligand head groups and hTLR2ED, indicating that the binding pocket of hTLR2ED might accommodate all of these ligands without major structural rearrangements. In addition, our models suggest that the hTLR2ED could also accommodate ligands with longer acyl chains than C16 without displacement of the ligand head group and only slight conformational adjustments of some of the loops on the outer face of hTLR2ED.
Figure 9. Modeling of hTLR2ED in complex with A) Pim2, B) Pim4, C) LTA, and D) LTA 19C, and E) FSL-1.
Modeling is based on the crystal structures of hTLR2ED in complex with Pam3CSK4 (Protein Data Bank 2Z7X) [12] and the Nogo receptor (PDB 1OZN) [19]. The most abundant N-linked glycans from the glycan analysis were added to the hTLR2ED models: Man3GlcNAc2 (Asn114), Man9GlcNAc2 (Asn199), Man3Fuc1GlcNAc2 (Asn414), Man9Glc1GlcNAc2 (Asn442). All ligands have two acyl chains with 16 carbon atoms, except for LTA 19C, which has two acyl chains with 19 carbon atoms and Pim4 which has one acyl chain of 19 carbon atoms and another of 16 carbon atoms.
Figure 10. Binding sites of hTLR2ED.
Close up views of the modeled binding sites for hTLR2ED in complex with A) Pim2, B) Pim4, C) LTA, and D) LTA 19C, and E) FSL-1.
Discussion
TLRs are one of the most important groups of receptors responsible for the recognition of bacteria, virus, parasites and initiation of a host immune response. TLRs are pathogen pattern-recognition receptors (PPRs) because they recognize conserved structures in macromolecules in these microorganisms. The crystal structures of hTLR2ED-Pam3CSK4, mTLR2ED-Pam2CSK4, and mTLR2ED-Pam3CSK4 showed that the two acyl chains connected to the cysteinyl S-glycerol occupy the hydrophobic binding pocket of TLR2ED[12].
Our aim was to investigate binding of other relevant lipopeptides and glycopeptides to hTLR2ED. To address this issue, we expressed and purified the complete ectodomain of human TLR2 (hTLR2ED) using a baculovirus expression system. The purified hTLR2ED is a monomer in solution with intramolecular disulfide bonds and N-linked glycosylation of the high-mannose type. It is likely that, besides disulfide bonds in the N-terminal cap and LRR domain, two disulfide bonds are also likely to be present in the C-terminal cap domain (Figure 1). This assumption is based on the observation of disulfide bonds in similar positions in hTLR3ED [35–37] and the Nogo receptor [19]. Employing CD spectroscopy we followed the thermal stability of hTLR2ED. We employed different algorithms to determine the remaining secondary structure, but did not obtain a satisfactory result, as the theoretical curves were statistically far away from the experimental curves. Although some secondary structure remains at 50ºC, the majority of the secondary structure appears to be lost at 95ºC as the curve shifts to a peak of 208 nm (random coil). Furthermore, although we did not measure the aggregation state, it is likely that the loss of secondary structure shown as the curve shifts to 208 nm would expose to the solvent the hydrophobic residues of the LRR motifs and result in aggregation. The relatively low value of ~45–55°C for the first melting point of hTLR2ED suggests that its secondary structure is thermally not very stable and might also indicate some structural flexibility. Asparagines (“asparagine ladder”) and phenylalanines (“phenylalanine spine”) in the consensus LRR repeats are known to be relevant for stabilization of LRR proteins through hydrogen bond formation and hydrophobic interactions, respectively[38]. The lack of those particular features in the central LRR repeats of hTLR2ED [12] (Figure 1) may render this domain more flexible and able to accommodate ligands with long acyl chains, thereby contributing to an explanation of its low melting point. We have previously tried binding methods such as Surface Plasmon Resonance to evaluate binding of ligands to TLR2, but did not achieve satisfactory results. Thus, we decided to employ native PAGE, which we have also successfully employed to study the binding to TLRs and accessory molecules such as TLR3 with dsRNA [37], CD36 with LTA, Pims, FSL-1 [10], MD-1 with LPS [39], and TLR5 with flagellin [40]. Electrophoretic mobility shift assays (EMSA) have also been successfully used to study the binding between LTA-CD14 [41] and TLR4-MD-2 with LPS [42]. Ligand binding alters the hydrodynamic size and/or charge of the protein, which then manifests itself in different electrophoretic mobilities of the liganded and unliganded forms. Here, we have demonstrated that hTLR2ED directly binds synthetic, diacyl- (FSL-1 and Pam2CSK4) and triacyl-lipopeptides (Pam3CSK4), purified LTA from S. aureus, and synthetic phosphatidylinositol mannosides (Pim2 and Pim4), in absence of TLR1 or TLR6. Soluble isoforms of hTLR2ED present in plasma [43], breast milk [43–44], amniotic fluids [45], saliva [46] and expressed in HEK cells [47] can act as a decoys of TLR2 and inhibit NFκB activation by transmembrane TLR2 [48]. The hTLR2ED expressed in insect cells should have the same inhibitory effect as the hTLR2ED from natural sources as the only difference with them would be in the types of glycan structures, which should not be relevant for binding as the glycosylation sites are not located near the binding site of hTLR2ED. Therefore, our ligand-binding studies under physiological conditions suggest that ligand competition between secreted isoforms of hTLR2ED and the transmembrane hTLR2 receptor is likely to be responsible for the down-regulation of TLR2 activation. Soluble TLR2 has also been implicated in the pathogenesis of HIV [28, 44]. Thus, we investigated whether hTLR2ED could interact with HIV-1 gp120. In native PAGE experiments, we did not observe binding between hTLR2ED and gp120 from HIV-1, consistent with gp120 not being a lipoprotein.
One of the main classes of TLR2 ligands are microbial lipoproteins. Synthetic lipopeptides can mimic the activity of microbial lipoproteins and, therefore, we investigated interaction of these peptides with hTLR2ED. CSK4 represents the amino-acid moiety of synthetic lipopeptides, such as Pam1CSK4, Pam2CSK4, and Pam3CSK4. No appreciable differences were observed in the electrophoretic mobility of hTLR2ED with and without CSK4, demonstrating that hTLR2ED does not interact with the amino-acid moiety of lipopeptides. Although no shift was seen for the hTLR2ED-Pam1CSK4 complex, a gel shift is observed when hTLR2ED binds Pam2CSK4. Although we did not detect binding of Pam1CSK4 to hTLR2ED, it is still possible that hTLR2ED may bind Pam1CSK4 with low affinity. The absence of the diacylglycerol moiety in Pam1CSK4 explains its low biological activity compared to Pam2CSK4 [49].
The crystal structure of the hTLR2ED/hTLR1ED complex reveals that TLR2 binds the two acyl chains linked to the cysteinyl S-glycerol of Pam3CSK4, and not the N-acyl chain which is bound by TLR1. The two cysteinyl S-glycerol fatty acid chains can establish a greater number of hydrophobic interactions and, thus, form a more stable complex with hTLR2ED.
Fibroblast-stimulating lipopeptide-1 (FSL-1) is a synthetic diacyl-lipopeptide that mimics the N-terminal sequence of the 44-kDa lipoprotein LP44 of Mycoplasma salivarium [50–52]. FSL-1 is a potent activator of antigen-presenting cells and is a useful tool for treatment of cancer and viral infections [53]. In native PAGE experiments, we observed that hTLR2ED binds Pam2CSK4 and FSL-1 fluorescent ligands. FSL-1 shares the cysteinyl S-glycerolipid of Pam2CSK4, but differs in the amino acids that are bound to the cysteine. Therefore, as in the case of Pam2CSK4, hTLR2ED is likely to interact with the cysteinyl S-diacyl glycerol moiety of FSL-1.
Peptidoglycan was thought to be a ligand for TLR2, but a recent report clearly showed that a purified peptidoglycan shows no activity, and that earlier reported activity was caused by the presence of contaminants, such as LTA[54]. Recent reports [55–58] also challenge the concept of LTA as a ligand for TLR2, suggesting that its reported activity is actually caused by contaminating lipoproteins. On the other hand, synthetic S. aureus LTA was reported to induce secretion of TNF-α from whole blood cells and macrophages in a TLR2-dependent manner[59–60].
