SUMMARY
The NLRP3 inflammasome assembles in response to danger signals, triggering self-cleavage of procaspase-1 and production of the proinflammatory cytokine IL-1β. Although virus infection activates the NLRP3 inflammasome, the underlying events remain incompletely understood. We report that virus activation of the NLRP3 inflammasome involves the 2′,5′-oligoadenylate (2-5A) synthetase (OAS)/RNase L system, a component of the interferon-induced antiviral response that senses double stranded RNA and activates endoribonuclease RNase L to cleave viral and cellular RNAs. The absence of RNase L reduces IL-1β production in influenza A virus-infected mice. RNA cleavage products generated by RNase L enhance IL-1β production but require the presence of 2′,3′-cyclic phosphorylated termini characteristic of RNase L activity. Additionally, these cleavage products stimulate NLRP3 complex formation with the DExD/H-box helicase, DHX33, and mitochondrial adapter protein, MAVS, which are each required for effective NLRP3 inflammasome activation. Thus, RNA cleavage events catalyzed by RNase L are required for optimal inflammasome activation during viral infections.
INTRODUCTION
The innate immune system provides a rapid protective response against a wide range of cellular insults sensed by the host as danger signals, including those associated with viral infections. Nucleic acid motifs produced by viruses often function as a type of danger signal known as pathogen associated molecular patterns (PAMPs) that are absent in uninfected cells (Schroder and Tschopp, 2010). Viral nucleic acids that function as PAMPs include double-stranded (ds) RNA, 5′-triphosphorylated (triP) single-strand (ss) RNA containing segments rich in polyuridine (Saito et al., 2008), and cytoplasmic genomic DNA (Schroder and Tschopp, 2010). These nucleic acids are sensed by germline-encoded pathogen-recognition receptors (PRRs) that initiate innate immune signaling (Takeuchi and Akira, 2010). The PRRs for viral nucleic acids include membrane-associated Toll-like receptors (TLRs) for double-stranded (ds)RNA (TLR3), single-stranded (ss)RNA (TLR7/8) and dsDNA (TLR9) (Takeuchi and Akira, 2010), cytoplasmic DNA sensors (cGAS, DAI/Zbp1, IFI16, and AIM2) (Unterholzner, 2013) and cytoplasmic sensors of dsRNA and/or 5′-triP single-stranded (ss) RNA [RIG-I-like receptors (RLRs) RIG-I and MDA5 (Takeuchi and Akira, 2010), DExD/H-box helicases (DDX1, DDX21, DHX33, and DHX36) (Liu et al., 2014; Mitoma et al., 2013; Zhang et al., 2011), protein kinase R (PKR) (Yim and Williams, 2014), and 2′,5′-oligoadenylate synthetases (OAS) (Chakrabarti et al., 2011)].
The NOD-like receptor (NLR) family includes additional PRRs that sense microbial PAMPs in the cytosol (Schroder and Tschopp, 2010). Different NLR members (including NLRP1, NLRP3, and NLRC4) or non-NLR proteins (notably AIM2) function in large molecular machines known as inflammasomes. The NLRP3 inflammasome is implicated in the host response to many different types of RNA viruses [including influenza A virus (IAV) (Allen et al., 2009; Ichinohe et al., 2009; Kanneganti et al., 2006a; Thomas et al., 2009), hepatitis C virus (Negash et al., 2013), Sendai virus (Kanneganti et al., 2006a), encephalomyocarditis virus (Rajan et al., 2011), vesicular stomatitis virus (VSV) (Rajan et al., 2011), West Nile virus (Ramos et al., 2012) and HIV (Guo et al., 2014)] and DNA viruses [(adenovirus (Muruve et al., 2008), varicella zoster (Nour et al., 2011), and herpes virus (Nour et al., 2011)]. In addition, the NLRP3 inflammasome has been linked to inherited autoinflammatory diseases, known as cryopyrin-associated periodic syndromes (Lamkanfi and Dixit, 2014; Ozkurede and Franchi, 2012).
Activation of the NLRP3 inflammasome requires two types of signals. The first signal (priming) occurs when microbial ligands or endogenous cytokines induce transcription by NF-κB of the NLRP3 and proIL-1β genes. Signal 2 (direct activation from host damage) releases auto-inhibition of NLRP3 allowing interaction with ASC through pyrin domains (PYD) present in both proteins. NLRP3 then nucleates prion-like filaments of ASC providing a platform for the zymogen, procaspase-1 binding through caspase-activation-and-recruitment-domains (CARD) (Cai et al., 2014; Lu et al., 2014). Assembly of the inflammasome results in self-cleavage of procaspase-1 (p45), into its activated form (a heterodimer of two cleavage products, p10 and p20). Caspase-1 is a cysteine protease that cleaves proIL-1β and proIL-18 at aspartic residues to generate the mature cytokines that are secreted and function in host responses to infection. Inflammasome activation leads to pyroptosis or inflammatory cell death with pore formation and cell swelling, thereby eliminating virus-infected cells (Bergsbaken et al., 2009). Despite considerable progress in understanding the biochemistry and biology of inflammasomes, precisely how viruses induce activation of the NLRP3 inflammasome remains largely unresolved. Here we investigated a possible role for RNase L in regulation of the NLRP3 inflammasome during viral infections.
RNase L is an ubiquitous endoribonuclease for single-stranded (ss) RNA that is often activated in virus-infected cells (Chakrabarti et al., 2011). IFNs induce transcription of a family of oligoadenylate synthetase (OAS) genes that encode proteins which produce 2′,5′-linked oligonucleotides of the formula px5′A (2′p5′A)n; x = 1–3; n≥2 (2-5A) from ATP when stimulated by viral dsRNA (Kerr and Brown, 1978). 2-5A binds to monomeric, inactive RNase L causing dimers to form (Dong and Silverman, 1995) which, in the presence of ADP or ATP, become catalytically active (Huang, 2014). RNase L mediates its antiviral activity by cleaving viral and cellular ssRNAs predominantly after UpAp and UpUp dinucleotides (Wreschner et al., 1981). Activation of RNase L induces autophagy (Chakrabarti et al., 2012) and apoptosis (Zhou et al., 1997), both of which contribute to its antiviral effects. In addition to the direct antiviral effects of RNase L from degradation of viral and cellular RNA, RNase L also initiates signaling events that regulate type I IFN production. RNase L either positively or negatively regulates viral-induction of type I IFNs depending on basal levels of OAS and RNase L in different cell types (Banerjee et al., 2014; Malathi et al., 2007). The RNA cleavage products generated by RNase L often have double-stranded regions (because RNase L is a ssRNA-specific endoribonuclease), a 5′-hydroxyl and a 2′,3′-cyclic phosphoryl group characteristic of metal ion-independent ribonucleases (Cooper et al., 2014). Signaling to the IFN-β gene by viral or cellular RNA cleavage products from RNase L action requires RIG-I and/or MDA5 and MAVS (Malathi et al., 2007). The role of RNase L in regulating type I IFN production led us to examine the possible effects of RNase L on inflammasome activation during viral infections. Our findings indicate that RNase L enhances NLRP3 inflammasome activation during viral infections in a signaling pathway that includes both DHX33 and MAVS. Activation of the NLRP3 inflammasome in bone marrow-derived dendritic cells (BMDC) and in human THP-1-derived macrophages occurred in response to direct activation of RNase L with 2-5A or transfection with cellular or viral RNA cleaved by RNase L. Our findings suggest a role for RNase L in inflammatory signaling that enhances survival from viral infections.
