SUMMARY
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related death worldwide1. Chemotherapies such as the topoisomerase II inhibitor (TopoIIi) etoposide effectively reduce disease in a minority of NSCLC patients2,3; therefore, alternative drug targets, including epigenetic enzymes, are under consideration for therapeutic intervention4. A promising potential epigenetic target is the methyltransferase EZH2, which in the context of the Polycomb Repressive Complex 2 (PRC2) is well known to tri-methylate Histone H3 at lysine 27 (H3K27me3) and elicit gene silencing5. Here, we demonstrate that EZH2 inhibition (EZH2i) had differential effects on TopoIIi response of NSCLCs in vitro and in vivo. EGFR and BRG1 mutations were genetic biomarkers that predicted enhanced sensitivity to TopoIIi in response to EZH2i. BRG1 loss-of-function mutant tumors responded to EZH2i with increased S phase, anaphase bridging, apoptosis, and TopoIIi sensitivity. Conversely, EGFR and BRG1 wild-type tumors up-regulated BRG1 in response to EZH2i and ultimately became more resistant to TopoIIi. EGFR gain-of-function mutant tumors were also sensitive to dual EZH2i and TopoIIi, due to genetic antagonism between EGFR and BRG1. These findings suggest an exciting opportunity for precision medicine in the genetically complex disease of NSCLC.
Keywords: Lung cancer, EZH2 inhibition, BRG1, EGFR, Chemotherapy, Etoposide
To validate that EZH2 is an important target for NSCLC, we generated a 116-gene lung cancer EZH2 co-expression gene signature (SI Table 1). This signature had predictive power for cancer progression using the Director’s Challenge dataset of 416 human lung adenocarcinomas6, partially due to stratification of later stage tumors to the EZH2 high group (Extended Data Fig. 1a). To control for this covariate, exclusively Stage 1 and moderately differentiated tumors were examined, confirming that the signature could robustly further stratify patients into risk groups (Fig. 1a). Gene ontology analysis revealed that the EZH2 co-expression signature was highly enriched for cell cycle, DNA synthesis and DNA repair genes (SI Table 2). One of the genes highly co-expressed with EZH2 in primary tumors was Topoisomerase 2A (TOP2A), which encodes the TopoII helicase targeted by etoposide.
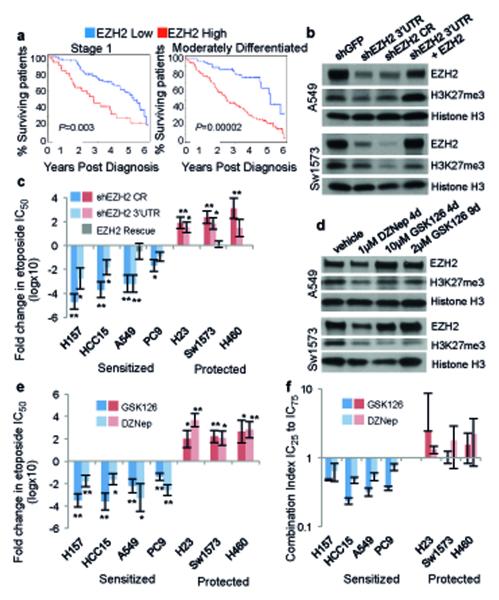
Figure 1. EZH2i sensitizes BRG1 or EGFR mutants to TopoIIi.
a, Director’s Challenge samples were hierarchically clustered into two risk groups using the EZH2 co-expression signature (Table S1). The Kaplan-Meier curves for only Stage 1 (n=94) or only moderately differentiated tumors (n=142) to 6 years post diagnosis are shown. b, Western Blot for EZH2 and H3K27me3 on indicated transduced lines, total Histone H3 is shown as loading control. CR indicates a coding region targeting hairpin. c, Fold change +/− s.e.m. in etoposide IC50 between transduced lines, n=3 biological replicates for HCC15, A549, PC9, H23 and Sw1573, n=4 biological replicates for HCC15 and H460, rescues n=3 biological replicates, * P<0.04, ** P<0.01. d, Western Blot for EZH2 and H3K27me3 on lines treated with indicated drugs. e, Fold change +/− s.e.m. in etoposide IC50 between vehicle treated and drug treated lines, n=3 biological replicates for all except n=4 biological replicates for H157 + DZNep, * P<0.04, ** P<0.01. f, Average Chou-Talalay combination index (CI) values +/− s.e.m. (also see SI Table 3) for fractions affected equivalent to IC25 through IC75; n=3 biological replicates.
To test EZH2 inhibition as a therapy for NSCLC, EZH2 expression was stably knocked-down with one of two different small hairpins in a panel of NSCLC cell lines. Western Blot confirmed that EZH2 protein and catalytic mark, H3K27me3, were decreased in each transduced cell line and could be rescued by EZH2 expression from a second lentivirus (Fig. 1b, Extended Data Fig. 1b). We then determined etoposide IC50 at 4 days. Of the 7 lines, HCC15, A549, H157 and PC9, termed ‘sensitized’ lines, had lower etoposide IC50 when EZH2 was knocked down. Conversely, H460, H23 and Sw1573 cell lines, termed ‘protected’ lines, had higher etoposide IC50 as shEZH2 lines (Fig. 1c). Rescue of EZH2 levels completely abrogated the change in etoposide IC50 driven by the 3’UTR targeting hairpin (A549 and Sw1573, Fig. 1c, grey bars). The sensitized and protected phenotypes were not due to differential degree of EZH2 knock-down (Extended Data Fig. 1b-c).
Next, we used pharmacological EZH2 inhibition via the S-adenosylhomocystein hydrolase inhibitor, DZNep, which causes proteosomal degradation of PRC2 components including EZH27,8 and the specific EZH2 methyltransferase inhibitor, GSK1269. Western Blot confirmed that 4 days of 1μM DZNep effectively reduced EZH2 protein and H3K27me3, and 10μM GSK126 for 4 days or 2μM GSK126 for 9 days caused decrease in H3K27me3 levels yet EZH2 remained unchanged (Fig. 1d, Extended Data Fig. 2a). 14 of 26 NSCLC cell lines were more sensitive to 4-day etoposide in the presence of 1μM DZNep, while the other lines were less sensitive to the chemotherapy in the presence of DZNep (Fig. 1e, Extended Data Fig. 2b). For the sensitized lines, pretreatment with 2μM GSK126 for 9 days sensitized the lines to 4-day etoposide with continued GSK126 treatment (14 days total). For the protected lines, 10μM of GSK126 for 4 days best recapitulated the etoposide protection caused by DZNep and shEZH2 (Fig. 1e, Extended Data Fig. 2c). IC50 shift results were validated with the Chou-Talalay Combination Index (CI)10, demonstrating strong synergism (CI<0.48) between DZNep and etoposide as well as synergism (CI<0.64) between GSK126 and etoposide (Fig. 1f, SI Table 3). The CI assay also confirmed drug antagonism (CI>1) in the protected lines.
We examined the mutational annotation available for the NSCLC lines and found that 12 of 14 sensitized cell lines harbored inactivating mutations in BRG1 (SMARCA4) or activating mutations in EGFR, while 10 of 12 protected cell lines were wild-type for the two genes (SI Table 4, Extended Data Fig. 2c, Fisher’s exact test p=0.001). Cell lines segregated into the same genotype-specific protected and sensitized classes when a different TopoII inhibitor, doxorubicin11, was combined with DZNep (Extended Data Fig. 2d).
