Abstract
For tRNA-dependent protein biosynthesis, amino acids are first activated by aminoacyl-tRNA synthetases (aaRSs) yielding the reaction intermediates aminoacyl-AMP (aa-AMP). Stable analogues of aa-AMP, such as aminoacyl-sulfamoyl-adenosines, inhibit their cognate aaRSs. Glutamyl-sulfamoyl-adenosine (Glu-AMS) is the best known inhibitor of Escherichia coli glutamyl-tRNA synthetase (GluRS). Thermodynamic parameters of the interactions between Glu-AMS and E. coli GluRS were measured in the presence and in the absence of tRNA by isothermal titration microcalorimetry. A significant entropic contribution for the interactions between Glu-AMS and GluRS in the absence of tRNA or in the presence of the cognate tRNAGlu or of the non-cognate tRNAPhe is indicated by the negative values of –TΔSb, and by the negative value of ΔCp. On the other hand, the large negative enthalpy is the dominant contribution to ΔGb in the absence of tRNA. The affinity of GluRS for Glu-AMS is not altered in the presence of the non-cognate tRNAPhe, but the dissociation constant K d is decreased 50-fold in the presence of tRNAGlu; this result is consistent with molecular dynamics results indicating the presence of an H-bond between Glu-AMS and the 3’-OH oxygen of the 3’-terminal ribose of tRNAGlu in the Glu-AMS•GluRS•tRNAGlu complex. Glu-AMS being a very close structural analogue of Glu-AMP, its weak binding to free GluRS suggests that the unstable Glu-AMP reaction intermediate binds weakly to GluRS; these results could explain why all the known GluRSs evolved to activate glutamate only in the presence of tRNAGlu, the coupling of glutamate activation to its transfer to tRNA preventing unproductive cleavage of ATP.
Introduction
Aminoacyl-tRNA synthetases (aaRS) play a front-line role in protein biosynthesis; they are responsible for the attachment of specific amino acids to their cognate tRNAs. This two-step reaction begins with the amino acid activation by condensation with an ATP molecule, creating an aminoacyl-adenylate (aa-AMP) which then reacts on the aaRS with the 3’-terminal adenosine of the tRNA’s acceptor stem, giving the aminoacyl-tRNA (aa-tRNA) which participates in protein biosynthesis on the ribosome. The essential function of aaRSs in translation makes them promising targets for inhibitors that could be used as antibiotics, such as pseudomonic acid A [1], an inhibitor of isoleucyl-tRNA synthetase (IleRS) used as a topical antibacterial treatment. Recent reviews [2] on the subject show that there are new developments in this field, including pharmacological patents [3].
Several types of stable analogues of aa-AMP inhibit aaRSs (reviewed by Chênevert et al., 2003, and by Finn and Tao, 2005) [4,5]. Aminoacyl-sulfamoyl-adenosines are amongst the most potent ones. Glutamyl-sulfamoyl-adenosine (Glu-AMS) (Fig 1) is a competitive inhibitor of Escherichia coli glutamyl-tRNA synthetase (GluRS) with a K i of 2.8 nM [6] and is 25 times less efficient against murine hepatic GluRS. This result suggests that the structures of the active sites of bacterial and mammalian GluRSs differ significantly, and indicates that Glu-AMS derivatives with bactericidal properties and low toxicity for humans could be developed.
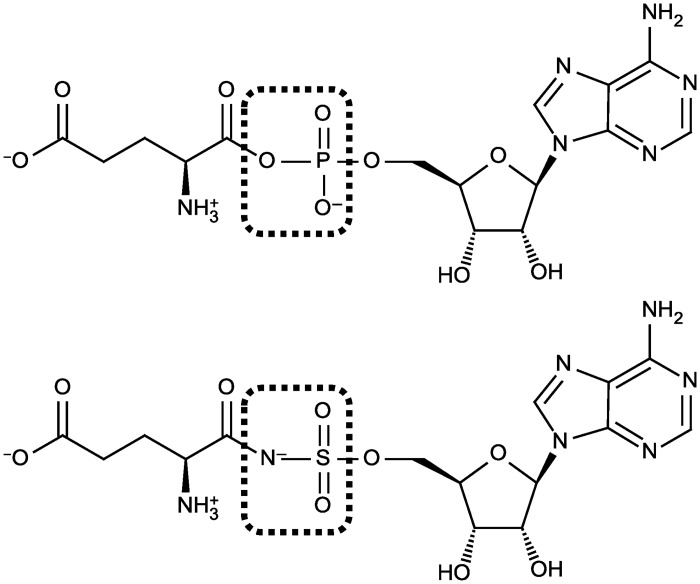
Fig 1. Structures of Glu-AMP and Glu-AMS.
Structures of the reaction intermediate Glu-AMP (top), and of its non-hydrolysable analogue Glu-AMS (bottom), which is an inhibitor of E. coli GluRS [6].
Most aaRS can activate their amino acid substrate in the absence of tRNA; the aa-AMP synthetized by these enzymes are relatively stable, which allows the characterization of their binding to their cognate aaRS (for instance, see Fersht (1977) [7] for isoleucyl-adenylate (Ile-AMP) and valyl-adenylate (Val-AMP)). On the other hand, all known GluRSs, glutaminyl-tRNA synthetases (GlnRSs), arginyl-tRNA synthetases (ArgRSs), and class 1 lysyl-tRNA synthetases (LysRSs) (closely linked to GluRSs) [8] do not activate their amino acid substrate in the absence of tRNA, but still catalyze the aminoacylation reaction via a two-step mechanism involving a very unstable aa-AMP intermediate [8–11] (reviewed by Schimmel and Söll, 1979, and by First et al., 2005) [12,13].
