Abstract
The Rim101/PacC transcription factor acts in a fungal-specific signaling pathway responsible for sensing extracellular pH signals. First characterized in ascomycete fungi such as Aspergillus nidulans and Saccharomyces cerevisiae, the Rim/Pal pathway maintains conserved features among very distantly related fungi, where it coordinates cellular adaptation to alkaline pH signals and micronutrient deprivation. However, it also directs species-specific functions in fungal pathogens such as Cryptococcus neoformans, where it controls surface capsule expression. Moreover, disruption of the Rim pathway central transcription factor, Rim101, results in a strain that causes a hyper-inflammatory response in animal infection models. Using targeted gene deletions, we demonstrate that several genes encoding components of the classical Rim/Pal pathway are present in the C. neoformans genome. Many of these genes are in fact required for Rim101 activation, including members of the ESCRT complex (Vps23 and Snf7), ESCRT-interacting proteins (Rim20 and Rim23), and the predicted Rim13 protease. We demonstrate that in neutral/alkaline pH, Rim23 is recruited to punctate regions on the plasma membrane. This change in Rim23 localization requires upstream ESCRT complex components but does not require other Rim101 proteolysis components, such as Rim20 or Rim13. Using a forward genetics screen, we identified the RRA1 gene encoding a novel membrane protein that is also required for Rim101 protein activation and, like the ESCRT complex, is functionally upstream of Rim23-membrane localization. Homologs of RRA1 are present in other Cryptococcus species as well as other basidiomycetes, but closely related genes are not present in ascomycetes. These findings suggest that major branches of the fungal Kingdom developed different mechanisms to sense and respond to very elemental extracellular signals such as changing pH levels.
Author Summary
Microorganisms that cause human disease use various cues to determine when they encounter a host. One of these signals is the slightly alkaline pH of human tissues. We have defined components of a pH-responsive signaling pathway in the human fungal pathogen Cryptococcus neoformans. This particular signaling pathway is specific for fungi and important for the virulence of several plant and animal pathogens. We have identified a previously unknown protein that activates this pH-sensing pathway. This protein and its downstream signaling pathway play a major role in the interaction of this fungal pathogen and the host immune system.
Introduction
To survive the harsh environment of the infected host, microorganisms must be able to sense their external environment and adaptively alter their cellular processes. For example, microorganisms often encounter changes in extracellular pH when moving between different microenvironments. For fungal pathogens, the alkaline-response transcription factor Rim101 plays a central role in adapting to changing environmental and host conditions.
Rim101 was first implicated in fungal pathogenesis in the human fungal pathogen Candida albicans in which the rim101Δ mutant was demonstrated to be defective in the yeast to hyphal transition, and therefore unable to cause disseminated infection [1]. More recently, Bertuzzi and Schrettl et al. demonstrated that the opportunistic pathogen Aspergillus fumigatus requires the Rim101 ortholog PacC for virulence in a murine model of infection [2]. Since then, several investigators have demonstrated that other human, insect, and plant fungal pathogens require this conserved alkaline response pathway to cause disease [3–7].
More recently, the physiological role of Rim101 has been defined in the opportunistic fungal pathogen, Cryptococcus neoformans. C. neoformans is a basidiomycete yeast that causes life-threatening meningitis in immunocompromised individuals. Due to the emergence of HIV and the increased use of immunosuppressive therapies, this once-rare pathogen has become a significant global health problem, with over 1 million new infections estimated each year [8]. In C. neoformans, Rim101 is required for the proper formation of the protective polysaccharide capsule, as well as growth under several stress conditions such as low iron, elevated salt concentrations, and alkaline pH [9,10]. In addition, Rim101 mediates cell wall modifications that allow for capsule attachment and the masking/repression of cell surface pathogen associated molecular patterns (PAMPs). In the rim101Δ mutant strain, failed cell wall masking of immunogenic PAMPs results in a dramatic hyper-inflammatory response and immune-mediated host damage in a mouse inhalation model of cryptococcal infection [11].
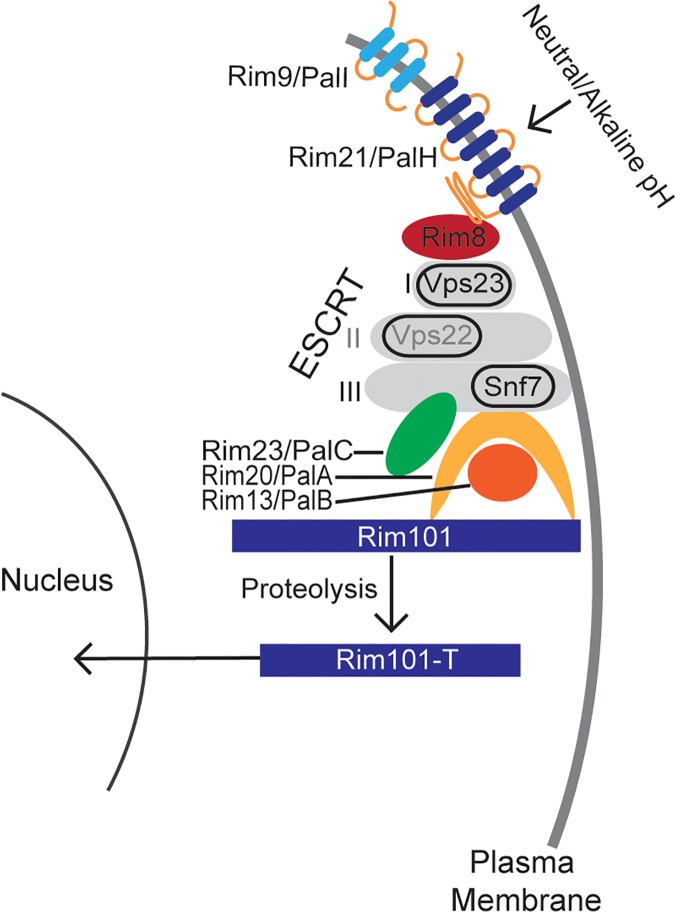
The signaling pathway responsible for sensing and activating Rim101 has been defined in model ascomycetes, such as Aspergillus nidulans and Saccharomyces cerevisiae, and it is highly conserved throughout other members of this phylum (Fig 1). In these species, initial activation of the Rim/Pal pathway involves extracellular sensing of pH by the 7-transmembrane domain receptor, Rim21/PalH [12–14]. This protein is a component of the membrane sensing complex, also comprised of the arrestin-like proteins Rim8/PalF, as well as the chaperone Rim9/PalI proteins [12,13,15]. Several ascomycete fungi require an additional component of the membrane signaling complex, Dfg16, which may serve as a Rim21/PalH chaperone [16]. Rim8/PalF recruitment to the sensing complex results in activation of part of the ESCRT complex, proteins also involved in endosome function [17–19]. Rim8/PalF binding to the ESCRT-I protein Vps23 induces the assembly of the ESCRT-II and—III complexes as a scaffolding platform for Rim pathway activation [19,20]. On this platform, the Rim proteolysis complex forms, composed of Rim23/PalC, Rim20/PalA, and the Rim13/PalB protease [21]. The intact Rim proteolysis complex catalyzes cleavage and activation of its target, the Rim101/PalC transcription factor [22,23]
Fig 1. A model of the canonical Rim pathway elucidated in ascomycete fungi [21,24].
This Rim/Pal signaling pathway has only recently been analyzed in basidiomycetes, a diverse group of fungi including some human and animal pathogens [9,25]. In the maize pathogen Ustilago maydis, the ESCRT-interacting complex (Rim23/PalC Rim20/PalA, and Rim13/PalB) was conserved and required for Rim101/PacC activation. However, elements of the membrane receptor complex (Rim9/PalI, Rim21/PalH, and Rim8/PalF) were either missing from the genome or not required for Rim101 processing [26]. In C. neoformans, there are several differences in the mechanism of Rim101 activation from established signaling paradigms in other fungal species. First, similar to U. maydis, the Rim/Pal membrane sensing complex proteins are not evident by homology searches. Also, we previously observed a novel connection between Rim101 and the cAMP/Protein Kinase A pathway [3,9]. The connection between these two conserved pathways reveals that the processes leading to Rim101 activation in this fungal species may occur in a somewhat distinct manner from that in distantly related ascomycetes. However, C. neoformans Rim101 activation also appears to require members of the canonical Rim activation pathway because deletion of the RIM20 ortholog resulted in phenotypes identical to a rim101Δ mutation [9]. This observation implicated the involvement of both the cAMP and the classical pH response pathway in the activation of C. neoformans Rim101 [3,9]. While the C. neoformans cAMP/PKA signaling pathway has been extensively characterized, comparatively little is known about the C. neoformans Rim pathway
In this study, we determined that C. neoformans encodes functional orthologs of the major components of the Rim101 proteolysis complex, Rim13/PalB, Rim23/PalC, and Rim20/PalA. In contrast to these highly conserved distal components of Rim101 activation, the surface pH-responsive proteins, Rim8/PalF and Rim21/PalH, could not be identified using sequence or domain-based searches. Utilizing a random mutagenesis screen specifically designed to find C. neoformans Rim101 activators, we identified a previously unrecognized membrane protein required for C. neoformans Rim101 activation, which we named RRA1 (Required for Rim101 Activation 1). While the Rra1 protein lacks significant sequence similarity to any component of the ascomycete Rim/Pal pathway, its predictive protein structure, containing 7-transmembrane domains, is similar to the Rim21/PalH sensors in ascomycetes. Moreover, predicted orthologs for Rra1 were found only in basidiomycetes. We also observe that Rim pathway signaling likely occurs in punctate regions on the plasma membrane. The recruitment of Rim23 to these puncta requires components of the ESCRT complex in addition to Rra1, suggesting that Rra1 functions upstream of other C. neoformans Rim pathway components. These findings suggest fundamental biological differences between the ways in which ascomycetes and basidiomycetes have developed cell surface signaling units to respond to common environmental signals.
Results
Neutral/alkaline pH induces Rim101 proteolytic cleavage and nuclear localization
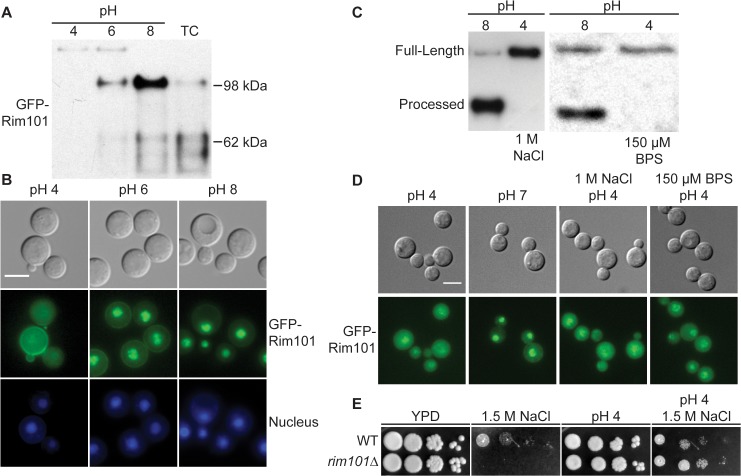
As modeled in Fig 1, the Ascomycete Rim101/PacC transcription factor is activated by Rim13/PalB-mediated proteolytic cleavage in response to elevated pH signals. Previously, we demonstrated that the GFP-Rim101 fusion protein, when overexpressed by the histone HIS3 promoter, was proteolytically cleaved from a 120 kDa form to a 70 kDa form in response to incubation in tissue culture medium [9]. To dissect the role of pH in Rim101 activation, we created a strain expressing GFP-Rim101 from its endogenous locus instead of the highly active histone3 promoter. The endogenously expressed GFP-RIM101 allele fully rescued all rim101Δ mutant phenotypes. Using this strain, we assessed GFP-Rim101 proteolysis in defined media at pH 4, 6, and 8. At pH 4 we exclusively observed the intact 140 kDa form of the GFP-Rim101 protein, suggesting that this is a non-inducing condition for Rim101 proteolysis and activation. In a dose-response relationship with increasing pH, we observed increased proteolytic processing of GFP-Rim101 resulting in a predominant 100 kDa processed form as well as lower molecular weight forms that either represent further processing or proteolytic degradation (Fig 2A). In addition, GFP-Rim101 proteolytic processing correlated with increased nuclear localization (Fig 2B). Together, this demonstrates that the C. neoformans Rim101 protein is activated by increasing pH.
Fig 2. Rim101 proteolysis and nuclear localization are dependent on pH.