To determine if the protected and sensitized phenotypes could be observed in vivo, we treated xenograft-bearing mice with etoposide and EZH2i. For the sensitized BRG1 mutant cell line H157, early treatment with dual etoposide and DZNep therapy prevented tumors from forming in 4/6 mice, proving more efficacious than etoposide or DZNep alone (Fig. 2a, Extended Data Fig. 3a-b). In contrast, the protected H23 xenografts that received early dual therapy grew significantly larger than those treated with either DZNep or etoposide alone (Fig. 2b, Extended Data Fig. 3b). Furthermore, in mice with established EGFR-driven PC9 xenografts the combination of GSK126 and etoposide prevented tumor growth (Fig. 2c).
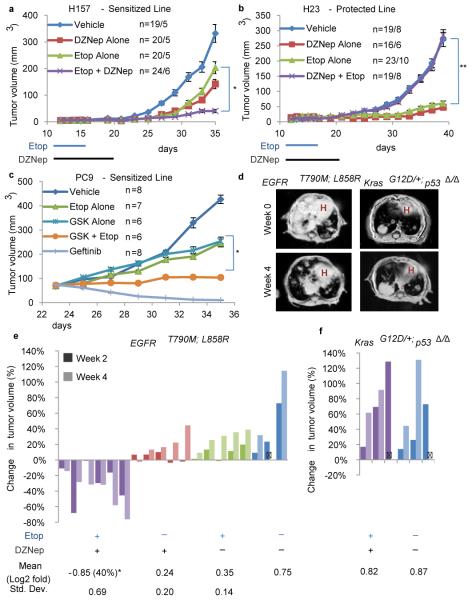
Figure 2. In vitro sensitivities to EZH2i + TopoIIi predict in vivo responses.
Either the H157 (a) or H23 (b) cell line was injected into the flanks of Nude mice and tumors were allowed to form. On day 12, mice were randomly segregated into cohorts that received either placebo, DZNep, etoposide or dual therapy for 2 weeks, and tumor size +/− s.e.m. were graphed, n for tumors/mice in each arm indicated on graphs, * P=0.002, ** P=0.0005 dual vs. etoposide. c, The PC9 cell line was injected into the flanks of Nude mice and tumors were allowed to grow to 70mm3. Mice were then treated with etoposide, GSK126, dual therapy or gefitinib (as a positive control) for 2 weeks, and tumor size +/− s.e.m. were graphed, n indicated on legend, mice with one tumor each, P<0.008 for dual vs etoposide or GSK126 alone. d, Representative MR images of mice of indicated genotypes on combination etoposide+DZNep treatment at 0 and 4 weeks post treatment initiation. H indicates heart area. e, Waterfall plot depicting tumor growth +/− s.e.m. of EGFRT790M;L858R tumors after 2 weeks and 4 weeks of treatment with vehicle (blue), etoposide (green), DZNep (red) and etoposide+DZNep (purple). The y-axis indicates % tumor growth vs. day 0. Each bar represents an individual mouse. Statistical analyses were performed on the 4 week log2 transformed data, P=0.008 dual vs. DZNep and P=0.004 dual vs. etoposide. f, Waterfall plot depicting tumor growth +/− s.e.m. of KrasG12D/+; p53Δ/Δ tumors after 2 weeks and 4 weeks of treatment with vehicle (blue) and etoposide+DZNep (purple).
Next, mouse models of lung cancer predicted to be sensitized (EGFRT790M; L858R transgenic; EGFR hereafter12) or protected (KrasG12D/+; p53Δ/Δ; Kras/p53 hereafter13) tumor types were treated with DZNep and etoposide. The Kras/p53 model, wild-type for Brg1 and Egfr, represents a predicted ‘protected’ cancer, whereas the EGFR model, driven by oncogenic EGFR, represents a predicted ‘sensitized’ cancer. Chemotherapy, DZNep, or combination therapy was then administered to randomized cohorts of mice with radiographically documented lung masses for 4 weeks (Fig. 2d). Marked tumor regression in the EGFR model was observed in response to 4 weeks of dual etoposide and DZNep treatment, while mice in the other treatment arms showed continued tumor growth (Fig. 2e, Extended Data Fig. 4a). In striking contrast, the Kras/p53 tumors proceeded to grow despite dual treatment (Fig. 2f). DZNep efficacy was confirmed by EZH2 immunohistochemistry for both models (Extended Data Fig. 4b,c).
To address the mechanism through which EZH2i changed sensitivity to TopoIIi, we considered the physical interaction between BRG1 and TopoII that allows for increased TopoII function14. Because BRG1 and EZH2 are known to be genetically antagonistic15, we hypothesized that protected cell lines up-regulated BRG1 in response to EZH2i and thereby had increased TopoII function. BRG1 transcript measured by RT-qPCR was reproducibly increased by DZNep treatment, though BRG1 levels were not significantly different when the cells were treated with GSK126 (Fig. 3a). To assess the function of BRG1 containing BAF complexes we quantified anaphase bridges, which are known to indicate a failure of TopoII to decatenate DNA prior to mitosis and can be attributable to BAF complex dysfunction (Extended Data Fig. 5a). DZNep or GSK126 treatment increased anaphase bridges in BRG1 mutant cell lines and decreased anaphase bridges in WT cells (Fig. 3b, Extended Data Fig. 5b). EGFR mutant cells, despite BRG1 up-regulation, also had increased anaphase bridging with DZNep or GSK126 treatment and showed high levels of EGFR in dividing cells (Extended Data Fig. 5c).
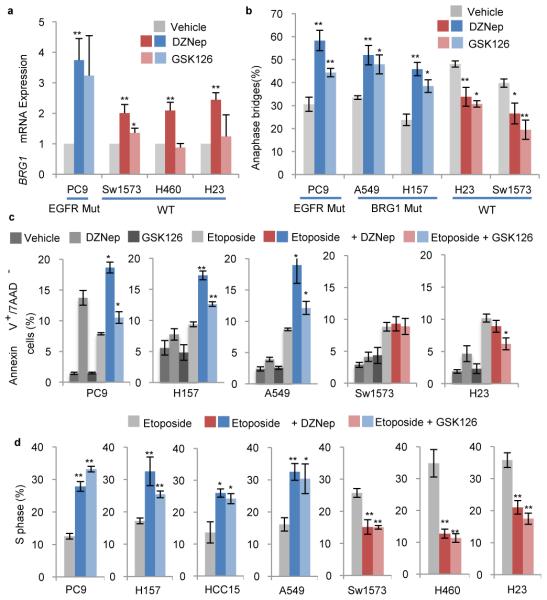
Figure 3. Dual EZH2i and TopoIIi differentially affects cell cycle, apoptosis and anaphase bridging.
a, RT-qPCR for BRG1 levels +/− s.e.m. in indicated cell lines in response to 4 days of 1μM DZNep or 10μM GSK126, n=6 biological replicates for DZNep, n=3 biological replicates for GSK126, * P<0.05, ** P<0.008. b, Percentage of anaphase structures with bridges +/− s.e.m. in vehicle-, DZNep- treated or GSK126-treated cell lines, n≥3 biological replicates and indicated in Methods, * P<0.04, ** P<0.02. c, Annexin V+/7AAD− cells +/− s.e.m. quantified by flow cytometry on cell lines cultured with indicated treatments for 3 days, n=4 biological replicates, * P<0.03, ** P<0.001 for Etop vs Dual treated or DZNep vs Dual treatment for PC9. d, 7-AAD cell cycle flow cytometry was performed on cell lines with or without 5μM etoposide or 1μM DZNep for 4 days. The average % S phase +/− s.e.m. of each culture is plotted, n=3 biological replicates for HCC15, A549, PC9 and H23, n=4 biological replicates for H157 and Sw1573, * P<0.05 ** P<0.009.
