Abstract
Posttranslational modification (PTM) of self-proteins has been shown to elicit clinically relevant immune responses in rheumatoid arthritis and celiac disease. Accumulating evidence suggests that recognition of modified self-proteins may also be important in type 1 diabetes. Our objective was to identify posttranslationally modified GAD65 peptides, which are recognized by subjects with type 1 diabetes, and to assess their disease relevance. We show that citrullination and transglutamination of peptides can enhance their binding to DRB1*04:01, a diabetes-susceptible HLA allele. These and corresponding modifications to amino acids at T-cell contact positions modulated the recognition of multiple GAD65 peptides by self-reactive T cells. Using class II tetramers, we verified that memory T cells specific for these modified epitopes were detectable directly ex vivo in the peripheral blood of subjects with type 1 diabetes at significantly higher frequencies than healthy controls. Furthermore, T cells that recognize these modified epitopes were either less responsive or nonresponsive to their unmodified counterparts. Our findings suggest that PTM contributes to the progression of autoimmune diabetes by eliciting T-cell responses to new epitope specificities that are present primarily in the periphery, thereby circumventing tolerance mechanisms.
Introduction
The importance of posttranslational modifications (PTMs) that modulate antigen recognition is established in human autoimmune disease. Citrullination of self-peptides in rheumatoid arthritis (RA) by peptidyl arginine deiminase enzymes converts arginine into citrulline, thereby enhancing peptide binding to susceptible HLA alleles (1) and increasing recognition by autoreactive T and B cells (2). Analogously in celiac disease, deamination of gliadin peptides by transglutaminase (transglutamination) converts glutamine into glutamic acid, enhancing their presentation and inducing robust T-cell activation (3). These T cells fail to recognize the corresponding unmodified peptides, underscoring the importance of PTM in loss of tolerance (4). Strikingly, the major HLA haplotypes that confer susceptibility for RA and celiac disease (DR4/DQ8 and DR3/DQ2) contribute the greatest genetic risk for type 1 diabetes (5). It is therefore compelling to investigate whether PTMs play an important role in type 1 diabetes.
Accumulating evidence suggests that modified self-antigens are recognized in type 1 diabetes. Mannering et al. (6) identified a modified insulin epitope that contains a vicinal disulfide bond. Delong et al. (7) described a chromogranin A epitope that is highly antigenic only after enzymatic modification with transglutaminase. More recently, a study by van Lummel et al. (8) demonstrated the binding of transglutaminated peptides to HLA-DQ8cis and DQ8trans molecules and isolated T cells that recognize a deamidated proinsulin peptide. In addition, published studies have documented transglutaminase-mediated cross linking and oxidative deamination of proteins within the islet (9,10) and conversion of the arginine within the insulin B chain to citrulline (11).
To further establish the importance of immune recognition of PTM epitopes in type 1 diabetes, we investigated the recognition of modified GAD65 epitopes by type 1 diabetes subjects. We report multiple modified GAD65 peptides that bind with high affinity to DRB1*04:01. T cells specific for these epitopes were present at higher ex vivo frequencies in subjects with type 1 diabetes than in healthy controls and preferentially recognized modified peptides.
Research Design and Methods
Human Subjects
Blood samples were collected from individuals with type 1 diabetes and healthy controls with HLA-DRB1*0401 haplotypes after obtaining written consent under an approved study by the Institutional Review Board at Benaroya Research Institute. Key attributes of these subjects are summarized in Supplementary Tables 1 and 2. Subjects with diabetes had an average age of 27.9 years, were sampled an average of 4 years after diagnosis, and were typically antibody positive for GAD and at least one other self-antigen. Healthy controls had an average age of 39.5. Autoantibody-positive subjects without diabetes had an average age of 37.5 years and were typically GAD and insulin autoantibody positive.
Peptides
Peptides (14–20 amino acids) representing modified versions of each GAD65 sequence with an arginine or glutamate (see Supplementary Table 3) were synthesized by Mimotopes. The biotinylated reference peptide HA306–318 (PKYVKQNTLKLAT) was synthesized by GenScript.
Peptide Binding Competition Assay
Peptide binding to HLA-DRB1*0401 was measured by incubating increasing concentrations of peptides in competition with 0.02 μmol/L biotinylated HA306–318 in wells coated with DRB1*0401 protein. After washing, residual biotin-HA306–318 was detected using europium-conjugated streptavidin (PerkinElmer) and quantified using a Victor2 D time-resolved fluorometer (PerkinElmer). Binding curves were simulated using Prism software (version 5.03, GraphPad Software Inc.), and IC50 values were calculated as the concentration needed to displace 50% of the reference peptide.
