Abstract
The reason that a certain subgroup of acute myeloid leukemia (AML) patients with t(8;21) translocation (generating the AML1/ETO fusion gene) displays a poor survival remains elusive. The proto-oncogene c-kit is expressed in approximately 80% of AML cases. The kinase domain mutation of the c-kit gene, one of the most common gain-of-function mutations associated with t(8;21) AML, predicts higher relapse risk and poor prognosis. However, the role of c-kit high expression in t(8;21) AML remains poorly understood. Here we evaluated the prognostic significance of c-kit expression levels in AML patients. The mRNA expression of c-kit was determined by real-time quantitative reverse transcription PCR in 132 adult AML patients. Patients were grouped into quartiles according to c-kit expression levels (Q1–Q4, each quartile containing 25% of patients) and divided into c-kit high (Q4; n = 33) and c-kit low (Q1–Q3; n = 99). High c-kit expression was associated with AML1/ETO-positive and with c-kit mutation. Of note, 35.8% of the AML1/ETO-positive AML patients carrying wild-type c-kit expressed high levels of c-kit, suggesting that other factors are involved in c-kit overexpression. High c-kit expression was associated with inferior overall and event-free survival in AML1/ETO-positive patients and was independently predictive for overall and event-free survival in multivariate analyses in a c-kit mutation-independent manner. Thus, high c-kit expression serves as a reliable molecular marker for poor prognosis, supporting a pathogenetic role of c-kit signaling in AML1/ETO-positive AML. AML1/ETO-positive patients with high c-kit expression might benefit from early treatment modifications and molecular target therapies.
Introduction
The translocation t(8;21)(q22;q22), generating the AML1/ETO fusion gene, is one of the most common structural chromosomal aberrations in patients with acute myeloid leukemia (AML). t(8;21) AML represents a favorable cytogenetic AML subgroup based on its excellent responsiveness to induction chemotherapy and high complete remission (CR) rate [1–3]. However, although the overall disease-free survival reaches about 60% in t(8;21) AML, about 30% to 40% of cases relapse after standard intensive chemotherapy, of which half become treatment resistant [4–7]. Therefore, t(8;21) AML is a heterogeneous disease with poor survival in a subset of patients. Multiple risk factors, including the white blood cell (WBC) count at initial examination, blood platelet count, sex chromosome abnormality and percentage of peripheral blood blasts, have been reported as prognostic factors in t(8;21) AML [5,8–10]. Even so, a certain subgroup of t(8;21) AML patients relapses without showing any above known risk factors. Therefore, stratification of the patients based on more universal risk factors may help identify patients who may benefit from more intensive therapies such as stem cell transplantation during the initial remission period.
Positivity for c-kit expression is present in 80% of AML cases [11], and the frequency of c-kit mutations ranges from 13% to 22% in AML with t(8;21) compared with less than 2% in AML cases overall [12,13]. It has been shown that the mutations of c-kit gene are a negative prognostic factor correlating higher incidence of relapse and a lower overall survival rate in adult patients as well as in children [13–15]. However, the clinical significance of c-kit high expression remains unclear in this subtype of leukemia. Given the pan-expression of c-kit in AML and the prognostic impact of c-kit mutations, we analyzed the mRNA expression levels of c-kit by quantitative real-time PCR (qPCR) in pretreatment bone marrow samples of 132 adults with AML and evaluated the prognostic significance of c-kit expression levels.
In the present study, we show that c-kit is highly upregulated in AML1/ETO-positive AML. The level of c-kit mRNA expression correlates perfectly to AML1/ETO. High expression of c-kit independently predicts more inferior overall and event-free survival in AML1/ETO-positive AML, regardless of c-kit mutations.
Methods
Ethics Statement
This study was carried out in accordance with principles of Declaration of Helsinki, and was approved by the Human Subject Ethics Committee in Chinese PLA General Hospital. Written informed consent was received from the participants or from the next of kin prior to inclusion in the study.
Patients and treatments
Bone marrow samples were analyzed from 132 patients with newly diagnosed, untreated AML from Chinese PLA General Hospital. Detection of t(8;21) was routinely accomplished by standard cytogenetic techniques and (or) by FISH using commercially available AML1/ETO probe (Vysis Inc.). Patients were treated with induction therapy consisted of idarubicin (10 mg/m2/day × 3)/ daunorubicin (60 mg/m2/day × 3)/ mitoxantrone (10 mg/m2/day × 3) and cytarabine (100 mg/m2/day × 7). Once CR was achieved, consolidation therapy was begun, consisting of intermediate/high dose cytarabine (1.5–2 g/m2/12 h on days 1–3) or standard-dose cytarabine-based chemotherapy (idarubicin/ daunorubicin/ mitoxantrone and cytarabine). Allogeneic and autologous hematopoietic stem-cell transplantations were performed in a risk-adapted and priority-based manner.