We next examined cell cycle and apoptosis dynamics of the lines. While the protected lines showed no difference in apoptotic levels in etoposide compared to dual treated cultures, the sensitized lines had significantly higher apoptotic fractions in dual treated cultures than in cultures treated with etoposide as a single agent (Fig. 3c). Furthermore, protected etoposide-treated lines had an average of 13.5% fewer cells in S phase in response to DZNep or shEZH2 compared to treatment with etoposide alone, suggesting these lines undergo cell cycle arrest sparing them from apoptosis. In contrast, sensitized etoposide-treated lines had an average of 16.5% more cells in S phase in response to DZNep or shEZH2 (Fig. 3d, Extended Data Fig. 6a), suggesting that apoptosis of G2/M cells that could not repair anaphase bridges contributed to enrichment for S phase.
While the links between EZH2, BRG1 and TopoII explained increased sensitivity of BRG1 mutants to TopoIIi, it was still unclear why EGFR mutants behaved similarly. In our panel of cell lines, 30% have a BRG1 mutation and 23% have an EGFR mutation, however none have both BRG1 and EGFR mutations (SI Table 4 and Extended Data Fig. 7a, Fisher’s exact test p=0.005). Similarly, in a panel of 412 sequenced primary adenocarcinomas16,17, 65 (15.7%) had mutation in EGFR, while 33 (8%) had mutations in BRG1. With these allele frequencies, 1.25% of the tumor samples were expected to have both BRG1 and EGFR mutations; however, only 1 (0.2%) was observed (Fisher’s exact test, p=0.019, p53RCCC7). The negative correlation of BRG1 and EGFR mutations suggests that they may be functionally redundant – BRG1 loss may be permissive for high EGFR expression. Supporting this idea, a strong negative correlation was found between EGFR and BRG1 expression in the Director’s Challenge Dataset (Fig. 4a). When comparing gene expression of EGFR and BRG1 mutant cell lines to WT lines, EGFR was the first of 21 shared up-regulated genes, over-expressed 3.1-fold in BRG1 mutant cell lines and 3.25-fold in EGFR mutant cell lines (Extended Data Fig. 7b-c, SI Table 5).
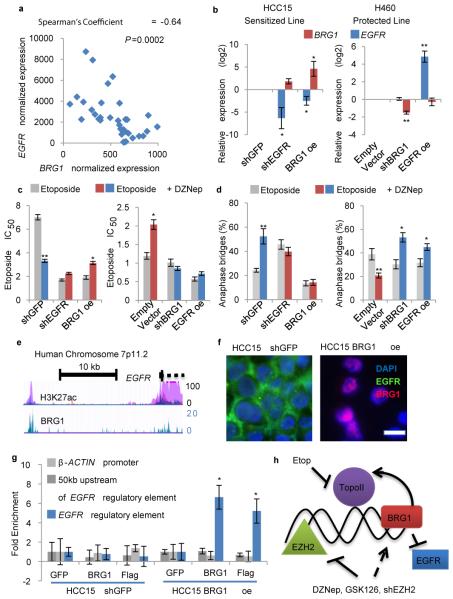
Figure 4. BRG1 and EGFR are genetically antagonistic and control the sensitized phenotype.
a, EGFR and BRG1 expression in tumors from the Director’s Challenge were plotted and correlation was assessed, n=36. b, RT-qPCR for expression of BRG1 and EGFR +/− s.e.m. in indicated transduced HCC15 and H460 cell lines, * P<0.04, **P<0.0001, n=5 biological replicates for H460, n=4 biological replicates for HCC15, oe indicates over-expression. c, The average etoposide IC50 +/− s.e.m. in the indicated vehicle- and DZNep-treated cells, n=3 biological replicates, * P<0.04, ** P<0.0001. d, Average percentage of anaphase structures with bridges +/− s.e.m. in indicated vehicle- or DZNep-treated cell lines, n=3 biological replicates, * P<0.04, ** P<0.02. e, Depiction of EGFR regulatory element including H3K27ac and BRG1 binding peaks and from ENCODE database. f, Immunofluorescence for EGFR and BRG1 on HCC15 shGFP (control) and BRG1 over-expressing cell lines, scale bar = 30μm. g, Chromatin immunoprecipitation for GFP (control), BRG1 or the FLAG-tag on the exogenously expressed BRG1 in the HCC15 shGFP (control) and BRG1 expressing cell lines. qPCR was performed with primers for indicated genomic sites and enrichment over GFP +/− s.e.m. was plotted, n=4 biological replicates, One-way ANOVA * P=0.02. h, In response to EZH2i: WT cells show increased BAF complex function and subsequent decrease in etoposide sensitivity; BRG1 mutant cells have increased etoposide sensitivity; and EGFR mutant cells act like BRG1 mutant cells due to genetic antagonism of EGFR by BRG1.
To better understand their genetic relationship, EGFR and BRG1 expression were manipulated (Fig. 4b, Extended Data Fig. 8a). BRG1 re-expression in the HCC15 lines converted the line from a sensitized to protected phenotype, and the HCC15 shEGFR line was no longer sensitized to etoposide by DZNep (Fig. 4c). Similarly, DZNep had no effect on etoposide IC50 when EGFR was over-expressed or BRG1 was knocked-down in H460 cells (Fig. 4c). These changes in etoposide sensitivity were consistent with changes in anaphase bridging, apoptosis and S phase accumulation (Fig. 4d, Extended Data Fig. 8b-c).
The ENCODE database18 shows a chromatin immunoprecipitation (ChIP) peak for BRG1 upstream of EGFR (Fig. 4e). HCC15 cells re-expressing BRG1 had lower levels of EGFR by RT-qPCR and immunofluorescence (Fig. 4b,f). We hypothesized that BRG1-containing BAF complexes bind to the EGFR regulatory element to disrupt EGFR transcription, as observed in other systems19. We confirmed the exogenously expressed BRG1 was being incorporated into BAF complexes (Extended Data Fig. 9a), and then performed ChIP assays. BRG1, immunoprecipitated with an antibody recognizing the protein itself or the FLAG-tag, was significantly associated with the EGFR regulatory element (Fig. 4g). This genetic antagonism explains why dividing cells in EGFR-driven cultures may not sufficiently up-regulate BRG1, and respond to etoposide+EZH2i with increased anaphase bridging as BRG1 mutant cells do (Fig. 4h).
Our results suggest that dual EZH2i+TopoIIi represent a treatment option for EGFR mutant tumors, even those that invariably develop resistance to EGFR TKIs12 . Furthermore, combination EZH2i+TopoIIi offers the first specific therapeutic for BRG1 mutant lung cancers. Importantly, the strong antagonism of etoposide and EZH2i warns against using this drug combination for EGFR and BRG1 WT tumors. Also notable is the fact that EZH2i either sensitized to etoposide (synergism) or protected from it (antagonism), but very rarely was additivity observed. We and others20,21 have found that BRG1 and EGFR mutations are significantly anti-correlated in NSCLC. Moreover, BRG1 mutant NSCLCs had elevated EGFR levels, raising the possibility that BRG1 mutant tumors represent a subset of patients with wild-type EGFR that respond to EGFR TKI20,21. Unlike Brg1-deficient MEFs14, BRG1 mutant lung cancer cells did not exhibit TopoII dysfunction unless EZH2 was inhibited, suggesting a novel connection between EZH2 and TopoII function.