MHC Class II Protein and Tetramer Reagents
Recombinant HLA- DRB1*04:01 was purified from insect cell culture supernatants by affinity chromatography and dialyzed against phosphate buffer, pH 6.0. To prepare tetramers, DRB1*0401 was biotinylated in vitro and subsequently incubated with 0.2 mg/mL of peptide at 37°C for 72 h in the presence of 0.2% n-octyl-β-D-glucopyranoside (Sigma-Aldrich) and 1 mmol/L Pefabloc SC. Monomers were conjugated into tetramers using RPE streptavidin (Invitrogen) at a molar ratio of 8:1.
In Vitro Tetramer Assays
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll underlay, resuspended in T-cell media (RPMI, 10% pooled human serum, 1% penicllin-streptomycin, 1% l-glutamine) at 4 × 106 cells/mL, and stimulated with groups of five peptides (20 mg/mL total) in 48-well plates for 14 days, adding medium and interleukin (IL)-2 as needed starting on day 7. To visualize responses, cells were stained with tetramers for 75 min at 37°C and then with CD4-PerCP (BD Biosciences), CD3-APC (eBioscience), and CD25-FITC (BioLegend) for 15 min at 4°C and analyzed on a FACSCalibur (BD Biosciences) using FlowJo software (TreeStar Inc.).
T-Cell Clone Isolation and Proliferation Assays
T-cell clones were isolated by staining in vitro cultures of PBMC from subjects with type 1 diabetes as described and sorting single tetramer-positive CD4+ T cells using a FACS Aria (BD Biosciences) and expanding in 96-well plates in the presence of 1 × 105 irradiated PBMC from an unrelated donor, 2 µg/mL phytohemagglutinin (Remel Inc.), adding T-cell media and IL-2 starting on day 10. Expanded cells were restained with tetramer. To assess specificity and preferential recognition of modified peptides, clones were plated at 104 cells/well with 1 × 105 irradiated PBMCs from an unrelated DRB1*04:01 donor and stimulated in triplicate with 10 μg/mL of either modified or unmodified peptide or with 150 µg/mL of GAD protein. After incubating for 48 h at 37°C, wells were pulsed with 3[H]-thymidine (1 µCi/well), and 3[H]-thymidine incorporation was measured 18 h later with a scintillation counter. To assess natural processing and enzymatic modification of GAD65 epitopes, GAD65 protein (Diamyd Medical) was unfolded by heating at 70°C for 30 min and then treated with PAD4 enzyme (Cayman Chemical) at pH 7.2 in PBS plus 2.5 mmol/L CaCl2 or transglutaminase (MyBioSource) at pH 5.0 in 50 mmol/L Tris plus 1 mmol/L CaCl2.
Intracellular Cytokine Analysis
To assess their functional phenotype, 106 T cells were activated with 50 ng/mL phorbol myristic acid and 1 µg/mL ionomycin for 30 min followed by incubation for 3 h at 37°C in the presence brefeldin A (eBioscience). Cells were fixed in fixation/permeabilization buffer (eBioscience), washed in permeabilization buffer (eBioscience), stained using anticytokine antibodies (IL-4 AF488 from eBioscience and interferon-γ AF700, IL-10 BV421, IL-17A APC/Cy7, and tumor necrosis factor (TNF)-α PerCP-Cy5.5 from BioLegend) and analyzed on an LSR II (BD Biosciences) and using FlowJo software (TreeStar Inc.).
Ex Vivo Tetramer Analysis
Direct analysis of T-cell frequency was accomplished using our previously published enrichment approach (12). Briefly, 30–50 × 106 PBMCs were resuspended in 200 µL of T-cell media, incubated with 50 nmol/L dasatinib for 10 min at 37°C, and stained with 20 μg/mL phycoerythrin (PE)-labeled tetramer at room temperature for 100 min, followed by antibody staining with CD4-APC (eBioscience), CD45RO-FITC (eBioscience), and a combination of CD14-PerCP and CD19-PerCP (BD Biosciences) for 15 min at 4°C. Cells were washed, incubated with PE magnetic beads (Miltenyi Biotec) for 20 min at 4°C, and enriched with a magnetic column, retaining 1% of the cells as a nonenriched sample. The PE-enriched and precolumn samples were labeled with Via-Probe (BD Biosciences) and analyzed on a FACSCalibur (BD Biosciences), gating on CD4+CD14−CD19−Via-Probe− and plotting tetramer versus CD45RO. Frequencies were calculated as previously described (12). Statistical analysis was performed using unpaired t tests with Welch correction with Prism software (version 5.03, GraphPad Software Inc.).