(A) GFP-Rim101 is proteolytically processed from 140 kDa to ~100 kDa in response to increasing pH. GFP-Rim101 was immunoprecipitated from wild-type cells after incubating for 5 hr at the indicated pH 8 (SC medium buffered with McIlvaine’s buffer). Protein processing was determined by western blotting using an α-GFP antibody. (B) GFP-Rim101 nuclear localization increases in response to increasing pH. Cells were cultured in the same way as in (A). GFP signal was assessed by epifluorescence microscopy. Nuclei were stained using Hoechst 33342 live nuclei stain. Scale bar = 5 μm. (C) GFP-Rim101 proteolysis is not induced by 1 M NaCl or 150 μM BPS. Cells were cultured in each indicated condition for 3 hr. GFP-Rim101 was analyzed by western blot. (D) GFP-Rim101 localization in response to pH 7 SC (McIlvaine’s) or pH 4 SC (McIlvaine’s) with 1 M NaCl or 150 μM BPS. Cell assessed epifluorescence microscopy after 30 min incubation in each condition. Scale bar = 5 μm (E) rim101Δ is not NaCl sensitive at pH 4. Strains spotted onto YPD, YPD 150mM HEPES pH 4 1.5 M NaCl, and YPD 1.5 M NaCl.
Rim101 is not activated by high NaCl or iron limitation
In addition to responding to alkaline pH, C. neoformans Rim101 is required for growth in the virulence-relevant conditions of iron limitation and high salt concentrations [9]. These stress responses are linked with increasing pH because alkaline pH both reduces the bioavailability of iron and disrupts the membrane charge gradient necessary for ion channels to properly function [27].To determine whether Rim101 could also be activated by high salt concentrations and/or iron limitation independent of pH, we analyzed Rim101 activation in response to high salt and low iron conditions at pH 4. Neither GFP-Rim101 proteolytic processing nor nuclear localization was induced by the addition of 1 M NaCl (Fig 2C and 2D). Similarly, iron deprivation due to the addition of the iron chelator BPS only slightly increased GFP-Rim101 nuclear localization, but it did not induce Rim101 proteolytic processing (Fig 2C and 2D). In addition, the rim101Δ mutant did not have a growth defect on 1.5 M NaCl at pH 4 (Fig 2E), whereas it displays a salt sensitive phenotype at more alkaline pH. Therefore, C. neoformans Rim101 appears to be activated specifically by neutral/alkaline pH, rather than by other cell stress conditions such as salt stress or limiting iron availability.
Identification and characterization of C. neoformans Rim13 and Rim23 orthologs
The signaling pathway responsible for Rim101 activation is highly conserved throughout fungi in the Ascomycota phylum (Fig 1). These proteins are typically recognizable by sequence conservation, even among fungi as divergent as the budding yeast S. cerevisiae and the hyphal fungus A. nidulans. In a previous study, we identified the gene encoding a functional ortholog of the Rim101 scaffolding protein Rim20 in the basidiomycete fungus C. neoformans and confirmed its role in capsule regulation, alkaline pH adaptation, and response to low iron [9]. Using S. cerevisiae and A. nidulans amino acid sequences for comparisons, we identified single genes encoding other components of the C. neoformans Rim101 proteolysis complex, including the Rim13 protease (CNAG_05601) and the Rim23 ESCRT-interacting protein (CNAG_02205) (S1 Fig).
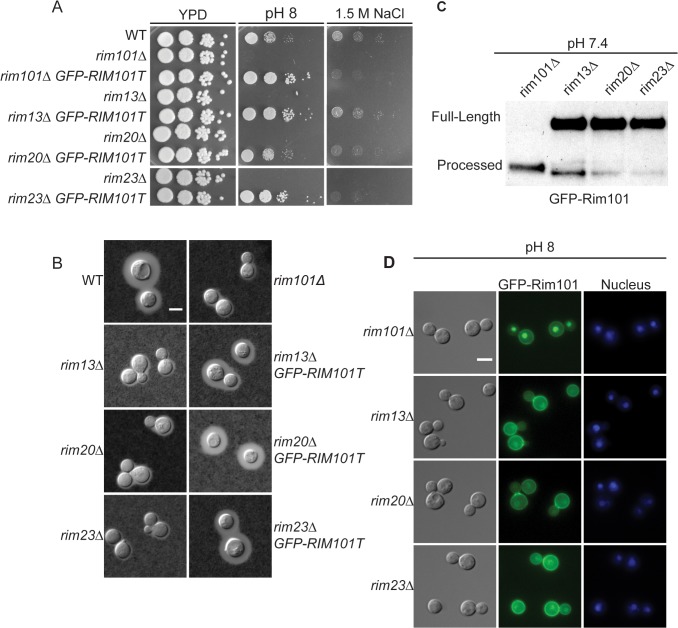
As expected, none of these genes were essential. Disruption of the RIM13 and RIM23 genes resulted in decreased growth on pH 8 and 1.5 M NaCl, similar to the rim101Δ mutant (Fig 3A). Notably, the rim13Δ and rim23Δ mutants also had a rim101Δ-like capsule defect, confirming a role for the pH-responsive signaling pathway in capsule regulation (Fig 3B). These rim13Δ and rim23Δ mutant phenotypes were rescued by reintroduction of the respective wild types alleles (S1 Fig).
Fig 3. Role of Rim13 and Rim23 orthologs in Rim101-regulated phenotypes.
(A) The C. neoformans RIM13 and RIM23 orthologs are required for pH 8 and NaCl tolerance. 10-fold serial dilutions of the indicated strains were spotted onto YPD, YPD 150mM HEPES pH 8, YPD 1.5M NaCl and incubated at 30°C for 48 hr -72 hr (B) The rim13Δ and rim23Δ mutants have a rim101Δ-like capsule defect. Cells were incubated in CO2-independent media for 48hr at 37°C. Capsule was visualized by counterstaining with India ink. (C) Rim101 proteolysis and localization are disrupted in rim13Δ, rim20Δ, and rim23Δ mutant strains. GFP-Rim101 was immunoprecipitated from each strain after 5 hr incubation in pH 7.4 YPD buffered with 150 mM HEPES. (D) GFP-Rim101 localization was assessed in the indicated strains after culturing for 5 hr in SC medium buffered with McIlvaine’s buffered to pH 8. Nuclei were stained with Hoechst 33342 live nuclei stain. Scale bar = 5 μm.
To directly determine whether Rim13, Rim23, and Rim20 are upstream activators of Rim101, we assessed Rim101 proteolytic activation and nuclear localization in each mutant. All three genes were required for both GFP-Rim101 proteolysis and nuclear localization, demonstrating that Rim101 activation requires each predicted Rim pathway component (Fig 3C and 3D). In A. nidulans and S. cerevisiae, expression of an active Rim101/PacC protein, lacking its inhibitory C-terminal domain rescues mutations in upstream Rim pathway components [28,29]. In a similar manner, we expressed of a truncated form of C. neoformans Rim101, lacking the C-terminal 603 amino acids, in the rim13Δ¸rim20Δ, and rim23Δ mutant strains. In these “upstream” mutants, this constitutively active form of Rim101 (GFP-Rim101T) restores capsule formation, as well as growth on pH 8 and in the presence of 1.5 M NaCl (Fig 3A and 3B).
ESCRT involvement in C. neoformans Rim pathway
In other fungi, the endosomal sorting complex required for transport (ESCRT) serves as a docking platform for Rim23, Rim20, and Rim13 proteins. Deletion of components of the ESCRT-I,-II, or-III complexes disrupts Rim101 activation [14,18,30–33]. In ascomycetes, the Rim pathway-dependent assembly of the ESCRT machinery is initiated by an interaction between the Rim8 arrestin-like protein and the ESCRT-I complex protein, Vps23. This recruits the ESCRT-II and then ESCRT-III complexes, eventually resulting in Rim101 proteolysis and activation [13,19,20].
Based on previous studies, there are several lines of evidence supporting a role for the ESCRT machinery in C. neoformans Rim101 activity. First, Hu et al., previously demonstrated that vps23Δ mutant phenotypes include sensitivity to pH 8 and 1.5M NaCl, along with displaying a capsule defect [34]. In addition, Chun and Madhani found that Vps25, a component of ESCRT-II, was required for several Rim101-dependent phenotypes [35]. More recently, Godinho et al demonstrated that the ESCRT-III component, Snf7, was required for capsule formation and full RIM101 transcription [36]. Rim101 induces its own transcription, which may partially explain the reason that disruption of a Rim pathway component would decrease RIM101 transcript levels [3]. Finally, both C. neoformans Rim23 and Rim20 contain predicted Snf7-interacting BRO1 domains (S1 Fig).
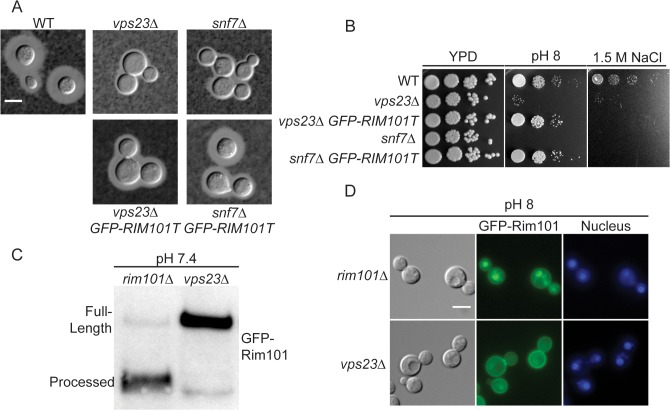
To directly determine whether the ESCRT complex is required for Rim101 activation in C. neoformans, we analyzed both the vps23Δ and snf7Δ mutants for defects in Rim pathway-associated phenotypes and Rim101 activation. Consistent with previously published data, both mutants displayed a marked capsule defect (Fig 4A). Both mutants were also sensitive to high salt and alkaline pH, similar to the rim101Δ mutant (Fig 4B) We also determined that GFP-Rim101 nuclear localization and proteolysis were disrupted in the vps23Δ mutation (Fig 4C and 4D). Similar to the rim13Δ, rim20Δ, and rim23Δ mutants, expression of the constitutively active, truncated GFP-Rim101T protein fully rescued the vps23Δ and snf7Δ mutant defects in growth at pH 8 and partially rescued their capsule defects (Fig 4A and 4B). Unlike the other Rim pathway mutants, GFP-Rim101T did not rescue the vps23Δ and snf7Δ growth defects on high salt concentrations. Therefore, both ESCRT-I and ESCRT-III components play conserved roles in C. neoformans Rim101 activation and function, although they have pleiotropic roles in regulating salt tolerance and complete capsule formation.
Fig 4. ESCRT complex proteins, Vps23 and Snf7, are required for Rim101 activation.
(A) snf7Δ and vps23Δ capsule defects are partially rescued by GFP-RIM101T expression. Strains cultured for 24 hr in tissue culture media. India ink used to visualize capsule. (B) GFP-RIM101T expression rescues the vps23Δ and snf7Δ growth defects on pH 8 but not 1.5 M NaCl. (C) GFP-Rim101 was immunoprecipitated from the indicated strains after 5 hr incubation in YPD with 150mM HEPES at pH 7.4. (D) GFP-Rim101 (full-length) nuclear localization is disrupted in the vps23Δ mutant. Localization was assessed after culturing for 5 hr in SC with McIlvaine’s buffer at pH 8. Nuclei were stained with Hoechst 33342 live nuclear stain. Scale bar = 5 μm.
Rim pathway membrane sensing complex components are not conserved in C. neoformans
In addition to the ESCRT machinery and the signaling components directly involved in Rim101 proteolytic activation, the classical ascomycete Rim pathway includes the integral membrane pH sensor Rim21/PalH, the integral membrane protein Rim9/PalI, and the arrestin-like Rim8/PalF. S. cerevisiae and C. albicans have an additional component of the Rim membrane sensing complex, Dfg16, which helps mediate Rim21/PalH localization and is also required for Rim pathway activation [16]. In ascomycetes, these plasma membrane Rim signaling components are identifiable based on highly conserved sequences and protein motifs. However, Rim21/PalH, Rim8/PalF, and Dfg16 orthologs could not be easily identified in C. neoformans using this approach. Similar searches did not identify predicted orthologs in any basidiomycete genome [26].
We also searched for predicted proteins with arrestin-like motifs that could be acting as Rim8 orthologs in C. neoformans. There were two predicted C. neoformans proteins (CNAG_02857, and CNAG_02341) that contained both the N- and C-terminal arrestin domains present in Rim8 proteins from other fungi. These were designated ALI1 and ALI2 (Arrestin Like 1 and 2). Disruption of these genes individually or in combination did not affect the Rim101-dependent phenotypes of capsule formation, growth at pH 8, or growth 1.5 M NaCl (S2 Fig) suggesting that these genes are not required for Rim101 activation.