Although peptidoglycan does not have fatty acid chains, LTA has a diacylglycerol structure similar to FSL-1, Pam2CSK4, and Pam3CSK4. Here, we show a considerable increase in the electrophoretic mobility of hTLR2ED after incubation with LTA from S. aureus. LTA from S. aureus consists of up to 50 negatively charged glycerophosphate units linked to a gentiobiosyl diacylglycerolipid that anchors the LTA to the plasma membrane [59], thus explaining the increased electrophoretic mobility of the hTLR2ED-LTA complex compared to uncomplexed hTLR2ED. Similarly increased electrophoretic mobility was shown for CD14-LTA and LBP-LTA complexes as compared with unliganded proteins in native PAGE experiments [41]. We also found that LTA can form aggregates with synthetic Pam2CSK4f, but that these complexes are not recognized by hTLR2ED. In fact, the electophoretic mobility of hTLR2ED incubated with LTA and Pam2CSK4f, is identical to that obtained when hTLR2ED is incubated with LTA alone. The acyl chains of the diacylglycerol anchor, as in the case of synthetic lipopeptides, are likely to be the motif responsible for interaction with hTLR2ED. The crystal structure of mTLR2ED/VLR in complex with LTA from S. pneumoniae also demonstrates the relevance of the diacylglycerol of LTA as the ligand binding moiety recognized by TLR2[13]. However LTA from S. pneumoniae has been shown to have an activity independent of TLR2 [58], showing that binding to the hydrophobic pocket of TLR2 is necessary but not sufficient for a microbial ligand to activate TLR2.
The acyl chains of LTA from S. aureus have a length between C14–C19[59], while those in Pam2CSK4 have only 16 carbon atoms (palmitates). Our modeling based on the published crystal structure of TLR2 [12] shows that the hydrophobic pocket can accommodate longer fatty acid chains than palmitic acid with minimal conformational changes to the outer loops of hTLR2ED. Thus, the extra carbon residues of some LTA molecules could establish additional hydrophobic interactions compared to Pam2CSK4 and could thus explain the preferential binding of hTLR2ED for LTA from S. aureus. Therefore, the binding of Pim4, which has one of the two acyl chains composed of 19 carbon atoms, confirms that TLR2 can accommodate ligands with acyl chains longer than 16 carbon atoms. Mycobacterium is an intracellular pathogen that lives inside macrophages, inhibiting macrophage activation and phagosome maturation [61–63]. Mycobacterium tuberculosis (M. tuberculosis) causes around two million deaths per year worldwide [64]. TLR2 has been shown to be important for controlling M. tuberculosis [65] and Mycobacterium leprae(M. leprae) [66] infections. Mycobacterium possess a unique cell wall structure rich in long fatty acids (mycolic acid), lipoglycans (lipoarabinomannan) and polysaccharides. Lipoarabinomannan is composed of three structures: a phosphatidyl-mannosyl-myo-inositol anchor (MPI), a polysaccharide backbone composed of D-arabinan and D-mannan, and chemical structures attached to the arabinan core termed ‘caps’. According to the type of cap motif, it is possible to classify lipoarabinomannans into three classes: if the cap is an oligomannose type, it is termed ManLAM; if phosphatidylinositol, it is termed PILAM; and, if no cap, it is termed AraLAM. PILAM is characteristic of fast growing mycobacteria, such as Mycobacterium. fortuitum and Mycobacterium smegmatis, while slow growing mycobacteria, such as M. tuberculosis and M. leprae, have ManLAM structures in the cell wall. Mycobacterium chelonae has araLAM in its cell wall [67]. Here, we show that the hTLR2ED is able to interact directly with synthetic phosphatidylinositol mannosides, demonstrating that Pim2 is the minimal lipoarabinomannan structure required for binding to the hTLR2ED. The phosphate group from myo-inositol phosphate enables the hTLR2ED-Pim2 complex to run with increased electrophoretic mobility compared to unliganded hTLR2ED. Changes in mobility of hTLR2ED-Pim2 by oligomerization of hTLR2ED are unlikely, as an oligomer should decrease the electrophoretic mobility, as for example TLR4 MD2 in complex with lipid A [42]. For Pim4, the extra two mannose residues and extra three carbon residues in one of its fatty acid chains seem to compensate for the negative charge of the phosphate group and make the complex hTLR2ED-Pim4 run with decreased electrophoretic mobility as compared to uncomplexed hTLR2ED. We have seen similar decreases in mobility by addition of one acyl chain in the case of the complex of hTLR2ED-Pam3CSK4 compared with hTLR2ED-Pam2CSK4. Although oligomerization of hTLR2ED could be another explanation for the decrease in mobility of hTLR2ED in complex with Pim4, Pam3CSK4 and Pam2CSK4, gel filtration of TLR2 ectodomain in complex with lipopeptides and glycolipids has been reported before showing that there is no oligomerization of TLR2 ectodomain [13].