Results
GAD65 Peptides Containing Modified Residues Bind to HLA-DRB1*0401
Conversion of arginine into citrulline has been shown to enhance the binding of peptides to certain HLA-DR proteins (1,2). Deamidation of glutamine to glutamic acid may also create potentially favorable interactions that enhance binding. To examine the influence of these modifications on HLA binding, we synthesized a peptide library containing every possible citrullinated residue (39 peptides with R→cit substitutions) and every possible deamidated glutamine residue (18 peptides with Q→E substitutions) within GAD65 and quantified the binding of each to DRB1*04:01. Fifteen peptides bound with appreciable affinity and were selected for further study (Peptide IC50 values and sequences are summarized in Supplementary Table 3).
Modified Self-Peptides Elicit In Vitro CD4+ T-Cell Responses
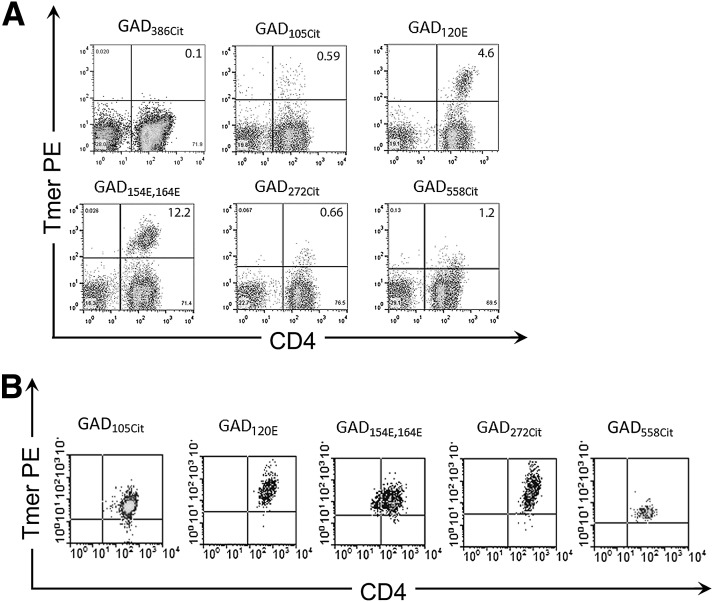
We then assessed the ability of these modified peptides to elicit T-cell responses by stimulating PBMCs from subjects with type 1 diabetes in vitro with modified peptides and staining with DRB1*04:01 tetramers. Five modified GAD65 peptides elicited positive responses in multiple subjects, including two peptides with Q→E substitutions and three peptides with R→cit substitutions (Fig. 1A). Sequences for these epitopes are summarized in Table 1, and their binding to DR0401 is shown in Supplementary Fig. 1. To further demonstrate that these were bona fide responses, T-cell clones were isolated and their specificity confirmed by tetramer staining. As shown in Fig. 1B, positive clones were obtained for all five of these epitope specificities. However, only three of the five epitopes were subsequently shown to be preferentially recognized in their modified form: GAD115–127 transglutaminated at position 120 (GAD120E), GAD153–172 transglutaminated at positions 154 and 164 (GAD154E,164E), and GAD265–284 citrullinated at position 272 (GAD272cit).
Figure 1.
Identifying immunogenic modified self-peptides. A: CD4+ T-cell responses to modified peptides from patients with type 1 diabetes (staining results are combined from multiple representative subjects). Responses were visualized by staining with DRB1*04:01 tetramers after 2 weeks of in vitro stimulation with modified peptides. Cells were gated on CD3 and CD4, and tetramer staining that was more than twice background was considered positive. The first panel (GAD386Cit) depicts a typical negative staining result. The remaining panels are typical positive results for each modified GAD peptide that was shown to be immunogenic. B: T-cell clones were sorted, expanded, and restained with tetramer to verify their specificity. In this way, three modified peptides were confirmed as bona fide epitopes. A representative staining for one clone specific for each modified GAD peptide is shown. In total, we isolated 4 GAD105Cit-specific clones from two different subjects, 14 GAD120E-specific clones from four different subjects, 10 GAD154E,164E-specific clones from four different subjects, 13 GAD272Cit-specific clones from two different subjects, and 9 GAD558Cit-specific clones from two different subjects. Tmer, tetramer.
Table 1.
Sequences and binding affinity for antigenic modified GAD65 peptides
| GAD65 peptide | Modification | Amino acid sequencea | Modified IC50 (µmol/L)b | Wild-type IC50 (µmol/L)b | RBAc |
|---|---|---|---|---|---|
| 89–108 | 105Cit | YAFLHATDLLPACDGEXPTL | 2.9 | 3.2 | 1.1 |
| 115–127 | 120E | MNILLEYVVKSFD | 0.2 | 0.7 | 3.5 |
| 153–172 | 154E,164E | PENLEEILMHCETTLKYAIK | 12.8 | 5.6 | 0.4 |
| 265–284 | 272Cit | KGMAALPXLIAFTSEHSHFS | 0.8 | 0.3 | 0.3 |
| 553–572 | 558Cit | KVNFFXMVISNPAATHQDID | 0.04 | 0.01 | 0.2 |
aThe residue that modulates recognition when modified is bolded in each sequence. X indicates citrulline.
bIC50 represents the peptide concentration that displaces half of the reference peptide.
cRBA represents the relative binding affinity of the modified peptide as compared with the unmodified peptide. A relative binding affinity >1 indicates superior binding.