The structures of Thermus thermophilus GluRS and of its complexes with several substrates and inhibitors [14] revealed that ATP binding by GluRS is switched to the productive mode by tRNAGlu binding [15], and that in the presence of tRNAGlu, GluRS recognizes specifically L-glutamate [16], excluding the non-cognate amino acids L-glutamine and D-glutamate which interact with GluRS in the absence of tRNA [17]. The structure of the T. thermophilus tRNAGlu•GluRS•Glu-AMS complex, which may represent the post-transition state of the glutamate-activation reaction, was determined at a resolution of 2.69 Å (PDB ID 2CV2) [16]. The reason for the tRNA-requirement in the activation reaction catalyzed by GluRSs and the three other above-mentioned aaRSs throughout evolution is not yet known.
We report here the values of thermodynamic parameters of the E. coli GluRS Glu-AMS interaction in the presence of the cognate tRNAGlu or of a non-cognate tRNAPhe, or in the absence of tRNA. These values suggest that all the known GluRSs evolved to activate glutamate only in the presence of tRNAGlu to prevent unproductive cleavage of ATP [18]. Moreover, this thermodynamic characterization of the GluRS Glu-AMS interaction (see equation below) could complement structural data for the design of less polar derivatives of Glu-AMS that could have bactericidal activity.
Materials and Methods
Enzyme and tRNA
Overproduction and purification of E. coli GluRS were performed as previously described [19] with the following modifications. A C-terminal histidine-tagged GluRS was used instead of the N-terminal tagged one. The overproduction was induced overnight at 30°C with 1 mM IPTG. The GluRS was purified to homogeneity, as revealed by SDS-PAGE analysis (result not shown).
Overproduction and purification of E. coli tRNAGlu-enriched total tRNA was done as described [20]. The aminoacylation plateau indicated that the final product contained 26% tRNAGlu. Saccharomyces cerevisiae tRNAPhe, used as a negative control, was purchased from Sigma-Aldrich (cat No: R4018).
GluRS inhibitor
Glu-AMS (5’-O-[N-(L-glutamyl)sulfamoyl]adenosine, Trilink Lot #A1004-060606), a stable analogue of the GluRS reaction intermediate Glu-AMP, and a potent inhibitor of E. coli GluRS with respect to glutamic acid [6] was purchased from RNA-TEC (Leuven, Belgium). A 10 mM stock solution was prepared in Tris-HCl buffer (50 mM, pH 7.9, 10 mM MgCl2).
Isothermal Titration Microcalorimetry
The GluRS solution with or without tRNA was dialyzed overnight in a D-tube dialyzer (Novagen) against 2 L of dialysis buffer (50 mM HEPES-KOH, pH 7.2, 10 mM MgCl2) at 4°C with light stirring. The next morning, the dialyzed solution was recovered and the volume adjusted by adding dialysis buffer to obtain the desired concentration (typically 9 μM GluRS and 11 μM tRNAGlu). This solution was kept on ice until use. Glu-AMS was diluted in the dialysis buffer to obtain a final concentration of 90 μM. All buffers and solutions were degased with stirring under vacuum. The microcalorimetry experiments were carried out in a VP-ITC 100 microcalorimeter (MicroCal, GE Healthcare) using deionized water as an internal reference for all assays. VPViewer ITC 2000 and Origin v 5.0 softwares (Microcal Software, Inc) were used for data collection and analysis, respectively.
To measure the thermodynamic parameters of the interaction between Glu-AMS and GluRS, in the presence and in the absence of tRNA, a solution containing 9 μM GluRS with or without a stoichiometric excess of tRNAGlu or S. cerevisiae tRNAPhe (Table 1) was placed in the sample cell of the microcalorimeter. The following conditions were used for all tests: reference power was set at 12 μcal/s and stirring at 300 rpm, the “high” feedback mode and “No check Temp”, “Fast Equil” and “Auto” ITC equilibration options were selected. The 90 μM Glu-AMS solution was loaded in the injection syringe. After a first injection of 1 μL over 2 s, a series of 39 injections (7.4 μL over 14.8 s) with 240 s between injections was performed. Injections of Glu-AMS in GluRS alone, GluRS•tRNAGlu or GluRS•tRNAPhe, were performed at 30°C (303 K), and done in duplicate. Injections of Glu-AMS in GluRS were also performed at 20°C and 37°C (293 and 310 K): 14 injections (20 μL over 40 s) followed the first injection of 1 μL. Each temperature was tested in duplicate, and in triplicate at 37°C.
Table 1. Influence of tRNA on GluRS/Glu-AMS binding at 30°C.
| E. coli GluRS | E. coli GluRS + E. coli tRNAGlu | E. coli GluRS + S. cerevisiae tRNAPhe | |
|---|---|---|---|
| na | 0.9998 ± 0.0094 | 1.0018 ± 0.0041 | 0.9902 ± 0.0410 |
| ΔHb (cal/mol) | -5041 ± 63 | -7990 ± 62 | -5173 ± 279 |
| ΔSb (cal/mol·K) | 13.19 ± 0.16 | 10.52 ± 0.08 | 7.92 ± 0.46 |
| ΔGb (cal/mol) | -9056 ± 38 | -11304 ± 209 | -8544 ± 309 |
| -TΔSb (cal/mol) | -3996 ± 50 | -3186 ± 25 | -2398 ± 140 |
n = stoichiometry coefficient (number of moles of Glu-AMS bound per mole of GluRS monomer), ΔHb = reaction enthalpy, ΔSb = reaction entropy, ΔGb = reaction energy (calculated with the formula ΔGb = -RT Ln K b, where R = 1.987 cal/mol·K).
