Abstract
The highly complex structure of the human brain is strongly shaped by genetic influences1. Subcortical brain regions form circuits with cortical areas to coordinate movement2, learning, memory3 and motivation4, and altered circuits can lead to abnormal behaviour and disease2. To investigate how common genetic variants affect the structure of these brain regions, here we conduct genome-wide association studies of the volumes of seven subcortical regions and the intracranial volume derived from magnetic resonance images of 30,717 individuals from 50 cohorts. We identify five novel genetic variants influencing the volumes of the putamen and caudate nucleus. We also find stronger evidence for three loci with previously established influences on hippocampal volume5 and intracranial volume6. These variants show specific volumetric effects on brain structures rather than global effects across structures. The strongest effects were found for the putamen, where a novel intergenic locus with replicable influence on volume (rs945270; P = 1.08 × 10−33; 0.52% variance explained) showed evidence of altering the expression of the KTN1 gene in both brain and blood tissue. Variants influencing putamen volume clustered near developmental genes that regulate apoptosis, axon guidance and vesicle transport. Identification of these genetic variants provides insight into the causes of variability inhuman brain development, and may help to determine mechanisms of neuropsychiatric dysfunction.
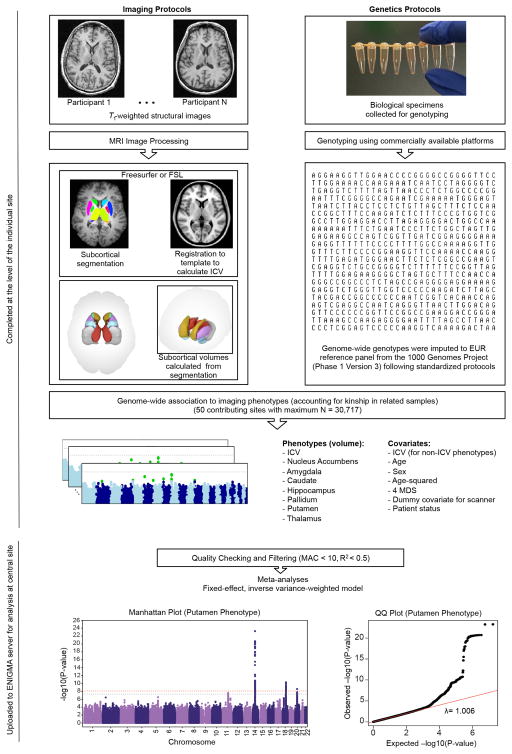
At the individual level, genetic variations exert lasting influences on brain structures and functions associated with behaviour and predisposition to disease. Within the context of the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) consortium, we conducted a collaborative large-scale genetic analysis of magnetic resonance imaging (MRI) scans to identify genetic variants that influence brain structure. Here, we focus on volumetric measures derived from a measure of head size (intracranial volume, ICV) and seven subcortical brain structures corrected for the ICV (nucleus accumbens, caudate, putamen, pallidum, amygdala, hippocampus and thalamus). To ensure data homogeneity within the ENIGMA consortium, we designed and implemented standardized protocols for image analysis, quality assessment, genetic imputation (to 1000 Genomes references, version 3) and association (Extended Data Fig. 1 and Methods).
After establishing that the volumes extracted using our protocols were substantially heritable in a large sample of twins (P < 1 × 10−4; see Methods and Extended Data Fig. 11a), with similar distributions to previous studies1, we sought to identify common genetic variants contributing to volume differences by meta-analysing site-level genome-wide association study (GWAS) data in a discovery sample of 13,171 subjects of European ancestry (Extended Data Fig. 2). Population stratification was controlled for by including, as covariates, four population components derived from standardized multidimensional scaling analyses of genome-wide genotype data conducted at each site (see Methods). Site-level GWAS results and distributions were visually inspected to check for statistical inflation and patterns indicating technical artefacts (see Methods).