METHODS
Cell Lines
Cell lines used are listed in Supplementary Table 4. All cell lines were maintained in RPMI 1640 media with 10% fetal bovine serum, 4mM L-glutamine and penicillin/streptomycin at 37°C, 5% CO2. Cell lines were obtained from the Meyerson Lab at Dana-Farber Cancer Institute. No mycoplasma was detected in cultures by either routine mycoplasma PCR or perinuclear DAPI staining. Cell line genotypes (SI Table 4) were obtained from published studies22,23, COSMIC database24 and CCLE database25.
Vectors
The pLKO.1 EZH2 shRNA construct clones TRCN0000040076 and TRCN0000040073 were purchased from SIGMA and the shGFP plasmid 12273 is available on Addgene26. Both shBRG1 and the matched empty vector were provided by the Smale lab27 and are available on Addgene, the BRG1 over-expression plasmid 19148 from the Massagué lab28 was purchased through Addgene, and the shEGFR and EGFR WT over-expression constructs were provided by the Jänne lab29. The EZH2 over-expression construct was derived by cloning human EZH2 cDNA into pLenti7.3/V5-DEST (Invitrogen). Lentivirus was packaged in HEK293T cells using established protocols30, and retrovirus was packaged in PlatE cells again using established protocols31. Cell lines were infected with viral-containing supernatant containing 6μg/mL polybrene (SIGMA) for a period of 10-18 hours. Infected cultures were selected with 1μg/mL puromycin (all sh constructs and EGFR oe, SIMGA), 200μg/mL hygromycin (BRG1 oe, Invitrogen), or by flow cytometry for GFP (EZH2 oe) 5 days post infection.
Small hairpin sequences:
GFP: GCCC(GCAAGCTGACCCTGAAGTTCAT)TCAAGAG(ATGAACTTCAGGGTCAGCTTGC)TTTT
EZH2 coding region:
CCGG(CGGAAATCTTAAACCAAGAAT)CTCGAG(ATTCTTGGTTTAAGATTTCCG)TTTTT
EZH2 3’ UTR:
CCGG(TATTGCCTTCTCACCAGCTGC)CTCGAG(GCAGCTGGTGAGAAGGCAATA)TTTTT
EGFR:
CCGG(GCTGAGAATGTGGAATACCTA)CTCGAG(TAGGTATTCCACATTCTCAGC)TTTTT
BRG1:
TTTG(TGGATAAGCAGCACAAGATT)TCAAGAG(AATCTTCTGCTGCTTCTCCA)TTTTT
Drugs
Etoposide and doxorubicin (SIGMA) were diluted to a stock of 100mM in DMSO for all cell culture experiments. DZNep was a kind gift from Dr. Marquez at NCI and was diluted in DMSO to a stock of 10mM. GSK126 was purchased from Xcess Bio as a 10mM stock in DMSO. All stocks were diluted in DMSO to 1000× concentration prior to addition into media at 2× concentration and final dilution onto plated cells 1:1.
Cytotox Assays
Cell lines were dissociated, counted and plated at 5,000 cells per well in flat bottom opaque tissue culture treated 96 well plates (CytoOne). Edge wells were filled with PBS. The following day, 2× drug diluted in media was added to each well such that the well then contained 100μl media with 1× drug concentration at the following doses: etoposide; 0, 0.1, 1, 3, 5, 7, 10, 50, 100, 500μM or doxorubicin; 0, 0.01, 0.1, 0.5, 1, 3, 5, 7, 10, 50μM, with or without additional 1μM DZNep, 10μM GSK126 or a continuation of 2μM GSK126 from a 9 day 2μM GSK126 pretreated culture. After 4 days, CellTiter-Glo (Promega) was added and luminescence was read on a BioTec plate reader to determine relative cell number in each well. Data were averaged for triplicate or quadruplicate technical replicates and normalized to the untreated wells, and whole runs (vehicle and EZH2i) were excluded if vehicle treated wells did not reach the threshold luminescence of 3000. Results from independent biological replicate experiments were input into GraphPad Prism software to extrapolate IC50 and s.e.m. of IC50 for a given cell line using the nonlinear regression analysis of log(inhibitor) vs. normalized response with a variable slope. For 1μM DZNep for 4 days with etoposide: n=3 biological replicates for H1975, H2030, HCC4006, A549, HCC2450, Calu1, H1650, H522, H2126, H1299, HCC15, H322, H2009, HCC95, H520, H460, Calu3, H2122, H23 and H3255, n=4 biological replicates for PC9, H157, HCC827, Sw1573, Calu6 and H441. For both 2μM GSK126 pretreated for 9 days and continued for 4 days with etoposide, and 10μM GSK126 treated for 4 days with etoposide n=3 biological replicates. The logIC50s were compared using GraphPad Prism software and P values reported are sum-of-squares F statistics. For graphing, logIC50 of vehicle control cells was subtracted from logIC50 of EZH2i treated cells and multiplied by ten to be depicted as log fold change × 10. Errors were estimated by calculating possible upper bound and lower bound of log fold changes based on GraphPad reported standard error of the mean for each logIC50 calculated. For Chou-Talalay combination index (CI) assays10, doses for etoposide, DZNep, or GSK126 were 0, 0.1, 0.5, 1, 3, 5, 7, 10, 30, 50, and 1:1 combinations of etoposide and DZNep or etoposide and GSK126. Survival percentages for 3 independent biological replicate experiments were averaged and input into CompuSyn software to extrapolate CI values. Any drug dose with mean survival over 100% was excluded because the CompuSyn software does not allow for values over 1. In addition, visual inspection led us to remove the highest doses of GSK126 from the analysis for every cell line, as is suggested by Chou32,33, leading to much better matching of the data-points to the median effect plot. Notably, removal of these data-points both increased synergy seen in the sensitized lines and increased antagonism seen in H460 and Sw1573 lines.
Flow Cytometry
For 7AAD-cell cycle analysis, cell lines were plated at 1.5×106 cells per 10cm plate and treated with drug for 4 days. Cells were then dissociated, fixed with 100% ice cold Ethanol for at least 2 hours, incubated for 30 minutes with 1mg/mL DNase-free RNase A (Thermo) and resuspended in 20μg/mL 7-Aminoactinomycin D (7AAD; Invitrogen). 30,000 events were collected on the BD Fortessa and analyzed with the ModFit LT software and results were averaged for 3 or 4 biological replicates (n indicated in legend).
For Annexin V/7AAD apoptosis analysis, cell lines were plated at 5×104 cells per well of 6 well plate and treated with drug for 3 days. Supernatant was retained and added to trypsinized suspensions of adherent cells. Cells were stained with Annexin V-FITC (BD Biosciences) according to manufacturer’s instructions, and resuspended with 1μg/mL 7AAD prior to analysis on BD Fortessa. For sensitized lines 2μM GSK126 pretreated for 9 days and continued for 3 days with etoposide was used, while for protected line 10μM GSK126 treated for 3 days with etoposide was used. Data was analyzed with FlowJo (Treestar) software and percentage Annexin V+/7AAD− cells were averaged for 4 biological replicate experiments.
Quantitative RT-PCR
RNA from treated cell lines was extracted using Absolutely RNA kits (Agilent) and cDNA was made using the SuperScript III kit (Invitrogen). Relative gene expression was assayed with Sybr green on the StepOnePlus™ Real-Time PCR System (Applied Biosystems). Relative expression was calculated by Gene of Interest(Ctreference-Ctexprimental)-CYPA(Ctreference-Ctexperimental) and graphed on the log2 scale or converted to linear scale. Statistics were performed on log2 data. For all experiments, the reference sample was a matched vehicle treated or control transduced cell line.