We identified one predicted ortholog (CNAG_05654) of the Rim9 component of the sensing complex, which has a 24% sequence similarity to the S. cerevisiae Rim9 protein. We also identified 2 other genes (CNAG_02114 and CNAG_04953) predicted to encode proteins with homology to the SUR7/PalI domain, a defining feature of Rim9 proteins. Disruption of any of these potential RIM9 orthologs, either separately or in combination in a triple mutant strain, did not affect Rim101-dependent phenotypes (S2 Fig).
Identification of novel C. neoformans Rim pathway component
As we were unable to identify components of the C. neoformans Rim pathway membrane sensing complex using sequence comparisons or searches for conserved protein domains, we designed a forward genetics screen to specifically identify novel Rim101 activators. The defining characteristic of a Rim pathway mutant is that its mutant phenotypes are rescued by expression of the constitutively active form of Rim101. Therefore, we created a strain encoding the truncated GFP-RIM101T allele under the control of the galactose-inducible GAL7 promoter, and we performed Agrobacterium tumefaciens-mediated random insertional mutagenesis in this strain. We then selected for mutants with rim101Δ-like phenotypes (sensitivity to alkaline pH and 1.5 M NaCl) in the presence of glucose (repressing conditions for GFP-Rim101T expression) and wild-type phenotypes on galactose-containing media (inducing conditions for GFP-Rim101T expression).
Using this random mutagenesis approach, we identified 40 mutants with galactose-suppressible alkaline and/or NaCl sensitivities. After identifying the site of each mutation, we recognized several disrupted genes among these mutants that were known to be required for Rim101 expression or activation. We identified one insertion in HAPX, which encodes a transcription factor required for full induction of RIM101 expression [37]. In this case, galactose-induced expression of GFP-Rim101T likely rescued the defect in RIM101 transcription, as opposed to a defect in Rim101 protein activation. We also identified an insertion in the RIM13 gene encoding the Rim101 activating protease. These data demonstrate that this screening strategy was successful in identifying genes known to be required for Rim101 activity.
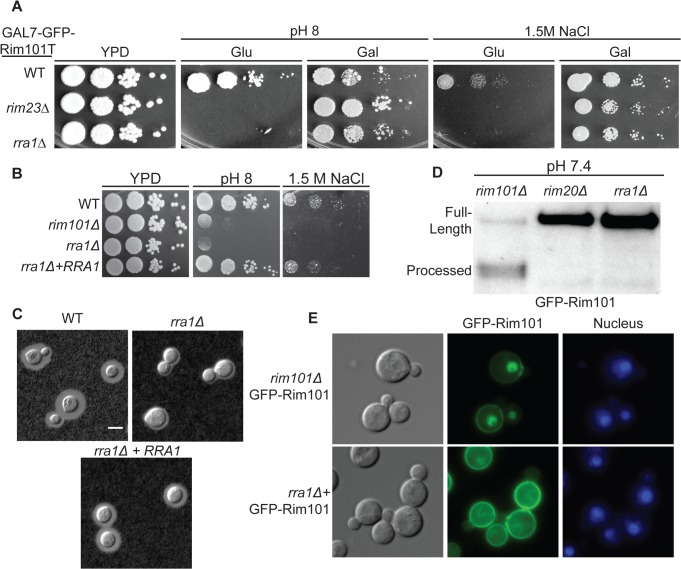
Among the additional mutants, we focused on a strain with an insertion in the CNAG_03488 locus due to its striking rim101Δ-like phenotypes on both 1.5 M NaCl and pH 8 (Fig 5A and 5B). Furthermore, these mutant phenotypes were completely rescued by the expression of GFP-Rim101T on galactose (Fig 5A). We confirmed these mutant phenotypes by independently disrupting the CNAG_03488 gene in the wild type H99 background (Fig 5B). In addition to pH 8 and 1.5M NaCl sensitivity, the CNAG_03488 mutant also displayed a rim101Δ-like capsule defect (Fig 5C). We complemented all mutant phenotypes by introduction of the wild type allele (Fig 5).
Fig 5. The rra1Δ mutant is phenotypically identical to other Rim pathway mutants.
(A) rra1Δ insertional mutant has pH 8 and 1.5 M NaCl growth defects that are rescued by GFP-RIM101T expression. 10-fold serial dilutions were spotted onto YPD, YPD with 150 mM HEPES pH 8, and YPD with 1.5 M NaCl. (B) Independent rra1Δ mutant has a growth defect pH 8 and 1.5 M NaCl. (C) The rra1Δ strain has a capsule defect. Cells were cultured for 48 hr in CO2-independent media at 37°C to induce capsule. Capsule was visualized by India ink staining. Scale bar = 5 μm. (D) The rra1Δ mutation disrupts GFP-Rim101 proteolysis. GFP-Rim101 was immunoprecipitated from the indicated mutant strains after 5 hr incubation in YPD with 150 mM HEPES at pH 7.4. (E) GFP-Rim101 nuclear localization is disrupted in the rra1Δ mutant. GFP-Rim101 was assessed after 5 hr incubation in pH 8 SC McIlvaine’s buffer.
To determine if the protein encoded by the CNAG_03488 gene was a component of the Rim pathway, we assessed GFP-Rim101 localization and proteolysis in the CNAG_03488 mutant. Like the verified activators of Rim101, CNAG_03488 was required for both Rim101 cleavage and nuclear localization (Fig 5D and 5E). Based on these results, we concluded that, like RIM13, RIM20, RIM23, and the ESCRT complex, the novel protein encoded by CNAG_03488 is required for Rim101 activation. To reflect its role in the Rim pathway, we named this gene RRA1 (Required for Rim101 Activation 1).
Rra1 is conserved throughout Basidiomycota
To determine whether Rra1 is a C. neoformans-specific or conserved component of the Rim pathway, we used the predicted Rra1 amino acid sequence to search for possible Rra1 orthologs in other fungal species. We only identified Rra1 homologs in other members of the basidiomycete phylum; however, these predicted proteins demonstrated a high degree of homology to Rra1 (Fig 6A and S3 Fig), even in distantly related members of this phylum such as U. maydis and P. graminis. We also demonstrated that Rra1 function in Rim pathway activation is functionally conserved in the divergent sibling species Cryptococcus gattii. Disruption of the C. gattii RRA1 (CNBG_2126) ortholog resulted in alkaline sensitivity and a capsule defect identical to the C. gattii rim101Δ mutant (CNBG_4424) (Fig 6B and 6C). These results demonstrate that Rra1 orthologs function in Rim101-related processes in different basidiomycete species.
Fig 6. Rra1 is a membrane protein and is conserved through Basidiomycete fungi.
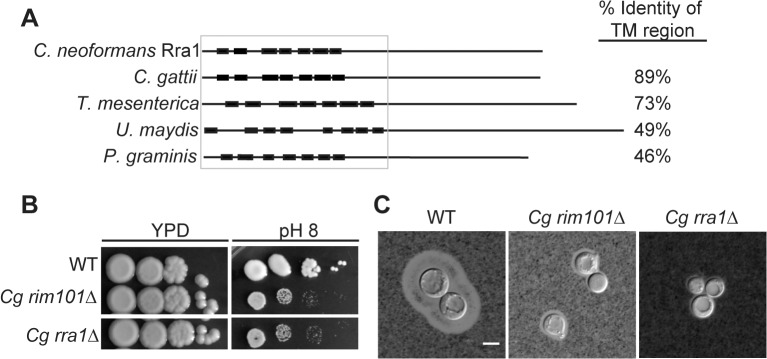
(A) C. neoformans Rra1 and its predicted basidiomycete orthologs contain 7–8 predicted transmembrane domains (closed boxes). C. neoformans CNAG_03488 (Rra1), C. gattii CGB_G5320C, Tremella mesenterica TREME_69388, Ustilago maydis um00299, Puccinia graminis PGTG_03106 predicted protein sequences are modeled. The TM domains were predicted using The TMPred TM prediction server [38] The % identity to the transmembrane region of C. neoformans Rra1 was determined by comparing 1–290 of the predicted Rra1 amino acid sequence to each of the predicted orthologs. (B) C. gattii rra1Δ and rim101Δ mutants are sensitive to pH 8. (C) Cg rra1Δ and rim101Δ have a capsule defect. Capsules visualized with India Ink after 24 hr incubation in tissue culture media. Scale bar = 5 μm.
C. neoformans Rra1 and its predicted basidiomycete orthologs all contain 7–8 predicted transmembrane domains (Fig 6A and S3 Fig), suggesting that they may be structurally similar to the Rim21/PalH pH sensors, which also contain 7 transmembrane domains [21] However, Rra1 does not share significant amino acid sequence homology with Rim21/PalH proteins, and direct BLAST comparisons between Rra1 and S. cerevisiae Rim21 and A. nidulans PalH produced Expected values greater than 1.
The Rim pathway does not require Gα protein activity
With 7 predicted transmembrane domains, Rra1 could also be structurally similar to GPCR proteins (G-protein Coupled Receptors). When activated, GPCRs interact with the Gα subunit of trimetric G proteins, leading to alteration in downstream signaling. C. neoformans encodes three characterized Gα proteins, Gpa1, Gpa2, and Gpa3 [39,40]. To begin to explore whether Rra1 is a GPCR and signaling through any of these Gα proteins, we analyzed the gpa1Δ, gpa2Δ, and gpa3Δ mutants for rim101Δ-like phenotypes. The gpa1Δ and gpa3Δ mutants grew like wild type on both pH 8 and 1.5 M NaCl, inconsistent with defective Rim pathway signaling. The gpa2Δ mutant was slightly sensitive to both pH 8 and 1.5 M NaCl, but these growth defects were not as severe as those displayed by a rim101Δ mutant or other Rim pathway mutants (S4 Fig). Moreover, Gpa2 plays a prominent role in C. neoformans mating, a cellular process that does not appear to be dependent on Rim signaling in this species [40]. Therefore, Gpa1, Gpa2, and Gpa3, were not required for Rim101 activation in C. neoformans and these proteins do not appear to be components of the C. neoformans Rim pathway.
pH-regulated Rim23-GFP localization requires Rra1 and ESCRT
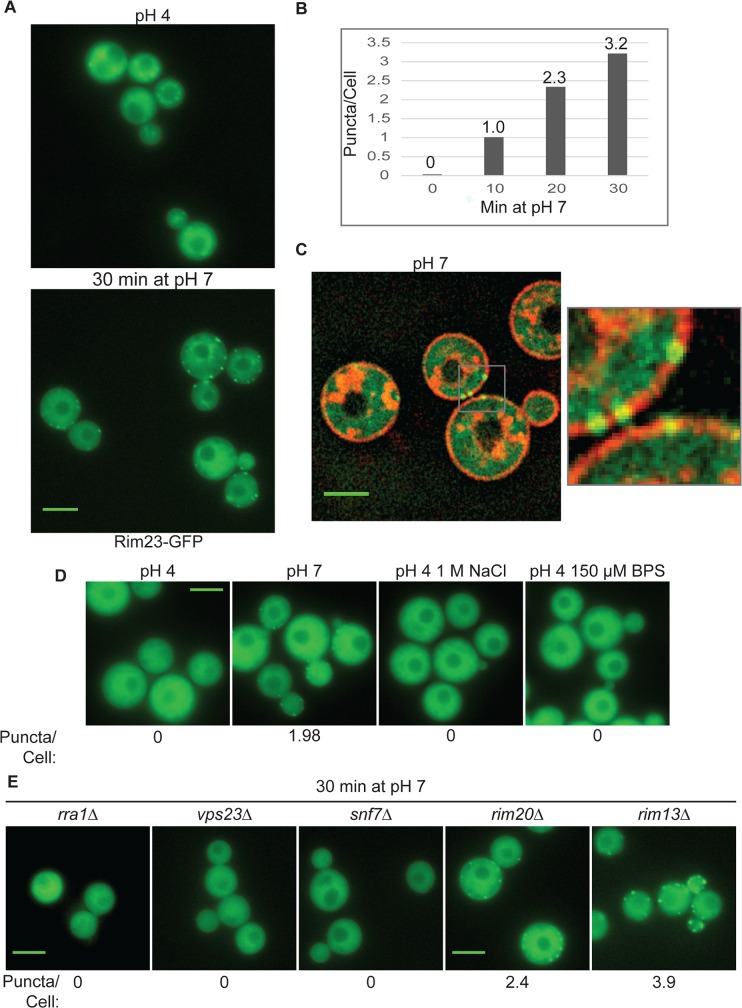
When activated, the ascomycete Rim21/PalH sensor recruits downstream Rim pathway components to punctate structures at the plasma membrane [12,14,19,21]. The order in which these components localize to this complex was used to establish the sequence of Rim/Pal protein activation (Fig 1). To determine if similar pH-regulated localization changes occur in C. neoformans, we expressed a Rim23-GFP fusion protein, regulated by the endogenous RIM23 promoter, in a rim23Δ mutant. This fusion protein was functional, fully rescuing the rim23Δ defects in pH 8 growth and capsule formation, and partially rescuing the 1.5 M NaCl growth defect. When incubated at pH 4, Rim23-GFP localized diffusely throughout the cell (Fig 7A). After shifting the cells to pH 7, Rim23-GFP migrates to distinct, cell surface-associated puncta. The number of puncta/cell increased from 1 to 3.2 between 10 min and 30 min of incubation at neutral pH (Fig 7B). These puncta appear to be on the plasma membrane or very close to the plasma membrane, co-localizing with non-endocytosed FM4-64 staining of the plasma membranes (Fig 7C). These punctate structures formed specifically in response to elevated pH, and not to other stress conditions associated with Rim101 function, such as high salt or low iron: the GFP-Rim23-containing puncta were not observed in cells shifted to pH 4+1 M NaCl, or pH 4+150 μM BPS (Fig 7D). Together, these data indicate that Rim23 cell surface puncta are specifically induced in response to pH signals, consistent with a role for neutral/alkaline pH in inducing Rim101 activation.