It is likely that hTLR2ED also binds to the acyl chains linked to the diacylglycerol moieties, as for lipopeptides. Although synthetic Pim2 and Pim4 were able to bind TLR2, they were not able to support TNF-α secretion from C57BL/6 mice[10]. Pims also have been shown to inhibit the activation of TLR2 in a CD14 independent way [68], decrease allergic response [69–70] and affect dendritic cell maturation [71]. Lipoarabinomannan and phosphatidylinositol mannosides were reported to be sensed by TLR1 and TLR2[41]. The failure to activate the TLR2 signalling pathway by these synthetic Pims likely results from the lack of a third acyl chain, which is necessary for TLR1 binding [72–73]. Therefore, the capacity of Pims to bind but not activate the TLR2 signalling pathway could explain their inhibitory properties. The binding site of hTLR2ED is situated on its convex face, while the concave face of hTLR2ED is covered by glycosylation, making it an unlikely site for binding ligands. The hydrophobic binding pocket of hTLR2ED, situated in the transition between the central LRR domain and the C-terminal LRR domain (LRR9-12), is the only binding site big enough to accommodate diacylglycerol structures [12] making it unlikely that ligands bind to a secondary site even at the high concentration of ligands employed. However, to rule out any possible nonspecific binding, future ligand binding experiments employing hTLR2ED with mutations that abrogate ligand binding, such as the mutation of Isoleucine 319 to Aspartate in the LRR 11 domain of human TLR2 [74] (Figure 1), could be performed. We have used in vitro conditions to test the binding of hTLR2ED with its ligands, where ligand concentrations are higher than in vivo conditions so that the experiments can be performed in the absence of accessory molecules such as CD36 or CD14. In vivo CD14 and CD36 are relevant to efficiently load ligands into TLR2 and activate the TLR2-dependent signaling [10]. Because of differences in the hydrophobic pocket of TLR2 between human and other species, such as murine [33] and equine[75], mammals may not be suitable models for the development of inhibitors for human TLR2. Instead, ligand binding assays such as those using native PAGE may be a better approach for evaluating inhibitors for human TLR2.
In summary, we showed that hTLR2ED is able to bind synthetic FSL-1, Pim2, Pim4, and LTA from S. aureus without the presence of its co-receptors TLR1 and TLR6. Although the structures of ligands recognized by hTLR2ED are all different, they all share the presence of two fatty acid chains linked to glycerol that is recognized by hTLR2ED (Figures 6, 9, 10). Overall, our data indicate the presence of a single binding site in hTLR2ED for microbial glycolipids and lipoproteins. Inhibitors with longer acyl chains than 16 carbon atoms, which bind but do not activate TLR2 signaling, could represent a novel strategy for the treatment of septic shock caused by Gram-positive bacteria.
Acknowledgments
We thank Dr. Terry Butters for providing the α-glucosidase II as well as helpful discussions, Julie Vanhnasy for technical support, and Drs. Robyn Stanfield and Adam Corper for comments and suggestions for the manuscript. This study was supported by NIH grant AI42266 (IAW), and by a Joint Scripps/Oxford Graduate Scholarship. We also thank the Wellcome Trust for grants to purchase the TofSpec and Q-TOF mass spectrometers and the Oxford Glycobiology Bequest for additional funding.