Primer sequences:
CYPA: F TCATCTGCACTGCCAAGACTG R CATGCCTTCTTTCACTTTGCC
EZH2: F AGGAGTTTGCTGCTGCTCTC R CCGAGAATTTGCTTCAGAGG
BRG1: F agcgatgacgtctctgaggt R gtacagggacaccagccact
EGFR: F TAACAAGCTCACGCAGTTGG R GTTGAGGGCAATGAGGACAT
Xenograft Experiments
For DZNep experiments, H157 or H23 cells were dissociated into single cells, counted and resuspended at 1×106 cells per 250μL of 1:1 media/matrigel (BD). 8- to 16-week-old female Foxn1nu (Nude) mice (Harlan) were injected subcutaneously with 1×106 cells in 2-4 spots on flanks. Etoposide and DZNep were administered from day 12 to day 17 post injections; etoposide: 20 mg/kg/day i.p. in corn oil once per day for 5 consecutive days, and DZNep 2mg/kg/day i.p. in corn oil twice per week for 1 week, or 1 mg/kg/day i.p. in corn oil twice per week for 2 weeks. Tumor growth was measured every other day by caliper in a non-blinded fashion. For GSK126 experiments, PC9 cells were dissociated into single cells, counted and resuspended at 1×106 cells per 250μL of 1:1 media/matrigel (BD). 8- to 16-week-old female Foxn1nu mice (Harlan) were injected subcutaneously with 1×106 cells in 1 spot on left flank. Tumors were allowed to grow for 23 days to a mean size of 70mm3. Mice were then randomized into groups that received etoposide, GSK126, both etoposide and GSK126, gefitinib or vehicle: Etoposide 10mg/kg/day i.p. in corn oil 3 times per weeks, GSK126 300mg/kg/day i.p. in 1:1 v/v GSK126/Captisol® mixture resuspended in sterile water with acetic acid to pH 4.8, gefitinib (LC Laboratories) 150mg/kg/day in 1% Tween-80 (SIGMA). All mouse experiments were approved by the CHB Animal Care and Use Committee and by the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee, both accredited by AAALAC, and were performed in accordance with relevant institutional and national guidelines and regulations.
Generation of the EZH2 Co-Expression Gene Signature
We used Oncomine34 to query the top 20 genes co-expressed with EZH2 in all datasets containing human non-small cell lung cancer samples and co-expression data6,35-41. 20 was the number of probes chosen to examine from each study in order to yield a list between 100-200 genes, which allowed for robust hierarchal clustering of samples similar to that in previous studies. Of the 180 probes, 64 were redundant, leading to a list of 116 genes highly co-expressed with EZH2 (SI Table 1). Because these data sets were from various microarray platforms, the gene list was then used to generate a probe list for the 116 genes corresponding to probes on the U133A Affymetrix array using the batch query function on the NetAffx website (http://www.affymetrix.com/analysis/index.affx). Gene Ontology analysis was performed on the EZH2 co-expression signature with dChip software (http://biosun1.harvard.edu/complab/dchip).
Microarray Analysis
All array data are publically available on Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and correspond to array files available from GSE482442 for all lines except A549, H522 and PC9, GSE5457 for A549 (2 replicates), GSE572043 for H522 and an additional A549, GSE767044 for an additional H1299, GSE1008945 for PC9 and H1650 and GSE3162546 for an additional PC9. Arrays were chosen based on availability in September 2012. Arrays were analyzed using R/Bioconductor (http://www.bioconductor.org/). Raw CEL files from U133A Affymetrix arrays were processed using the robust multiarray average (rma) algorithm47. To identify genes correlating with the phenotypic groups, we used limma48 to fit a statistical linear model to the data and then tested for differential gene expression in the three groups using the vennSelect package, WT: H460, H441, H2122, H2009, Calu6, HCC95, EGFR mutant: H1650, HCC827, HCC4006, H1975, H3255, PC9, Brg1 mutant: A549, H1299, H157, H2126, H522, HCC15. Results were adjusted for multiple testing using the Benjamini and Hochberg (BH) method49, and significance was determined using a False-Discovery-Rate cutoff of less than 5%. For correlation between EGFR and BRG1 expression in the Director’s Challenge dataset, the arrays were processed using RMA and limma as described above. Tumors with an rma normalized expression of more than 2000 for EGFR or 575 for BRG1 were plotted and correlation was assessed – because these data were nonparametric, Spearman’s correlation coefficient was used. Using this same method of selecting for highest expressing tumors we could visualize positive correlations between EZH2 and other EZH2 co-expression gene signature members (data not shown).
Kaplan-Meier Analysis
Raw gene expression data from the Director’s Challenge human lung adenocarcinoma samples6 were obtained (https://caintegrator.nci.nih.gov/caintegrator/). Probe intensities from the Affymetrix U133A platform used in these studies were normalized and modeled using dChip software50 (http://biosun1.harvard.edu/complab/dchip). Kaplan-Meier survival analyses were implemented after the samples were hierarchically clustered using centroid linkage into two risk groups using the EZH2 co-expression gene signature. Survival differences between the two risk groups were assessed using the Mantel-Haenszel log rank test. The large area between the two risk groups and its associated small P value from the Mantel-Haenszel log rank test implicate a robust survival classification model.
Statistical Analysis
Except where indicated, a 2-tailed Student’s t-test with equal variance was used to compare measurements between 2 conditions with at least 3 biological replicates per condition. Normal distribution was checked with the Kolmogorov-Smirnov Test – data that failed test at α=0.05 were considered normally distributed. If the data were nonparametric, then a 2-tailed Mann Whitney U test was used instead of a T-test to assess the P value. Unless noted otherwise, pooled data is represented by the mean and standard error. P values are indicated in figure legends, and P-values less than 0.05 were considered significant.
Western Blot
Whole cell extracts were made in RIPA buffer (0.5% Deoxycholate, 1% IGEPAL-CA630, 0.1% sodium dodecyl sulfate, 150mM NaCl, 50mM Tris-8.1), lysates were cleared by centrifugation, and protein concentrations were quantified with the Pierce BCA Protein Assay Kit (Thermo). For Western blotting, 25 μg of protein extract per sample was denatured with heat and reducing agents, separated on a 4-12% acrylamide gel (BioRad) and transferred to nitrocellulose (GE Healthcare). Antibodies used for Western blotting were: EZH2 (clone D2C9; Cell Signaling; 1:200), Histone H3 (polyclonal; AbCAM ab1791; 1:2000) and H3K27me3 (polyclonal; Millipore 07-449; 1:1000) all incubated overnight at 4°C. All antibodies have detailed species validation available online from vendors. Secondary antibody, anti-rabbit-HRP (Santa Cruz sc-2313; 1:10,000), was incubated for 1 hour at room temperature. After washing, chemiluminescence was visualized with Western Lightning Plus-ECL (PerkinElmer) and exposure onto KODAK BioMax XAR film.