RESULTS
RNase L enhances IL-1β induction and improves animal survival during IAV infections
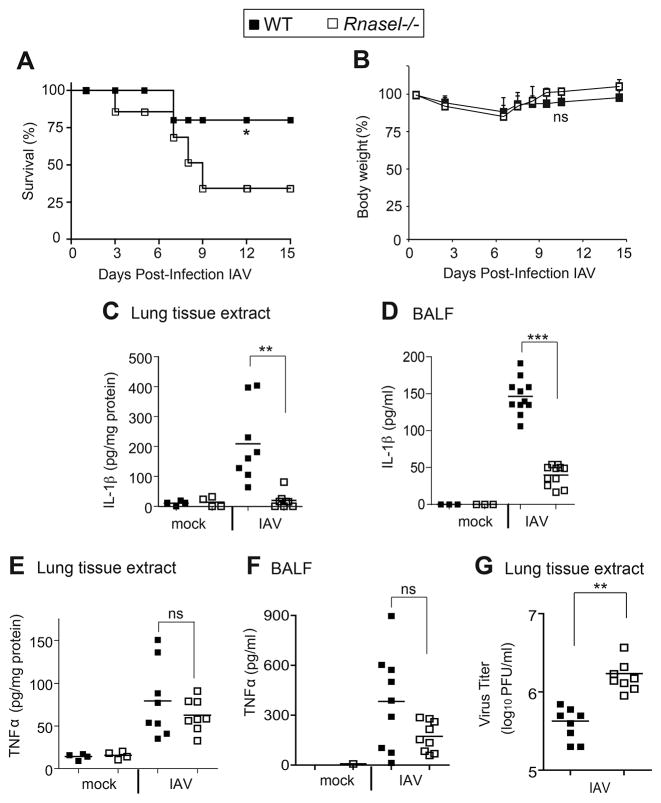
To determine the possible effect of RNase L on IL-1β induction during viral infections in vivo, wild type (wt) and Rnasel−/− mice were infected with the mouse-adapted influenza A virus (IAV), strain A/PR/8/H1N1, by the intranasal (i.n.) route. At 15 days post-infection (dpi), 80% of the wt animals survived infection compared with only 34% survival for the Rnasel−/− mice (Figure 1A). In contrast, there were no significant differences in body weights between the infected wt and Rnasel−/− mice (Figure 1B). Levels of IL-1β were measured both in lung tissue extracts and in brochoaveolar lavage fluid (BALF) at 2 dpi (Figure 1C,D). Deletion of RNase L decreased mean levels of IL-1β obtained after IAV infections by 90% and 75% of wt levels in lung tissue and BALF, respectively. IAV-induced levels of TNFα were moderately reduced by deletion of RNase L in lung tissue and BALF, possibly due to secondary induction of TNFα by IL-1β (Thomas et al., 2009) (Figure 1E,F). Previously, we reported decreased IFN-β production in Rnasel−/− mice after infection with the picornavirus, encephalomyocarditis virus, or with the rhabdovirus, vesicular stomatitis virus (VSV) (Malathi et al., 2007). Similarly, after IAV infection there was significantly less IFN-β in BALF from Rnasel−/− mice compared with wt mice (Figure S1). At 7 dpi, viral yields were modestly increased (by 4-fold) in the lungs of Rnasel−/− mice compared to wt mice (Figure 1G). These results show that, in IAV-infected mice, RNase L has prosurvival and antiviral effects that correlate with increased production of IL-1β and IFN-β
Figure 1. RNase L deficiency in mice enhances the lethality of IAV while decreasing the induction of IL-1β.
(A) Survival of wt and Rnasel−/− mice infected i.n. with IAV. (B) Mean body weights corresponding to panel (A). (C,D) IL-1β and (E,F) TNF-α levels determined by ELISAs at 2 days post-infection (dpi) with IAV in (C,E) lung homogenates and (D,F) BALF. (G) Viral titers from lungs harvested at 7 dpi, determined by plaque assays. horizontal lines, mean ± standard deviation (SD). Significance was determined by (A,B) Kaplan-Meier analysis or (C–G) two-tailed Student’s t tests. *** p<0.001; **p< 0.01; *p<0.05; ns, not significant. Numbers of mice used: (A,B) n= 14; (C,E) n=8; (D) n=11; (F) n=9; and (G) n=8. See also Figure S1.
RNase L enhances inflammasome activation during viral infections
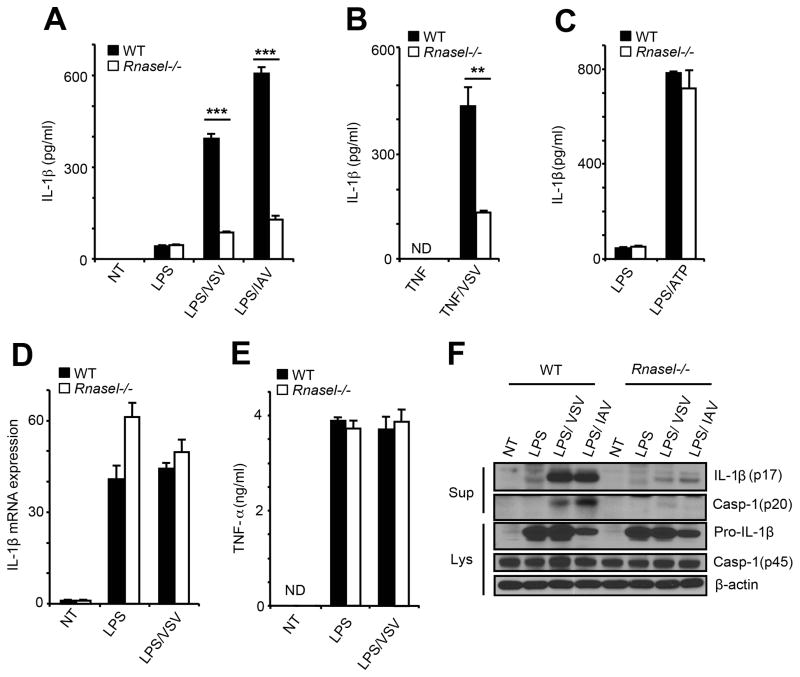
Secretion of IL-1β following LPS priming and VSV or IAV infections was decreased by 80% in Rnasel−/− BMDC (Figure 2A). LPS treatment alone primed inflammasomes, but only minimally induced IL-1β. In addition, in the absence of LPS, VSV or IAV alone failed to induce IL-1β (Figure S2). The RNase L effect on IL-1β induction was apparent between 8 and 18 hours post-infection with VSV, and between 24 and 36 hours of infection with IAV (Figure S2). Similarly, deletion of RNase L reduced IL-1β induction by 70% after priming with TNFα followed by VSV infection (Figure 2B). ATP activates the NLRP3 inflammasome through the P2X7 ion channel leading to potassium efflux (Franchi et al., 2007). RNase L had no effect on IL-1β induction by LPS and ATP, suggesting that RNase L is not a general regulator of inflammasome activation (Figure 2C). To determine if RNase L affects inflammasome priming (signal 1), levels of IL-1β mRNA were measured by qRT-PCR. However, RNase L had no effect on IL-1β mRNA induction in BMDC by LPS alone or by LPS followed by VSV infection (Figure 2D). Also, there was no effect of RNase L on TNFα induction by LPS or by LPS/VSV (Figure 2E). To assess the effect of RNase L on inflammasome activation (signal 2), processing of procaspase-1 (45 kDa) to one of its self-cleavage products (the 20 kDa subunit of mature active caspase-1), and of proIL-1β (31 kDa) to mature IL-1β (17 kDa), was monitored by immunoblotting (Figure 2F). LPS treatment by itself failed to produce either activated caspase-1 p20 or mature IL-1β (p17). In contrast, LPS plus either VSV or IAV infection of wt BMDC caused a large increase in levels of mature IL-1β (p17) and a clearly detectable increase in caspase-1 p20. However, in Rnasel−/− BMDC, LPS treatment followed by VSV or IAV infections produced only minimal amounts of IL-1β p17 or caspase-1 p20 (Figure 2F). These findings are consistent with a role for RNase L in inflammasome activation (signal 2), but not in priming (signal 1), during these viral infections.
Figure 2. Inflammasome activation in response to viral infections is reduced in the absence of RNase L.