T Cells Specific for Modified GAD65 Epitopes Are Present at Higher Frequencies in Subjects With Type 1 Diabetes
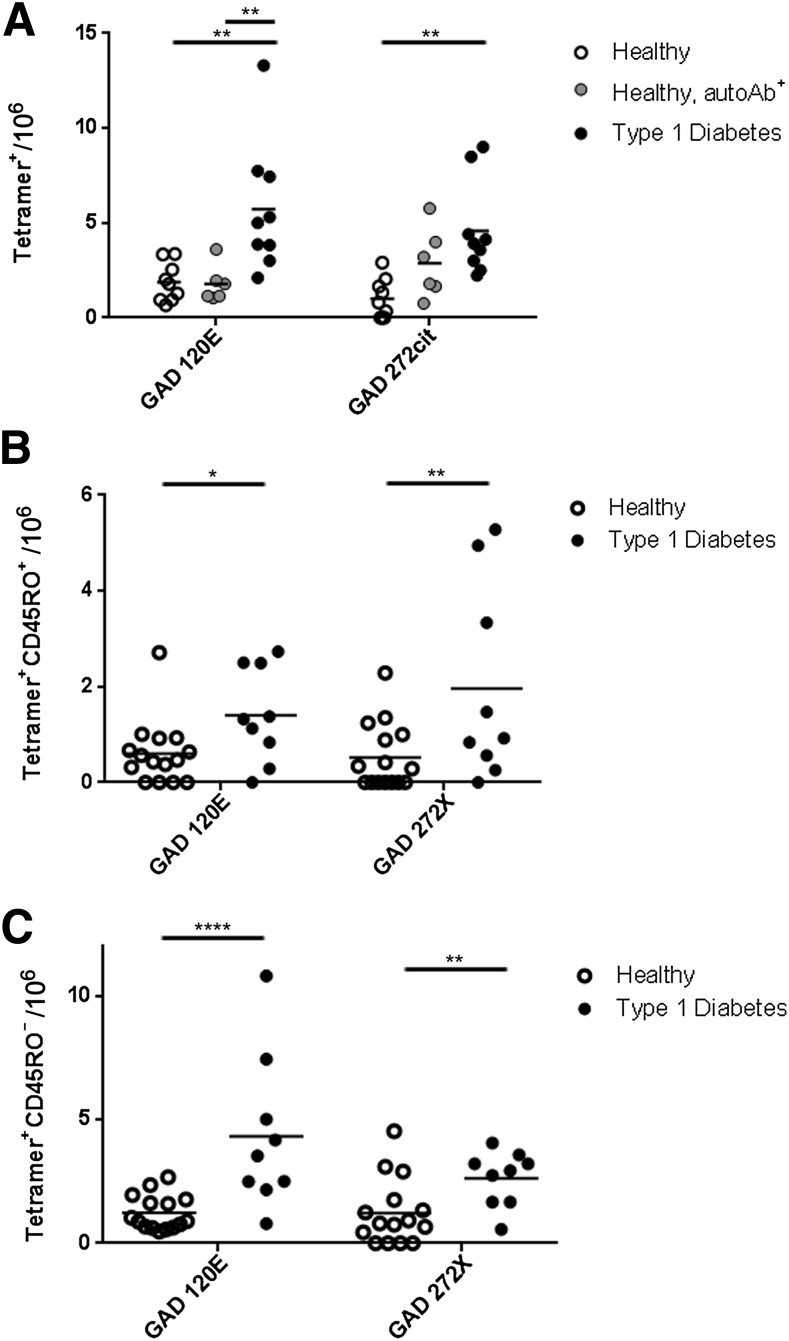
To address the disease relevance of these modified epitopes, we directly assessed the frequencies of T cells that recognize modified GAD65 epitopes in peripheral blood samples from nine subjects with type 1 diabetes, six autoantibody-positive HLA-matched controls, and nine autoantibody-negative controls. Because the total frequency of GAD65-reactive CD4+ T cells in peripheral blood is less than 1 in 30,000 cells (13), we used an anti-PE magnetic enrichment strategy to accurately detect these cells (Supplementary Fig. 2). GAD120E-specific T cells were present at significantly higher frequencies in subjects with type 1 diabetes than in antibody-positive or antibody-negative controls (P < 0.01 for both comparisons) (Fig. 2A). GAD272cit specific T cells were more frequent in subjects with type 1 diabetes than in antibody-negative controls (P < 0.01). Antibody-positive controls had intermediate frequencies of GAD272cit-specific T cells, which were not significantly different from subjects with type 1 diabetes or antibody-negative controls. GAD154E,164E-specific T-cell frequencies were relatively low in all subjects (≤4 cells per million), and no significant differences were seen between subjects with type 1 diabetes and controls (data not shown). Similarly, frequencies of GAD105cit- and GAD558cit-specific T cells were measured in a limited number of subjects, but these cells were present at very low frequencies (≤3 cells per million; data not shown) suggesting that these cells may be of limited importance.