Fig 7. Rim23-GFP forms plasma membrane-associated puncta under neutral/alkaline pH conditions.
(A) Rim23-GFP was visualized at pH 4 SC McIlvaine’s buffer and after 30 min after shift to pH 7 SC McIlvaine’s buffer. (B) The number of Rim23-GFP puncta increase with time after shifting from pH 4 to pH 7. Quantification Rim23-GFP puncta/cell at pH 4 (0 min) and after 10, 20, 30 min incubation at pH 7. (C) Rim23-GFP puncta are closely associated with the plasma membrane. Cells were stained with FM-464 after 1 hr incubation at pH 7. (D) 1 M NaCl and 150 μM BPS does not induce Rim23-GFP puncta formation. Cells images after 30 min incubation in indicated culture media. (E) Rim23-GFP puncta formation was disrupted by rra1Δ, vps23Δ, and snf7Δ mutants. Cells were imaged after 30 min incubation at pH 7. The number of puncta/cell was quantified for 62–100 cells/strain. All strains in this figure were cultured in SC McIlvaine’s buffer media. All scale bars = 5 μm.
We next determined which components of the Rim pathway were required for the observed Rim23-GFP plasma membrane localization. Neither Rim20 nor Rim13, other members of the Rim proteolysis complex, were required for Rim23-GFP cell surface puncta formation. However, pH-driven Rim23 localization to surface punctate structures did not occur in the rra1Δ, vps23Δ and snf7Δ mutants (Fig 7E). Therefore, Rim20 and Rim13 are not required for Rim23 localization, while both the ESCRT pathway proteins and Rra1 are functionally upstream of Rim23-puncta formation.
Rim pathway mutants induce an increased inflammatory response during infection
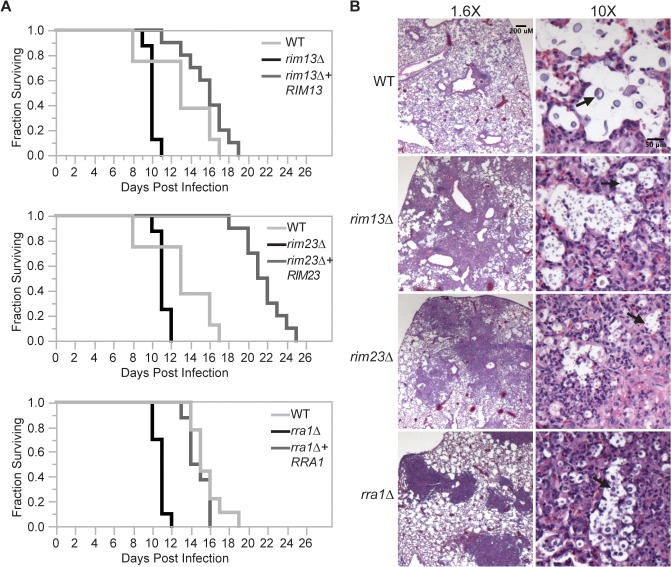
During infection, C. neoformans Rim101 regulates the expression of several genes involved in cell wall synthesis and maintenance by directly binding to elements in their promoters [3,11]. In the absence of Rim101-directed cell wall maintenance, the rim101Δ strain induces a hyper-inflammatory response in the lungs of infected mice. In this way, infection with this attenuated strain results in the paradoxical death of the host. We previously demonstrated that Rim101 activity is also regulated by the PKA/cAMP pathway, and pka1Δ mutant displays cell wall changes similar to the rim101Δ mutant [3,11]. As Rim101 activation is regulated by both the cAMP/PKA pathway and the Rim pathway, we examined whether mutations in other components of the Rim pathway were sufficient to alter virulence in a murine inhalation model of cryptococcosis. For each strain, 8–10 A/J mice were intranasally inoculated with 5x104 C. neoformans cells. Mice were monitored daily and sacrificed when they displayed symptoms of disease consistent with imminent death. In each experiment, all mice infected with the wild type died between 18–20 days. In contrast to the mice infected with the wild type and reconstituted strains, all mice infected with the rim13Δ, rim23Δ, or rra1Δ mutant cells succumbed to the infection by day 12 (Fig 8A). This statistically significant decrease in survival (p>0.001) was consistent with the mortality of mice infected with a rim101Δ mutant [9,11]. Both the rim13Δ+RIM13 and rra1Δ+RRA1 reconstituted strains resulted in animal mortality at a rate very similar to WT. The rim23Δ+RIM23 reconstituted strain displayed a mild delay in virulence, which we subsequently found was likely due to a temperature sensitive phenotype specific only to this isolate. An independent rim23Δ+RIM23 isolate without a temperature-sensitive phenotype was used for the rest of these experiments.
Fig 8. Effects of Rim pathway mutants on virulence.
(A) rim13Δ, rim23Δ, and rra1Δ mutants are hypervirulent in a murine model of cryptococcosis. 8–10 A/Jcr female mice were intranasally inoculated with 1X105 cryptococcal cells and monitored daily for survival. (B) Histopathological analysis revealed increased inflammatory cell infiltration in Rim-pathway mutants. Infected A/Jcr mouse lungs were harvested on day 7 post inoculation and H & E stain. Black arrows mark C. neoformans cells.
Like the rim101Δ mutant, the rim13Δ, rim23Δ, and rra1Δ mutants induced an increased infiltration of inflammatory cells in the lungs of infected mice. Histologic examination of infected mouse lungs 7 days after infection revealed that, unlike the lungs infected with wild type, those infected with Rim pathway mutants were characterized by diffuse regions of intense inflammatory cell infiltration (Fig 8B). This type of inflammation is consistently observed in mice infected with the rim101Δ mutant strain [11]. These results suggest that the immune system recognizes other Rim pathway mutants in a similar manner to a rim101Δ mutant.
The histopathological images also allowed us to analyze size and morphology of the C. neoformans cells. In the wild type infected lungs, there were numerous titan-sized cells [41,42]. In contrast, the rim13Δ, rim23Δ, and rra1Δ mutant cells were noticeably smaller in size (Fig 8B). Therefore, like rim101Δ, these mutants also have a defect in titan cell formation in vivo [43]. Together, these results are consistent with a model in which Rim13, Rim23, and Rra1 act as mediators of Rim101 activation in the context of infection.
Discussion
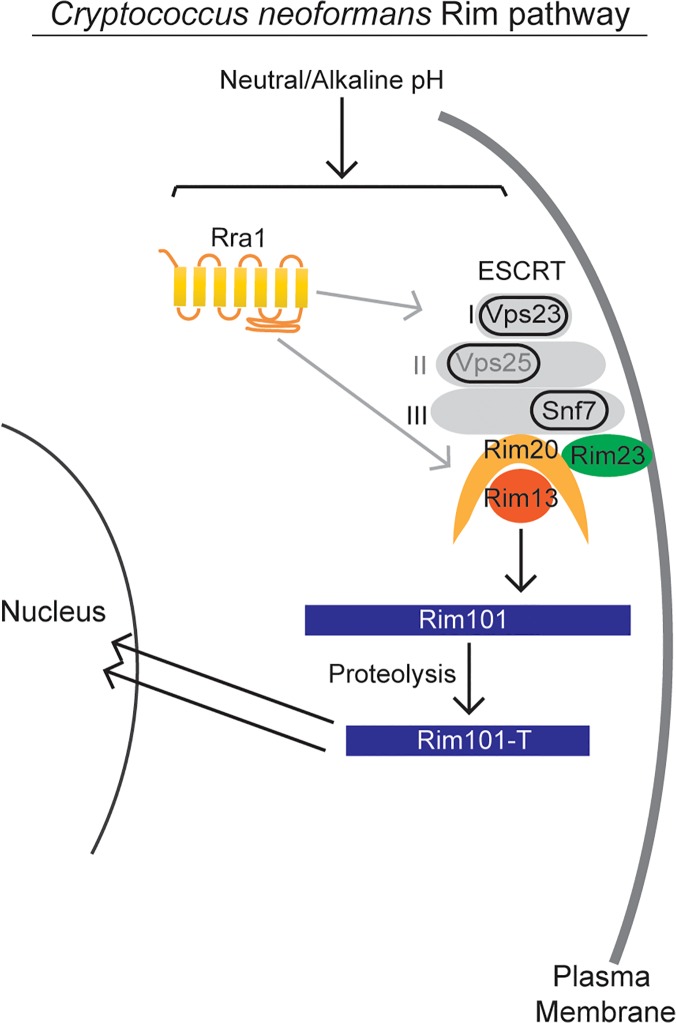
In this study, we identified both conserved and novel components of the C. neoformans Rim pathway. Based on our results as well as investigation of this pathway in other fungi, we propose the model in Fig 9 for Rim101 activation in C. neoformans. In response to neutral/alkaline pH, both the newly discovered Rra1 membrane protein and the ESCRT—I,-II,-III complex proteins control Rim23 relocation to punctate structures on the plasma membrane. In addition to Rim23, the other components of the Rim101 proteolysis complex are also functionally conserved in C. neoformans. Proteolysis enhances Rim101 nuclear localization, where it acts as both a transcriptional activator and repressor of a large group of genes, some of which are necessary for adaption to neutral/alkaline pH [3,11].
Fig 9. Model of C. neoformans Rim pathway.
Our data supports a model in which the Rim pathway is activated by neutral/alkaline pH. In this pathway, the newly discovered Rra1 membrane protein and the ESCRT complex, including Vps23 (ESCRT-I) likely Vps25 (ESCRT-II) and Snf7 (ESCRT-III), induce Rim23 puncta formation and activate the Rim101-proteolysis complex. The Rim13 protease cleaves the Rim101 transcription factor, inducing its nuclear localization. Active Rim101 then transcriptional changes necessary for adapting to alkaline pH.
While the C. neoformans Rim pathway is required for responding to several stressful conditions, our results suggest that it is specifically activated by increased pH. We found that neither iron limitation nor high NaCl concentrations could alone induce Rim101 processing. In contrast, increasing pH resulted in a dose-dependent increase in Rim101 proteolytic processing and nuclear localization. The marginally increased intensity of GFP-Rim101 nuclear staining under iron limitation was likely due to increased levels of full-length Rim101; which may possess some intrinsic signaling activity. Rim101 transcription is activated by iron-responsive transcription factors, such as HapX, which themselves are activated by this low-iron growth condition [37] The exact mechanism of Rim pathway pH sensing is still largely unknown in any system, although recent research in S. cerevisiae Rim signaling suggests that altered membrane polarity serves as at least one of the pathway activating signals [12,44].