All values and errors in this table were obtained by weighting by inverse variance [32], except for ΔGb values and errors, obtained by simple average and standard error calculations.
Raw data and calculated values for each separated ITC runs are shown in S1 Table.
The following nomenclature was used to describe the interaction at equilibrium between Glu-AMS and either GluRS or a GluRS•tRNA complex:
Homology modeling
The primary sequence of E. coli GluRS (471 residues) was obtained from the UniProt Consortium (2012) (UniProt P04805). Two structures were identified as templates for homology modeling from a standard protein Blast (BLASTP) query using the Protein Data Bank (PDB) database on the NCBI/Blast web server [21]. The two GluRSs identified are from Burkholderia thailandensis [22] and Thermosynechococcus elongatus [23], with Uniprot Q2SX36 and Q8DLI5, respectively. A multiple sequence alignment of the sequences using the default parameters of T-Coffee v10.00.r1613 Build 432 [24] showed similarities of 64.7% and 54.3% between the E. coli GluRS and the B. thailandensis and T. elongatus GluRSs, and identities of 49.7% and 41.3%, respectively (the multiple alignment is given in S1 Fig). However, several sections of the B. thailandensis GluRS crystal structure are missing. Consequently, the T. elongatus GluRS structure (PDB 2CFO) was chosen as the template for homology modeling.
The models were built using the T-Coffee alignment and MOE [25] with default parameters, as described previously [26], with the exception that 10 different side chain positions were explored for each model. The best models were validated using MolProbity [27], and the model with the highest score was used for system preparation and docking simulations. Then, three important water molecules (residues 2001, 2007 and 2009 of 2CFO) were added to the model according to 2CFO and 2CV2 crystal structures. In addition, new conformations of Met-250 were generated using the rotamer explorer tool in MOE, as this residue was pointing toward the solvent and would clash with the tRNA. The lowest energy conformation pointing toward the active site was chosen. Then, the Glu-AMS from 2CV2 was added to the modeled structure. This receptor was prepared with the MOE LigX tool to adjust the hydrogen atoms, the rotamers and to minimize the system's energy as previously described [28]. Two models were then built, with and without the tRNA (hereafter referred to as E. coli GluRS and E. coli GluRS•tRNAGlu, respectively). The E. coli GluRS•tRNAGlu model was built by adding the tRNAGlu from PDB 2CV2 to the E. coli GluRS model. The tRNAGlu conformation was energy minimized while keeping fixed all the other atoms. For both models, Open Babel 2.3.2 [29,30] was used to convert pdb files in pdbqt and assign Gasteiger charges.
Docking simulations
Glu-AMS has been docked to both E. coli GluRS and E. coli GluRS•tRNAGlu models. RMSDs between the best ranked conformation of each system and PDB 2CV2 were calculated for the Glu-AMS heavy atoms. All docking calculations were carried out with Autodock VINA 1.1.2 [31] using a rigid receptor for the protein and some flexibility between nucleotides C74 and C75 of tRNAGlu.
Results
Influence of tRNA on the interaction of GluRS with Glu-AMS
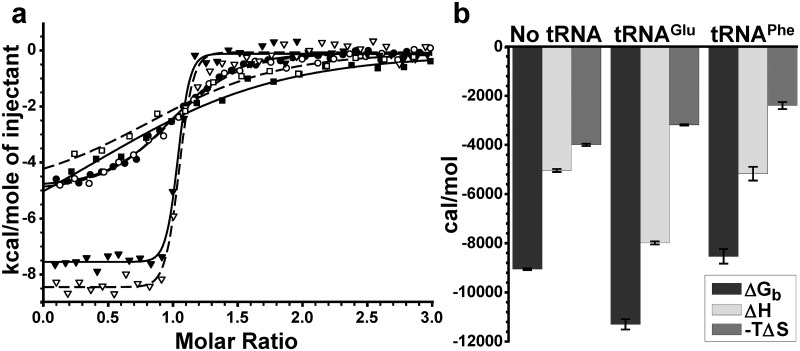
The thermodynamic parameters ΔGb, ΔHb and -TΔSb of the interaction of GluRS with Glu-AMS were measured at 30°C in the absence of tRNA and in the presence of a small excess of E. coli tRNAGlu and, as a negative control, of the non-cognate tRNAPhe from S. cerevisiae (Fig 2 and Table 1; all values and errors in this table were obtained by weighting by inverse variance [32], except for ΔGb values and errors, obtained by simple average and standard error calculations).
Fig 2. Glu-AMS binding to GluRS with/without tRNA.
(a) Integrated ITC curves of Glu-AMS binding to GluRS with/without tRNA. Binding of Glu-AMS: to GluRS alone (circles), to GluRS with saturating concentration of tRNAGlu in enriched total tRNA from E. coli (upside-down triangles), to GluRS with 11.23 µM tRNAPhe from brewer’s yeast (squares). A duplicata of each tested condition is shown. The values shown in (b) were calculated from means of two distinct experiments reported in Table 1, weighting by inverse variance [32].