RNA isolation and real-time quantitative reverse transcription PCR
Mononuclear cells from bone marrow samples of patients were prepared by Ficoll-Hypaque (Sigma-Aldrich, St Louis, MO) gradient centrifugation. Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, USA). Reverse transcription for obtaining cDNA was performed SuperScript III First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. The expression of AML1/ETO and c-kit was detected by qPCR using TaqMan Gene Expression Assay (Applied Biosystems, Foster City, CA). Expression of the target genes was determined by absolute quantification method using ABL1 levels for normalization [16]. The primers and probes specific for c-kit and ABL1 used have been previously described [17]. The following sets of primers and probes were used for AML1/ETO detection: Forward: CAAGTCGCCACCTACCACAGA; Reverse: AGCCTAGATTGCGTCTTCACATC; Probe: FAM-CCATCAAAATCACAGTGGAT-NFQ-MGB.
Statistical analysis
CR was defined as recovery of morphologically normal bone marrow and normal peripheral blood cell count (absolute neutrophil count >1,000/mm3 and platelet count >100,000/mm3) and no signs or symptoms of the disease or evidence of central nervous system leukemia or other extramedullary infiltration [18]. Relapse was defined by >5% bone marrow blasts, circulating leukemic blasts or development of extramedullary leukemia. Overall survival (OS) was measured from the beginning of therapy until date of death or last follow-up. Event-free survival (EFS) was defined as the time from study entry to first event. An event was defined as failure to achieve a CR, relapse after achieving a CR, or death.
Wilcoxon signed-rank test was selected to determine the difference of c-kit expression levels between groups of samples. Spearman’s correlation coefficient (r) was used to access the correlation of mRNA levels between AML1/ETO and c-kit. To compare clinical outcome of patients with different c-kit expression levels, the cohort was stratified using the quartile grouping method described previously [19]. Patients were grouped into quartiles according to c-kit expression levels (Q1–Q4, each quartile containing 25% of patients) and divided into high c-kit (Q4; n = 33) and low c-kit (Q1–Q3; n = 99) based on the trend observed in clinical outcome after performing a Cox regression analysis for EFS with c-kit quartile grouping as the independent variable. c-kit expression ranged between 0.1965 and 10.7182 with the following median expression for each quartile: 0.3604 (Q1), 0.7595 (Q2), 1.0637 (Q3), and 2.9547 (Q4). In this model, AML1/ETO-positive patients in the highest c-kit quartile showed a significant difference of EFS, as compared to the remaining patients with lower c-kit expression levels. The differences in regression coefficients with SE for each quartile were as follows: Q1 versus Q4, -13.692 (SE 405.935), P = 0.973; Q2 versus Q4, -0.615 (SE 0.399), P = 0.123; Q3 versus Q4, = -0.859 (SE 0.386), P = 0.026. Survival curves were generated using the Kaplan-Meier method and the log-rank test was used to compare survival between groups. Clinical features across groups were compared using the 2-sided Fisher exact test for categorical data and the nonparametric Mann-Whitney U test for continuous variables. The Cox proportional hazards model with stepwise forward selection were constructed to determine whether c-kit expression was associated with outcome when adjusting for other prognostic variables. The full multivariate model used the variables significant at a 10% level in univariate analysis, including c-kit expression (low vs. high), c-kit mutation status (mutation vs. wild-type), white blood count (10 × 109/L increase), bone marrow blasts (10% increase), age (10-year increase), cytarabine-based chemotherapy (high- vs. standard-dose), hematopoietic stem-cell transplantations (Allogeneic- vs. no, autologous- vs. no) and CR achievement (1 vs. ≥ 2 courses). The possible influence of sample bias on the results and the stability of the model were examined by bootstrap resampling method [20]. A total of 1000 bootstrap samples were generated for each analysis. Cox regression was run separately on these 1000 samples to obtain robust estimates of the standard errors of coefficients, and hence the P values and 95% confidence intervals of the model coefficients. SPSS 20.0 software was used to process the data. A P value of less than 0.05 was chosen as a threshold for statistical significance.