Figure 2.
CD4+ T cells specific for modified autoantigens are detected ex vivo more frequently in patients than in controls. PBMCs were isolated and costained ex vivo with tetramer and anti-CD45RO to quantify antigen-specific cells. A: The total frequency of GAD65-specific CD4+ T cells as measured for nine subjects with diabetes, nine antibody-negative controls, and six antibody-positive controls (all HLA matched). GAD120E frequencies were significantly higher in subjects with type 1 diabetes (black circles, 5.7 ± 1.1 cells per million) than in autoantibody-negative controls (white circles, 1.9 ± 0.3 cells per million) or autoantibody-positive controls (gray circles, 1.8 ± 0.4 cells per million). GAD272Cit frequencies also differed significantly between subjects with type 1 diabetes (black circles, 4.6 ± 0.8 cells per million) than in autoantibody-negative controls (white circles, 1.0 ± 0.3 cells per million). B: The frequency of GAD65-specific memory CD4+ T cells as measured for 9 subjects with type 1 diabetes and 15 healthy subjects (9 autoantibody negative and 6 autoantibody positive). GAD120E memory frequencies were significantly higher in subjects with type 1 diabetes (black circles, 1.4 ± 0.3 cells per million) than in HLA-matched controls (white circles, 0.6 ± 0.2 cells per million). GAD272Cit memory frequencies also differed significantly between subjects with type 1 diabetes (black circles, 2.0 ± 0.7 cells per million) and HLA-matched controls (white circles, 0.5 ± 0.2 cells per million). C: The frequency of GAD65-specific naïve CD4+ T cells as measured for 9 subjects with type 1 diabetes and 15 healthy subjects (nine autoantibody negative and six autoantibody positive). Naïve GAD120E frequencies were significantly higher in subjects with type 1 diabetes (black circles, 4.3 ± 1.0 cells per million) than in HLA-matched controls (white circles, 1.2 ± 0.2 cells per million). Naïve GAD272Cit frequencies also differed significantly between subjects with type 1 diabetes (black circles, 2.6 ± 0.4 cells per million) and HLA-matched controls (white circles, 1.2 ± 0.3 cells per million). *P < 0.05; **P < 0.01; ****P < 0.0001. autoAb+, autoantibody positive; X, citrulline.
Relevant epitopes would be expected to elicit a memory response in subjects with type 1 diabetes. We assessed the ex vivo memory phenotype of GAD-specific CD4+ T cells by costaining with tetramer and anti-CD45RO (Fig. 2B). GAD120E-specific memory T cells and GAD272cit-specific memory T cells were significantly more frequent in subjects with type 1 diabetes than in controls (P = 0.013 and P = 0.01, respectively). Therefore, CD4+ memory T cells specific for two GAD65 epitopes generated by PTM are present at significantly higher frequencies in subjects with type 1 diabetes than in healthy controls. Interestingly, naïve (CD45RO−) T cells specific for GAD120E and GAD272cit were also present at significantly higher frequencies in subjects with type 1 diabetes (P = 0.0005 and P = 0.008, respectively) (Fig. 2C).