IL-1β levels by ELISAs from (A) wt and Rnasel−/− BMDC primed with LPS for 6 hrs and infected with VSV for 12 hrs or IAV for 24 hrs, (B) wt and Rnasel−/− BMDC primed with TNFα for 6 hrs and infected with VSV for 12 hrs and (C) BMDC primed with LPS for 6 hrs and treated with extracellular ATP (0.5 mM) for 1 hr. (D) IL-1β mRNA levels by qRT-PCR and (E) TNFα levels by ELISAs from LPS primed wt and Rnasel−/− BMDCs without or with VSV infection. (F) Cleavage of caspase-1 and pro-IL-1β in LPS primed wt and Rnasel−/− BMDCs without or with VSV or IAV infection determined in immunoblots. Cell lysates (Lys); cell supernatants (Sup); error bars, SD; **, p<0.01; ***, p<0.001 by two-tailed Student’s t tests. See also Figure S2.
Involvement of MAVS but not RIG-I and/or MDA5 in viral activation of the NLRP3 inflammasome
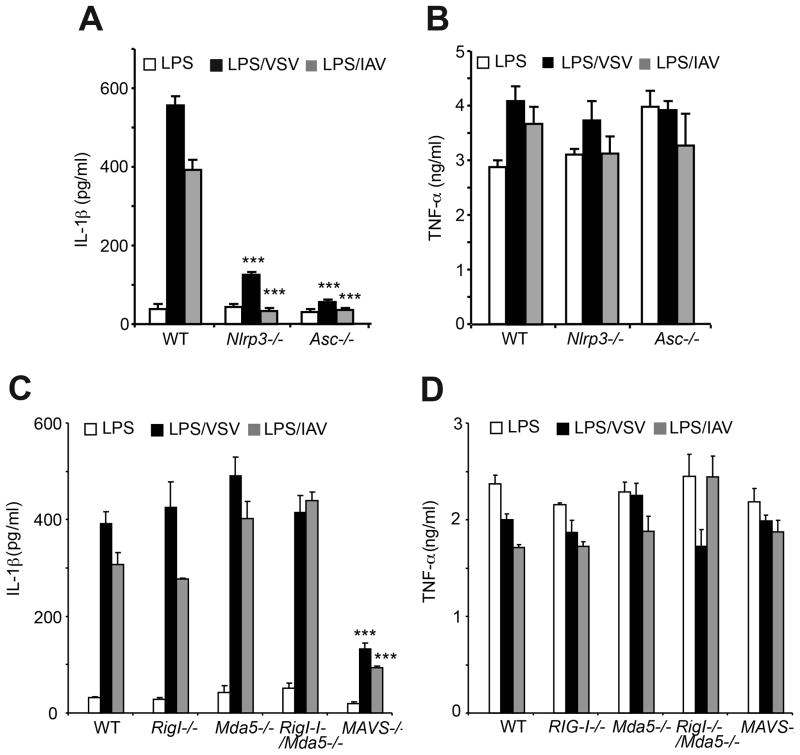
In BMDC, viral induction of IL-1β, but not TNF-α, was dependent on both NLRP3 and ASC (Figure 3A&B). In addition, MAVS, but not RIG-I or MDA5 either alone or in combination (double knockout), was required for optimal induction of IL-1β by VSV or IAV (Figure 3C). TNF-α levels were unaffected by RIG-I and/or MDA5 or by MAVS (Figure 3D). In contrast to effects of viruses, IL-1β induction by ATP was independent of MAVS (Figure S3). In control experiments, we ruled out differences in cell death rates as a cause of the reduction in IL-1β production observed in LPS-treated, virus-infected Mavs−/− BMDM (data not shown). These results show that activation of the NLRP3 inflammasome during infection with either VSV or IAV is dependent on MAVS for optimal activation, but not on RIG-I and/or MDA5.
Figure 3. Involvement of NLRP3, ASC, and MAVS in IL-1β induction but not TNFα induction in response to viral infections.
(A,B) LPS primed wt, Nlrp3−/− and Asc−/− BMDC were infected with VSV or IAV. (C,D) LPS primed wt, Rig I−/−, Mda5−/−, RigI−/−/Mda5−/−, and Mavs−/− BMDC were infected with VSV or IAV. IL-1β and TNFα levels were measured by ELISAs. Error bars, SD; ***, p<0.001 by two-tailed Student’s t tests. See also Figure S3.
Activation of RNase L stimulates the NLRP3 inflammasome
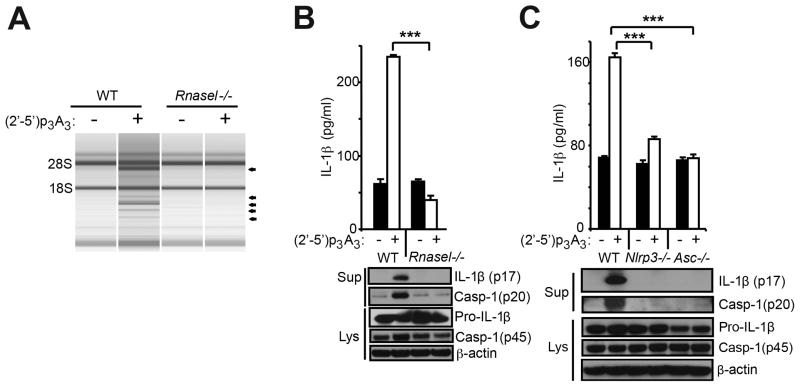
To determine the effect of RNase L activation on the NLRP3 inflammasome, BMDC from wt and Rnasel−/− mice were transfected with (2′-5′)p3A3 (2-5A), a potent and highly specific activator of RNase L (Chakrabarti et al., 2011). Characteristic RNase L-mediated rRNA cleavage products (Silverman et al., 1983) were observed following (2′-5′)p3A3 transfection of wt but not Rnasel−/− BMDC (Figure 4A). Similarly, IL-1β induction was observed after (2′-5′)p3A3 transfection of wt but not Rnasel−/− BMDC (Figure 4B, upper panel). In addition, immunoblots of cell supernatant (Sup) showed (2′-5′)p3A3 induction of mature IL-1β (p17) and of cleaved caspase-1 (p20) in wt but not in Rnasel−/− BMDC (Figure 4B, lower panel). (2′-5′)p3A3 induction of IL-1β was dependent on both NLRP3 and ASC (Figure 4C). These results show direct activation of RNase L with (2′-5′)p3A3 causes IL-1β production in BMDC.
Figure 4. IL-1β production by direct activation of RNase L is dependent on both NLRP3 and ASC.
BMDC were primed with Pam3Csk4 (200 ng/ml) for 16 hrs and mock transfected or transfected with (2′-5′)p3A3 for additional 8 hrs as indicated. (A) rRNA cleavage in response to (2′-5′)p3A3 transfection was monitored in RNA chips. (B,C) IL-1β and cleaved caspase-1 was measured in cell supernatants (Sup), whereas proIL-1β, procaspase-1 and β-actin were measured in cell lysates (Lys). Upper panels, ELISAs; lower panels, immunoblots. Error bars, SD; ***; p<0.001 by two-tailed Student’s t tests.
RNA cleavage products activate the NLRP3 inflammasome
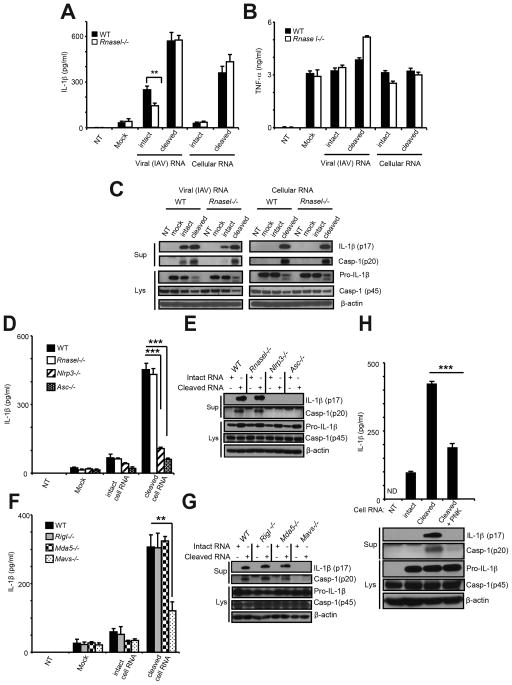
We next investigated effects of RNase L-cleaved RNA on the NLRP3 inflammasome. Either IAV genomic RNA or cellular RNA were cleaved in vitro with purified, recombinant human RNase L activated by (2′-5′)p3A3. Kinetics of RNA cleavage was monitored with a FRET probe in parallel reactions containing cellular or IAV genomic RNA (Figure S4A,B). In addition, cleavage of cellular and IAV genomic RNA was monitored in RNA chips (Agilent) (Figure S4C,D, respectively).