Immunoprecipitation
Cultured cells (10×106 per line) were collected by trypsinization and pelleting, followed by PBS wash and pelleting. On ice, cell pellets were resuspended in 750μL Hypotonic Buffer (10mM Tris-HCl, pH 7.4, 10mM NaCl, 3mM MgCl2) and incubated for 15 minutes. 37.5μL of 10% NP-40 was added to each tube followed by vortexing for 10 seconds and centrifugation for 10 minutes at 3,000 rpm at 4°C. Nuclear pellets were then resuspended in RIPA buffer for 30 minutes on ice with vortexing, lysates were cleared by centrifugation, and protein concentrations were quantified with the Pierce BCA Protein Assay Kit (Thermo). Antibodies directed against Flag (M2; SIGMA; 1:50), BAF155 (R-18; Santa Cruz; 1:50), or ARID1a (PSG3; Santa Cruz; 1:10) were incubated with 300μg of each nuclear extract with 1:1 Protein A and Protein G agarose beads (GE Healthcare) rotating at 4°C overnight. All antibodies have detailed species validation available online from vendors. Beads were washed 3× in 1mL RIPA buffer with the second wash being rotated at 4°C for 30 minutes, then resuspended in ~35μL 1× reducing buffer and boiled for 5 minutes prior to loading 10μL per lane. Proteins were separated on a 4-12% acrylamide gel (BioRad) and transferred to nitrocellulose (GE Healthcare). Antibodies for Western blotting were: Flag (M2; SIGMA; 1:1000), BAF155 (R-18; Santa Cruz; 1:1000), and ARID1a (polyclonal; Bethyl Laboratories A310-040A; 1:1000). Secondary antibody, anti-rabbit-HRP (Santa Cruz sc-2313; 1:10,000), anti-mouse-HRP (Santa Cruz sc-2314; 1:10,000), or anti-goat-HRP (Santa Cruz sc-2020; 1:10,000), was incubated for 1 hour at room temperature. After washing, chemiluminescence was visualized with Western Lightning Plus-ECL (PerkinElmer) and exposure onto KODAK BioMax XAR film.
Anaphase Bridge Analysis
To quantify anaphase bridges, cells were grown on 4-well cultures slides (Lab Tek II). Adherent cells were fixed with 4% paraformaldehyde for 20 min, washed and stained with Vectashield with 4′,6-diamidino-2-phenylindole (DAPI; Vector Labs). Images were taken of each anaphase structure, and the number of anaphases with bridges over the total number of anaphases (between 11 and 34 total anaphases per well of a 4-well chamber slide) was recorded for each of three or more independent biological replicate experiments in a blinded fashion. Exact biological replicate n for Fig. 3b are: PC9 vehicle=7 DZNep=4 GSK126=4; A549 vehicle=3 DZNep=3 GSK126=3; H157 vehicle=7 DZNep=5 GSK126=3; H23 vehicle=6 DZNep=4 GSK126=3; Sw1573 vehicle=4 DZNep=4 GSK126=3 and for Extended Data Fig. 5b are H441 vehicle=3 DZNep=3; H2009 vehicle=3 DZNep=3; H522 vehicle=3 DZNep=3; and H1650 vehicle=4 DZNep=4. Imaging was performed with a Nikon 90i camera with 100× objective and oil emersion and NIS-Elements software and processed with NIS-Elements and Adobe Photoshop.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde and permeabilized with 10% Normal Donkey Serum (NDS; Jackson ImmunoResearch), 0.25% Triton-X (SIGMA), both in PBS. Primary antibodies, Brg1 (clone G-7, Santa Cruz), EGFR (polyclonal; Cell Signaling 2232), were incubated over-night at 1:100 dilution in PBS, 10% NDS. All antibodies have detailed species validation available online from vendors. Slides were washed 3× and secondary antibodies, anti-mouse-AlexaFluor594 and anti-rabbit-AlexaFluor488 (Invitrogen) were incubated at 1:500 for 1 hour. After washing, cover slips were mounted with Vectashield with 4′,6-diamidino-2-phenylindole (DAPI; Vector Labs). Imaging was performed with a Nikon 90i camera and NIS-Elements software and processed with NIS-Elements and Adobe Photoshop. All treatment groups were imaged with the same exposure time and equivalent processing. Images were chosen to highlight the difference between BRG1high interphase cells and EGFRhigh dividing cells in EZH2i treated PC9 cultures.
Treatment and MRI of Endogenous Mouse Models
Doxycycline inducible EGFRT790M;L858R transgenic mice12, and Lox-Stop-Lox-KrasG12D/+;p53fl/fl (KrasG12D/+; p53Δ/Δ)13,51 mice were maintained on a mixed background and housed in a pathogen-free environment at the Harvard School of Public Health and were handled in strict accordance with Good Animal Practice as defined by the Office of Laboratory Animal Welfare. All animal work was done with Dana-Farber Cancer Institute IACUC approval. Cohorts of male and female EGFRT790M;L858R ; CCSP-rtTA were put on a doxycycline diet at 6 weeks of age to induce the expression of mutant EGFR, while male and female KrasG12D/+; p53Δ/Δ mice received intranasal adeno-Cre between 6 and 8 weeks of age. Mice were evaluated by Magnetic resonance imaging (MRI) 12 to 16 weeks after doxycycline diet or adeno-Cre infection to document and quantify the lung cancer burden before being randomized to various treatment study cohorts. Treated mice in all cohorts have the similar initial tumor burden. Tumor bearing mice were randomized into cohorts treated either with vehicle (corn oil), etoposide 10mg/kg i.p. 3× per week for 4 weeks; DZNep 4mg/kg i.p. 2× per week for 4 weeks, or both etoposide and DZNep. The mice were imaged by MRI biweekly to determine the reduction in tumor volume during the respective treatments as described previously in a non-blinded fashion12. The tumor burden volume and quantification were reconstructed on 3D slicer software (http://www.slicer.org). Immunohistochemistry was performed as described with anti-EZH2 (clone D2C9; Cell Signaling) or anti-pEGFR (Y1068; clone D7A5; Cell Signaling) and developed using Vectastain Elite ABC kit (Vector Labs). Imaging was performed with a Nikon 90i camera and NIS-Elements software and processed with NIS-Elements and Adobe Photoshop.
Chromatin Immunoprecipitation (ChIP)
5×106 cells were fixed in 1% formaldehyde for 10 minutes prior to addition of glycine to a concentration of 1mM. Cells were pelleted, washed and resuspended in ChIP sonication buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mM Tris 8.1, 150 mM NaCl, 5 mM EDTA) containing protease and phosphastase inhibitors (Roche). Samples were sonicated for a total of 3 minutes in 30 second cycles with 1 minute breaks. Sonicated samples were centrifuged for 15 minutes to clear the lysates, and resulting whole cell extracts were used for pull-downs. Antibodies directed against GFP (Ab-1; Neomarkers), Brg1 (G-7; Santa Cruz) and Flag (M2; SIGMA) were incubated with equal proportions of whole cell extracts at 1:30 dilution overnight, rotating at 4°C. 1:1 Protein A and Protein G agarose beads (GE Healthcare) were added and incubated for 2 hours at 4°C. Beads were then pelleted and washed with high salt wash buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mM Tris-8.1, 500 mM NaCl, 5 mM EDTA), followed by LiCl immune complex buffer (250mM LiCl, 0.5% IGEPAL-CA630, 0.5% Deoxycholate, 10mM Tris-8.1, 1mM EDTA), and TE (10mM Tris-8.1, 1mM EDTA) prior to suspension in Elution buffer (1% SDS, 0.1 M NaHCO2, 0.01 mg/ml salmon sperm DNA (GE Healthcare)). Crosslinks were reversed at 65°C overnight, beads were pelleted, and resulting supernatant was incubated with 0.4mg/mL Proteinase K (SIGMA) for 2 hours at 37°C. DNA from each sample was purified using Qiagen PCR purification columns following the manufacturer’s instructions. Samples were resuspended in 100μL 10mM Tris-8.1 and 2μL were used for each Sybr green PCR reaction (Applied Biosystems). Enrichment was calculated by 2^average(Ctreference-Ctexprimental) for each genomic region of interest. Reference samples were the GFP, both Brg1 and FLAG samples were experimental. Statistical analyses (one way ANOVA) were performed on log2 transformed data for 3 independent biological replicates.