The abbreviations used are
- TLRs
toll-like receptors
- hTLR2ED
human toll-like receptor 2 ectodomain
- HIV
human immunodeficiency virus
- LTA
lipoteichoic acid
- gp120
glycoprotein 120
- Pim2
phosphatidyl inositol mannoside 2
- GPI
glycosylphosphatidylinositol
- C19
19 carbon atoms
- FSL-1
fibroblast stimulating lipopeptide-1
- FSL-1f
fibroblast stimulating lipopeptide-1 fluorescein
- EMSA
electrophoretic mobility shift assay
- ESI
electrospray ionization
- CD
cluster of differentiation
- VLR
variable lymphocyte receptor (VLR)
- 2-AB
2- aminobenzamide
- ODN
oligodeoxynucleotide
- Pam3CSK4
tripalmitoyl-cysteinyl-seryl-(lysyl)3-lysine
- Pam2CSK4
dipalmitoyl-cysteinyl-seryl-(lysyl)3-lysine
- Pam2CSK4f
dipalmitoyl-cysteinyl-seryl-(lysyl)3-lysine fluorescein
- PamCSK4
palmitoyl-cysteinyl-seryl-(lysyl)3-lysine
- CSK4
cysteinyl-seryl-(lysyl)3-lysine
- GFP
green fluorescent protein
- PBS
phosphate buffer saline
- PPRs
pattern-recognition receptors
- Man
mannose
- Glc
glucose
- Fuc
fucose
- GlucNac
N-acetylglucosamine
- A1
mono-antennary
- Gal
galactose
- Ni-NTA
nickel-nitriloacetic acid
- LRR
leucine-rich repeats
- HPLC
high-performance liquid chromatography
- FPLC
fast protein liquid chromatography
- MS
mass spectrometry
- MALDI TOF
matrix-assisted laser desorption and ionization time of flight
- S. aureus
Staphylococcus aureus
- S. pneumoniae
Streptococcus pneumoniae
- M
Mycobacterium
Footnotes
Competing interest: The authors have declared that they have no competing interests.
References
- 1.Akira S. TLR signaling. Curr Top Microbiol Immunol. 2006;311:1–16. doi: 10.1007/3-540-32636-7_1. [DOI] [PubMed] [Google Scholar]
- 2.Moresco EM, LaVine D, Beutler B. Toll-like receptors. Curr Biol. 2011;21:R488–493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 3.Vasselon T, Detmers PA, Charron D, et al. TLR2 recognizes a bacterial lipopeptide through direct binding. J Immunol. 2004;173:7401–7405. doi: 10.4049/jimmunol.173.12.7401. [DOI] [PubMed] [Google Scholar]
- 4.Murakami S, Iwaki D, Mitsuzawa H, et al. Surfactant protein A inhibits peptidoglycan-induced tumor necrosis factor-alpha secretion in U937 cells and alveolar macrophages by direct interaction with toll-like receptor 2. J Biol Chem. 2002;277:6830–6837. doi: 10.1074/jbc.M106671200. [DOI] [PubMed] [Google Scholar]
- 5.Massari P, Visintin A, Gunawardana J, et al. Meningococcal porin PorB binds to TLR2 and requires TLR1 for signaling. J Immunol. 2006;176:2373–2380. doi: 10.4049/jimmunol.176.4.2373. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Lee DS, Madrenas J. Evolving Bacterial Envelopes and Plasticity of TLR2-Dependent Responses: Basic Research and Translational Opportunities. Front Immunol. 2013;4:347. doi: 10.3389/fimmu.2013.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozinsky A, Underhill DM, Fontenot JD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahringer U, Lindner B, Inamura S, et al. TLR2 - promiscuous or specific? A critical reevaluation of a receptor expressing apparent broad specificity. Immunobiology. 2008;213:205–224. doi: 10.1016/j.imbio.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Hoebe K, Georgel P, Rutschmann S, et al. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez-Dalmaroni MJ, Xiao N, Corper AL, et al. Soluble CD36 ectodomain binds negatively charged diacylglycerol ligands and acts as a co-receptor for TLR2. PLoS One. 2009;4:e7411. doi: 10.1371/journal.pone.0007411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Bergenhenegouwen J, Plantinga TS, Joosten LA, et al. TLR2 & Co: a critical analysis of the complex interactions between TLR2 and coreceptors. J Leukoc Biol. 2013;94:885–902. doi: 10.1189/jlb.0113003. [DOI] [PubMed] [Google Scholar]
- 12.Jin MS, Kim SE, Heo JY, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Kang JY, Nan X, Jin MS, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Monie TP, Moncrieffe MC, Gay NJ. Structure and regulation of cytoplasmic adapter proteins involved in innate immune signaling. Immunol Rev. 2009;227:161–175. doi: 10.1111/j.1600-065X.2008.00735.x. [DOI] [PubMed] [Google Scholar]
- 15.Radcliffe CM, Arnold JN, Suter DM, et al. Human follicular lymphoma cells contain oligomannose glycans in the antigen-binding site of the B-cell receptor. J Biol Chem. 2007;282:7405–7415. doi: 10.1074/jbc.M602690200. [DOI] [PubMed] [Google Scholar]