Transfection of intact IAV genomic RNA into Pam3Csk4-primed wt and Rnasel−/− BMDC induced mean averages of 249 and 143 pg/ml of IL-1β, respectively (Figure 5A). However, prior cleavage of IAV RNA by RNase L increased IL-1β induction to mean averages of 572 and 576 pg/ml in wt and Rnasel−/− BMDC, respectively (Figure 5A). The similar levels of IL-1β in these two cell types were consistent with effects of the cleaved RNAs acting in a pathway downstream of RNase L. Intact cellular RNA failed to induce IL-1β in either wt or RNasel−/− BMDC. In contrast, cleaved cellular RNA induced mean averages of 360 and 435 pg/ml of IL-1β in wt and Rnasel−/− BMDC, respectively. There was no effect of IAV genomic or cellular RNA, either intact or cleaved, on TNF-α levels following Pam3Csk4 treatment (Figure 5B). Immunoblot analysis showed enhanced inflammasome activation when cleaved IAV RNA or cleaved cellular RNA were transfected compared with intact RNAs (Figure 5C). The effect was more evident in the case of cellular RNA in that there was no detectable processing of proIL-1β or procaspase-1 p45 when transfecting intact RNA, but pronounced processing of both proteins in response to cleaved RNA. Nlrp3−/− and Asc−/− BMDC were defective for IL-1β induction and for procaspase-1 and proIL-1β processing in response to cleaved RNA (Figure 5D,E). In addition, there was a partial dependence on MAVS (a decrease of 62% in Mavs−/− BMDC), but not RIG-I or MDA5, for induction of IL-1β and for NLRP3 activation (Figure 5F,G). Also, there was no effect of a double knockout of RIG-I and MDA5 on IL-1β induction by RNase L-cleaved cellular RNA, indicating that these RLRs were non-redundant in their lack of effect (Figure S4E). TNF-α levels were unaffected by either cleaved or uncleaved cellular RNA (Figure S4F,G).
Figure 5. RNase L generated RNA cleavage products bearing 2′,3′-cyclic phosphates induce IL-1β production.
(A–C) wt and Rnasel−/− BMDC were primed with Pam3Csk4 (200 ng/ml) for 16 hrs and transfected with intact or RNase L-cleaved IAV RNA or cellular RNA for 8 hrs. (D–G) BMDC of different genotypes (as indicated) were primed with Pam3Csk4 and transfected with uncleaved or RNase L-cleaved cellular RNAs for 8 hrs. (H) Intact, cleaved, or cleaved and PNK-treated cellular RNA were transfected into Pam3Csk4 treated BMDC. (A,B,D,F,&H upper) ELISAs; (C,E,G, and H lower) immunoblots. Cell supernatants (Sup); cell lysates (Lys). Error bars, SD;. **p<0.01;***, p<0.001 by two-tailed Student’s t tests. See also Figure S4.
To determine which moieties of the cleaved RNAs might contribute to inflammasome activation, the 2′,3′-cyclic phosphoryl termini were removed with T4 polynucleotide kinase (PNK) (Figure S4H,I) (Nedialkova et al., 2009). The efficacy of PNK in removing 2′,3′-cyclic phosphates was determined by cleaving the synthetic RNA substrate, C11U2C7 (Carroll et al., 1996), with RNase L following by incubation of the cleavage product, C11U2>2′,3′p, with PNK, or as a control shrimp alkaline phosphatase (SAP) which requires longer incubation to remove cyclic 2′,3′-phosphates (Nedialkova et al., 2009). Removal of 2′,3′-cyclic phosphates was monitored by ligation of the free 3′-OH end to [32P]pCp with T4 RNA ligase (Nedialkova et al., 2009). PNK removed the 2′,3′-cyclic phosphates within 2 min, whereas SAP required at least 60 min of incubation. IL-1β induction in BMDC was decreased from mean averages of 422 to 188 pg/ml when the 2′,3-cyclic phosphates were removed with PNK from RNase L-cleaved cellular RNA (Figure 5H, upper panel). Furthermore, processing of proIL-1β and procaspase-1 was reduced sharply when the dephosphorylated cleaved RNA was used (Figure 5H, lower panel). These findings support a role for the 2′,3-cyclic phosphoryl groups in recognition of the cleaved RNA during signaling to the NLRP3 inflammasome.
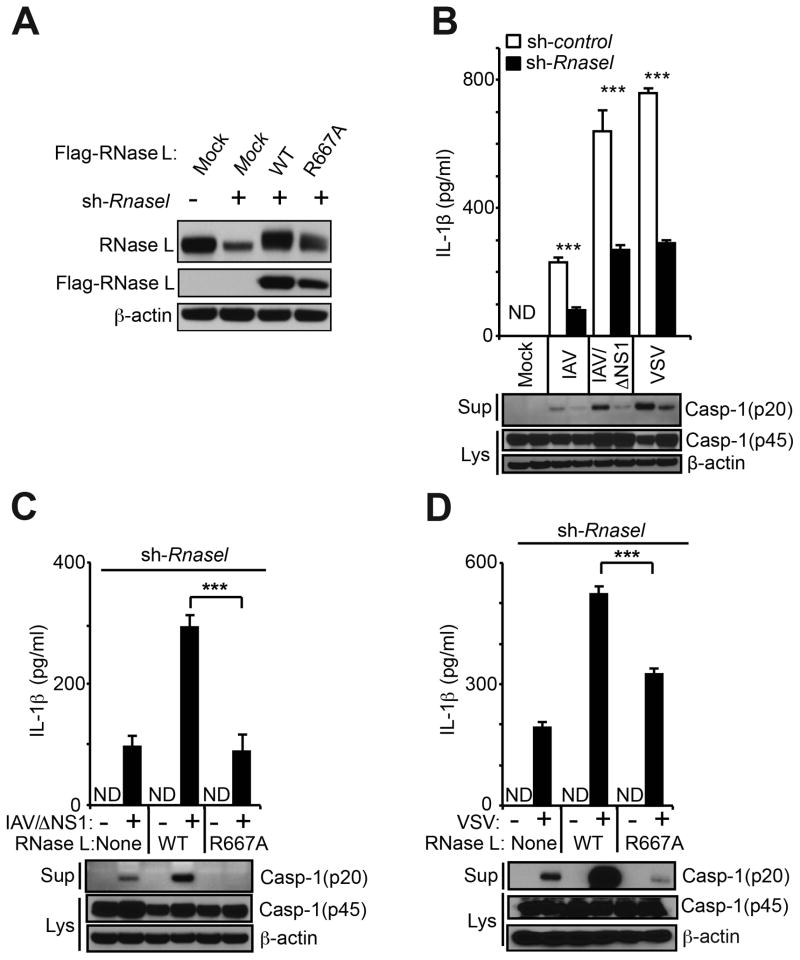
The enzymatic activity of RNase L is required to stimulate the NLRP3 inflammasome