A substantial entropic contribution for the interactions between Glu-AMS and GluRS in the absence of tRNA or in the presence of the cognate tRNAGlu or of the non-cognate tRNAPhe is indicated by the negative values of-TΔSb under these three conditions (Fig 2). The importance of the entropic contribution is also revealed by the negative value of ΔCp in the absence of tRNA (see below). On the other hand, the large negative enthalpy is the dominant contributor to the free energy of the Glu-AMS GluRS interaction in the absence of tRNA; it is not altered in the presence of the non-cognate tRNAPhe, but is strongly increased in the presence of the cognate tRNAGlu, resulting in an increase of the negative value of ΔGb from -9050 to -11300 cal/mol) (Table 1). These results indicate that there are many favorable hydrogen bonds and/or van der Waals interactions between Glu-AMS and GluRS.
Temperature-dependence of the GluRS/Glu-AMS interaction
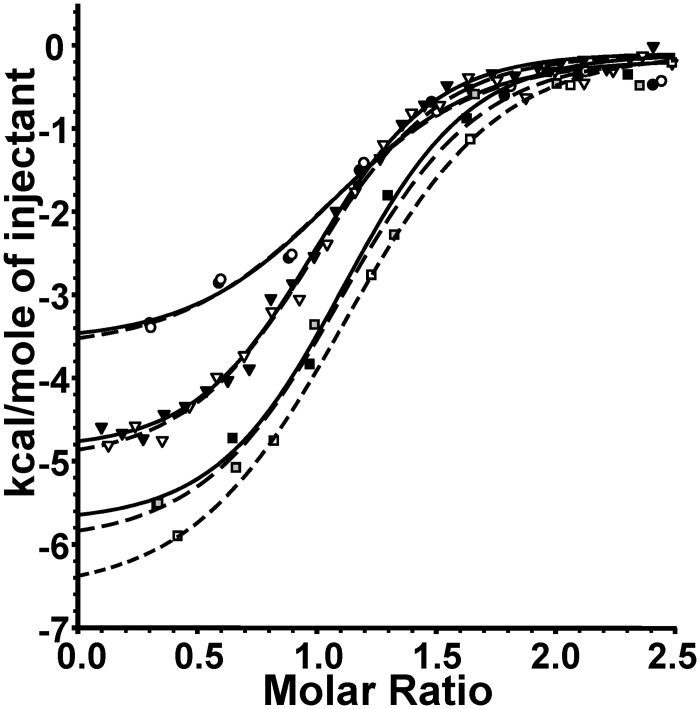
The influence of temperature on Glu-AMS binding to GluRS was also investigated by microcalorimetry at 20, 30 and 37°C (i.e. 293, 303 and 310 K), in the absence of tRNAGlu (Fig 3). The ΔGb values are similar at these temperatures, but ΔHb values increase with temperature (Table 2).
Fig 3. Temperature-dependence of Glu-AMS binding to GluRS.
Integrated ITC curves of Glu-AMS (90 μM) binding to GluRS (9 μM) at different temperatures; 20°C (circles), 30°C (upside-down triangles), 37°C (squares).
Table 2. Temperature-dependance of the GluRS Glu-AMS interaction.
| T (K) | 293 | 303 | 310 |
|---|---|---|---|
| n | 1.0097 ± 0.0369 | 0.9998 ± 0.0094 | 1.0016 ± 0.0095 |
| ΔHb (cal/mol) | -3929 ± 191 | -5041 ± 63 | -6395 ± 81 |
| ΔSb (cal/mol·K) | 15.29 ± 0.74 | 13.19 ± 0.16 | 9.146 ± 0.117 |
| ΔGb (cal/mol) | -8396 ± 1.6 | -9056 ± 38 | -9326 ± 100 |
| -TΔSb (cal/mol) | -4479 ± 218 | -3996 ± 50 | -2835 ± 36 |
n = stoichiometry coefficient (number of moles of Glu-AMS bound per mole of GluRS monomer), ΔHb = reaction enthalpy, ΔSb = reaction entropy, T = temperature, ΔGb = reaction energy (calculated with the formula ΔGb = -RT Ln K b, where R = 1.987 cal/mol·K).
Injections of Glu-AMS at 90 μM were done in a starting concentration of 9 μM of GluRS.
Raw data and calculated values for each separated ITC runs are shown in S2 Table.
By plotting these ΔHb values as a function of temperature (S2 Fig), we calculated the change in heat capacity (ΔCp) using the following equation: ΔCp = (ΔHT2 - ΔHT1)/(T2—T1) [33]. The calculated value is -143 ± 23 cal/mol·K.