The ascomycete Rim101/PalC proteolysis complex consists of Rim23/PalC, Rim20/PalA, and Rim13/PalB. Upon pathway activation, these proteins are recruited by the ESCRT complex to plasma membrane puncta [12,14,19]. We demonstrate here demonstrate that the Rim101 proteolysis complex is functionally conserved in C. neoformans and is likely assembled and localized similarly to the ascomycete Rim101/PacC proteolysis complex. In addition to the required role of Rim23, Rim20 and Rim13 in Rim101 activation, we demonstrated that Rim23 is recruited to plasma membrane puncta when the pathway is activated. Rim23 is likely recruited to these punctate structures through an interaction with the assembled ESCRT machinery. The majority of the Rim23 protein sequence consists of the conserved Snf7-interacting domain, which likely facilitates an ESCRT-III/Rim23 interaction. Further supporting this model, both Vps23 (ESCRT-I) and Snf7 (ESCRT-III) were required for Rim23 puncta formation. The same Snf7-interacting domain is also present in the Rim20 amino acid sequence, indicating that, like in ascomycetes, both Rim23 and Rim20 interact with the assembled ESCRT-III complex in C. neoformans. [14,19,21]. Taken together, our results suggest that the C. neoformans Rim101 proteolysis complex is recruited to the plasma membrane after Rim pathway activation and organized in a similar manner to the ascomycete Rim101 proteolysis complex.
The role of the C. neoformans ESCRT complex in Rim101 activation has been suggested by several studies on ESCRT components [34,45,46]. We demonstrated that at least two documented ESCRT-mutant phenotypes, alkaline pH sensitivity and a capsule formation defect, result from failed Rim101-activation. This suggests that other ESCRT-dependent processes, such as heme acquisition [34], may also be dependent on the Rim pathway. However, we also observed that the ESCRT complex regulates multiple processes independent of Rim101 activation. First, the vps23Δ and snf7Δ mutants displayed NaCl sensitivity that was more severe than other Rim pathway mutants, and this was not rescued by expression of the constitutively active Rim101T. Furthermore, constitutively active Rim101T expression only partially rescued the vps23Δ and snf7Δ capsule defects. Perhaps the most striking difference between the ESCRT mutants and other Rim pathway mutants is their differing roles in pathogenesis. Unlike the Rim pathway mutants, which are hypervirulent in a murine inhalation infection model, both snf7Δ and vps23Δ mutants are avirulent [34,36]. These more severe ESCRT mutant phenotypes reflect the multiple roles the ESCRT machinery serves in C. neoformans biology and pathogenesis.
Prior to this study, the only identified basidiomycete Rim pathway components were identified based on sequence similarity to components of the ascomycete Rim/Pal pathway [25,26]. This strategy failed to identify the most upstream Rim pathway components in both U. maydis and C. neoformans or other members of this phylum. In this study, we performed a forward genetics screen and identified a novel component of the C. neoformans Rim pathway. This novel protein, Rra1, is functionally conserved in at least one related basidiomycete, C. gattii, and is conserved at a sequence level in more distantly related basidiomycetes. Rra1 is required for all Rim101-dependent phenotypes and is functionally upstream of the pH-regulated subcellular localization of the Rim101 proteolysis complex protein, Rim23. Unlike the ESCRT complex components, disruption of RRA1 did not lead to additional, non-Rim101 dependent phenotypes, suggesting that Rra1 functions specifically in Rim pathway activation.
While the exact function of Rra1 is unknown, it has two characteristics that suggest that it may function as a Rim pathway pH sensor. Like the ascomycete Rim21/PalH pH sensor, Rra1 was required for neutral/alkaline pH induced Rim23 puncta formation [14]. In addition, Rra1, in C. neoformans and other basidiomycetes, is predicted to contain 7 transmembrane domains. However, Rra1 does not share significant amino acid sequence homology with any Rim21/PalH protein. Since the exact Rim21/PalH amino acids responsible for sensing pH are unknown, we are as yet unable to determine if Rra1 also contains similar pH sensing domains. Rra1 may also sense pH through histidine or paired glutamic acid/aspartic acid residues that are common in other pH sensing GPCRs or ion transporting proteins, and there are several of these amino acids that appear within the basidiomycete Rra1 amino acid sequence [47–50]. While the predicted Rra1 secondary structure suggests a functional similarity to Rim21/PalH, it is also possible that Rra1 serves a different purpose in Rim pathway activation. Like Dfg16 in S. cerevisiae, Rra1 may serve a supportive role in the proper localization of the Rim pathway pH sensor [12,16,19]. Rra1 may also function in a different cellular location, such as the vacuolar membrane or ER, which are known sites of pH-sensing and pH homeostasis [51,52]. Further studies on this novel protein will significantly advance our understanding of how C. neofromans, and other basidiomycetes sense pH.
Other components of the C. neoformans Rim membrane sensing complex remain elusive. None of the three potential Rim9/PalI orthologs nor the two candidate Rim8/PalF arrestin-like proteins were required for Rim pathway activation. Given the Rra1 structural similarity to GPCRs, we also examined whether the Rim pathway could signal through a canonical Gα protein, but none of the three known Gα proteins were required for Rim pathway activation. More extensive genetic screens will likely identify additional components for the C. neoformans Rim pathway, including cell surface chaperones and linking proteins.
The role of the Rim pathway in C. neoformans pathogenesis is complex. First, this pathway is required for C. neoformans survival in the host and rim101Δ mutants are rapidly and efficiently cleared in animal models of infection. Mechanistically, we have demonstrated that Rim101 is required for survival in the face of specific conditions found in micro-niches of the host; alkaline pH, iron deprivation, salt stress. Paradoxically, while these attenuated strains are poorly viable, they simultaneously induce an overly exuberant immune response. This is likely due to failed organization of the cell wall-masking phenotype and exposure of highly immunogenic epitopes. Interestingly, in human cases of cryptococcal meningitis, transcriptional studies of the pathogens at the site of infection demonstrate that RIM101 transcripts are among the most highly induced during infection [53]. These previous studies suggested that Rim101-regulated cell wall changes in C. neoformans may be induced by the cAMP/Pka1 pathway. Here, we show that other elements of the Rim pathway are also required for Rim101-mediated interactions with the host.
In this study, we have provided the most thorough investigation of the Rim pathway in any basidiomycete fungus. Together, these data demonstrate the importance of microbial adaptation to environmental and host-specific signals for pathogen survival. Additionally, they further enrich our understandings of pathogen adaptation and host immune recovery to minimize host damage during infections.
Materials and Methods
Strains, media, and growth conditions
Gene loci analyzed in this study are shown in Table 1.The strains used in this study are shown in Table 2. The C. neoformans H99 Matα genetic background was used to create each mutant and fluorescent strain [54]. Unless otherwise stated, cells were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) [55]. To create the eGFP-RIM101 strains (KS88-2, KS87-2, and KS118) each mutant strain was crossed with KN99 Mata on MS mating media [56]. Spores were isolated by microdissection and recombinant spores were identified by PCR and nourseothricin (NAT) or neomycin (NEO) resistance. The resulting Mata mutant strains were crossed with rim101Δ + GFP-RIM101 (TOC105) and recombinant spores were isolated by microdissection. The rim90Δ rim91Δ rim92Δ (KS136) strain was created by first creating rim91Δ rim92Δ MATa KS117 and crossing it with the KS63 strain. The GAL7-RIM101 70T strains were created by crossing rim23Δ + GAL7-RIM101 70T (KS159) with KN99 Mata and isolating recombinant progeny. The KS161 strain was identified by PCR and phenotype.
Table 1. Gene loci analyzed in this study.
| Genes | CNAG number | GeneBank |
|---|---|---|
| RIM13 | CNAG_05601 | AFR99029.2 |
| RIM20 | CNAG_03582 | AFR96803.1 |
| RIM23 | CNAG_02205 | AFR95615.1 |
| RRA1 | CNAG_03488 | AFR96713.2 |
| RIM90 | CNAG_05654 | AFR99085.1 |
| RIM91 | CNAG_02114 | AFR94219.1 |
| RIM92 | CNAG_04953 | AFR94219.1 |
| ALI1 | CNAG_02857 | AFR93834.1 |
| ALI2 | CNAG_02341 | AFR95488.1 |
| CgRIM101 | CNGB_4424 | |
| CgRRA1 | CNGB_2126 |
Table 2. Strain list.
| Strains | Genotype | Source |
|---|---|---|
| H99 | MATα | [57] |
| KN99 | MAT a | [58] |
| KS33 | rim13Δ::NEO MATα | this study |
| KS110 | rim13Δ::NEO MAT a | this study |
| TOC66 | rim13Δ::NEO + RIM13 + pCH233 (NAT) MATα | this study |
| KS88-2 | rim13Δ::NEO + eGFP-RIM101 pCH233 (NAT) MATα | this study |
| KS140 | rim13Δ::NEO +GFP-RIM101T + pCH233 (NAT) MATα | this study |
| KS94 | rim23Δ::NEO MATα | this study |
| KS82-2 | rim23Δ::NEO MAT a | this study |
| KS81-2 | rim23Δ::NEO + RIM23 + pCH233 (NAT) MATα | this study |
| KS87-2 | rim23Δ::NEO + eGFP-RIM101 + pCH233 (NAT) MATα | this study |
| KS141 | rim23Δ::NEO +GFP-RIM101T + pCH233 (NAT) MATα | this study |
| KS159 | rim23Δ::NEO + pTO22 (GAL7-GFP-RIM101T NAT) MATα | this study |
| TOC17 | rim20Δ::NAT MATα | [8] |
| KS118-2 | rim20Δ::NAT + eGFP-RIM101 pJAF (NEO) MATα | this study |
| KS77-2 | rim20Δ::NAT +GFP-RIM101T + pJAF (NEO) MATα | this study |
| TOC2 | rim101Δ::NAT MATα | [8] |
| TOC105 | rim101Δ::NAT + eGFP-RIM101 MATα | [2] |
| KS208 | rim101Δ::NAT + eGFP-RIM101 MAT a | this study |
| TOC65 | rim101Δ::NAT + GFP-RIM101T + pJAF (NEO) MATα | this study |
| vps23-9 | vps23Δ::NEO MATα | [20] |
| KS211 | vps23Δ::NEO + eGFP-RIM101 + pCH233 (NAT) MATα | this study |
| KS75-2 | vps23Δ::NEO +GFP-RIM101T + pCH233 (NAT) MATα | this study |
| snf7Δ | snf7Δ::NEO | [36] |
| KS151 | snf7Δ::NEO +GFP-RIM101T + pCH233 (NAT) MATα | this study |
| KS63 | rim90Δ::NEO MATα | this study |
| KS57 | rim91Δ::NEO MATα | this study |
| KS53 | rim92Δ::NEO MATα | this study |
| KS117 | rim91Δ::NEO rim92Δ::NEO MAT a | this study |
| KS136 | rim90Δ::NEO rim91Δ::NEO rim92Δ::NEO MATα | this study |
| KS118 | ali1Δ::NEO MATα | this study |
| KS94-2 | ali2Δ::NAT MATα | this study |
| KS97-2 | ali1Δ::NEO ali2Δ::NAT MATα | this study |
| KS161 | H99 + pTO22 (GAL7-GFP-RIM101T NAT) MATα | this study |
| KS216 | AGRO MUTANT rra1Δ::NEO + (GAL7-GFP-RIM101T NAT) MATα | this study |
| KS183 | rra1Δ::NEO MATα | this study |
| KS202 | rra1Δ::NEO + pKS38 (RRA1 NAT) MATα | this study |
| KS185 | rra1Δ::NEO + eGFP-RIM101 MATα | this study |
| KS214 | rra1Δ::NEO + pKS34 (pHIS3-GFP-RRA1 NAT) MATα | this study |
| KS289 | rim23Δ::NEO + GFP-RIM23 + NAT MATα | this study |
| KS290 | rim23Δ::NEO + GFP-RIM23 + NAT MAT a | this study |
| KS292 | snf7Δ::NEO rim23Δ::NEO + GFP-RIM23 + pCH233 (NAT) | this study |
| KS296 | vps23Δ::NEO rim23Δ::NEO + GFP-RIM23 + pCH233 (NAT) | this study |
| KS298 | rim13Δ::NEO rim23Δ::NEO + GFP-RIM23 + pCH233 (NAT) | this study |
| KS299 | rim20Δ::NEO rim23Δ::NEO + GFP-RIM23 + pCH233 (NAT) | this study |
| KS301 | rra1Δ::NEO rim23Δ::NEO + GFP-RIM23 + pCH233 (NAT) | this study |
| YSB25 | gpa2Δ::NEO | [40] |
| YPH105 | gps3Δ::NEO | [40] |
| YSB83 | gpa1Δ::NAT | [59] |
| R265 | C. gattii | [60] |
| KS260 | C. gattii rra1Δ::NEO | this study |
| KS261 | C. gattii rim101::NEO | this study |
YP-Gal media contained 1% yeast extract, 2% peptone, and 3% galactose. The pH 4 and pH 8 media was made by adding 150 mM HEPES buffer to YPD or YP-Gal liquid media, adjusting the pH with concentrated HCl (for pH 4) or NaOH (for pH 8), prior to autoclaving. The NaCl plates were made by adding NaCl to YPD to a concentration of 1.5 M. To induce capsule, strains were incubated for 48 hr in CO2-independent medium (Gibco) with 150 rpm shaking at 37°C.