Results
Correlations of c-kit expression with t(8;21) AML
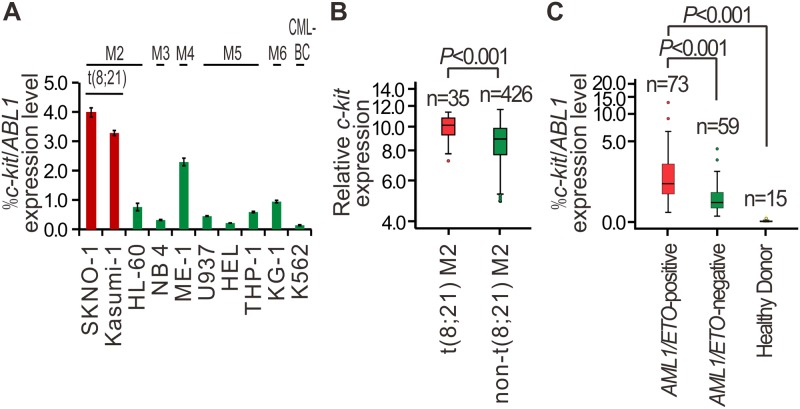
We initially detected c-kit mRNA expression levels in 10 myeloid leukemia cell lines using qPCR (Fig 1A) and then analyzed c-kit expression pattern in a previously published microarray dataset GSE6891 [21], which included gene expression profiles of 461 blood or bone marrow samples from AML patients (Fig 1B). The result showed c-kit was significantly upregulated in t(8;21) AML cell lines and patients, as compared to t(8;21)-negative subtypes. To further confirm the selectively high expression of c-kit in t(8;21) AML cells, using qPCR analysis, we measured c-kit levels in bone marrow mononuclear cells from 132 newly diagnosed AML patients and 15 healthy donors (S1 Dataset). As shown in Fig 1C, c-kit mRNA levels were significantly elevated in bone marrow samples from AML1/ETO-positive patients (n = 73), as compared to those from AML1/ETO-negative (n = 59) and healthy controls (n = 15; both P = 0.000).
Fig 1. Selective high expression of c-kit in AML1/ETO-positive AML cell lines and patients.
(A) qPCR showing c-kit expression in myeloid leukemia cell lines. CML-BC: chronic myeloid leukemia in blast crisis. Bars indicate the mean±SEM from three independent experiments. ABL1 levels were measured for normalization. (B) Normalized c-kit expression in pretreatment samples of 461 patients with de novo AML (GEO database, GSE6891). The gene expression was determined using gene-expression arrays (Affymetrix HGU133 Plus 2.0 GeneChips), which reflected by the intensity of hybridization of labeled mRNA to the gene chip. Median values are depicted by the horizontal lines. The Mann-Whitney U test was used to compare expression levels between groups. (C) qPCR showing c-kit expression level in bone marrow samples from untreated AML patients at diagnosis and healthy donors. ABL1 levels were measured for normalization. Median values are depicted by the horizontal lines. The Mann-Whitney U test was used to compare expression levels between groups.
c-kit expression with respect to clinical and biologic characteristics
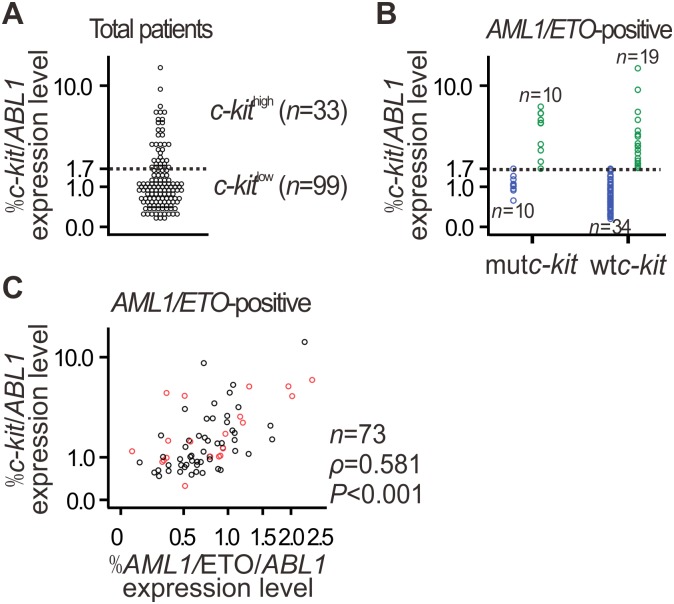
For further determination of the correlation between c-kit and AML1/ETO expressions, patients were divided into two groups, high and low, according to c-kit mRNA levels (Fig 2A). Correlation analyses between c-kit levels and the clinical and biologic features of the patients revealed that high c-kit expression was significantly associated with AML1/ETO-positive (P = 0.000) or c-kit mutation (mutc-kit) (P = 0.019) in the overall cohort (Table 1). Of note, high expression of c-kit is independent of mutc-kit (P>0.05; Table 1) in the AML1/ETO-positive cohort. Actually, in AML1/ETO-positive patients carrying wild-type c-kit (wtc-kit), 19 of 53 (35.8%) presented high level of c-kit expression (Fig 2B). Further, there was a strong positive correlation between AML1/ETO and c-kit expression levels in the AML1/ETO-positive patients, especially in those carrying wtc-kit (n = 53, ρ = 0.601, P<0.001) (Fig 2C).
Fig 2. Positive correlation between AML1/ETO and c-kit expression levels in AML1/ETO-positive AML.
(A) The patients described in Fig 1C were divided into high and low c-kit expression groups, which described in detail in statistical analysis. The threshold is depicted as a dashed line. Patient characteristics are described in Table 1. (B) Stratification of AML1/ETO-positive AML patients with high and low c-kit expression according to c-kit mutation status. (C) Correlation between c-kit and AML1/ETO levels in AML1/ETO-positive AML patients was assessed by the Spearman rank correlation coefficient. Gene expression was detected by qPCR. ABL1 levels were measured for normalization. Black circles indicate AML patients carrying wtc-kit.