Structural analysis of the GluRS Glu-AMS interaction
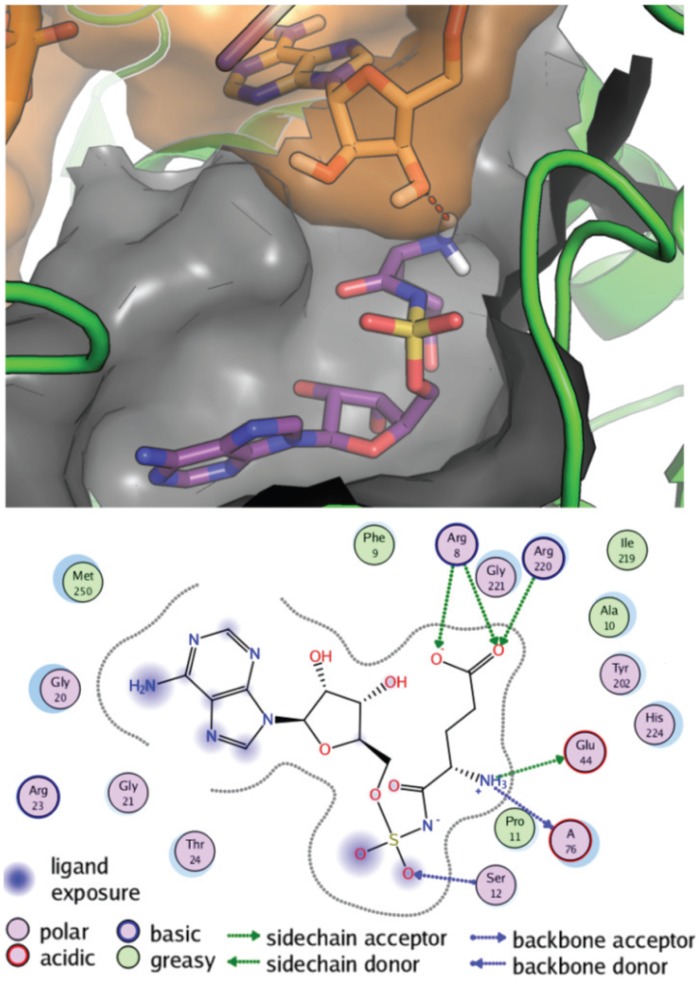
Homology models for E. coli GluRS, both in the absence and in the presence of E. coli tRNAGlu were built from the T. elongatus GluRS crystal structure. Docking of Glu-AMS showed that the binding mode was conserved in both models. The RMSDs between the docked Glu-AMS conformations and the T. thermophilus crystal structures (PDB 2CV2) are 0.79 and 0.82 Å in the absence and in the presence of tRNAGlu, respectively. The docking result for E. coli GluRS•tRNAGlu model is presented in Fig 4 for the E. coli GluRS•tRNAGlu model, where the Glu-AMS NH3 moiety interacts with both GluRS Glu44 and with the 3’-OH group of tRNAGlu A76, leaving free the vicinal 2’-OH group on which GluRS transfers the glutamyl group from Glu-AMP [34].
Fig 4. Molecular docking of Glu-AMS and E. coli tRNAGlu on E. coli GluRS.
Structural representation of the Glu-AMS and E. coli tRNAGlu in the E. coli GluRS binding site from the docking simulations. Top: Glu-AMS is in purple sticks, E. coli tRNAGlu A76 is in orange sticks and a transparent orange surface, E. coli GluRS is in green cartoon, and the binding site is depicted as a grey surface. The H-bond formed between Glu-AMS and A76 is shown as a dotted red line. Only the hydrogen atoms of the NH3 group involved in this H-bond are shown for clarity. Bottom: 2D representation of the Glu-AMS docked conformation bound to E. coli GluRS. The binding pocket is represented with a grey dotted line, polar and non-polar residues are represented as magenta and green circles, respectively, and polar interactions are shown as green and blue lines.
Discussion
Aminoacyl adenylates (aa-AMP) are one of the products of the activation reaction catalyzed by aaRSs, and one of the substrates of the subsequent transfer reaction (reviewed by Giegé and Springer, 2012) [35]. Most aa-AMP are relatively stable when bound to their activating enzyme, and can be isolated in complex with a corresponding aaRS [36]. In 1963, Meister reported the synthesis of 16 aa-AMP, not including glutamyl-AMP [37]. The instability and transient existence of Glu-AMP in the formation of Glu-tRNA were revealed in kinetic studies of the reaction mechanism of E. coli GluRS [38]. Glu-AMS, an analogue of Glu-AMP, is the best known inhibitor of E. coli GluRS [6] probably because the dimensions of the sulfamoyl group are nearly the same as those of the phosphate group, and because it can exist in the anionic form, both in solution and in the solid state, due to the acidity of the NH function; the negative charge is delocalized over several atoms, and the anion is a good mimic of the phosphate ion [4].
The microcalorimetric study reported here reveals a substantial entropic contribution for the interactions between Glu-AMS and GluRS in the absence of tRNA or in the presence of the cognate tRNAGlu or of a non-cognate tRNAPhe, indicated by the negative values of-TΔSb. The entropy term for Glu-AMS binding at 30°C (Table 1) to the free GluRS (the apo-enzyme) is greater (13.2 cal/mol·K) than for its binding to the GluRS/tRNAGlu complex (the holoenzyme) (10.5 cal/mol·K). This result indicates that the active site is less crowded in the apoenzyme than in the holoenyme; the fit in the holoenzyme would have to be better than in the apoenzyme. In other words, the Glu-AMS would be more constrained in the holoenzyme, thus restricting motion. This would translate into a smaller entropy term. Such favorable entropic contribution is in agreement with the values reported for the binding of ATP, whose polarity is similar to that of Glu-AMS, to MEK1 [39] and to F1-ATPase [40]. A favorable entropic contribution was reported to arise from the desolvation of the polar binding site [41], as the polar residues forming the cavity constrain the water molecules in a stiff H-bond network. The negative value of ΔCp during the binding of Glu-AMS to GluRS (S2 Fig) also suggests the breaking of a tight H-bond network, similarly to the hydrophobic interactions. Indeed, in the case of the hydrophobic effect, such a change in heat capacity of binding is thought to arise from the accommodation of non-polar groups by water [42,43].