- 16.Ainge GD, Parlane NA, Denis M, et al. Phosphatidylinositol mannosides: Synthesis and adjuvant properties of phosphatidylinositol di- and tetramannosides. Bioorg Med Chem. 2006;14:7615–7624. doi: 10.1016/j.bmc.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Zajonc DM, Ainge GD, Painter GF, et al. Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. J Immunol. 2006;177:4577–4583. doi: 10.4049/jimmunol.177.7.4577. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 19.He XL, Bazan JF, McDermott G, et al. Structure of the Nogo receptor ectodomain: a recognition module implicated in myelin inhibition. Neuron. 2003;38:177–185. doi: 10.1016/s0896-6273(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 20.Petrescu AJ, Milac AL, Petrescu SM, et al. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 21.Weber AN, Morse MA, Gay NJ. Four N-linked glycosylation sites in human toll-like receptor 2 cooperate to direct efficient biosynthesis and secretion. J Biol Chem. 2004;279:34589–34594. doi: 10.1074/jbc.M403830200. [DOI] [PubMed] [Google Scholar]
- 22.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 23.Rudd PM, Elliott T, Cresswell P, et al. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 24.Zapun A, Darby NJ, Tessier DC, et al. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- 25.Harrison RL, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell expression system and engineering of insect cells to produce “mammalianized” recombinant glycoproteins. Adv Virus Res. 2006;68:159–191. doi: 10.1016/S0065-3527(06)68005-6. [DOI] [PubMed] [Google Scholar]
- 26.Marchal I, Jarvis DL, Cacan R, et al. Glycoproteins from insect cells: sialylated or not? Biol Chem. 2001;382:151–159. doi: 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenfield NJ. Methods to estimate the conformation of proteins and polypeptides from circular dichroism data. Anal Biochem. 1996;235:1–10. doi: 10.1006/abio.1996.0084. [DOI] [PubMed] [Google Scholar]
- 28.Heggelund L, Flo T, Berg K, et al. Soluble toll-like receptor 2 in HIV infection: association with disease progression. AIDS. 2004;18:2437–2439. [PubMed] [Google Scholar]
- 29.Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 30.Kerkmann M, Rothenfusser S, Hornung V, et al. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J Immunol. 2003;170:4465–4474. doi: 10.4049/jimmunol.170.9.4465. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Palmer GH, Abbott JR, et al. CpG ODN 2006 and IL-12 are comparable for priming Th1 lymphocyte and IgG responses in cattle immunized with a rickettsial outer membrane protein in alum. Vaccine. 2003;21:3307–3318. doi: 10.1016/s0264-410x(03)00176-2. [DOI] [PubMed] [Google Scholar]
- 32.Oliveira-Nascimento L, Massari P, Wetzler LM. The Role of TLR2 in Infection and Immunity. Front Immunol. 2012;3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabiec A, Meng G, Fichte S, et al. Human but not murine toll-like receptor 2 discriminates between tri-palmitoylated and tri-lauroylated peptides. J Biol Chem. 2004;279:48004–48012. doi: 10.1074/jbc.M405311200. [DOI] [PubMed] [Google Scholar]
- 34.Buwitt-Beckmann U, Heine H, Wiesmuller KH, et al. Lipopeptide structure determines TLR2 dependent cell activation level. FEBS J. 2005;272:6354–6364. doi: 10.1111/j.1742-4658.2005.05029.x. [DOI] [PubMed] [Google Scholar]
- 35.Bell JK, Askins J, Hall PR, et al. The dsRNA binding site of human Toll-like receptor 3. Proc Natl Acad Sci U S A. 2006;103:8792–8797. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell JK, Botos I, Hall PR, et al. The molecular structure of the Toll-like receptor 3 ligand- binding domain. Proc Natl Acad Sci U S A. 2005;102:10976–10980. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 38.Bell JK, Mullen GE, Leifer CA, et al. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 39.Yoon SI, Hong M, Han GW, et al. Crystal structure of soluble MD-1 and its interaction with lipid IVa. Proc Natl Acad Sci U S A. 2010;107:10990–10995. doi: 10.1073/pnas.1004153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon SI, Kurnasov O, Natarajan V, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song DH, Lee JO. Sensing of microbial molecular patterns by Toll-like receptors. Immunol Rev. 2012;250:216–229. doi: 10.1111/j.1600-065X.2012.01167.x. [DOI] [PubMed] [Google Scholar]