Table 1. Comparison of clinical and biologic variables of AML patients according to c-kit expression.
| Characteristic | Overall cohort (n = 132) | P value | AML1/ETO-positive (n = 73) | P value | AML1/ETO-positive and wtc-kit (n = 53) | P value | |||
|---|---|---|---|---|---|---|---|---|---|
| c-kit high | c-kit low | c-kit high | c-kit low | c-kit high | c-kit low | ||||
| Patients no. | 33 | 99 | 29 | 44 | 19 | 34 | |||
| Age a | 33 (11–60) | 36 (12–84) | 0.245 | 33.0 (11–60) | 26 (12–82) | 0.875 | 34 (11–59) | 26 (12–62) | 0.676 |
| Sex, no. (%) | 0.534 | 0.655 | 0.910 | ||||||
| Male | 22 (66.7) | 60 (60.6) | 18 (62.1) | 25 (56.8) | 12 (63.2) | 22 (64.7) | |||
| Female | 11 (33.3) | 39 (39.4) | 11 (37.9) | 19 (43.2) | 7 (36.8) | 12 (35.3) | |||
| WBC (× 109/L) a | 12.7 (2.7–290.0) | 13.1 (0.3–362.0) | 0.870 | 12.2 (2.7–55.9) | 14.5 (1.5–76.3) | 0.423 | 11.0 (3.5–37.4) | 14.6 (2.1–76.3) | 0.349 |
| Bone marrow blasts (%) a | 64.0 (21.2–92.0) | 55.0 (7.4–97.0) | 0.084 | 65.0 (36.8–92.0) | 53.1 (7.4–95.0) | 0.038 | 64.0 (36.8–92.0) | 52.1 (7.4–95.0) | 0.040 |
| FAB subtypes, no. (%) | 0.546 | ||||||||
| M2 | 30 (26.1) | 85 (73.9) | 29 (39.7) | 44 (60.3) | 19 (35.8) | 34 (64.2) | |||
| M3 | 0 (0) | 2 (100) | |||||||
| M4 | 3 (33.3) | 6 (66.7) | |||||||
| M5 | 0 (0) | 4 (100) | |||||||
| M6 | 0 (0) | 2 (100) | |||||||
| AML1/ETO status b , no. (%) | 0.000 | ||||||||
| AML1/ETO positive | 29 (87.9) | 44 (44.4) | |||||||
| AML1/ETO negative | 4 (12.1) | 55 (55.6) | |||||||
| c-kit mutation status c , no. (%) | 0.019 | 0.270 | |||||||
| mutc-kit | 10 (34.5) | 10 (13.9) | 10 (34.5) | 10 (22.7) | |||||
| wtc-kit | 19 (65.5) | 62 (86.1) | 19 (65.5) | 34 (77.3) | |||||
| Chemotherapy c , no. (%) | 0.575 | 0.752 | 0.376 | ||||||
| High-dose cytarabine-based | 9 (37.5) | 19 (31.1) | 9 (45.0) | 10 (50.0) | 6 (37.5) | 8 (53.5) | |||
| Standard-dose cytarabine-based | 15 (62.5) | 42 (68.9) | 11 (55.0) | 10 (50.0) | 10 (62.5) | 7 (46.7) | |||
| HSCT, no. (%) | 0.895 | 0.899 | 0.972 | ||||||
| Allo-HSCT | 8 (24.2) | 27 (27.3) | 6 (20.7) | 8 (18.2) | 3 (15.8) | 6 (17.6) | |||
| Auto-HSCT | 3 (9.1) | 7 (7.1) | 3 (10.3) | 6 (13.6) | 2 (10.5) | 4 (11.8) | |||
| No HSCT | 22 (66.7) | 65 (65.7) | 20 (69.0) | 30 (68.2) | 14 (73.7) | 24 (70.6) | |||
| CR c , no. (%) | 0.297 | 0.606 | 0.611 | ||||||
| 1 course | 15 (62.5) | 31 (50.0) | 13 (65.0) | 12 (57.1) | 11 (68.8) | 9 (60.0) | |||
| ≥ 2 courses | 9 (37.5) | 31 (50.0) | 7 (35.0) | 9 (42.9) | 5 (31.3) | 6 (40.0) | |||
a Values represent median (range).
b The AML/ETO status was confirmed using qPCR analysis.
c Information is not available in some cases. HSCT, hematopoietic stem cell transplantation.