To analyze GFP-Rim101 localization, strains were grown overnight (~18 hr) at 30°C with 150 rpm shaking. Cells were then pelleted and resuspended in either pH 4 or pH 8 Synthetic Complete media buffered with McIlvaine’s buffer. Strains were shaken at 150 rpm, 30°C for 5 hr
Molecular biology
The primers used to create each mutant and fluorescent strain are listed in Table 3.
Table 3. Primers.
| Primer name | 5'-3' Primer sequences |
|---|---|
| AA1792 | GTCATAGCTGTTTCCTGGGGCGGATGATGCAGAGTTA |
| AA1793 | ACTGGCCGTCGTTTTACTAACAAAGGTAACCGTCGGT |
| AA1794 | ACCGACGGTTACCTTTGTTAGTAAAACGACGGCCAGT |
| AA1795 | AAGTCAGCGGTCTTGAGGAA |
| AA1796 | TAAGGGCTAAAGTCGGAGCA |
| AA1797 | TAACTCTGCATCATCCGCCCCAGGAAACAGCTATGAC |
| AA1812 | ATCTTGCCATTGATGATAG |
| AA1813 | GTCGGAAGATTAAAAAGTG |
| AA3226 | GGATTGGTCTAGGGCCTCTT |
| AA3227 | GTCATAGCTGTTTCCTGTATGTATAATGATTATATCTG |
| AA3228 | CAGATATAATCATTATACATACAGGAAACAGCTATGAC |
| AA3229 | CGTATATCATATTCAACTTTCTCGTTTTCCCAGTCACGAC |
| AA3230 | GTCGTGACTGGGAAAACGAGAAAGTTGAATATGATATACG |
| AA3231 | ATTTAGCCCCGTCGTCTTCT |
| AA3241 | CCGATGTAGTGGCCAAGTCT |
| AA3242 | TGGACATACCAGACGATCCA |
| AA3254 | GAGGACTACTTGGGCGTCAA |
| AA3255 | GTCATAGCTGTTTCCTGCTGTCGGACCGTGTTTATCG |
| AA3256 | CGATAAACACGGTCCGACAGCAGGAAACAGCTATGAC |
| AA3257 | ATATTATAAGTTAGAGGTTAGGTTTTCCCAGTCACGAC |
| AA3258 | GTCGTGACTGGGAAAACCTAACCTCTAACTTATAATAT |
| AA3259 | GGACGGGAGTGTAATGAGGA |
| AA3260 | TGTGCATTCTGCATGGTTTT |
| AA3261 | GTCATAGCTGTTTCCTGGTTTTATAGTTCCGAAGTTGAC |
| AA3262 | GTCAACTTCGGAACTATAAAACCAGGAAACAGCTATGAC |
| AA3263 | GGATGGAATTATAGAATGGCGTTTTCCCAGTCACGAC |
| AA3264 | GTCGTGACTGGGAAAACGCCATTCTATAATTCCATCC |
| AA3265 | CTTCGCCCTTTGATCTTGAG |
| AA3267 | AAGATGTGATCGCGTGAATG |
| AA3268 | GTCATAGCTGTTTCCTGGATGGCAGTTTAGTTGTGAG |
| AA3269 | CTCACAACTAAACTGCCATCCAGGAAACAGCTATGAC |
| AA3270 | GATGAAGATGGCAAAATATATTGTTTTCCCAGTCACGAC |
| AA3271 | GTCGTGACTGGGAAAACAATATATTTTGCCATCTTCATC |
| AA3272 | TGAAGAAAGGGGAGGTGATG |
| AA3292 | GCTTTTGATGACCCTGTCGT |
| AA3293 | CCAAAGACGTGTGATTGTGG |
| AA3294 | TGGTGATCCATGCTTGTTGT |
| AA3295 | AATTTATCCGGGAGGAATCG |
| AA3296 | ATTTCTTACGGCCGGAACTT |
| AA3361 | GACTGGGCCTATGTTGAGGA |
| AA3362 | GTCATAGCTGTTTCCTGCGTGAGGTGTAGGGAAGGAGCAG |
| AA3363 | CTGCTCCTTCCCTACACCTCACGCAGGAAACAGCTATGAC |
| AA3364 | GCAAAATAAAAAGAATGTATCAAGTTTTCCCAGTCACGAC |
| AA3365 | GTCGTGACTGGGAAAACTTGATACATTCTTTTTATTTTGC |
| AA3366 | ACGAATAATAGGGGGCATCC |
| AA3401 | ACGAATAATAGGGGGCATCC |
| AA3402 | ACATCGCATCTCGAGGTTTC |
| AA3505 | CTGAGCGGTGTCCTTTTCTC |
| AA3506 | GTCATAGCTGTTTCCTGGGTGTGGGTGTGGTTGTCGTGGT |
| AA3507 | ACCACGACAACCACACCCACACCCAGGAAACAGCTATGAC |
| AA3508 | GTATATCTAGATTGAACAACTAAGTTTTCCCAGTCACGAC |
| AA3509 | GTCGTGACTGGGAAAACTTAGTTGTTCAATCTAGATATAC |
| AA3510 | TTTCAGTTCCGAGGTGCTCT |
| AA3672 | GATTCGCACCATTGGTCTTT |
| AA3673 | TAACGCGGAGCTCTGATCTT |
| AA4011 | CTCCCTCCACCAGATACCAA |
| AA4012 | CTGCCACAAAGTTGAACGTC |
| AA3970 | ACCACCACCATCCTAACCAG |
| AA3971 | ACGAGGAAGAAGGGTAAGGC |
| AA4031 | GGATCCATGGATGCAGGGACT |
| AA4032 | GGATCCAAGGCCAAGAAGGGAAA |
| AA4033 | GGATCCAAGGGATTGCAAGTGGTCAG |
| AA1551 | AGCTGTGCGTATCCAATAAT |
| AA3358 | CTCCTCGCCCTTGCTCACCATCTTGGCCTTGCTGTTAAC |
| AA3357 | GTTAACAGCAAGGCCAAGATGGTGAGCAAGGGCGAGGAG |
| AA1879 | GCAAGAATTGGCTGCCCTCTAGGCATACCTGCCAAACCTAA |
| AA1880 | TTAGGTTTGGCAGGTATGCCTAGAGGGCAGCCAATTCTTGC |
| AA1752 | ACTGATAGATCTGAGGAAAGCGTCAAGGATATG |
| AA1463 | AGTTCCGCATCAGTCTTGCT |
| AA1657 | GAGGAAAGCGTCAAGGATATG |
| AA1489 | CCTGAGGACGCTTGAAAGTC |
| AA3305 | GGAGTTCGTGACCGCCGC |
| AA1753 | AGTTAAGATCTATGGCTTACCCAATTCTCCC |
| AA3078 | CAAGAATTGGCTGCCCTCTAAGTCGAGTTGGAAGAGAGTG |
| AA3079 | AAGAGGGTCCACGCCTCCCTAGAGGGCAGCCAATTCTTGC |
| AA3491 | CCTCGCCCTTGCTCACCATGAAGTAGTTGCCCTTGCCGGC |
| AA3492 | GCCGGCAAGGGCAACTACTTCATGGTGAGCAAGGGCGAGG |
| AA3493 | CAAAATAAAAAGAATGTATCAATCAGTACAGCTCGTCCATG |
| AA3494 | CATGGACGAGCTGTACTGATTGATACATTCTTTTTATTTTG |
| AA4206 | GCAGTACAGGTGGAGATTGC |
| AA4207 | GTCATAGCTGTTTCCTGGTTGGCCTTGTTGCTTCA |
| AA4208 | ACTGGCCGTCGTTTTACGGAGATAACCAACTCTTG |
| AA4209 | CCCTCACTCTCAGATCGGTC |
| AA4210 | GCCTCATCCAACGTCCTTTC |
| AA4211 | GTCATAGCTGTTTCCTGGACTGTAATGGCCTTATG |
| AA4212 | ACTGGCCGTCGTTTTACGGATCATGATGAAGGAGA |
| AA4213 | TAAAGAGCTGGGTGTCTGGG |
| AA3934 | TCGATGCGATGTTTCGCT |
| AA3935 | CCTGAATGAACTGCAGGA |
| M13R | CAGGAAACAGCTATGAC |
| M13F -20 | GTAAAACGACGGCCAGT |
The rim13Δ (KS33), rim23Δ (KS94), ali1Δ (KS118), ali2Δ (KS94-2), rim90 (KS63), rim91 (KS57), rim92Δ (KS53) strains were created by replacing the entire open reading frame with the dominant selectable NAT or NEO resistance genes. Overlap PCR was used to create each KO cassette as previously described [61], which was introduced into the recipient strain by biolistic transformation [62]. The following primer combinations were used to create each KO construct: RIM13 5’ fragment: AA1796 and AA1792, RIM13 3’ fragment: AA1793 and AA1795, NEO resistance marker: AA1797 and AA1794. AA1796 and AA1795 were used to amplify the final deletion construct. RIM23 5’ fragment: AA3361 and AA3362, RIM23 3’ fragment: AA3365 and AA3366, NEO resistance marker: AA3363 and AA3364, AA3361 and AA3366 were used to amplify the final deletion construct. ALI1 5’ fragment: AA3254 and AA3255, ALI1 3’ fragment: AA3258 and AA3259, NEO resistance marker: AA3256 and AA3257. AA3254 and AA3259 were used to amplify the final deletion construct. ALI2 5’ fragment: AA3505 and AA3506 ARR2 3’ fragment: AA3509 and AA3510, NEO resistance marker: AA3507 and AA3508. AA3505 and AA3510 were used to amplify the final deletion construct. RIM90 5’ fragment: AA3267 and AA3268, RIM90 3’ fragment: AA3271 and AA3272, NEO resistance marker: AA3269 and AA3270. AA3267 and AA3272 were used to amplify the final deletion construct. RIM91 5’ fragment: AA3226 and AA3227, RIM91 3’ fragment: AA3230 and AA3231, NEO resistance marker: AA3228 and AA3229. AA3226 and AA3231 were used to amplify the final deletion construct. RIM92 5’ fragment: AA3260 and AA3261, RIM92 3’ fragment: AA3264 and AA3265, NEO resistance marker: AA3262 and AA3263. AA3260 and AA3265 were used to amplify the final deletion construct. The ali1Δ ali2Δ (KS97-2) strain was created by disrupting the ALI2 gene in the KS118 strain. The rra1Δ (KS183) and rra1Δ + eGFP-RIM101 (KS185) strain was created by using primers AA3970 and AA3971 to PCR amplify the NEO resistance marker from KS216 and introduce it into wild type H99. Each mutant strain was confirmed by Southern blot. The primers used to create each Southern probe were: RIM13: AA1812 and AA1813, RIM23: AA3401 and AA3402, RIM90: AA3292 and AA3267, RIM91: AA3241 and AA3242. RIM92 AA3293 and AA3294, ALI1: AA3295 and AA3296. ALI2: AA3672 and AA3673, RRA1: AA4011 and AA4012.
The C. gattii rim101Δ and rra1Δ mutants were created in the R265 background using the split marker method previously described [63]. For the Cgrim101Δ mutant the following primers were used with R265 gDNA as the template. CgRIM101 5’ fragment: AA4206 and AA4207, CgRIM101 3’ fragment: AA4208 and AA4209. The 5’ region of the NEO cassette was amplified from pJAF using the M13R primer and AA3935 and the 3’ region with M13F -20 and AA3934. The Cgrim101Δ 5’ KO cassette was created by combining the 5’ CgRIM101 and NEO PCR PCR products by overlap PCR using AA4206 and AA3935. The 3’ KO cassette was similarly made using AA3932 and AA4209. These overlap PCR products were combined and transformed into R265. The Cgrra1Δ mutant was created in the same way, using the following primer pairs. CgRRA1 5’ fragment: AA4210 and AA4211 and CgRRA1 3’ fragment: AA4212 and AA4213. Southern blot analysis was used to confirm a insertion at the correct locus.