To investigate the involvement of the endoribonuclease activity of RNase L in NLRP3 inflammasome activation, RNase L was first stably-depleted in human THP-1 macrophages (Tsuchiya et al., 1980) with lentivirus expressing shRNA against the 3′-UTR of RNase L mRNA (sh-Rnasel) followed by drug selection (Figure 6A). Subsequently, the RNase L knockdown cells were reconstituted by expressing wt RNase L or mutant R667A RNase L [which completely inactivates the ribonuclease function (Dong et al., 2001)] from constructs that lack the natural 3′-UTR, including the shRNA target site (Figure 6A). Viral infections were performed on the THP-1 cells expressing control shRNA (sh-control) or sh-Rnasel. The parental IAV strain (A/PR/8/H1N1) produced relatively low levels of IL-1β in the sh-control cells that were further reduced by 65% in the sh-Rnasel cells (Figure 6B). An IAV/ΔNS1 mutant virus, lacking NS1 protein that counteracts OAS-RNase L (Min and Krug, 2006), produced a more robust induction of IL-1β in the THP-1 cells. Treatment of THP-1 cells with sh-Rnasel reduced IL-1β induction by IAV/ΔNS1 by 58% compared to the sh-control levels (Figure 6B). Similar partial dependency of IL-1β induction on RNase L in THP-1 cells was obtained with VSV infection. Also, IAV/ΔNS1 produced robust production of IL-1β in wt but not in Rnasel−/− mouse peritoneal macrophages (Figure S5A). Reconstitution of the sh-Rnasel THP-1 cells with wt RNase L enhanced IL-1β induction to 308% of the sh-control levels in response to IAV/ΔNS1 (Figure 6C, upper panel). In contrast, the R667A mutant of RNase L, which lacks ribonuclease activity (Dong et al., 2001), was deficient in IAV/ΔNS1 induction of IL-1β. In addition, IAV/ΔNS1 induction of procaspase-1 cleavage was enhanced by wt RNase L, but not by R667A mutant RNase L (Figure 6C, lower panel). In VSV-infected THP-1 cells, wt RNase L enhanced IL-1β induction by 271% whereas the R667A mutant RNase L was partially deficient for IL-1β induction (Figure 6D, upper panel). In these cells, procaspase-1 processing was stimulated by wt but not by R667A RNase L (Figure 6D, lower panel). To further validate the involvement of RNase L in inflammasome activation in THP-1 cells, RNase L levels were depleted with siRNA. Procaspase-1 processing in response to IAV/ΔNS1 infection was reduced in cells depleted for RNase L (Figure S5B). IL-1β induction was decreased in the si-Rnasel treated cells in response to IAV/ΔNS1 infection, but not in response to LPS/ATP (Figure S5C). These findings demonstrate that the NLRP3 inflammasome is dependent on the nuclease function of RNase L. In addition, these results extend the stimulatory effect of RNase L on inflammasome activation to human cells.
Figure 6. A functional nuclease domain in RNase L is required for inflammasome stimulation.
(A) RNase L levels in THP-1 cells or expressing sh-Rnasel alone or with Flag-tagged wt or R667A mutant RNase L cDNAs. Immunoblots were probed with (upper panel) monoclonal antibody to human RNase L, (middle panel) antibody to Flag epitope, and (bottom panel) antibody to β-actin. (B) IL-1β and caspase-1 (p20) in cell supernatants (Sup) and procaspase-1 (p45) and β-actin in cell lysates (Lys) from THP-1 cells expressing sh-control or sh-Rnasel and infected with IAV, IAV/ΔNS1 or VSV. (C,D) THP-1 cells expressing Rnasel shRNA or reconstituted with wt or mutant R667A RNase L infected with (C) IAV/ΔNS1 or (D) VSV. (B–D), upper panels, ELISAs; lower panels, immunoblots. Error bars, SD; ***, p<0.001 by two-tailed Student’s t tests. See also Figure S5.
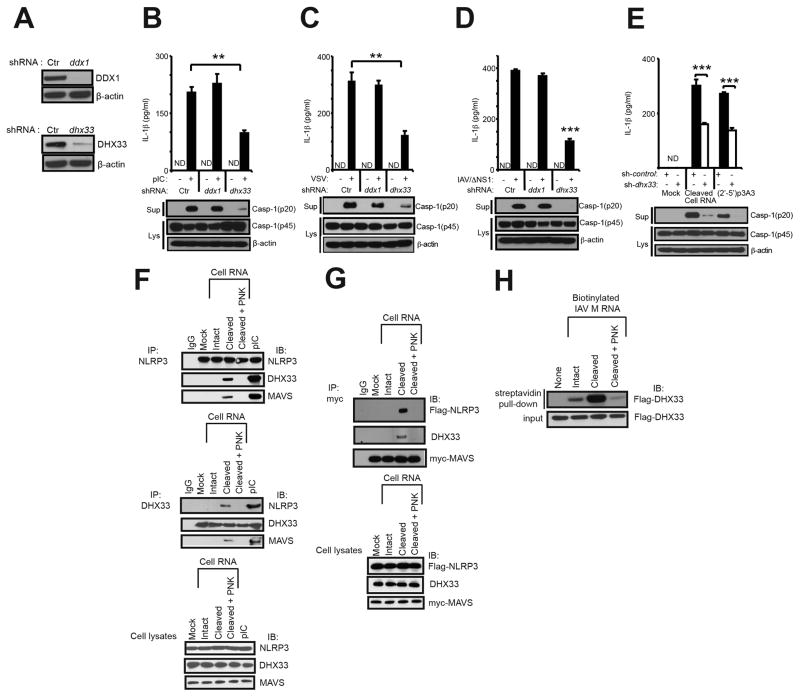
Involvement of DHX33 in RNase L signaling to the NLRP3 inflammasome
The DExD/H-box RNA helicase, DHX33, was recently shown to bind dsRNA and to interact with and activate NLRP3 (Mitoma et al., 2013). To determine whether DHX33 was a sensor for RNA cleavage products of RNase L, we depleted either DHX33, or as a control the helicase DDX1, in THP-1 cells by means of shRNA expressed from recombinant lentiviruses. DDX1 is a helicase family member implicated as a dsRNA sensor and as an inducer of type-I IFN (Zhang et al., 2011). Efficient knockdown of both proteins was obtained as determined in immunoblots (Figure 7A). Transfection with the synthetic dsRNA, poly (rI):poly (rC) (pIC), potently induced IL-1β in control shRNA (sh-control) cells and in DDX1-depleted (sh-Ddx1) cells. In contrast, IL-1β induction by pIC was reduced by 52% in the DHX33-depleted (sh-Dhx33) cells (Figure 7B, upper panel), similar to previously reported results by others (Mitoma et al., 2013). Induction of IL-1β by either VSV or IAV/ΔNS1 was unaffected by reducing expression of DDX1, whereas depleting DHX33 reduced IL-1β levels by 61% and 71%, respectively, of the sh-control levels (Figure 7C,D, upper panels). Cleavage of procaspase-1 (p45) to p20 was also deficient in DHX33-depleted, pIC-treated or virus-infected cells (Figure 7B–D, lower panels). Similarly, IL-1β induction following transfection of either cleaved cellular RNA or (2′-5′)p3A3 was reduced by 47% and by 51% of the sh-control levels in DHX33-depleted cells (Figure 7E, upper panel). Procaspase-1 cleavage was also deficient in the DHX33-depleted THP-1 cells transfected with either cleaved RNA or (2′-5′)p3A3 (Figure 7E, lower panel).
Figure 7. Involvement of DHX33 in RNase L-mediated inflammasome activation.