c-kit expression and outcome in overall cohort of AML patients
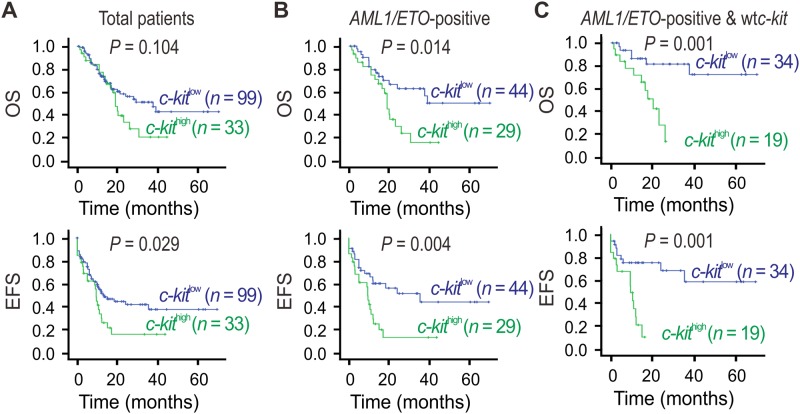
Prognostic significance of c-kit expression analysis was first performed in the whole population (Fig 3A). High c-kit expression was not a poor prognostic factor for OS. In contrast, EFS were significantly shorter in the patients with high c-kit expression (99 patients) compared with the patients with low c-kit expression (33 patients). The estimated 3-year OS rates of c-kit high patients were 16.6% plus or minus 7.7% (median = 9.4 months) versus 38.2% plus or minus 6.8% (median = 13.4 months) for c-kit low patients (P = 0.029; Fig 3A).
Fig 3. Higher expression of c-kit in AML1/ETO-positive AML predicts more inferior prognosis.
(A-C) Kaplan-Meier estimate for OS and EFS in indicated patient subgroups. Survival curves were compared using log-rank test.
c-kit expression and survival in AML1/ETO-positive AML patients
A total of 78 patients with AML1/ETO expression were separately analyzed. Patients expressing high c-kit levels had a shorter OS (16.1%±9.2% at 3 years, median = 19.0 months vs. 63.0%±8.4% at 3 years, median = not reached, P = 0.014), and EFS (13.5%±7.8% at 3 years, median = 9.4 months vs. 51.7%±8.8% at 3 years, median = 35.3 months, P = 0.004) than patients expressing low c-kit levels (Fig 3B). Notably, further stratification of patient groups based on c-kit mutation status improved the predictive capability of c-kit for both OS and EFS (Fig 3C). In the AML1/ETO-positive AML patients carrying wtc-kit [n = 53, about 73% (53/73) of the entire AML1/ETO-positive patients], those expressing high c-kit levels had a shorter OS (14.2%±12.4% at 3 years, median = 20.3 months vs. 81.6%±7.6% at 3 years, median = not reached, P = 0.001) and EFS (10.5%±9.1% at 3 years, median = 9.5 months vs. 58.6%±12.2% at 3 years, median = not reached, P = 0.001) than those expressing low c-kit levels.
Multivariate analysis and c-kit expression
According to the prognostic value of c-kit expression in AML patients, c-kit expression was entered into a multivariate model in addition to factors significantly associated with prognosis in univariate analysis (Table 2). In the overall cohort, c-kit expression was not independently predictive for OS or EFS. In contrast, in the AML1/ETO-positive patients, c-kit expression was an independent prognostic parameter for both OS (P = 0.049) and EFS (P = 0.033). Remarkably, in the AML1/ETO-positive and wtc-kit subgroup, high c-kit expression was the only parameter with prognostic significance for OS (P = 0.003) and EFS (P = 0.002). To further investigate the stability of the classical multivariate Cox models shown in Table 2, we conducted a bootstrap resampling procedure to calculate the standard errors of regression coefficients along with corresponding P values and 95% confidence intervals of the model. We found no significant differences in P values of regression coefficients between the classical Cox regression and bootstrap Cox regression models (Table 2).
Table 2. Multivariate analysis for OS and EFS and bootstrap resampling for variable in the multivariate model.