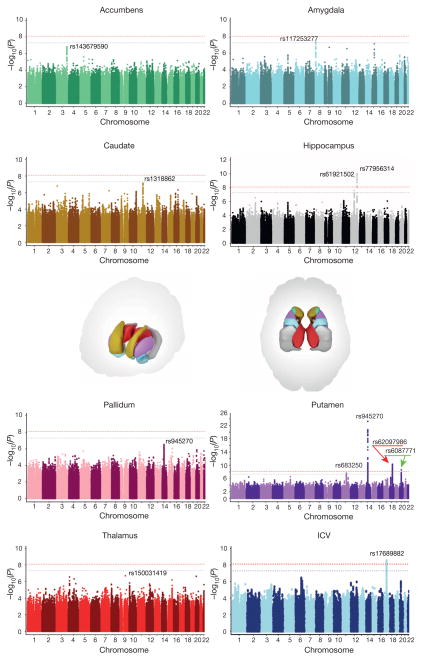
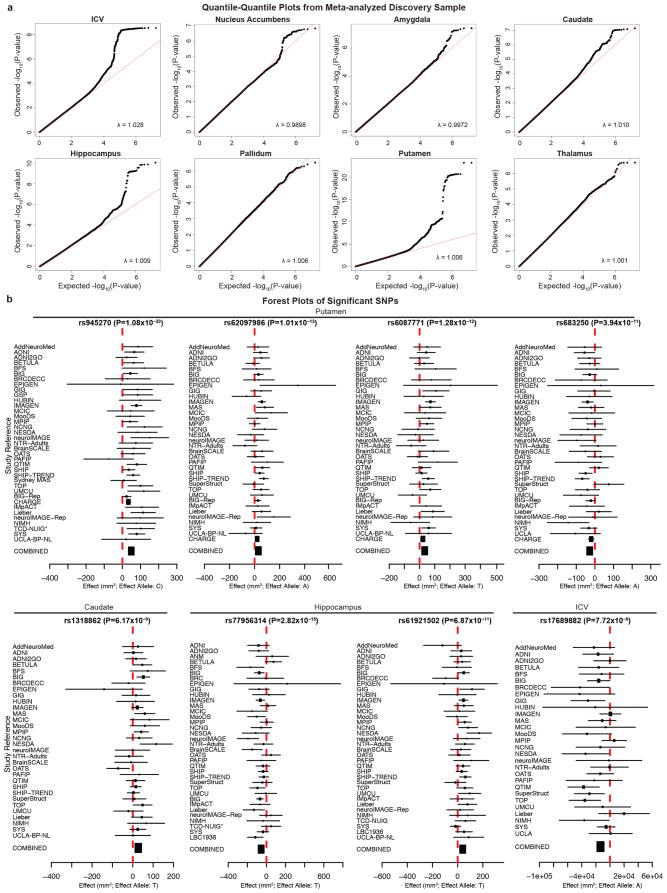
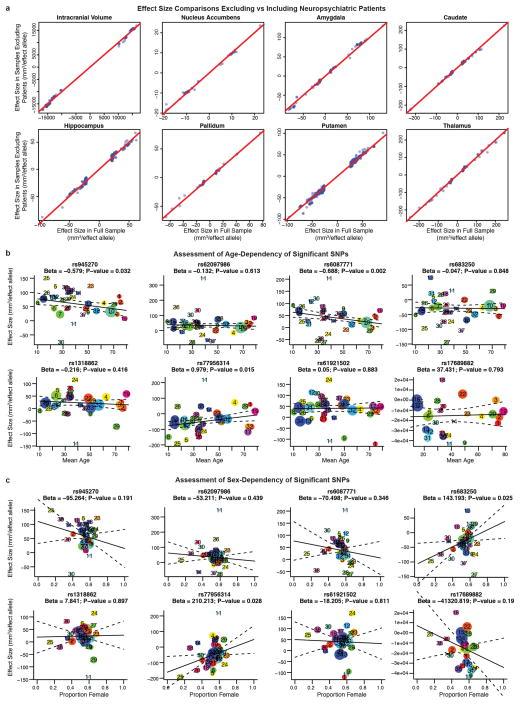
Meta-analysis of the discovery sample identified six genome-wide significant loci after correcting for the number of variants and traits analysed (P < 7.1 × 10−9; see Methods): one associated with the ICV, two associated with hippocampal volume, and three with putamen volume. Another four loci showed suggestive associations (P < 1 × 10−7) with putamen volume (one locus), amygdala volume (two loci), and caudate volume (one locus; Table 1, Fig. 1 and Supplementary Table 5). Quantile–quantile plots showed no evidence of population stratification or cryptic relatedness (Extended Data Fig. 4a). We subsequently attempted to replicate the variants with independent data from 17,546 individuals. All subcortical genome-wide significant variants identified in the discovery sample were replicated (Table 1). The variant associated with the ICV did not replicate in a smaller independent sample, but was genome-wide significant in a previously published independent study6, providing strong evidence for its association with the ICV. Moreover, two suggestive variants associated with putamen and caudate volumes exceeded genome-wide significance after meta-analysis across the discovery and replication data sets (Table 1). Effect sizes were similar across cohorts (P > 0.1, Cochran’s Q test; Extended Data Fig. 4b). Effect sizes remained consistent after excluding patients diagnosed with anxiety, Alzheimer’s disease, attention-deficit/hyperactivity disorder, bipolar disorder, epilepsy, major depressive disorder or schizophrenia (21% of the discovery participants). Correlation in effect size with and without patients was very high (r > 0.99) for loci with P < 1 × 10−5, indicating that these effects were unlikely to be driven by disease (Extended Data Fig. 5a). The participants’ age range covered most of the lifespan (9–97 years), but only one of the eight significant loci showed an effect related to the mean age of each cohort (P = 0.002; rs6087771 affecting putamen volume; Extended Data Fig. 5b), suggesting that nearly all effects are stable across the lifespan. In addition, none of these loci showed evidence of sex effects (Extended Data Fig. 5c).