CD4+ T-Cell Clones Preferentially Recognize Modified Peptides
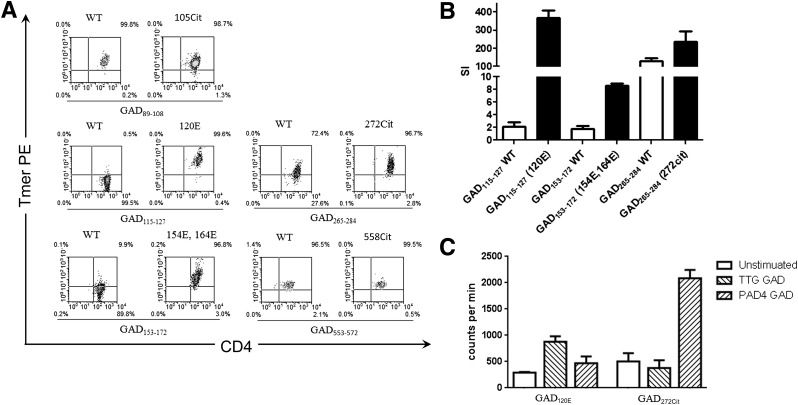
Although T cells from subjects with type 1 diabetes recognized modified GAD peptides, these could represent T cells that recognize wild-type peptide and cross react with modified peptide. We assessed the preferential recognition of modified epitopes using two approaches. First, T-cell clones isolated from subjects with type 1 diabetes were stained with tetramer loaded with either modified peptide or the corresponding wild-type peptide (Fig. 3A). CD4+ T-cell clones specific for GAD120E or GAD154E,164E were effectively stained only by tetramers loaded with the Q→E substituted peptide. Clones specific for GAD272cit could be stained by either tetramer, but staining had a consistently higher mean fluorescence intensity (MFI) for tetramers loaded with the R→cit-substituted peptide. In contrast, T-cell clones specific for GAD105cit had a similar MFI when stained by either the wild-type or R→cit-substituted peptide, and T-cell clones specific for GAD558cit had a higher MFI when stained by the wild-type peptide. Because the unmodified GAD120Q peptide was previously reported as a DRB1*04:01-restricted epitope (14), we also isolated GAD120Q-specific clones. GAD120Q-specific clones were effectively stained only by tetramers loaded with the unmodified peptide (data not shown), suggesting that although these peptides differ only by one modified amino acid, they are distinct epitopes. These results were further confirmed by measuring the proliferation of T-cell clones in response to either modified or wild-type peptide (Fig. 3B). GAD120E- and GAD154E,164E-specific clones proliferated only in response to modified peptide. GAD272cit-specific clones proliferated in response to both peptides; however, consistent with tetramer staining, the citrullinated peptide induced more proliferation than its wild-type counterpart.
Figure 3.
CD4+ T-cell clones preferentially recognize modified peptides and protein. A: Representative tetramer-positive CD4+ T-cell clones specific for modified GAD65 epitopes were stained using tetramers loaded with modified peptide or the corresponding unmodified version (wild-type peptide) and the staining MFI compared. Clones specific for GAD120E, GAD154E,164E, and GAD272Cit were preferentially stained by modified peptide tetramers while clones specific for GAD105Cit and GAD558Cit were not. B: Preferential recognition was further confirmed by measuring the proliferative responses of clones following peptide stimulation with either modified peptide (black bars) or the corresponding unmodified peptide (white bars), as determined by [3H] thymidine incorporation. Data are represented as stimulation index normalizing the proliferation of each clone based on [3H] thymidine incorporation of unstimulated wells. C: To determine whether epitope modifications can occur via enzymatic treatment and that these epitopes can be processed and presented from intact GAD protein, we measured the proliferative responses of clones following stimulation with in vitro transglutaminated (TTG, right diagonal pattern) or citrullinated (PAD4, left diagonal pattern) GAD protein as compared with unstimulated T cells (white bars). GAD120E-specific T cells proliferated in response to TTG-treated GAD (stimulation index = 3.1) but not PAD4-treated GAD (stimulation index = 1.6). GAD272Cit-specific T cells proliferated in response to PAD4-treated GAD (stimulation index = 4.1) but not TTG-treated GAD (stimulation index = 1.7). Data are represented as counts per minute (proportional to the proliferation of each clone based on [3H] thymidine incorporation). Tmer, tetramer; WT, wild-type peptide; SI, stimulation index.
CD4+ T Cells Specifically Proliferate in Response to Modified GAD Protein and Secrete Inflammatory Cytokines
If T-cell responses to modified GAD epitopes play an important role in type 1 diabetes, the corresponding peptides should occur naturally and responding T cells should have a phenotype that contributes to autoimmunity. To assess whether modified GAD peptides can be processed and presented from intact protein and are accessible for enzymatic modification, we assessed the proliferation of T-cell clones specific for GAD120E, GAD154E,164E, and GAD272cit in response to in vitro citrullinated and transglutaminated GAD65 protein. As shown in Fig. 3C, GAD120E-specific T cells proliferated in response to tissue transglutaminase (TTG)-treated GAD protein but not PAD4-treated GAD. Correspondingly, GAD272cit-specific T cells proliferated in response to PAD4-treated GAD protein but not TTG-treated GAD. GAD154E,164E-specific T cells were not able to proliferate in response to TTG-treated GAD (data not shown), suggesting either that this epitope is difficult to process from intact protein or that these residues are not readily accessible for modification.
To assess their functional phenotype, we characterized the cytokine profiles of T-cell clones specific for GAD120E, GAD154E,164E, and GAD272cit by intracellular cytokine staining. As shown in Supplementary Fig. 3, these T cells predominantly secreted TNF-α and interferon-γ, with some clones also producing IL-4 and/or IL-10.