On the other hand, the large negative enthalpy is the dominant contribution to the free energy of the Glu-AMS binding to GluRS in the absence of tRNA; it suggests that there are several favorable hydrogen bonds and/or van der Waals interactions between Glu-AMS and GluRS. The free energy is not altered in the presence of the non-cognate tRNAPhe, but is strongly increased in the presence of the cognate tRNAGlu. This increased binding of Glu-AMS to GluRS in the presence of the cognate tRNAGlu results from the reorientation of the tRNAGlu 3’ end, observed from PDB 2CV2 for the T. thermophilus GluRS [16], leading to an additional H-bond between the residue A76 and Glu-AMS (see S3 Fig), also confirmed for E. coli GluRS from docking simulations (Fig 4). Indeed, the free energy of binding difference of about 2.2 kcal/mol observed in the presence of tRNAGlu (Table 1) is in the range of the known values for the strength of H-bonds [41]. The Glu-AMS•GluRS•tRNAGlu complex is a posttransition-state mimic [16], i.e. a state following the glutamate activation reaction and preceding the attack by tRNAGlu on Glu-AMP; in this posttransition-state, the stronger binding of Glu-AMS to GluRS in the presence than in the absence of tRNAGlu revealed by our results, is due to a direct involvement of tRNAGlu to the binding of Glu-AMS to the GluRS•tRNAGlu complex.
Because of the high instability of the aminoacylation reaction intermediate Glu-AMP [44], it is very difficult to characterize its interaction with GluRS. The very high structural similarity between Glu-AMP and Glu-AMS allowed us to use the latter in structural studies which revealed that the presence of tRNAGlu bound to GluRS is required for the correct positioning of the α-phosphate of ATP and of the α-COOH of glutamate for the catalysis of the activation reaction [16]. The 50-fold decrease in the affinity of GluRS for Glu-AMS in the absence of tRNAGlu (Table 1 and Fig 2) suggests that the Glu-AMP GluRS interaction in the absence of tRNAGlu is much weaker than that between other aaRSs and their cognate aa-AMP, and has the same order of magnitude as the interaction between a non-cognate aa-AMP and an aaRS, such as tyrosyl-AMP (Tyr-AMP) and phenylalanyl-tRNA synthetase (PheRS) (Table 3).[7,45–48] The released intermediate would likely be hydrolyzed very fast by one of the mechanisms of pre-transfer editing [49] (reviewed by Ling et al., 2009) [50]. This putative low affinity of GluRS for Glu-AMP could explain why this enzyme evolved to require the presence of its cognate tRNA to activate glutamate, allowing the immediate transfer of glutamate from Glu-AMP to the acceptor end of tRNA, and thus preventing unproductive cleavage of ATP. The fact that all the known GluRSs share this property [18] supports this model. The generality of this model could be tested by determining the influence of cognate and non-cognate tRNAs on the binding of each of the three other aaRSs, whose activation reaction is tRNA-dependent (GlnRS, ArgRS and class I LysRS), to the corresponding aminoacyl-sulfamoyl adenosine.
Table 3. Affinities of several aaRSs for cognate and non-cognate aa-AMP.
| aaRS | aa-AMP or analogues | K d aa-AMP aaRS | Reference |
|---|---|---|---|
| GluRS from E. coli | Glu-AMS | 309 nM in the absence of tRNAGlu 7 nM in the presence of tRNAGlu | This work a |
| PheRS from baker’s yeast | Phe-AMP | 5 nM | [45] |
| Tyr-AMP | 1000 nM | [46] | |
| ThrRS from yeast mitochodria | Threonyl-sulfamoyl-adenosine (Thr-AMS) | 4.5 nM | [47] |
| Seryl-sulfamoyl-adenosine (Ser-AMS) | 450 nM | [47] | |
| IleRS from E. coli | Ile-AMP and Val-AMP | This IleRS binds Val-AMP with a 150-fold weaker affinity than Ile-AMP. | [7]; reviewed by Fersht, 1998 [48] |
a: These K d values (dissociation constant) were calculated with the formula K d = 1/K b.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Prof. Pierre Lavigne (Biochemistry, Université de Sherbrooke, Canada) for stimulating discussions. This work was supported by grant #CG051791 to J.L. and grant #9988 to J.A.K. from the Natural Sciences and Engineering Research Council of Canada (NSERC), and by the grant #PR-133605 to J.L. and R.C., and a Ph.D. fellowship to X.B. from the “Fonds de Recherche du Québec—Nature et Technologies, (FRQNT)”.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grant #CG051791 to JL and grant #9988 to JAK from the Natural Sciences and Engineering Research Council of Canada (NSERC) (http://www.nserc-crsng.gc.ca), and by the grant #PR-133605 to JL and RC from the Fonds de Recherche du Québec - Nature et Technologies, (FRQNT) (http://www.frqnt.gouv.qc.ca). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fuller AT, Mellows G, Woolford M, Banks GT, Barrow KD, Chain EB (1971) Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens . Nature 234: 416–417. [DOI] [PubMed] [Google Scholar]
- 2. Vondenhoff GHM, Van Aerschot A (2011) Aminoacyl-tRNA synthetase inhibitors as potential antibiotics. European Journal of Medicinal Chemistry 46: 5227–5236. 10.1016/j.ejmech.2011.08.049 [DOI] [PubMed] [Google Scholar]