| Patients | Variable a | Cox regression | Bootstrap resampling | ||||
|---|---|---|---|---|---|---|---|
| P value | Hazard Ratio (95.0% CI) | Regression Coefficient | Std. Error | Bootstrap b | |||
| P value | 95.0% CI | ||||||
| overall cohort | |||||||
| OS | |||||||
| I/HD vs. SD cytarabine-based chemotherapy | 0.002 | 0.261 (0.110–0.618) | -1.311 | 0.439 | 0.001 | -2.294 – -0.562 | |
| EFS | |||||||
| I/HD vs. SD cytarabine-based chemotherapy | 0.002 | 0.321 (0.155–0.666) | -1.068 | 0.361 | 0.002 | -1.857 – -0.467 | |
| AML1/ETO-positive patients | |||||||
| OS | |||||||
| c-kit high vs. c-kit low | 0.049 | 2.810 (1.003–7.872) | 1.033 | 0.995 | 0.048 | -0.039–2.483 | |
| I/HD vs. SD cytarabine-based chemotherapy | 0.012 | 0.233 (0.075–0.721) | -1.458 | 1.214 | 0.008 | -3.157 – -0.411 | |
| EFS | |||||||
| c-kit high vs. c-kit low | 0.033 | 2.739 (1.086–6.910) | 1.008 | 0.809 | 0.030 | 0.054–2.346 | |
| I/HD vs. SD cytarabine-based chemotherapy | 0.013 | 0.298 (0.115–0.771) | -1.211 | 0.609 | 0.005 | -2.483 – -0.400 | |
| AML1/ETO-positive and wtc-kit patients | |||||||
| OS | |||||||
| c-kit high vs. c-kit low | 0.003 | 5.086 (1.732–14.933) | 1.626 | 0.651 | 0.005 | 0.637–3.222 | |
| EFS | |||||||
| c-kit high vs. c-kit low | 0.002 | 4.093 (1.695–9.888) | 1.409 | 0.490 | 0.004 | 0.599–2.527 | |
a Variables considered for model inclusion were: c-kit expression (high vs. low), c-kit mutation status (mutation vs. wild-type), WBC count (10×109/L increase), bone marrow blasts (10% increase), age (10-year increase), cytarabine-based chemotherapy (intermediate/high dose- vs. standard-dose), HSCT (allo- vs. no, auto- vs. no) and CR achievement (1 vs. ≥ 2 courses). Only variables significantly associated with outcomes in univariate analysis were included in the multivariate model.
b bootstrap results are based on 1000 bootstrap samples. SD, standard dose; I/HD, intermediate/high dose.
Discussion
Although AML1/ETO-positive AML patients achieve an initial complete remission, most of them relapse for undefined reasons. The present investigation was undertaken to address why a certain subgroup of AML1/ETO-positive AML patients displays a poor survival. Our findings establish c-kit as a reliable molecular marker, which identify patients with an inferior prognosis in an otherwise prognostically favorable AML1/ETO-positive AML.
In analyzing c-kit expression in AML patients, we found that c-kit was highly elevated in AML1/ETO-positive patients as compared to AML1/ETO-negative ones. When AML1/ETO-positive patients were divided into two groups regarding c-kit expression levels, we evidenced the positive correlation between AML1/ETO and c-kit. As a leukemia-initiating transcriptional factor, AML1/ETO is not sufficient in itself to induce leukemogenesis [22]. The c-kit proto-oncogene plays a very important role in the occurrence and development of multiple tumor types through regulating cellular differentiation and proliferation [23–26]. Mutations of the activation loop of c-kit kinase domain, leading to constitutive activation of the c-kit receptor kinase and thereby its downstream signaling transduction pathways, co-operate with AML1/ETO to cause leukemia as a critical genetic event [27]. Mutations of c-kit were associated with a higher relapse risk and a lower overall survival rate in t(8;21) AML patients [13–15,28]. Especially, AML1/ETO9a, a spliced isoform of AML1/ETO [29], correlates with c-kit overexpression/mutations and indicates poor disease outcome in the FAB-M2 subtype of t(8;21) AML [30]. However, the correlation between AML1/ETO and c-kit high expression and their biological importance and functional properties in t(8;21) AML remains poorly understood.
In the present study, the fact that high c-kit expression was also detected in a considerable part of AML1/ETO-positive AML patients carrying wtc-kit, indicating that other factors, except for mutc-kit, might contribute to the elevated expression of c-kit. Actually, the statistically significant positive correlation between AML1/ETO and c-kit mRNA levels in AML patients, especially in those carrying wtc-kit, suggests a potential regulatory interaction between AML1/ETO and c-kit. According to the data of our group, AML1/ETO can inhibit the expression of tumor suppressor gene microRNA-193a by inducing DNA hypermethylation in the promoter region of microRNA-193a [31]. The microRNA-193a can inhibit the overexpression of c-kit oncogene in AML1/ETO-positive leukemia cells [32]. Therefore, it is very likely that c-kit is a target of AML1/ETO. Actually, a previous study has proved that induced expression of AML1/ETO in leukemia cells significantly up-regulates both mRNA and protein levels of c-kit gene [33]. Together, these findings support the notion that c-kit could be a downstream target of AML1/ETO. Further mechanistical analysis should be done to unveil the molecular regulation pathway between AML1/ETO and c-kit.
While t(8;21) AML have a relatively good prognosis, about 30% to 40% of relapse rate has been observed for unknown reasons [4–7]. In this study, we have identified high c-kit expression as an independent prognostic factor associated with an inferior EFS and OS in AML1/ETO-positive AML patients. It is worth noting that c-kit-associated inferior prognosis in AML1/ETO-positive AML patients is c-kit mutation-independent because the predictive capability of c-kit for both OS and EFS was further improved in the subgroup of patients carrying AML1/ETO and wtc-kit. Certain reports have suggested that the number of AML1/ETO transcripts could serve as an indicator for relapse, since higher AML1/ETO transcript at diagnosis and a limited reduction of AML1/ETO transcript at the time of achieving complete remission predict a higher risk of relapse [34–35]. However, the underlying mechanisms are still not known. In the present study, the significant correlation between AML1/ETO and c-kit expressions or between the high c-kit expression and the poor outcome in AML1/ETO-positive patients provides an alternative explanation for why high AML1/ETO transcript predicts high relapse risk and supports the critical contribution of c-kit to AML1/ETO-driven leukemia.