The GFP-RIM101T strains were created by first creating TOC65 (rim101Δ + GFP-RIM101T). The following PCR primer combinations were used to amplify the RIM101 promoter and terminator (from H99 gDNA) and a truncated GFP-RIM101 fusion (from pTO2): RIM101 promoter: AA1551 and AA3358, GFP-RIM101 truncation:AA3357 and AA1879, RIM101 terminator: AA1880 and AA1752. Overlap PCR was then performed and was transformed, along with pJAF (NEO), into TOC2. Replacement of the rim101Δ NAT disruption construct was confirmed by PCR, and the expression and functionality of GFP-RIM101T was verified by reconstitution of the rim101Δ mutant phenotypes. Primers AA1463 and AA1657 were then used to amplify the promoter, ORF, and terminator of the GFP-70T from the TOC65 gDNA. The GFP-RIM101T was then co-transformed with either pJAF (NEO) or pCH233 (NAT) into the following mutant strains: vps23Δ (vps23-9), rim13Δ (KS33), rim20Δ (TOC17), rim23Δ (SK94). Transformants were screened using PCR (AA1489 and AA3305) and epifluorescence microscopy to detect the presence and expression of the GFP-RIM101 fusion respectively.
KS289 (rim23Δ::NEO + GFP-RIM23 + NAT MATα) was created by creating a RIM23-GFP fusion using the following PCR reactions. AA3361 and AA3491 were used to amplify RIM23 and its promoter from H99 gDNA. AA3492 and AA3493 were used to amplify GFP from a GFP-containing vector. AA3494 and AA3166 were used to amplify that RIM23 terminator region. These PCR products were mixed and overlap PCR was performed using AA3361 and AA3366. The overlap piece was mixed with pCH233 (NAT) and transformed into KS94 (rim23Δ). Transformants that had rescued growth on pH 8 all displayed GFP fluorescence. The isolate chosen for this study was rescued for 1.5 M NaCl growth and capsule formation. The other RIM23-GFP strains were created by crossing KS289 or KS290 (Mata) to the each mutant strain. Progeny were selected based on pH 8 and 1.5M NaCl sensitivity and the genotypes were confirmed using PCR. GFP-RIM23 was the only RIM23 allele in each of these strains.
To reconstitute the rim13Δ mutant, primers AA1796 and AA1795 were used to amplify the RIM13 promoter, gene, and terminator sequence and then co-transformed with pCH233 (NAT) into the KS33 strain. PCR and rescued phenotype were used to verify restored expression of RIM13. The rim23Δ mutant was similarly reconstituted using primers AA3361 and AA3366. The rra1Δ mutant was reconstituted by amplifying the RRA1 promoter, ORF, and terminator using primers AA4033 and AA4032 and then TA cloning into pCR2.1. The insert was digested from the TA vector with BamHI cloned into the single BamHI site in pJAF to create pKS38. The KS183 strain (rra1Δ) was then transformed with pKS38 and transformants were screened for rescued mutant phenotypes.
The pKS34 vector was created by using AA4031 and AA4032 to amplify the RRA1 gene from H99 genomic DNA and then TA cloned into pCR2.1. RRA1 was then cloned into the single BamHI site in pCN19 [64].To create rra1Δ + GFP-RRA1 (KS214), KS183 was transformed with pKS34.
The pTO22 construct was created by amplifying and truncating the RIM101 gene using the following primer pairs: 5’ flank: AA1753 and AA1879, 3’ flank: AA1880 and AA1752. The entire piece was amplified using AA1753 and AA1752, TA cloned into pCR2.1, and then cloned into the single BamHI site in pCN70 (pGAL7-GFP-NEO).
Insertional mutagenesis and assessment of mutants
Agrobacterium tumefaciens-mediated random mutagenesis was carried out as previously described [65]. Briefly, A. tumefaciens expressing the pPZP-NEOcc plasmid was incubated with shaking at room temperature for 48 hr in Luria-Bertani (LB) medium containing kanamycin. Cells were washed and resuspended to an OD600 of 0.15 in liquid induction media (IM) containing 200 μM acetosyringone (AS) and incubated for 6 hr An overnight YPD culture of KS161 was harvested, washed with IM and resuspended in IM at a concentration of 107 cells/mL. 200 μL of the A. tumefaciens and KS161 suspensions were mixed and aliquoted on IM agar containing acetosyringone. Cells were incubated for 3 days and then scraped onto selection media (YPD + Neo + 100 μg/mL cefotaxime). Colonies passaged 1X on the selection plates and then inoculated into liquid selection media in 96-well plates. Duplicates of each isolate were inoculated. Mutants were then inoculated onto YP Dextrose or YP Galactose media containing 1.5M NaCl or 150mM HEPES adjusted to pH 8. A total of 3500 mutants were screened in duplicate. Mutants with growth defects on either condition were selected and rescreened. The genetic location of each insertion was identified by deep sequencing using MiSeq deep sequencing.
Protein extraction, immunoprecipitation, and western blot
Protein extracts were prepared as previously described [66]. To initially assess GFP-Rim101 proteolytic processing at multiple pH’s, the rim101Δ + GFP-RIM101 (TOC105) strain was grown overnight (~18 hr) in 50ml YPD, 30°C, 150 rpm shaking. Cells were pelleted and resuspended in SC McIlvaine’s buffer at pH 4, pH 6, or pH 8 to an OD600nm of 1. Samples were further incubated at 30°C 150 rpm for 3 or 5 hr Cells were harvested, flash frozen on dry ice, and lysed in 0.4 mL lysis buffer containing 2x protease inhibitors (Complete, Mini, EDTA-free; Roche) and 1x phosphatase inhibitors (PhosStop; Roche) and 1 mM phenylmethanesulfonylfluoride (PMSF). Lysis was performed by bead beating (0.5 mL of 3 μM glass beads in a Mini-BeadBeater-16 (BioSpec), 10 cycles for 30 s each). Supernatants were transferred to new tubes and glass beads washed 2 times with 0.4 mL lysis buffer. Lysates were cleared by centrifugation at 15,000 rpm, 4°C, for 10min. 5 μLGFP-TRAP resin (Chemotek), which was equilibrated in lysis buffer, was added to the cleared lysate and inverted at 4°C for 4 hr GFP-TRAP resin was washed 3 times with 1 mL lysis buffer and protein was eluted in 30 μL NuPage sample buffer with 1X NuPage Reducing Agent by boiling for 5 min. Western blots were performed as described previously using 4–8% NuPage BisTris gels to separate the samples. To detect GFP-Rim101, blots were incubated in anti-GFP primary antibody (Roche) (using a 1/10,000 dilution) and then in secondary anti-mouse peroxidase-conjugated secondary antibody (using a 1/25,000 dilution, Jackson Labs).
Assessment of GFP-Rim101 proteolysis in the upstream Rim pathway mutants was performed as described above but with several alterations. First, YPD 150 mM HEPES adjusted to a pH 7.4 was used instead of SC McIlvaine’s buffer. Second, GFP-Rim101 was immunoprecipitated with anti-GFP antibody (Roche) for 1 hr and then incubated 2 hr with 80 μL Protein G Sepharose (Thermo Scientific). Finally, a 3–8% NuPage Tris-Acetate gel was used to separate samples.
Microscopy
The high-resolution fluorescent images of Rim23-GFP were captured using a Delta Vision Elite deconvolution microscope equipped with a Coolsnap HQ2 high resolution charge-coupled-device (CCD) camera. All other differential interference (DIC) and fluorescent images were captured using a Zeiss Axio Imager A1 fluorescence microscope equipped with an AxioCam MRM digital camera.
Virulence studies
Virulence studies were carried out according to [67]. 8–10 female A/Jcr mice were anesthetized with 100–140 μL of 12 mg/ml ketamine HCl and 1 mg/ml Xylazine in PBS. Each mouse was intranasally inoculated with 1X105 cells in a volume of 25 μL. Mice were monitored and weighed daily and sacrificed based on predetermined symptoms that predict imminent death. Kaplan-Meyer curves were analyzed by log-rank test (JMP software; SAS Institute, Cary, NC). All studies were performed in compliance with Duke University institutional guidelines for animal experimentation.
Ethics statement
All animal work was performed according to the Duke University Institutional guidelines for animal experimentation. All mice were anesthetized using an intraperitoneal injection of Ketamine/Xylazine mixture and were sacrificed by CO2. Approved secondary methods of ensuring animal death were also used. These techniques are consistent with IACUC-approved animal handling protocol- A217-11-08.
Supporting Information
(A) Model of C. neoformans Rim13, Rim20, and Rim23 orthologs. Conserved protein domains were predicted using Pfam and Super Family databases. The E-values for each domain prediction: Rim23 PF03097: 5.50E-9; Rim20 PF03097: 1.90E-97, PF13949: 1.10E-71; Rim13 SSF54001: 1.16E-42, SSF49758 (2 domains): 1.57E-19 and 2.09E-14. (B) Expression of RIM13 and RIM23 wild type alleles rescues rim13Δ and rim23Δ pH 8s and 1.5 M NaCl growth defects. (C) Expression of the wild type alleles also rescues the rim13Δ and rim23Δ mutant capsule defects. Cells were cultured for 48 hr in CO2-independent media 37°C to induce capsule formation. Capsule was visualized by India ink stain.
(TIF)
(A) The ali1Δ, ali2Δ, ali1Δ ali2Δ, and rim90Δ rim91Δ rim92Δ mutants grow like WT on YPD with 150 mM HEPES at pH 8 and YPD + 1.5 M NaCl. 10-fold serial dilutions of each sample were spotted onto the indicated plates. (B) The ali1Δ, ali2Δ, ali1Δ ali2Δ, and rim90Δ rim91Δ rim92Δ mutants do not have a capsule formation defect. Cells were cultured for 48 hr in CO2-independent media 37°C to induce capsule formation. Capsule was visualized by India ink stain.
(TIF)
The following orthologs are represented: C. neoformans CNAG_03488 (Rra1), C. gattii CGB_G5320C, Tremella mesenterica TREME_69388, Puccinia graminis PGTG_03106, and Ustilago maydis um00299. The gray shaded regions mark predicted transmembrane helices. The similarity between sequences is represented by color, with red representing the most similar and blue representing the least similar. Alignment was created using the T-Coffee multiple sequence alignment server [68]. The numbers indicate amino acid position.
(TIF)
The indicated strains were spotted onto YPD, YPD 150mM HEPES pH 8, and YPD 1.5M NaCl and incubated for 2–4 days at 30°C.