- 3. Gadakh B, Van Aerschot A (2012) Aminoacyl-tRNA synthetase inhibitors as antimicrobial agents: a patent review from 2006 till present. Expert Opinion on Therapeutic Patents 22: 1453–1465. 10.1517/13543776.2012.732571 [DOI] [PubMed] [Google Scholar]
- 4. Chênevert R, Bernier S, Lapointe J (2003) Inhibitors of aminoacyl-tRNA synthetases as antibiotics and tools for structural and mechanistic studies In: Lapointe J, Brakier-Gingras L, editors. Translation Mechanisms. Georgetown, Texas: Eurekah.com/Landes Bioscience; pp. 416–428. [Google Scholar]
- 5. Finn J, Tao J (2005) Aminoacyl-tRNA synthetases as anti-infective drug targets In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Georgetown, Texas: Eurekah.com/Landes Bioscience; pp. 405–413. [Google Scholar]
- 6. Bernier S, Dubois DY, Habegger-Polomat C, Gagnon L- P, Lapointe J, Chênevert R (2005) Glutamylsulfamoyladenosine and pyroglutamylsulfamoyladenosine are competitive inhibitors of E. coli glutamyl-tRNA synthetase. Journal of Enzyme Inhibition and Medicinal Chemistry 20: 61–67. [DOI] [PubMed] [Google Scholar]
- 7. Fersht AR (1977) Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry 16: 1025–1030. [DOI] [PubMed] [Google Scholar]
- 8. Ibba M, Losey HC, Kawarabayasi Y, Kikuchi H, Bunjun S, Söll D (1999) Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement. Proceedings of the National Academy of Sciences of the United States of America 96: 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ravel JM, Wang SF, Heinemeyer C, Shive W (1965) Glutamyl and Glutaminyl Ribonucleic Acid Synthetases of Escherichia coli W. Separation, Properties, and Stimulation of Adenosine Triphosphate-Pyrophosphate Exchange by Acceptor Ribonucleic Acid. Journal of Biological Chemistry 240: 432–438. [PubMed] [Google Scholar]
- 10. Mehler AH, Mitra SK (1967) The activation of arginyl transfer ribonucleic acid synthetase by transfer ribonucleic acid. Journal of Biological Chemistry 242: 5495–5499. [PubMed] [Google Scholar]
- 11. Kern D, Lapointe J (1979) Glutamyl transfer ribonucleic acid synthetase of Escherichia coli. Study of the interactions with its substrates. Biochemistry 18: 5809–5818. [DOI] [PubMed] [Google Scholar]
- 12. Schimmel PR, Söll D (1979) Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annual Review of Biochemistry 48: 601–648. [DOI] [PubMed] [Google Scholar]
- 13. First EA (2005) Catalysis of the tRNA aminoacylation reaction In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Georgetown, Texas: Eurekah.com/Landes Bioscience; pp. 328–352. [Google Scholar]
- 14. Nureki O, Vassylyev DG, Katayanagi K, Shimizu T, Sekine S-i, Kigawa T, et al. (1995) Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science 267: 1958–1965. [DOI] [PubMed] [Google Scholar]
- 15. Sekine S-i, Nureki O, Dubois DY, Bernier S, Chênevert R, Lapointe J, et al. (2003) ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO Journal 22: 676–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sekine S-i, Shichiri M, Bernier S, Chênevert R, Lapointe J, Yokoyama S (2006) Structural bases of transfer RNA-dependent amino acid recognition and activation by glutamyl-tRNA synthetase. Structure 14: 1791–1799. [DOI] [PubMed] [Google Scholar]
- 17. Hara-Yokoyama M, Yokoyama S, Miyazawa T (1986) Conformation change of tRNAGlu in the complex with glutamyl-tRNA synthetase is required for the specific binding of L-glutamate. Biochemistry 25: 7031–7036. [DOI] [PubMed] [Google Scholar]
- 18. Freist W, Gauss DH, Söll D, Lapointe J (1997) Glutamyl-tRNA sythetase. Biological Chemistry 378: 1313–1329. [PubMed] [Google Scholar]
- 19. Dubois DY, Blais SP, Huot JL, Lapointe J (2009) A C-truncated glutamyl-tRNA synthetase specific for tRNAGlu is stimulated by its free complementary distal domain: mechanistic and evolutionary implications. Biochemistry 48: 6012–6021. 10.1021/bi801690f [DOI] [PubMed] [Google Scholar]
- 20. Madore E, Florentz C, Giegé R, Sekine S-i, Yokoyama S, Lapointe J (1999) Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34. European Journal of Biochemistry 266: 1128–1135. [DOI] [PubMed] [Google Scholar]
- 21.NCBI/Blast. Available: http://blast.ncbi.nlm.nih.gov/Blast.cgi
- 22. Baugh L, Gallagher LA, Patrapuvich R, Clifton MC, Gardberg AS, Edwards TE, et al. (2013) Combining functional and structural genomics to sample the essential Burkholderia structome. Plos One 8: e53851 10.1371/journal.pone.0053851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schulze JO, Masoumi A, Nickel D, Jahn M, Jahn D, Schubert WD, et al. (2006) Crystal structure of a non-discriminating glutamyl-tRNA synthetase. Journal of Molecular Biology 361: 888–897. [DOI] [PubMed] [Google Scholar]
- 24. Notredame C, Higgins DG, Heringa J (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology 302: 205–217. [DOI] [PubMed] [Google Scholar]
- 25.MOE. Available: www.chemcomp.com
- 26. Karlsson M, Strid Å, Sirsjö A, Eriksson LA (2008) Homology Models and Molecular Modeling of Human Retinoic Acid Metabolizing Enzymes Cytochrome P450 26A1 (CYP26A1) and P450 26B1 (CYP26B1). Journal of Chemical Theory and Computation 4: 1021–1027. [DOI] [PubMed] [Google Scholar]