- 42.Kim HM, Park BS, Kim JI, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 43.LeBouder E, Rey-Nores JE, Rushmere NK, et al. Soluble forms of Toll-like receptor (TLR)2 capable of modulating TLR2 signaling are present in human plasma and breast milk. J Immunol. 2003;171:6680–6689. doi: 10.4049/jimmunol.171.12.6680. [DOI] [PubMed] [Google Scholar]
- 44.Henrick BM, Nag K, Yao XD, et al. Milk matters: soluble Toll-like receptor 2 (sTLR2) in breast milk significantly inhibits HIV-1 infection and inflammation. PLoS One. 2012;7:e40138. doi: 10.1371/journal.pone.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dulay AT, Buhimschi CS, Zhao G, et al. Soluble TLR2 is present in human amniotic fluid and modulates the intraamniotic inflammatory response to infection. J Immunol. 2009;182:7244–7253. doi: 10.4049/jimmunol.0803517. [DOI] [PubMed] [Google Scholar]
- 46.Kuroishi T, Tanaka Y, Sakai A, et al. Human parotid saliva contains soluble toll-like receptor (TLR) 2 and modulates TLR2-mediated interleukin-8 production by monocytic cells. Mol Immunol. 2007;44:1969–1976. doi: 10.1016/j.molimm.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 47.Raby AC, Le Bouder E, Colmont C, et al. Soluble TLR2 reduces inflammation without compromising bacterial clearance by disrupting TLR2 triggering. J Immunol. 2009;183:506–517. doi: 10.4049/jimmunol.0802909. [DOI] [PubMed] [Google Scholar]
- 48.Liew FY, Xu D, Brint EK, et al. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 49.Meng G, Grabiec A, Vallon M, et al. Cellular recognition of tri-/di-palmitoylated peptides is independent from a domain encompassing the N-terminal seven leucine-rich repeat (LRR)/LRR-like motifs of TLR2. J Biol Chem. 2003;278:39822–39829. doi: 10.1074/jbc.M304766200. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura J, Shibata K, Hasebe A, et al. Signaling pathways induced by lipoproteins derived from Mycoplasma salivarium and a synthetic lipopeptide (FSL-1) in normal human gingival fibroblasts. Microbiol Immunol. 2002;46:151–158. doi: 10.1111/j.1348-0421.2002.tb02680.x. [DOI] [PubMed] [Google Scholar]
- 51.Okusawa T, Fujita M, Nakamura J, et al. Relationship between structures and biological activities of mycoplasmal diacylated lipopeptides and their recognition by toll-like receptors 2 and 6. Infect Immun. 2004;72:1657–1665. doi: 10.1128/IAI.72.3.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shamsul HM, Hasebe A, Iyori M, et al. The Toll-like receptor 2 (TLR2) ligand FSL-1 is internalized via the clathrin-dependent endocytic pathway triggered by CD14 and CD36 but not by TLR2. Immunology. 2010;130:262–272. doi: 10.1111/j.1365-2567.2009.03232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose WA, 2nd, McGowin CL, Pyles RB. FSL-1, a bacterial-derived toll-like receptor 2/6 agonist, enhances resistance to experimental HSV-2 infection. Virol J. 2009;6:195. doi: 10.1186/1743-422X-6-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Travassos LH, Girardin SE, Philpott DJ, et al. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 2004;5:1000–1006. doi: 10.1038/sj.embor.7400248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashimoto M, Tawaratsumida K, Kariya H, et al. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J Immunol. 2006;177:3162–3169. doi: 10.4049/jimmunol.177.5.3162. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto M, Furuyashiki M, Kaseya R, et al. Evidence of immunostimulating lipoprotein existing in the natural lipoteichoic acid fraction. Infect Immun. 2007;75:1926–1932. doi: 10.1128/IAI.02083-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer K, Stein K, Ulmer AJ, et al. Cytokine-inducing lipoteichoic acids of the allergy-protective bacterium Lactococcus lactis G121 do not activate via Toll-like receptor 2. Glycobiology. 2011;21:1588–1595. doi: 10.1093/glycob/cwr071. [DOI] [PubMed] [Google Scholar]