(TIF)
Acknowledgments
We acknowledge Dr. Sam Johnson and the Duke Microscopy Core for assistance with the Delta Vision microscopy. We thank Dr. James Kronstad for providing us with strains. We thank Sandra Breeding and the MBL Molecular Mycology Woods Hole Class of 2013 for valuable assistance with the random-mutagenesis screen.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
These studies were supported by National Institutes of Health grant AI074677 (to JAA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Davis D, Edwards JE, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000. Oct;68(10):5953–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertuzzi M, Schrettl M, Alcazar-Fuoli L, Cairns TC, Muñoz A, Walker LA, et al. The pH-responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 2014. Oct 16;10(10):e1004413 10.1371/journal.ppat.1004413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O’Meara TR, Xu W, Selvig KM, O’Meara MJ, Mitchell AP, Alspaugh JA. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol. 2014. Feb 9;34(4):673–84. 10.1128/MCB.01359-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang W, Shang Y, Chen P, Gao Q, Wang C. MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ Microbiol. 2014 Mar 10; [DOI] [PubMed]
- 5. Narayan S, Batta K, Colloby P, Tan CY. Cutaneous cryptococcus infection due to C. albidus associated with Sezary syndrome. Br J Dermatol. 2000;143(3):632–4. [DOI] [PubMed] [Google Scholar]
- 6. You B-J, Choquer M, Chung K-R. The Colletotrichum acutatum gene encoding a putative pH-responsive transcription regulator is a key virulence determinant during fungal pathogenesis on citrus. Mol Plant Microbe Interact. 2007. Sep;20(9):1149–60. [DOI] [PubMed] [Google Scholar]
- 7. Landraud P, Chuzeville S, Billon-Grande G, Poussereau N, Bruel C. Adaptation to pH and role of PacC in the rice blast fungus Magnaporthe oryzae . PLoS One. Public Library of Science; 2013. Jan 16;8(7):e69236 10.1371/journal.pone.0069236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–30 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 9. O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, et al. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. Public Library of Science; 2010. Feb;6(2):e1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chun CD, Madhani HD. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans . PLoS One. Public Library of Science; 2010;5(9):e12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O’Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio. 2013. Jan;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Obara K, Yamamoto H, Kihara A. Membrane protein Rim21 plays a central role in sensing ambient pH in Saccharomyces cerevisiae . J Biol Chem. 2012. Nov 9;287(46):38473–81. 10.1074/jbc.M112.394205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herranz S, Rodríguez JM, Bussink H-J, Sánchez-Ferrero JC, Arst HN, Peñalva MA, et al. Arrestin-related proteins mediate pH signaling in fungi. Proc Natl Acad Sci U S A. 2005. Aug 23;102(34):12141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galindo A, Calcagno-Pizarelli AM, Arst HN, Peñalva MÁ. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J Cell Sci. 2012. Apr 1;125(Pt 7):1784–95. 10.1242/jcs.098897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink H-J, Múnera-Huertas T, et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell. 2007. Dec 1;6(12):2365–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barwell KJ, Boysen JH, Xu W, Aaron P, Mitchell AP. Relationship of DFG16 to the Rim101p pH Response Pathway in Saccharomyces cerevisiae and Candida albicans Relationship of DFG16 to the Rim101p pH Response Pathway in Saccharomyces cerevisiae and Candida albicans †. 2005; [DOI] [PMC free article] [PubMed]
- 17. Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. Elsevier Inc.; 2011. Jul 19;21(1):77–91. [DOI] [PubMed] [Google Scholar]
- 18. Xu W, Smith FJ, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans . Mol Biol Cell. 2004. Dec;15(12):5528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Obara K, Kihara A. Signaling Events of the Rim101 Pathway Occur at the Plasma Membrane in a Ubiquitination-Dependent Manner. Mol Cell Biol. 2014. Sep 15;34 (18): 3525–34. 10.1128/MCB.00408-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herrador A, Herranz S, Lara D, Vincent O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol Cell Biol. 2010. Feb 15;30(4):897–907. 10.1128/MCB.00132-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peñalva MA, Lucena-Agell D, Arst HN. Liaison alcaline: Pals entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol. 2014. Nov 1;22C:49–59. [DOI] [PubMed] [Google Scholar]
- 22. Xu W, Mitchell AP. Yeast PalA / AIP1 / Alix Homolog Rim20p Associates with a PEST-Like Region and Is Required for Its Proteolytic Cleavage Yeast PalA / AIP1 / Alix Homolog Rim20p Associates with a PEST-Like Region and Is Required for Its Proteolytic Cleavage. 2001;183(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Díez E, Alvaro J, Espeso E a, Rainbow L, Suárez T, Tilburn J, et al. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002. Mar 15;21(6):1350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr Opin Microbiol. 2009/07/28 ed. 2009;12(4):365–70. 10.1016/j.mib.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 25. Aréchiga-Carvajal ET, Ruiz-Herrera J. The RIM101/pacC Homologue from the Basidiomycete Ustilago maydis Is Functional in Multiple pH-Sensitive Phenomena. Eukaryot Cell. 2005. Jun;4(6):999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Antonio C-CJ, Lucila O-C, Miriam T-S, Scott G, José R-H. Functional analysis of the pH responsive pathway Pal/Rim in the phytopathogenic basidiomycete Ustilago maydis . Fungal Genet Biol. 2010;47(5):446–57. 10.1016/j.fgb.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 27.Kosman DJ. Iron metabolism in aerobes: Managing ferric iron hydrolysis and ferrous iron autoxidation. Coordination Chemistry Reviews. 2013. p. 210–7. [DOI] [PMC free article] [PubMed]
- 28. Li W, Mitchell AP. Proteolytic Activation of Rim1p, a Positive Regulator of Yeast Sporulation and Invasive Growth. Genetics. 1997;145(1):63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Orejas M, Espeso EA, Tilburn J, Sarkar S, Arst HN Jr., Penalva MA. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995/07/01 ed. 1995;9(13):1622–32. [DOI] [PubMed] [Google Scholar]
- 30. Kullas AL, Li M, Davis DA. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans . Eukaryot Cell. 2004. Dec;3(6):1609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blanchin-Roland S, Da Costa G, Gaillardin C. ESCRT-I components of the endocytic machinery are required for Rim101-dependent ambient pH regulation in the yeast Yarrowia lipolytica . Microbiology. 2005. Nov;151(Pt 11):3627–37. [DOI] [PubMed] [Google Scholar]
- 32. Rodríguez-Galán O, Galindo A, Hervás-Aguilar A, Arst HN, Peñalva MA. Physiological involvement in pH signaling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J Biol Chem. 2009. Feb 13;284(7):4404–12. 10.1074/jbc.M808645200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Calcagno-Pizarelli AM, Hervás-Aguilar A, Galindo A, Abenza JF, Peñalva MA, Arst HN. Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT-0, I, II and III genes by inactivation of a salt-tolerance pathway allows examination of ESCRT gene roles in pH signalling. J Cell Sci. 2011. Dec 1;124(Pt 23):4064–76. 10.1242/jcs.088344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu G, Caza M, Cadieux B, Chan V, Liu V, Kronstad J. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect Immun. 2013;81(1):292–302. 10.1128/IAI.01037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chun CD, Madhani HD. Ctr2 Links Copper Homeostasis to Polysaccharide Capsule Formation and Phagocytosis Inhibition in the Human Fungal Pathogen Cryptococcus neoformans . PLoS One. Public Library of Science; 2010;5(9):e12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Godinho RM da C, Crestani J, Kmetzsch L, Araujo G de S, Frases S, Staats CC, et al. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci Rep. Nature Publishing Group; 2014. Jan 2;4:6198 10.1038/srep06198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, et al. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans . PLoS Pathog. 2010;6(11):e1001209 10.1371/journal.ppat.1001209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hofmann K, Stoffel W. TMBASE—a database of membrane spanning protein segments. Biol Chem HoppeSeyler. 1993;374:166. [Google Scholar]
- 39. Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 andcAMP. Genes Dev. 1997;11(23):3206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsueh Y-PP, Xue C, Heitman J. G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans . Mol Biol Cell. 2007. Sep 1;18(9):3237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, et al. Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity. PLoS Pathog. Public Library of Science; 2010;6(6):e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. Fungal Cell Gigantism during Mammalian Infection. PLoS Pathog. Public Library of Science; 2010;6(6):e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Okagaki LH, Wang Y, Ballou ER, O’Meara TR, Bahn Y-SS, Alspaugh JA, et al. Cryptococcal Titan Cell Formation Is Regulated by G-Protein Signaling in Response to Multiple Stimuli. Eukaryot Cell. 2011. Oct;10(10):1306–16. 10.1128/EC.05179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ikeda M, Kihara A, Denpoh A, Igarashi Y. The Rim101 Pathway Is Involved in Rsb1 Expression Induced by Altered Lipid Asymmetry. 2008;19(May):1922–31. 10.1091/mbc.E07-08-0806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Da C. Godinho RM, Crestani J, Kmetzsch L, de S. Araujo G, Frases S, Staats CC, et al. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci Rep. Macmillan Publishers Limited. All rights reserved; 2014. Sep 2;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chun CD, Madhani HD. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans . PLoS One. 2010;5(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ludwig M-G, Vanek M, Guerini D, Gasser JA, Jones CE, Junker U, et al. Proton-sensing G-protein-coupled receptors. Nature. Nature Publishing Group; 2003. Sep 4;425(6953):93–8. [DOI] [PubMed] [Google Scholar]
- 48. Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–43. 10.1038/nrmicro2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levin LR, Buck J. Physiological Roles of Acid-Base Sensors. Annu Rev Physiol. Annual Reviews; 2015. Feb 10;77(1):347–62. [DOI] [PubMed] [Google Scholar]
- 50. Harrison JS, Higgins CD, O’Meara MJ, Koellhoffer JF, Kuhlman BA, Lai JR. Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins. Structure. Elsevier Ltd; 2013. Jul 2;21(7):1085–96. 10.1016/j.str.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heath VL, Shaw SL, Roy S, Cyert S, Heath VL, Shaw SL, et al. Hph1p and Hph2p, Novel Components of Calcineurin-Mediated Stress Responses in Saccharomyces cerevisiae Hph1p and Hph2p, Novel Components of Calcineurin-Mediated Stress Responses in Saccharomyces cerevisiae. 2004;
- 52. Dechant R, Saad S, Ibáñez AJ, Peter M. Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Mol Cell. 2014. Aug 7;55(3):409–21. 10.1016/j.molcel.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, Toffaletti DL, Tenor JL. The Cryptococcus neoformans Transcriptome at the Site of Human. 2014; [DOI] [PMC free article] [PubMed]
- 54. Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, et al. Analysis of the Genome and Transcriptome of Cryptococcus neoformans var. grubii Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation. PLoS Genet. 2014. Apr;10(4):e1004261 10.1371/journal.pgen.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sherman F. Getting started with yeast. Methods Enzym. 1991;194:3–41. [DOI] [PubMed] [Google Scholar]
- 56. Murashige T, Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plant. Blackwell Publishing Ltd; 1962;15(3):473–97. [Google Scholar]
- 57. Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101(1):177–94. [PMC free article] [PubMed] [Google Scholar]
- 58. Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and Virulence of congenic a and α isolates. Infect Immun. 2003;71(9):4831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xue C, Bahn Y-SS, Cox GM, Heitman J. G Protein-coupled Receptor Gpr4 Senses Amino Acids and Activates the cAMP-PKA Pathway in Cryptococcus neoformans . Mol Biol Cell. 2006;17(2):667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004;101(49):17258–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the Sexual Cycle of the Primary Fungal Pathogen Cryptococcus neoformans var. gattii: Implications for an Outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2(5):1036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175(5):1405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim MS, Kim SY, Jung KW, Bahn YS. Targeted gene disruption in Cryptococcus neoformans using double-joint PCR with split dominant selectable markers. Methods Mol Biol. 2012;845:67–84. 10.1007/978-1-61779-539-8_5 [DOI] [PubMed] [Google Scholar]
- 64. Price MS, Nichols CB, Alspaugh JA. The Cryptococcus neoformans Rho-GDP Dissociation Inhibitor Mediates Intracellular Survival and Virulence. Infect Immun. 2008;76(12):5729–37. 10.1128/IAI.00896-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Idnurm A, Reedy JL, Nussbaum JC, Heitman J. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell. 2004. Apr 9;3(2):420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nichols CB, Ferreyra J, Ballou ER, Alspaugh JA. Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot Cell. 2009;8(2):181–9. 10.1128/EC.00351-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a Virulence Factor in Experimental Cryptococcosis. Infect Immun. 2000;68(2):443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Model of C. neoformans Rim13, Rim20, and Rim23 orthologs. Conserved protein domains were predicted using Pfam and Super Family databases. The E-values for each domain prediction: Rim23 PF03097: 5.50E-9; Rim20 PF03097: 1.90E-97, PF13949: 1.10E-71; Rim13 SSF54001: 1.16E-42, SSF49758 (2 domains): 1.57E-19 and 2.09E-14. (B) Expression of RIM13 and RIM23 wild type alleles rescues rim13Δ and rim23Δ pH 8s and 1.5 M NaCl growth defects. (C) Expression of the wild type alleles also rescues the rim13Δ and rim23Δ mutant capsule defects. Cells were cultured for 48 hr in CO2-independent media 37°C to induce capsule formation. Capsule was visualized by India ink stain.
(TIF)
(A) The ali1Δ, ali2Δ, ali1Δ ali2Δ, and rim90Δ rim91Δ rim92Δ mutants grow like WT on YPD with 150 mM HEPES at pH 8 and YPD + 1.5 M NaCl. 10-fold serial dilutions of each sample were spotted onto the indicated plates. (B) The ali1Δ, ali2Δ, ali1Δ ali2Δ, and rim90Δ rim91Δ rim92Δ mutants do not have a capsule formation defect. Cells were cultured for 48 hr in CO2-independent media 37°C to induce capsule formation. Capsule was visualized by India ink stain.
(TIF)
The following orthologs are represented: C. neoformans CNAG_03488 (Rra1), C. gattii CGB_G5320C, Tremella mesenterica TREME_69388, Puccinia graminis PGTG_03106, and Ustilago maydis um00299. The gray shaded regions mark predicted transmembrane helices. The similarity between sequences is represented by color, with red representing the most similar and blue representing the least similar. Alignment was created using the T-Coffee multiple sequence alignment server [68]. The numbers indicate amino acid position.
(TIF)
The indicated strains were spotted onto YPD, YPD 150mM HEPES pH 8, and YPD 1.5M NaCl and incubated for 2–4 days at 30°C.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.