- 27. Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography 66: 12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Corbeil CR, Williams CI, Labute P (2012) Variability in docking success rates due to dataset preparation. Journal of Computer-Aided Molecular Design 26: 775–786. 10.1007/s10822-012-9570-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: An open chemical toolbox. Journal of Cheminformatics 3: 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Open Babel. Available: http://openbabel.org
- 31. Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31: 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buckland ST, Burnham KP, Augustin NH (1997) Model Selection: An Integral Part of Inference. Biometrics 53: 603–618. [Google Scholar]
- 33. Thomson JA, Ladbury JE (2005) Isothermal Titration Calorimetry: a tutorial In: Ladbury JE, Doyle ML, editors. Biocalorimetry 2, Applications of calorimetry in the biological sciences. England: John Wiley and Sons, Ltd; pp. 37–58. [Google Scholar]
- 34. Moras D (1992) Structural and functional relationships between aminoacyl-tRNA synthetases. Trends in Biochemical Sciences 17: 159–164. [DOI] [PubMed] [Google Scholar]
- 35. Giegé R, Springer M (20 November 2012, posting date.) Aminoacyl-tRNA Synthetases in the Bacterial World In: Böck A, Curtiss III R, Kaper JB, Karp PD, Neidhardt FC, Nyström T et al. , editors. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. Available: http://wwwecosalorg. Washington, DC.: ASM Press; 10.1128/ecosalplus.ESP-0009-2013 [DOI] [Google Scholar]
- 36. Allende JE, Allende CC (1971) Detection and isolation of complexes between aminoacyl-tRNA synthetases and their substrates. Methods in Enzymology 20: 210–220. [Google Scholar]
- 37. Meister A (1963) Preparation and properties of amino acyl adenylates. Methods in Enzymology 6: 751–757. [Google Scholar]
- 38. Kern D, Lapointe J (1980) The catalytic mechanism of glutamyl-tRNA synthetase of Escherichia coli. Evidence for a two-step aminoacylation pathway, and study of the reactivity of the intermediate complex. European Journal of Biochemistry 106: 137–150. [PubMed] [Google Scholar]
- 39. Smith CK, Windsor WT (2007) Thermodynamics of nucleotide and non-ATP-competitive inhibitor binding to MEK1 by circular dichroism and isothermal titration calorimetry. Biochemistry 46: 1358–1367. [DOI] [PubMed] [Google Scholar]
- 40. Salcedo G, Cano-Sánchez P, de Gómez-Puyou MT, Velázquez-Campoy A, García-Hernández E (2014) Isolated noncatalytic and catalytic subunits of F1-ATPase exhibit similar, albeit not identical, energetic strategies for recognizing adenosine nucleotides. Biochimica et Biophysica Acta 1837: 44–50. 10.1016/j.bbabio.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 41. Yu H, Rick SW (2010) Free energy, entropy, and enthalpy of a water molecule in various protein environments. Journal of Physical Chemistry B 114: 11552–11560. [DOI] [PubMed] [Google Scholar]
- 42. O'Brien R, Haq I (2005) Application of Biocalorimetry: Binding, Stability and Enzyme Kinetics In: Ladbury JE, Doyle ML, editors. Biocalorimetry 2, Applications of calorimetry in the biological sciences. England: John Wiley and Sons, Ltd; pp. 3–34. [Google Scholar]
- 43. Blume A, Tuchtenhagen J (1992) Thermodynamics of ion binding to phosphatidic acid bilayers. Titration calorimetry of the heat of dissociation of DMPA. Biochemistry 31: 4636–4642. [DOI] [PubMed] [Google Scholar]
- 44. Kern D, Lapointe J (1980) The catalytic mechanism of the glutamyl-tRNA synthetase from Escherichia coli. Detection of an intermediate complex in which glutamate is activated. Journal of Biological Chemistry 255: 1956–1961. [PubMed] [Google Scholar]
- 45. Baltzinger M, Lin S-X, Remy P (1983) Yeast phenylalanyl-tRNA synthetase: symmetric behavior of the enzyme during activation of phenylalanine as shown by a rapid kinetic investigation. Biochemistry 22: 675–681. [DOI] [PubMed] [Google Scholar]
- 46. Lin S-X, Baltzinger M, Remy P (1983) Fast kinetic study of yeast phenylalanyl-tRNA synthetase: an efficient discrimination between tyrosine and phenylalanine at the level of the aminoacyladenylate-enzyme complex. Biochemistry 22: 681–689. [DOI] [PubMed] [Google Scholar]
- 47. Ling J, Peterson KM, Simonović I, Söll D, Simonović M (2012) The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase. Journal of Biological Chemistry 287: 28518–28525. 10.1074/jbc.M112.372920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fersht AR (1998) Sieves in sequence. Science 280: 541 [DOI] [PubMed] [Google Scholar]
- 49. Gruic-Sovulj I, Uter N, Bullock T, Perona JJ (2005) tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase. Journal of Biological Chemistry 280: 23978–23986. [DOI] [PubMed] [Google Scholar]
- 50. Ling JQ, Reynolds N, Ibba M (2009) Aminoacyl-tRNA Synthesis and Translational Quality Control. Annual Review of Microbiology 63: 61–78. 10.1146/annurev.micro.091208.073210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.