(A) shRNA mediated depletion of DDX1 or DHX33 in THP-1 macrophages. (B–E) THP-1 macrophages expressing different shRNAs [sh-control, sh-Ddx1 or sh-Dhx33] were (B) transfected with pIC (2 μg/ml for 8 hrs), (C) infected with VSV, (D) infected with IAV/ΔNS1, and (E) mock transfected or transfected with cell RNA cleaved with RNase L or transfected with (2′-5′)p3A3. Upper panels, ELISAs; lower panels, immunoblots (IB). (F) THP-1 macrophages were transfected with intact, cleaved, cleaved and PNK-treated cellular RNA or pIC. Immunoprecipitations (IP) were with anti-NLRP3, anti-DHX33, or an isotype control IgG. Cell lysates were used as controls (bottom panel). (G) HEK293T cells were co-transfected with myc-MAVS and flag-NLRP3 cDNAs. After 72 hrs, cells were transfected with intact, cleaved or cleaved and PNK treated cellular RNA. IPs were with anti-myc followed by immunoblotting (IB) with anti-flag, anti-DHX33 or anti-myc antibodies. Cell lysates were used as controls (lower panel). (H) Purified flag-DHX33 was incubated with biotinylated IAV M gene RNA (intact, cleaved, or cleaved and PNK-treated). Biotinylated RNAs were precipitated with streptavidin beads followed by IB with anti-flag antibody to detect DHX33. cell supernatants (Sup); cell lysates (lys). Error bars, SD; **p<0.01;***, p<0.001 by two-tailed Student’s t tests. See also Figure S6.
Interaction of DHX33, MAVS and NLRP3 in response to cleaved RNA
To determine whether RNase L-mediated RNA cleavage products cause DHX33, NLRP3 and MAVS to associate in intact cells, immunoprecipitations were performed (Figure 7F & G). Total cell RNA was left intact, cleaved with RNase L, or cleaved and dephosphorylated with PNK. THP-1 macrophages were then transfected with these RNAs or with pIC and immunoprecipitations (IP) performed on cell extracts followed by immunoblotting. After transfection with cleaved cell RNA or with pIC, IP of NLRP3 pulled-down both DHX33 and MAVS, while IP of DHX33 pulled-down NLRP3 and MAVS (Figure 7F). Similar results were obtained with pIC transfections as previously reported (Mitoma et al., 2013). In contrast, no complexes of DHX33-MAVS-NLRP3 were observed in mock transfected cells, or in cells transfected with intact cell RNA or cleaved and PNK-treated cell RNA. In addition, IP of myc-tagged MAVS pulled-down flag-tagged NLRP3 and DHX33 in HEK293T cells in response to cleaved cell RNA, but not with intact or cleaved and PNK-treated cell RNA (Figure 7G). These results show that a complex containing DHX33, MAVS and NLRP3 is formed in response to RNase L-cleaved RNA that was dependent on the PNK-sensitive 2′,3′-cyclic phosphoryl group.
RNase L-cleaved RNA directly interacts with DHX33
To determine if cleavage by RNase L directly affects the affinity of RNA for DHX33, biotinylated RNA was synthesized in vitro from the IAV M gene cDNA and incubated with purified flag-tagged DHX33. The biotinylated RNA was then pulled-down with streptavidin beads followed by immunoblotting for DHX33 [as described by (Mitoma et al., 2013)]. Interaction between RNA and DHX33 was greatly increased following cleavage by RNase L (Figure 7H). In contrast, PNK treatment resulted in a loss of affinity for DHX33. The interaction was blocked with unmodified cleaved RNA, whereas unlabeled intact RNA reduced but did not completely block binding and unlabeled RNA that was cleaved and PNK-treated was ineffective (Figure S6). These findings indicate RNase L-generated RNA cleavage products have enhanced affinity for DHX33, dependent on the PNK-sensitive cyclic phosphoryl groups at the 2′,3′-termini
DISCUSSION
Our results show that RNase L activation in immune cells enhances activation of NLRP3 inflammasomes resulting in increased secretion of IL-1β. Inflammasome activation during IAV infections was previously linked to a reduction in tissue damage due to effects on cell recruitment and tissue repair in the lung (Allen et al., 2009; Iwasaki and Pillai, 2014; Thomas et al., 2009). In particular, epithelial necrosis and pulmonary functions were more severely compromised in Nlrp3 deficient mice in response to IAV infections (Thomas et al., 2009). In addition, a prior study showed that IL-1β promotes adaptive immunity thus controlling IAV infections and is protective in a mouse model (Pang et al., 2013). We showed here that during IAV infections levels of IL-1β were significantly elevated in lung tissue and BALF from wt compared with Rnasel−/− mice. Therefore, it is likely that RNase L contributes to animal survival from IAV infections at least partly by enhancing IL-1β production. When RNase L was absent, infections with either VSV or IAV resulted in reduced cleavage of procaspase-1 and proIL-1β to their mature forms. Similar to some previous studies, we observed that IL-1β induction by either VSV or IAV requires both inflammasome proteins, NLRP3 and ASC (Allen et al., 2009; Ichinohe et al., 2009; Rajan et al., 2011; Thomas et al., 2009). A role for RLRs is more controversial in that contrasting studies showed either involvement (Poeck et al., 2010) or no involvement (Rajan et al., 2011) of RIG-I in caspase-1 dependent inflammasome activation in response to VSV. Our data agree with the latter study in that IL-1β secretion during VSV infections was completely independent of RIG-I. Indeed, single or combined deficiencies in RIG-I and/or MDA5 failed to impair IL-1β secretion in response to either VSV or IAV, indicating these RNA helicases are non-redundant in their lack of effect. In contrast to RLRs, the adapter MAVS was required for optimal activation of induction of IL-1β in response to either IAV or VSV infections, as well as for IL-1β secretion upon transfection with RNase L-generated RNA cleavage products. MAVS was previously implicated in NLRP3 inflammasome activation in response to different stimuli (Subramanian et al., 2013), including Sendai virus (Park et al., 2013). MAVS promotes localization of the NLRP3 inflammasome to the mitochondria (Park et al., 2013; Subramanian et al., 2013). However, there was no effect of NLRP3, ASC, or MAVS on viral induction of TNF-α, a cytokine that is not processed by inflammasomes. In addition, whereas a previously study show MAVS dependence for NLRP3 activation by ATP (Subramanian et al., 2013), we found no effect of MAVS deletion on NLRP3 activation by ATP [(Figure S3) and (Franchi et al., 2014)].
Prior studies investigated how priming (signal 1) and activation (signal 2) of the NLRP3 inflammasome occurs during IAV infections [reviewed in (Iwasaki and Pillai, 2014)]. Signal 1 (priming) was not observed in response to VSV or IAV as determined by a lack of IL-1β secretion unless an NF-κB stimulus such as TNF-α or LPS was used. However, after priming both VSV and IAV resulted in a robust signal 2 in wt but not in Rnasel−/− BMDC. Direct and highly specific activation of RNase L by the 2-5A triadenylate, (2′-5′)p3A3, triggered NLRP3 inflammasome activation as measured by processing of proIL-1β and procaspase-1 and by IL-1β secretion. However, RNase L activation by 2-5A did not induce IL-1β in BMDC lacking either NLRP3 or ASC. Evidence points to a role for the RNase L-generated RNA cleavage products in inflammatory signaling. Whereas intact IAV genomic RNA, but not intact cellular RNA, modestly stimulated NLRP3, RNA cleavage products from RNase L-mediated digestion of either type of RNA highly stimulated the NLRP3 inflammasome. Activation by the RNA cleavage products was dependent on NLRP3 and ASC, with a requirement for MAVS for optimal activity, whereas RIG-I and/or MDA5 had no effect. During IAV infections in vivo, commensal bacteria are believed to provide the priming step (signal 1) (Ichinohe et al., 2011), whereas signal 2 is provided by viral ssRNA (Thomas et al., 2009), proton flux through the viral-encoded matrix 2 (M2) trans-Golgi ion channel (Ichinohe et al., 2010), and amyloid-like structures of the IAV virulence protein, PB1-F2 (McAuley et al., 2013). Our results suggest that RNA cleavage products from RNase L activity during IAV infections are additional stimuli for NLRP3 inflammasome activation.