In conclusion, we identified high c-kit expression as an independent adverse prognostic factor in adult AML1/ETO-positive AML, thereby serving as a useful marker for poor prognosis. Our results demonstrate the role of c-kit, which is essential to identify therapeutic targets that are specific to AML1/ETO-positive AML.
Supporting Information
Individual clinical characteristics and c-kit mRNA expression levels of 132 newly diagnosed AML patients (A) and 15 healthy donors (B).
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was partially supported by grants from the National Natural Science Foundation of China (81170518 to LY; 81000221 and 81370010 to XNG; 81171820 to JL and 81370635 to YHL), the National Public Health Grand Research Foundation (201202017 to LY), the Capital Public Health Project (Z111107067311070 to LY), and the Beijing Natural Science Foundation (7122169 to XNG and 7112126 to JL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ferrara F, Del Vecchio L. Acute myeloid leukemia with t(8;21)/AML1/ETO: a distinct biological and clinical entity. Haematologica. 2002;87: 306–319. [PubMed] [Google Scholar]
- 2. Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92: 2322–2333. [PubMed] [Google Scholar]
- 3. Byrd JC, Dodge RK, Carroll A, Baer MR, Edwards C, Stamberg J, et al. Patients with t(8;21)(q22;q22) and acute myeloid leukemia have superior failure-free and overall survival when repetitive cycles of high-dose cytarabine are administered. J Clin Oncol. 1999;17: 3767–3775. [DOI] [PubMed] [Google Scholar]
- 4. Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116: 354–365. 10.1182/blood-2009-11-254441 [DOI] [PubMed] [Google Scholar]
- 5. Narimatsu H, Yokozawa T, Iida H, Tsuzuki M, Hayakawa M, Takeo T, et al. Clinical characteristics and outcomes in patients with t(8;21) acute myeloid leukemia in Japan. Leukemia. 2008;22: 428–432. [DOI] [PubMed] [Google Scholar]
- 6. Lin P, Chen L, Luthra R, Konoplev SN, Wang X, Medeiros LJ. Acute myeloid leukemia harboring t(8;21)(q22;q22): a heterogeneous disease with poor outcome in a subset of patients unrelated to secondary cytogenetic aberrations. Mod Pathol. 2008;21: 1029–1036. 10.1038/modpathol.2008.92 [DOI] [PubMed] [Google Scholar]
- 7. Gorin NC, Labopin M, Frassoni F, Milpied N, Attal M, Blaise D, et al. Identical outcome after autologous or allogeneic genoidentical hematopoietic stem-cell transplantation in first remission of acute myelocytic leukemia carrying inversion 16 or t(8;21): a retrospective study from the European Cooperative Group for Blood and Marrow Transplantation. J Clin Oncol. 2008;26: 3183–3188. 10.1200/JCO.2007.15.3106 [DOI] [PubMed] [Google Scholar]
- 8. Nguyen S, Leblanc T, Fenaux P, Witz F, Blaise D, Pigneux A, et al. A white blood cell index as the main prognostic factor in t(8;21) acute myeloid leukemia (AML): a survey of 161 cases from the French AML Intergroup. Blood. 2002;99: 3517–3523. [DOI] [PubMed] [Google Scholar]
- 9. Schlenk RF, Benner A, Krauter J, Büchner T, Sauerland C, Ehninger G, et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2004;22: 3741–3750. [DOI] [PubMed] [Google Scholar]
- 10. Marcucci G, Mrózek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23: 5705–5717. [DOI] [PubMed] [Google Scholar]
- 11. Ikeda H, Kanakura Y, Tamaki T, Kuriu A, Kitayama H, Ishikawa J, et al. Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood. 1991;78: 2962–2968. [PubMed] [Google Scholar]
- 12. Kuchenbauer F, Schnittger S, Look T, Gilliland G, Tenen D, Haferlach T, et al. Identification of additional cytogenetic and molecular genetic abnormalities in acute myeloid leukaemia with t(8;21)/AML1/ETO. Br J Haematol. 2006;134: 616–619. [DOI] [PubMed] [Google Scholar]
- 13. Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24: 3904–3911. [DOI] [PubMed] [Google Scholar]
- 14. Schnittger S, Kohl TM, Haferlach T, Kern W, Hiddemann W, Spiekermann K, et al. KIT-D816 mutations in AML1/ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107: 1791–1799. [DOI] [PubMed] [Google Scholar]
- 15. Shimada A1, Taki T, Tabuchi K, Tawa A, Horibe K, Tsuchida M, et al. KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): a study of the Japanese Childhood AML Cooperative Study Group. Blood. 2006;107: 1806–1809. [DOI] [PubMed] [Google Scholar]