- 58.Gisch N, Kohler T, Ulmer AJ, et al. Structural reevaluation of Streptococcus pneumoniae Lipoteichoic acid and new insights into its immunostimulatory potency. J Biol Chem. 2013;288:15654–15667. doi: 10.1074/jbc.M112.446963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deininger S, Figueroa-Perez I, Sigel S, et al. Use of synthetic derivatives to determine the minimal active structure of cytokine-inducing lipoteichoic acid. Clin Vaccine Immunol. 2007;14:1629–1633. doi: 10.1128/CVI.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morath S, Stadelmaier A, Geyer A, et al. Synthetic lipoteichoic acid from Staphylococcus aureus is a potent stimulus of cytokine release. J Exp Med. 2002;195:1635–1640. doi: 10.1084/jem.20020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deretic V, Fratti RA. Mycobacterium tuberculosis phagosome. Mol Microbiol. 1999;31:1603–1609. doi: 10.1046/j.1365-2958.1999.01279.x. [DOI] [PubMed] [Google Scholar]
- 62.Miller BH, Fratti RA, Poschet JF, et al. Mycobacteria inhibit nitric oxide synthase recruitment to phagosomes during macrophage infection. Infect Immun. 2004;72:2872–2878. doi: 10.1128/IAI.72.5.2872-2878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vergne I, Fratti RA, Hill PJ, et al. Mycobacterium tuberculosis phagosome maturation arrest: mycobacterial phosphatidylinositol analog phosphatidylinositol mannoside stimulates early endosomal fusion. Mol Biol Cell. 2004;15:751–760. doi: 10.1091/mbc.E03-05-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briken V, Miller JL. Living on the edge: inhibition of host cell apoptosis by Mycobacterium tuberculosis. Future Microbiol. 2008;3:415–422. doi: 10.2217/17460913.3.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Underhill DM, Ozinsky A, Smith KD, et al. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc Natl Acad Sci U S A. 1999;96:14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krutzik SR, Ochoa MT, Sieling PA, et al. Activation and regulation of Toll-like receptors 2 and 1 in human leprosy. Nat Med. 2003;9:525–532. doi: 10.1038/nm864. [DOI] [PubMed] [Google Scholar]
- 67.Jankute M, Grover S, Rana AK, et al. Arabinogalactan and lipoarabinomannan biosynthesis: structure, biogenesis and their potential as drug targets. Future Microbiol. 2012;7:129–147. doi: 10.2217/fmb.11.123. [DOI] [PubMed] [Google Scholar]
- 68.Court N, Rose S, Bourigault ML, et al. Mycobacterial PIMs inhibit host inflammatory responses through CD14-dependent and CD14-independent mechanisms. PLoS One. 2011;6:e24631. doi: 10.1371/journal.pone.0024631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ainge GD, Hudson J, Larsen DS, et al. Phosphatidylinositol mannosides: synthesis and suppression of allergic airway disease. Bioorg Med Chem. 2006;14:5632–5642. doi: 10.1016/j.bmc.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 70.Harper JL, Hayman CM, Larsen DS, et al. A PIM(2) analogue suppresses allergic airway disease. Bioorg Med Chem. 2011;19:917–925. doi: 10.1016/j.bmc.2010.11.058. [DOI] [PubMed] [Google Scholar]
- 71.Mazurek J, Ignatowicz L, Kallenius G, et al. Divergent effects of mycobacterial cell wall glycolipids on maturation and function of human monocyte-derived dendritic cells. PLoS One. 2012;7:e42515. doi: 10.1371/journal.pone.0042515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilleron M, Nigou J, Nicolle D, et al. The acylation state of mycobacterial lipomannans modulates innate immunity response through toll-like receptor 2. Chem Biol. 2006;13:39–47. doi: 10.1016/j.chembiol.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 73.Gilleron M, Quesniaux VF, Puzo G. Acylation state of the phosphatidylinositol hexamannosides from Mycobacterium bovis bacillus Calmette Guerin and mycobacterium tuberculosis H37Rv and its implication in Toll-like receptor response. J Biol Chem. 2003;278:29880–29889. doi: 10.1074/jbc.M303446200. [DOI] [PubMed] [Google Scholar]
- 74.Kajava AV, Vasselon T. A network of hydrogen bonds on the surface of TLR2 controls ligand positioning and cell signaling. J Biol Chem. 2010;285:6227–6234. doi: 10.1074/jbc.M109.063669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Irvine KL, Hopkins LJ, Gangloff M, et al. The molecular basis for recognition of bacterial ligands at equine TLR2, TLR1 and TLR6. Vet Res. 2013;44:50. doi: 10.1186/1297-9716-44-50. [DOI] [PMC free article] [PubMed] [Google Scholar]