RNase L is often activated in virus-infected cells where it cleaves single-stranded regions of host and viral RNAs, predominantly after UU or UA dinucleotides leaving as termini 5′-hydroxyls and 2′,3′-cyclic phosphates (Cooper et al., 2014; Wreschner et al., 1981). We showed here that optimal activation of the NLRP3 inflammasome by these RNA cleavage products depends on the 2′,3′-cyclic phosphate termini. IRE1, a kinase-endoribonuclease that functions in the unfolded protein response, also produces 2′,3′-cyclic phosphorylated termini which are immunostimulatory (Eckard et al., 2014). It remains to be determined whether other features of the RNA cleavage products from RNase L activity contribute to the effect, in particular double-stranded regions which are left intact by this enzyme.
Previously, the NLRP3 inflammasome was shown to be activated in macrophages by transfection of either viral RNA or the synthetic dsRNA, pIC (Kanneganti et al., 2006a). The mechanism whereby NLRP3 senses viral dsRNA is unknown, but it is believed to be indirect, as has been suggested for sensing microbial PAMPs by NLR-like proteins in plants (Mackey et al., 2003). DHX33 is a DExD/H-box RNA helicase with a helicase C type domain, similar to RIG-I and MDA5, but lacking a CARD domain (Liu et al., 2014). DHX33 interacts with dsRNA, MAVS and NLRP3 and its depletion rendered human macrophages resistant to caspase-1 activation and secretion of IL-1β and IL-18 (Liu et al., 2014; Mitoma et al., 2013). DHX33 is also implicated in dsRNA signaling to the type I IFN genes in response to either pIC or reovirus (Liu et al., 2014). Our data indicate involvement of RNase L-mediated RNA cleavage products generated in virus-infected cells in NLRP3 activation through DHX33 and MAVS. Cleaved RNA products generated by RNase L caused formation of a complex containing DHX33, MAVS and NLRP3. Current findings are consistent with a previous study showing that pIC stimulated a complex containing DHX33, NLRP3 and ASC (Mitoma et al., 2013). Moreover, the RNA cleavage products directly bind to DHX33. Both immune complex formation and affinity for DHX33 were ablated by PNK treatment of the cleaved RNA, which removes 2′,3′-cyclic phosphate from the RNA. Our results suggest a model in which direct binding of the cleaved RNA to DHX33 stimulates association with MAVS and NLRP3 inducing oligomerization and recruitment of procaspase-1 through ASC. Recently it was shown that dsRNA acts through NLRP3 and MAVS to induce membrane damage and potassium efflux to activate NLRP3 (Franchi et al., 2014). Perhaps the DHX33-MAVS-NLRP3 complex induced by cleaved RNA is also activated by potassium efflux. However, whereas RIG-I and MDA5 are redundant for pIC stimulation of the NLRP3 inflammasome (Franchi et al., 2014), that was not the case for RNA cleavage products generated by RNase L, suggesting at some level immune signaling by these different RNA species are distinct (Figure S4E).
RNase L is a relatively general antiviral enzyme capable of inhibiting a broad range of RNA and DNA viruses (Silverman, 2007). Future studies will determine the range of viruses that stimulate inflammasomes through RNase L. Also, structural or sequence requirements in the RNA cleavage products may be further defined. Additional co-factors that contribute to the immune signaling pathway may be explored, such as possible involvement other DExD/H-box helicases. It will also be interesting to know whether OAS and RNase L affect other types of NLR- or non-NLR-inflammasomes. Taken together, our findings suggest that during viral infections RNase L-generated RNA cleavage products are sensed by DHX33 leading to NLRP3 activation.
EXPERIMENTAL PROCEDURES
Cell culture
Bone marrow cells isolated from hind limbs of 8–12 weeks old C57BL6 wt, Rnasel−/− (Zhou et al., 1993), Nlrp3−/− (Kanneganti et al., 2006b), Asc−/− (Ozoren et al., 2006), RigI−/− (Kato et al., 2005), Mda5−/− (Gitlin et al., 2006), RigI−/−Mda5−/− (Errett et al., 2013), and Mavs−/− mice (see Supplemental Information) were grown and differentiated in Iscove’s modified Dulbecco’s medium (IMDM) containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 1% non-essential amino acids, 50 μM β-mercaptoethanol and 20 ng/ml of GM-CSF for 9 days before they were used for experiments. THP-1 cells (ATCC TIB-202) were maintained in RPMI-1640 medium containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 1% non-essential amino acids and 50 μM β-mercaptoethanol. HEK293T cells and MDCK cells were maintained in DMEM medium with 10% (v/v) heat inactivated FBS.
Viruses
IAV A/PR/8/34 (H1N1) grown in 11-day embryonated chicken eggs and IAV/ΔNS1 lacking the NS1 gene were kindly provided by Adolfo Garcia-Sastre (New York) (Gack et al., 2009). IAV/ΔNS1 was grown in MDCK-NS1 cells (Gack et al., 2009) with DMEM medium supplemented with 0.3 % bovine albumin, 1% penicillin streptomycin, and 1 μg/ml of L- (tosylamido-2-phenyl) ethyl chloromethyl ketone(TPCK)-treated trypsin (Sigma-Aldrich). Vesicular stomatitis virus (VSV, Indiana strain) (a gift from Dr. Amiya Banerjee, Cleveland, OH) was grown in CV1 cells.
Viral infections of mice
Male mice (6 wk of age) anesthetized with ketamine–xylazine were infected intranasally (i.n.) with 1×105 pfu of IAV (in 20 μl). BALF was collected by washing the trachea and lungs twice and injecting a total of 2 ml PBS containing 0.1% BSA (Ichinohe et al., 2009). BALF was used for cytokine measurements. Lungs of infected mice were excised on 7 days post infection and homogenized in 1 ml PBS using a mechanical homogenizer. The viral titers were quantified by plaque assay on MDCK cells as described previously (Manicassamy et al., 2010). Experiments involving mice were performed under an approved IACUC protocol from the Cleveland Clinic.
Viral infection of cells
BMDC were primed with LPS (100 ng/ml) or TNF-α (25 ng/ml) for 6 hrs, washed, and infected with VSV (moi=1), IAV (moi=1) for 12 and 24 hrs respectively in serum free IMDM. THP-1 cells were treated with PMA (80 ng/ml) for 16 hrs, washed and cultured in complete (with 10% FBS) RPMI media for an additional 48 hrs. Differentiated THP-1 cells were placed in serum-free RPMI followed by infection with VSV, IAV or IAVΔNS1 (each at moi=1) for 90 min. Subsequently the THP-1 cells were replenished with complete RPMI media for additional 12 and 24 hrs, respectively. Supernatants were collected for ELISAs and both supernatants and lysates were used for immunoblotting.
Transfections of BMDC
BMDCs were primed with Pam3Csk4 (200ng/ml) for 16 hrs. The cells were subsequently transfected with viral RNA (100 ng/ml), cellular RNA (250 ng/ml) or (2′-5′)p3A3 (10 μM) with lipofectamine 2000 (Life Technologies) for an additional 8 hrs.
ELISAs
ELISAs for mouse IL-1β and TNF-α and for human IL-1β were performed as described (BD Biosciences).
IL-1β mRNA levels
Mouse IL-1β mRNA expression was determined by qRT-PCR using the following primer pair with SYBR green. Fwd: 5′GCAACTGTTCCTGAACTCAACT 3′; Rev: 5′ATCTTTTGGGGTCCGTCAACT 3′ (Primer bank ID: 6680415a1 http://pga.mgh.harvard.edu/primerbank/index.html).
Co-immunoprecipitation assays
THP-1 cells (1 × 107) were differentiated with PMA (80 nM), and then transfected with 300ng of cellular RNAs (intact or RNase L cleaved or cleaved and PNK treated) or pIC with lipofectamine 2000. After 4h the cells were resuspended in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% [vol/vol] Nonidet-P40, 5 mM EDTA, and 10% [vol/vol] glycerol). Lysates were immunoprecipitated with control rabbit immunoglobulin G (IgG) or anti-DHX33 antibody or anti-NLRP3 antibody (Cell Signaling Technology, D2P5E) with Protein-A agarose (Sigma-Aldrich). Myc-tagged MAVS cDNA (0.5 μg) and Flag-tagged NLRP3 cDNA (0.5 μg) were co-transfected into HEK293T cells with lipofectamine 2000. After 72h the cells were transfected with 300 ng of cellular RNAs (intact, cleaved or cleaved and PNK-treated). At 4 hr after transfection cells were lysed in buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 1% [vol/vol] Triton-X, 5 mM EDTA, and 10% [vol/vol] glycerol). Lysates were immunoprecipitated with anti-myc antibody (Sigma-Aldrich) with Protein G-agarose (GE Healthcare Life Sciences).