Primer sequences:
β-Actin: F TCGAGCCATAAAAGGCAACT R TCTCCCTCCTCCTCTTCCTC
EGFR regulatory element: F CCTTGTAGATTGGGGCTGAG R AGTTTGGGGGTGGAAGAAAG
50kb upstream of regulatory element:
F GGCTGAGACAGAGGGAACAC R CCATCTCAGCCTCCCAAGTA
Extended Data
Extended Data Figure 1. Verification of EZH2 as a potential target for NSCLC a.
Survival of lung adenocarcinoma patients in the Director’s Challenge dataset. Samples were hierarchically clustered using the primary tumor generated EZH2 co-expression signature (SI Table 1) into two risk groups. The Kaplan-Meier curve for the whole dataset is shown, n=416, P<0.00001. b, Western Blot was performed on whole cell extracts from indicated lines for EZH2 and its catalytic mark H3K27me3, total Histone H3 is shown as loading control. CR indicates a coding region targeting hairpin. c, RT-qPCR for EZH2 mRNA in the indicated cell lines after plating at equal density and treating for 4 days with indicated treatments. Each cell line is normalized to its shGFP control, n=2 biological replicates.
Extended Data Figure 2. Pharmacological inhibition of EZH2 changes response of cells to TopoII inhibitors a.
Western Blot for EZH2 and H3K27me3 was performed on whole cell extracts after administration of 1μM DZNep for 4 days, 10μM GSK126 for 4 days, 2μM GSK126 for 9 days, or vehicle. Total Histone H3 is shown as a loading control. b, The fold change in etoposide IC50 +/− s.e.m. in response to DZNep is plotted, n=3 biological replicates for H1975, H2030, HCC4006, A549, HCC2450, Calu1, H1650, H522, H2126, H1299, HCC15, H322, H2009, HCC95, H520, H460, Calu3, H2122, H23 and H3255, n=4 biological replicates for PC9, H157, HCC827, Sw1573, Calu6 and H441, * P<0.02. Cell lines with mutations in BRG1 or EGFR are indicated. Note that the H23 cell line has a very late coding region mutation in BRG1 (K1533N) and is predicted to produce functional protein22, consistent with its protected phenotype in our assays. c, Average fold change +/− s.e.m. between vehicle treated and indicated EZH2i treated lines for etoposide IC50 is graphed, n=3 biological replicates, * P<0.03, ** P<0.01. d, Fold change in doxorubicin IC50 in response to DZNep, n=2 biological replicates.
Extended Data Figure 3. Xenograft experiments confirm sensitized and protected phenotypes a.
Representative image of mouse injected at 4 sites (arrows) with H23 tumor cells 12 days after cell injection. b, Representative images of mice injected at 4 sites with either H23 or H157 cells, and treated with indicated drugs, 35 days after cell injection. Palpable tumors that remain are indicated with arrows.
Extended Data Figure 4. Autochthonous mouse models confirm genotype specificity of dual EZH2i and TopoIIi a.
Representative images of H+E stained lung from EGFRT790M; L858R mice treated with indicated therapies for 4 weeks. Areas with tumors of similar sizes were chosen for comparison, scale bar=200μm. b, Histology from KrasG12D/+/p53Δ/Δ mouse lung tumors after 1 week of indicated treatments; top image is H+E, bottom image is EZH2 IHC, scale bar=100μm. c, Histology from EGFRT790M; L858R mouse lung tumors after 4 weeks of indicated treatments; top image is H+E, center image is phospho-EGFR IHC, and bottom image is EZH2 IHC, scale bar=100μm.
Extended Data Figure 5. EZH2i modulates anaphase bridging differentially by genotype a.
Representative images of nuclei undergoing a normal anaphase and of nuclei that scored positively for the presence of anaphase bridges. b, Percentage of anaphase bridging +/− s.e.m. in additional BRG1 wild-type H2009 and H441, BRG1 mutant H522, and EGFR mutant H1650 cell lines, n=3 biological replicates for all except H1650 n=4, *P<0.05. c, Immunofluorescence on PC9 cultures showing increase in BRG1 staining in interphase nuclei in response to EZH2i while anaphase nuclei retain strong EGFR stain, representative of 3 biological replicates, scale bar = 30μm.
Extended Data Figure 6. Cell cycle and apoptosis analysis of dual EZH2i and TopoIIi treated lines a.
7-AAD cell cycle flow cytometry on cultures corresponding showing the matched controls for each experiment shown in Fig. 3d. The average % S phase +/− s.e.m. of each culture is plotted, n=3 biological replicates for H460, H23, Calu6, PC9, HCC15, A549 and H522, n=4 biological replicates for Sw1573, H441 and H157.
Extended Data Figure 7. EGFR and BRG1 negatively correlate in NSCLC a.
Additional NSCLC cell lines with known EGFR and BRG1 mutations used to estimate mutually exclusivity of the two mutations b, Venn diagram of differential gene expression overlap between cell lines of various genotypes. c, Average probe intensity +/− s.e.m. of EGFR probe (201983_s_at) and EZH2 probe (203358_s_at) on the U133A Affymetrix array for cell lines with various EGFR and BRG1 mutational statuses, n=6/genotype, see Methods, * P=0.014.
Extended Data Figure 8. Modulation of EGFR and BRG1 influences sensitized and protected phenotypes a.
RT-qPCR for BRG1, EGFR and EZH2 expression +/− s.e.m. in the various indicated treated transduced cell lines, n=3 biological replicates. b, For the indicated HCC15 and H460 stably transduced etoposide treated cell lines, 7AAD flow cytometry was used to assess changes in S phase percentage +/− s.e.m. in response to DZNep, n=3 biological replicates, * P=0.02, ** P<0.001. c, Percentage sub-G1 fractions +/− s.e.m. of the indicated 4-day cultures were assessed during 7AAD cell cycle flow cytometry analysis. Critically, for these assays the supernatant of each culture was retained and combined with the trypsinized adherent cells to accurately reflect the total amount of apoptosis/necrosis in each culture, n=3 biological replicates, ** P=0.03.
Extended Data Figure 9. Confirmation that BRG1 re-expression leads to formation of BAF complex a.
Immunoprecipitation of BAF complex members from nuclear lysates of the (left) BRG1 mutant HCC15 shGFP control cell line and (right) the HCC15 line with BRG1 re-expressed shows that exogenously expressed FLAG-tagged BRG1 does result in BRG1-containing BAF complex formation. Blot is representative of 3 biological replicates.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Kim Lab, F. Luo, P. Louis, K. Harrington, X. Wang and J. Brainson for technical assistance and discussions, J. Crabtree, D. Hargreaves, C. Kadoch, L. Zon, K. Cichowski, M. Enos, S. Orkin, A. Gutierrez and C. Roberts for helpful discussions. This work was supported in part by the Ladies Auxiliary to the Veterans of Foreign Wars, PF-12-151-01-DMC from the American Cancer Society, and the Uniting Against Lung Cancer Young Investigator Award supported by Meryl Bralower (CMF), Boston University Undergraduate Research Opportunities Program (PTD), RO1 HL090136, U01 HL100402 RFA-HL-09-004, American Cancer Society Research Scholar Grant RSG-08-082-01-MGO, the V Foundation for Cancer Research, a Basil O’Conner March of Dimes Starter Award, the Harvard Stem Cell Institute, and the Lung Cancer Research Foundation (CFK), the NIH grants CA122794, CA140594, CA163896, CA166480, CA154303, and CA120964 (KKW), the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (VEM), and the NIH grant K08 CA163677 (PSH).