- 16. Viehmann S, Teigler-Schlegel A, Bruch J, Langebrake C, Reinhardt D, Harbott J. Monitoring of minimal residual disease (MRD) by real-time quantitative reverse transcription PCR (RQ-RT-PCR) in childhood acute myeloid leukemia with AML1/ETO rearrangement. Leukemia. 2003;17: 1130–1136. [DOI] [PubMed] [Google Scholar]
- 17. Gao XN, Lin J, Ning QY, Gao L, Yao YS, Zhou JH, et al. A histone acetyltransferase p300 inhibitor C646 induces cell cycle arrest and apoptosis selectively in AML1-ETO-positive AML cells. PLoS One. 2013;8: e55481 10.1371/journal.pone.0055481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheson BD, Cassileth PA, Head DR, Schiffer CA, Bennett JM, Bloomfield CD, et al. Report of the National Cancer Institute-sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol. 1990;8: 813–819. [DOI] [PubMed] [Google Scholar]
- 19. Kühnl A, Gökbuget N, Kaiser M, Schlee C, Stroux A, Mochmann LH, et al. Overexpression of LEF1 predicts unfavorable outcome in adult patients with B-precursor acute lymphoblastic leukemia. Blood. 2011;118: 6362–6367. 10.1182/blood-2011-04-350850 [DOI] [PubMed] [Google Scholar]
- 20. Sauerbrei W, Schumacher M. A bootstrap resampling procedure for model building: application to the Cox regression model. Stat Med. 1992;11: 2093–2109. [DOI] [PubMed] [Google Scholar]
- 21. de Jonge HJ, Valk PJ, Veeger NJ, ter Elst A, den Boer ML, Cloos J, et al. High VEGFC expression is associated with unique gene expression profiles and predicts adverse prognosis in pediatric and adult acute myeloid leukemia. Blood. 2010;116: 1747–1754. 10.1182/blood-2010-03-270991 [DOI] [PubMed] [Google Scholar]
- 22. Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer cell. 2002;1: 63–74. [DOI] [PubMed] [Google Scholar]
- 23. Heinrich MC, Blanke CD, Druker BJ, Corless CL. Inhibition of KIT tyrosine kinase activity: a novel molecular approach to the treatment of KIT-positive malignancies. J Clin Oncol. 2002;20: 1692–1703. [DOI] [PubMed] [Google Scholar]
- 24. Krystal GW, Hines SJ, Organ CP. Autocrine growth of small cell lung cancer mediated by coexpression of c-kit and stem cell factor. Cancer Res. 1996;56: 370–376. [PubMed] [Google Scholar]
- 25. Ali S, Ali S. Role of c-kit/SCF in cause and treatment of gastrointestinal stromal tumors (GIST). Gene. 2007;401: 38–45. [DOI] [PubMed] [Google Scholar]
- 26. Yasuda A, Sawai H, Takahashi H, Ochi N, Matsuo Y, Funahashi H, et al. The stem cell factor/c-kit receptor pathway enhances proliferation and invasion of pancreatic cancer cells. Mol Cancer. 2006;5: 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci U S A. 2001;98: 10398–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boissel N, Leroy H, Brethon B, Philippe N, de Botton S, Auvrignon A, et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia. 2006;20: 965–970. [DOI] [PubMed] [Google Scholar]
- 29. Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12: 945–949. [DOI] [PubMed] [Google Scholar]
- 30. Jiao B, Wu CF, Liang Y, Chen HM, Xiong SM, Chen B, et al. AML1-ETO9a is correlated with C-KIT overexpression/mutations and indicates poor disease outcome in t(8;21) acute myeloid leukemia-M2. Leukemia. 2009;23: 1598–1604. 10.1038/leu.2009.104 [DOI] [PubMed] [Google Scholar]
- 31. Li Y, Gao L, Luo X, Wang L, Gao X, Wang W, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121: 499–509. 10.1182/blood-2012-07-444729 [DOI] [PubMed] [Google Scholar]
- 32. Gao XN, Lin J, Li YH, Gao L, Wang XR, Wang W, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30: 3416–3428. 10.1038/onc.2011.62 [DOI] [PubMed] [Google Scholar]
- 33. Wang YY, Zhou GB, Yin T, Chen B, Shi JY, Liang WX, et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad Sci U S A. 2005;102: 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoo SJ, Chi HS, Jang S, Seo EJ, Seo JJ, Lee JH, et al. Quantification of AML1-ETO fusion transcript as a prognostic indicator in acute myeloid leukemia. Haematologica. 2005;90: 1493–1501. [PubMed] [Google Scholar]
- 35. Leroy H, de Botton S, Grardel-Duflos N, Darre S, Leleu X, Roumier C, et al. Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21). Leukemia. 2005;19: 367–372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual clinical characteristics and c-kit mRNA expression levels of 132 newly diagnosed AML patients (A) and 15 healthy donors (B).
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.