See Supplemental Information for additional experimental procedures.
Supplementary Material
Acknowledgments
We thank Adolfo Garcia-Sastre (New York) for IAVs, Amiya Banerjee (Cleveland) for VSV, Jason Weber (St. Louis) for DHX33 constructs, Babal Kant Jha (Cleveland) for RNase L and 2-5A, Millenium Pharmaceuticals for Nlrp3 and Asc mutant mice, Shizuo Akira (Osaka) for Rig-I mice, Marco Colonna (St. Louis) for Mda5 mutant mice, Aimee McMillan for mouse colony management and Irina Polyakova (Cleveland) and Christina Gaughan (Cleveland) for expert technical assistance,. The studies were supported by NIH grant (CA044059) and the Mal and Lea Bank Chair Fund (to R.H.S.), NIH grants R01AI063331 and R01DK091191 (to G. N.) and NIH grants AI074973, AI083019, and AI88778 (to M.G.).
Footnotes
CONFLICT OF INTEREST STATEMENT
Co-author Luigi Franchi is currently an employee of Lycera, a biotechnology company that develops drugs for inflammation. The other co-authors do not have a conflict of interest.
AUTHOR CONTRIBUTIONS
A.C., S.B., L.F., G.N. and R.H.S. wrote the paper. A.C., S.B., L.F., and R.H.S. designed the experiments. L.F., G.N., M.G., and YM Loo provided reagents.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Chakrabarti A, Jha BK, Weiss SR, Silverman RH. Cell-Type-Specific Effects of RNase L on Viral Induction of Beta Interferon. mBio. 2014:5. doi: 10.1128/mBio.00856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nature reviews Microbiology. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like Polymerization Underlies Signal Transduction in Antiviral Immune Defense and Inflammasome Activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SS, Chen E, Viscount T, Geib J, Sardana MK, Gehman J, Kuo LC. Cleavage of oligoribonucleotides by the 2′,5′-oligoadenylate- dependent ribonuclease L. The Journal of biological chemistry. 1996;271:4988–4992. doi: 10.1074/jbc.271.9.4988. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. RNase L triggers autophagy in response to viral infections. Journal of virology. 2012;86:11311–11321. doi: 10.1128/JVI.00270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DA, Jha BK, Silverman RH, Hesselberth JR, Barton DJ. Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic acids research. 2014 doi: 10.1093/nar/gku118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Niwa M, Walter P, Silverman RH. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA. 2001;7:361–373. doi: 10.1017/s1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. The Journal of biological chemistry. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, Crow YJ, Stetson DB. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nature immunology. 2014 doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. Journal of virology. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Munoz-Planillo R, Ozkurede U, Kim YG, Chakrabarti A, Gale M, Jr, Silverman RH, Colonna M, Akira S, Nunez G. Cytosolic Double-Stranded RNA Activates the NLRP3 Inflammasome via MAVS-Induced Membrane Permeabilization and K+ Efflux. J Immunol. 2014 doi: 10.4049/jimmunol.1400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kanneganti TD, Dubyak GR, Nunez G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. The Journal of biological chemistry. 2007;282:18810–18818. doi: 10.1074/jbc.M610762200. [DOI] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell host & microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Gao J, Taxman DJ, Ting JP, Su L. HIV-1 Infection Induces Interleukin-1beta Production via TLR8 Protein-dependent and NLRP3 Inflammasome Mechanisms in Human Monocytes. The Journal of biological chemistry. 2014;289:21716–21726. doi: 10.1074/jbc.M114.566620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zeqiraj E, Dong B, Jha BK, Duffy N, Orlicky S, Thevakumaran M, Pillon MC, Ceccarelli DF, Wan L, Juang YC, Mao DYL, Gaughan C, Brinton MA, Perelygin AA, Kourinov I, Guarne A, Silverman RH, Sicheri F. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon induced antiviral activity. Molecular Cell. 2014;53:221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. The Journal of experimental medicine. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nature immunology. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nature reviews Immunology. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. The Journal of biological chemistry. 2006a;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006b;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kerr IM, Brown RE. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Mechanisms and Functions of Inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lu N, Yuan B, Weng L, Wang F, Liu YJ, Zhang Z. The interaction between the helicase DHX33 and IPS-1 as a novel pathway to sense double-stranded RNA and RNA viruses in myeloid dendritic cells. Cellular & molecular immunology. 2014;11:49–57. doi: 10.1038/cmi.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Tate MD, MacKenzie-Kludas CJ, Pinar A, Zeng W, Stutz A, Latz E, Brown LE, Mansell A. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS pathogens. 2013;9:e1003392. doi: 10.1371/journal.ppat.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H, Hanabuchi S, Kim T, Bao M, Zhang Z, Sugimoto N, Liu YJ. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Nedialkova DD, Ulferts R, van den Born E, Lauber C, Gorbalenya AE, Ziebuhr J, Snijder EJ. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. Journal of virology. 2009;83:5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negash AA, Ramos HJ, Crochet N, Lau DT, Doehle B, Papic N, Delker DA, Jo J, Bertoletti A, Hagedorn CH, Gale M., Jr IL-1beta production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS pathogens. 2013;9:e1003330. doi: 10.1371/journal.ppat.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour AM, Reichelt M, Ku CC, Ho MY, Heineman TC, Arvin AM. Varicella-zoster virus infection triggers formation of an interleukin-1beta (IL-1beta)-processing inflammasome complex. The Journal of biological chemistry. 2011;286:17921–17933. doi: 10.1074/jbc.M110.210575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkurede VU, Franchi L. Immunology in clinic review series; focus on autoinflammatory diseases: role of inflammasomes in autoinflammatory syndromes. Clinical and experimental immunology. 2012;167:382–390. doi: 10.1111/j.1365-2249.2011.04535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozoren N, Masumoto J, Franchi L, Kanneganti TD, Body-Malapel M, Erturk I, Jagirdar R, Zhu L, Inohara N, Bertin J, et al. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- Pang IK, Ichinohe T, Iwasaki A. IL-1R signaling in dendritic cells replaces pattern-recognition receptors in promoting CD8(+) T cell responses to influenza A virus. Nature immunology. 2013;14:246–253. doi: 10.1038/ni.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Juliana C, Hong S, Datta P, Hwang I, Fernandes-Alnemri T, Yu JW, Alnemri ES. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J Immunol. 2013;191:4358–4366. doi: 10.4049/jimmunol.1301170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nature immunology. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Rajan JV, Rodriguez D, Miao EA, Aderem A. The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. Journal of virology. 2011;85:4167–4172. doi: 10.1128/JVI.01687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos HJ, Lanteri MC, Blahnik G, Negash A, Suthar MS, Brassil MM, Sodhi K, Treuting PM, Busch MP, Norris PJ, Gale M., Jr IL-1beta signaling promotes CNS-intrinsic immune control of West Nile virus infection. PLoS pathogens. 2012;8:e1003039. doi: 10.1371/journal.ppat.1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. Journal of virology. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. Journal of virology. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Thomas PG, Dash P, Aldridge JR, Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) International journal of cancer Journal international du cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- Unterholzner L. The interferon response to intracellular DNA: why so many receptors? Immunobiology. 2013;218:1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Interferon action--sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- Yim HC, Williams BR. Protein kinase R and the inflammasome. J Interferon Cytokine Res. 2014;34:447–454. doi: 10.1089/jir.2014.0008. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. Embo J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.