Footnotes
CONTRIBUTIONS
C.M.F., C.X., K.K.W. and C.K. designed the study; C.M.F., C.X., P.T.J., J.M.B. and Y.J.L performed the experiments; S.P.R. cloned the EZH2 cDNA vector; H.Z. performed EZH2 IHC, V.E.M. provided DZNep, P.S.H. analyzed primary tumor sequencing data; K.K.W. allowed autochthonous mouse models studies in his laboratory; C.M.F. and C.K. wrote the manuscript with comments from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Jemal A, et al. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61:69–90. doi: 10.3322/caac.20107. doi:10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Zornosa C, et al. First-Line Systemic Therapy Practice Patterns and Concordance With NCCN Guidelines for Patients Diagnosed With Metastatic NSCLC Treated at NCCN Institutions. Journal of the National Comprehensive Cancer Network. 2012;10:847–856. doi: 10.6004/jnccn.2012.0088. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, et al. Randomized phase II study of concurrent cisplatin/etoposide or paclitaxel/carboplatin and thoracic radiotherapy in patients with stage III non-small cell lung cancer. Lung Cancer. 2012;77:89–96. doi: 10.1016/j.lungcan.2012.02.011. doi:10.1016/j.lungcan.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. doi:10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. doi:10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Shedden K, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. doi:10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & Development. 2007;21:1050–1063. doi: 10.1101/gad.1524107. doi:10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury SR, et al. (-)-Epigallocatechin-3-gallate and DZNep reduce polycomb protein level via a proteasome-dependent mechanism in skin cancer cells. Carcinogenesis. 2011;32:1525–1532. doi: 10.1093/carcin/bgr171. doi:10.1093/carcin/bgr171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. doi:10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 10.Chou T, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs of enzyme inhibitors. Advanced Ezyme Regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 11.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Research. 2009;37:738–748. doi: 10.1093/nar/gkn937. doi:10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji H, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. doi:10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Jackson EL, et al. The Differential Effects of Mutant p53 Alleles on Advanced Murine Lung Cancer. Cancer Research. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. doi:10.1158/0008-5472.can-05-2193. [DOI] [PubMed] [Google Scholar]
- 14.Dykhuizen EC, et al. BAF complexes facilitate decatenation of DNA by topoisomerase II. Nature. 2013;497:624–627. doi: 10.1038/nature12146. doi:10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson BG, et al. Epigenetic Antagonism between Polycomb and SWI/SNF Complexes during Oncogenic Transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. doi:10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imielinski M, et al. Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. doi:10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.TCGA Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. doi:10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ENCODE An integrated encyclopedia of DNA elements in the human genome. Nature. 2013;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oike T, et al. A Synthetic Lethality-Based Strategy to Treat Cancers Harboring a Genetic Deficiency in the Chromatin Remodeling Factor BRG1. Cancer Research. 2013;73:5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. doi:10.1158/0008-5472.can-12-4593. [DOI] [PubMed] [Google Scholar]
- 21.Matsubara D, et al. Lung cancer with loss of BRG1/BRM, shows epithelial mesenchymal transition phenotype and distinct histologic and genetic features. Cancer Science. 2013;104:266–273. doi: 10.1111/cas.12065. doi:10.1111/cas.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina PP, et al. Frequent BRG1/SMARCA4–inactivating mutations in human lung cancer cell lines. Human Mutation. 2008;29:617–622. doi: 10.1002/humu.20730. doi:10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto H, et al. PIK3CA Mutations and Copy Number Gains in Human Lung Cancers. Cancer Research. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. doi:10.1158/0008-5472.can-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orimo A, et al. Stromal Fibroblasts Present in Invasive Human Breast Carcinomas Promote Tumor Growth and Angiogenesis through Elevated SDF-1/CXCL12 Secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. doi:http://dx.doi.org/10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 27.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2 nucleosome remodeling complexes during an inflammatory response. Genes & Development. 2006;20:282–296. doi: 10.1101/gad.1383206. doi:10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi Q, He W, Zhang XH-F, Le H-V, Massagué J. Genome-wide Impact of the BRG1 SWI/SNF Chromatin Remodeler on the Transforming Growth Factor β Transcriptional Program. Journal of Biological Chemistry. 2008;283:1146–1155. doi: 10.1074/jbc.M707479200. doi:10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engelman JA, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. The Journal of Clinical Investigation. 2006;116:2695–2706. doi: 10.1172/JCI28656. doi:10.1172/jci28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fillmore CM, et al. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proceedings of the National Academy of Sciences. 2010;107:21737–21742. doi: 10.1073/pnas.1007863107. doi:10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zacharek SJ, et al. Lung Stem Cell Self-Renewal Relies on BMI1-Dependent Control of Expression at Imprinted Loci. Cell Stem Cell. 2010;9:272–281. doi: 10.1016/j.stem.2011.07.007. doi:10.1016/j.stem.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou T-C. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Research. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. doi:10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 33.Chou T-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacological Reviews. 2006;58:621–681. doi: 10.1124/pr.58.3.10. doi:10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 34.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9 doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 36.Garber ME, et al. Diversity of gene expression in adenocarcinoma of the lung. Proceedings of the National Academy of Sciences. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. doi:10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon GJ, et al. Translation of Microarray Data into Clinically Relevant Cancer Diagnostic Tests Using Gene Expression Ratios in Lung Cancer and Mesothelioma. Cancer Research. 2002;62:4963–4967. [PubMed] [Google Scholar]
- 38.Landi MT, et al. Gene Expression Signature of Cigarette Smoking and Its Role in Lung Adenocarcinoma Development and Survival. PLoS ONE. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohrbeck A, et al. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. Journal of Translational Medicine. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su AI, et al. Molecular Classification of Human Carcinomas by Use of Gene Expression Signatures. Cancer Research. 2001;61:7388–7393. [PubMed] [Google Scholar]
- 41.Yu K, et al. A Precisely Regulated Gene Expression Cassette Potently Modulates Metastasis and Survival in Multiple Solid Cancers. PLoS Genet. 2008;4:e1000129. doi: 10.1371/journal.pgen.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lockwood WW, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. doi: http://www.nature.com/onc/journal/v27/n33/suppinfo/onc200898s1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shankavaram UT, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Molecular Cancer Therapeutics. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. doi:10.1158/1535-7163.mct-06-0650. [DOI] [PubMed] [Google Scholar]
- 44.Su L-J, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyanaga A, et al. Antitumor activity of histone deacetylase inhibitors in non-small cell lung cancer cells: development of a molecular predictive model. Molecular Cancer Therapeutics. 2008;7:1923–1930. doi: 10.1158/1535-7163.MCT-07-2140. doi:10.1158/1535-7163.mct-07-2140. [DOI] [PubMed] [Google Scholar]
- 46.Balko J, et al. Gene expression patterns that predict sensitivity to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer cell lines and human lung tumors. BMC Genomics. 2006;7:289. doi: 10.1186/1471-2164-7-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. doi:10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 48.Smyth GK, Michaud J. l., Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. doi:10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 49.Hochberg Y, Benjamini Y. More powerful procedures for multiple hypothesis testing. Statistical Medicine. 1990:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 50.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proceedings of the National Academy of Sciences. 2001;98:31–36. doi: 10.1073/pnas.011404098. doi:10.1073/pnas.98.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Curtis SJ, et al. Primary Tumor Genotype Is an Important Determinant in Identification of Lung Cancer Propagating Cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. doi:10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.