Discussion
The importance of immune recognition of modified antigens is established in human autoimmune diseases, including RA and celiac disease, and emerging evidence supports their possible importance in type 1 diabetes. This evidence includes unique recognition of a disulfide-modified insulin epitope by human T cells (6), increased immunogenicity of a transglutaminated chromogranin A epitope by T-cell clones isolated from the NOD mouse (7), and cross recognition of a deamidated proinsulin peptide (8). We believe that our present study provides important new evidence that CD4+ T cells that recognize modified epitopes play a role in type 1 diabetes. We have identified five peptides with PTM that bind to DRB1*04:01 and are recognized by T-cell clones from subjects with type 1 diabetes. We demonstrated that two of these modifications can occur via in vitro enzymatic treatment and that the corresponding epitopes can be processed and presented from intact GAD protein. T cells that recognized these two epitopes (GAD120E and GAD272cit) were present at significantly higher ex vivo frequencies in subjects with type 1 diabetes than in HLA-matched controls.
CD4+ T cells that recognize modified epitopes were not present at elevated frequencies in islet autoantibody–positive nondiabetic individuals, indicating that accumulation of these T cells is not directly linked to autoantibody development; rather these cells may be associated with a later stage of progression. Notably, subjects with type 1 diabetes had higher frequencies of memory (CD45RO+) cells specific for GAD65-modified epitopes than healthy controls. Subjects with type 1 diabetes also had higher frequencies of naïve (CD45RO−) cells specific for GAD65-modified epitopes, suggesting that subjects who progress to develop diabetes may select a larger repertoire of potentially autoreactive T cells that recognize these modified GAD65 epitopes. Alternatively, these could be antigen experienced CD4+ T cells that have lost CD45RO expression due to chronic antigen exposure. The mere presence of CD4+ T cells that recognize modified GAD65 epitopes does not guarantee that these cells play a role in diabetes. However, T-cell clones that recognize modified epitopes predominantly secreted TNF-α and interferon-γ. This Th1-like phenotype supports the relevance of T-cell recognition of modified antigens in the loss of tolerance in type 1 diabetes.
Out of the many modified self-peptides tested, the two that were present at elevated frequencies in subjects with type 1 diabetes have been previously reported as DRB1*04:01-restricted epitopes in their unmodified forms (14,15). This raises the possibility that wild-type peptide-specific T cells may simply cross react with modified peptide. Indeed, GAD272 is recognized by T cells in its unmodified form but interacts more strongly with T-cell receptor (TCR; based on MFI of tetramer staining) and provokes stronger proliferation when citrullinated. In contrast, distinct T cells recognize GAD120E and GAD120Q, as mismatched tetramers could not cross stain GAD120E- and GAD120Q- specific T cells, and clones specific for GAD120E failed to proliferate in response to wild-type peptide. Therefore, modified epitopes can not only provoke stronger responses by T cells that recognize wild-type peptide, but also elicit responses from T cells that only recognize modified peptide. However, our results also show that not every modification leads to increased recognition, as GAD105cit and GAD558cit were not preferentially recognized in comparison with the corresponding unmodified peptides.
Modified residues could enhance or alter peptide recognition by modulating HLA binding and/or TCR interactions. Indeed, altered peptide ligands are well-known to modulate immune recognition, enhancing or diminishing T-cell recognition. PTMs can be considered to be a specialized variety of naturally occurring altered peptide ligands. Our previously published work suggests some correlation between HLA binding affinity and T-cell recognition. For example, substituting a wild-type Leu with a disfavored Pro or Tyr residue within a DRB1*08:01-restricted epitope reduced its binding by twofold and 10-fold, respectively; clones specific for that epitope had twofold less proliferation in response to the Pro-substituted peptide and failed to respond to the Tyr-substituted peptide (16). Therefore, given that GAD120E binds to DRB1*04:01 with 3.5-fold higher affinity than GAD120Q, the enhanced stability of that HLA/peptide complex could alter its recognition by CD4+ T cells. In addition, the bulkier, negatively charged glutamic acid may alter the orientation of T-cell contact residues, enabling its recognition by a different population of T cells. GAD272cit binds DRB1*04:01 with a lower affinity than GAD272R. The predicted binding register for this sequence places the modified residue in a nonanchor position, suggesting that citrullination of this peptide increases recognition mainly by modulating TCR interactions.
The phenomenon of epitope spreading, in which different epitopes are recognized during different stages of disease, is well documented in the experimental autoimmune encephalomyelitis model (17). Our findings support the notion that modification of β-cell proteins by TTG and peptidyl arginine deiminase enzymes, which are upregulated in response to inflammation and stress (18,19), generates a wave of neoepitopes that are recognized within the islet. T cells that recognize such tissue-specific modified epitopes may evade normal mechanisms of central and peripheral tolerance, comprising a latent pool of autoreactive cells that are activated during secondary stages of disease progression. Looking ahead, it will be important to address the existence of modified epitopes present within other antigens and restricted by other disease-associated HLA alleles; to determine the stage of disease during which T cells specific for modified self-antigens appear; and to characterize their frequency and phenotype in at-risk individuals as they progress toward disease.
Supplementary Material
Article Information
Acknowledgments. The authors thank McKenzie Lettau (Benaroya Research Institute) and Jani Klein (Benaroya Research Institute) for assisting with subject recruitment and Diana Sorus (Benaroya Research Institute) for administrative support of this project.
Funding. Funding for this project was provided by JDRF International (17-2012-121 to E.A.J.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.W.M. performed experiments and drafted the manuscript. I-T.C. produced HLA-DR proteins, contributed to discussion, and reviewed the manuscript. C.G. and J.O. assisted with subject selection, contributed to discussion, and reviewed the manuscript. W.W.K. assisted with experimental design, contributed to discussion, and reviewed the manuscript. E.A.J. designed and performed experiments and edited the manuscript. E.A.J. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-1952/-/DC1.
References
- 1.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol 2003;171:538–541 [DOI] [PubMed] [Google Scholar]
- 2.James EA, Moustakas AK, Bui J, et al. HLA-DR1001 presents “altered-self” peptides derived from joint-associated proteins by accepting citrulline in three of its binding pockets. Arthritis Rheum 2010;62:2909–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Wal Y, Kooy Y, van Veelen P, et al. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J Immunol 1998;161:1585–1588 [PubMed] [Google Scholar]
- 4.Arentz-Hansen H, Körner R, Molberg O, et al. The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med 2000;191:603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erlich H, Valdes AM, Noble J, et al. Type 1 Diabetes Genetics Consortium . HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mannering SI, Harrison LC, Williamson NA, et al. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J Exp Med 2005;202:1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delong T, Baker RL, He J, Barbour G, Bradley B, Haskins K. Diabetogenic T-cell clones recognize an altered peptide of chromogranin A. Diabetes 2012;61:3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Lummel M, Duinkerken G, van Veelen PA, et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes 2014;63:237–247 [DOI] [PubMed] [Google Scholar]
- 9.Owen RA, Bungay PJ, Hussain M, Griffin M. Transglutaminase-catalysed cross-linking of proteins phosphorylated in the intact glucose-stimulated pancreatic beta-cell. Biochim Biophys Acta 1988;968:220–230 [DOI] [PubMed] [Google Scholar]
- 10.Akagawa M, Sasaki T, Suyama K. Oxidative deamination of lysine residue in plasma protein of diabetic rats. Novel mechanism via the Maillard reaction. Eur J Biochem 2002;269:5451–5458 [DOI] [PubMed] [Google Scholar]
- 11.Hayashi H, Morioka M, Ichimiya S, et al. Participation of an arginyl residue of insulin chain B in the inhibition of hemagglutination by Porphyromonas gingivalis. Oral Microbiol Immunol 1993;8:386–389 [DOI] [PubMed] [Google Scholar]
- 12.Kwok WW, Roti M, Delong JH, et al. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol 2010;125:1407–1409, e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oling V, Marttila J, Ilonen J, et al. GAD65- and proinsulin-specific CD4+ T-cells detected by MHC class II tetramers in peripheral blood of type 1 diabetes patients and at-risk subjects. J Autoimmun 2005;25:235–243 [DOI] [PubMed] [Google Scholar]
- 14.Wicker LS, Chen SL, Nepom GT, et al. Naturally processed T cell epitopes from human glutamic acid decarboxylase identified using mice transgenic for the type 1 diabetes-associated human MHC class II allele, DRB1*0401. J Clin Invest 1996;98:2597–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel SD, Cope AP, Congia M, et al. Identification of immunodominant T cell epitopes of human glutamic acid decarboxylase 65 by using HLA-DR(alpha1*0101,beta1*0401) transgenic mice. Proc Natl Acad Sci U S A 1997;94:8082–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow IT, James EA, Gates TJ, et al. Differential binding of pyruvate dehydrogenase complex-E2 epitopes by DRB1*08:01 and DRB1*11:01 Is predicted by their structural motifs and correlates with disease risk. J Immunol 2013;190:4516–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuohy VK, Yu M, Yin L, Kawczak JA, Kinkel RP. Spontaneous regression of primary autoreactivity during chronic progression of experimental autoimmune encephalomyelitis and multiple sclerosis. J Exp Med 1999;189:1033–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ientile R, Caccamo D, Griffin M. Tissue transglutaminase and the stress response. Amino Acids 2007;33:385–394 [DOI] [PubMed] [Google Scholar]
- 19.Foulquier C, Sebbag M, Clavel C, et al. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum 2007;56:3541–3553 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.