Table 1.
Genetic variants at eight loci were significantly associated with putamen, hippocampus, caudate nucleus and ICV
| Discovery cohort
|
Replication cohort
|
Discovery + replication cohorts
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Marker | A1 | A2 | Frq | Effect (se) | P value | Sample size |
Effect (se) | P value | Sample size |
Effect (se) | P value | Total sample size |
Variance explained (%) |
Diff./ allele (%) |
| Putamen | rs945270 | C | G | 0.58 | 60.64 (6.00) | 5.43 × 10−24 | 13,145 | 39.15 (5.46) | 7.81 × 10−13 | 15,130 | 48.89 (4.04) | 1.08 × 10−33 | 28,275 | 0.52 | 0.94 |
| Putamen | rs62097986 | A | C | 0.44 | 39.53 (6.01) | 4.86 × 10−11 | 13,145 | 22.46 (5.53) | 4.89 × 10−5 | 14,891 | 30.28 (4.07) | 1.01 × 10−13 | 28,036 | 0.20 | 0.58 |
| Putamen | rs6087771 | T | C | 0.71 | 40.72 (6.82) | 2.42 × 10−9 | 11,865 | 26.97 (6.57) | 4.02 × 10−5 | 13,675 | 33.58 (4.73) | 1.28 × 10−12 | 25,540 | 0.20 | 0.64 |
| Putamen | rs683250 | A | G | 0.63 | −33.97 (6.08) | 2.33 × 10−8 | 13,145 | −22.30 (5.89) | 1.50 × 10−4 | 13,113 | −27.95 (4.23) | 3.94 × 10−11 | 26,258 | 0.17 | 0.51 |
| Caudate | rs1318862 | T | C | 0.58 | 26.27 (4.89) | 7.54 × 10−8 | 13,171 | 31.82 (14.23) | 0.025 | 1,860 | 26.86 (4.62) | 6.17 × 10−9 | 15,031 | 0.22 | 0.74 |
| Hip. | rs77956314 | T | C | 0.91 | −54.21 (8.37) | 9.33 × 10−11 | 13,163 | −57.43 (12.69) | 6.04 × 10−6 | 4,027 | −55.18 (6.99) | 2.82 × 10−15 | 17,190 | 0.36 | 1.40 |
| Hip. | rs61921502 | T | G | 0.84 | 43.40 (6.89) | 2.92 × 10−10 | 13,163 | 26.81 (13.32) | 0.044 | 3,046 | 39.90 (6.12) | 6.87 × 10−11 | 16,209 | 0.26 | 1.01 |
| ICV | rs17689882 | A | G | 0.22 | −15,335.88 (2,582.20) | 2.87 × 10−9 | 10,944 | −5,202.15 (5,428.60) | 0.337 | 1,878 | −13,460.47 (2,331.05) | 7.72 × 10−9 | 12,822 | 0.26 | 0.96 |
The allele frequency (frq) and effect size are given with reference to allele 1 (A1). Effect sizes are given in units of mm3 per effect allele. Results are provided for the discovery samples and the combined meta-analysis of the discovery and replication cohorts (all European ancestry). Additional validation was attempted in non-European ancestry generalization samples (shown in Supplementary Table 6). The variance explained gives the percentage variance explained by a given SNP after correcting for covariates (see Methods for additional details). The percentage difference in volume per effect allele (Diff./allele) is based on the absolute value of the final combined effect divided by a weighted average of the brain volume of interest across all sites in the discovery sample and then multiplied by 100. Hip, hippocampus.
Figure 1. Common genetic variants associated with subcortical volumes and the ICV.
Manhattan plots coloured with a scheme that matches the corresponding structure (middle) are shown for each subcortical volume studied. Genome-wide significance is shown for the common threshold of P = 5 × 10−8 (grey dotted line) and also for the multiple comparisons-corrected threshold of P = 7.1 × 10−9 (red dotted line). The most significant SNP within an associated locus is labelled.
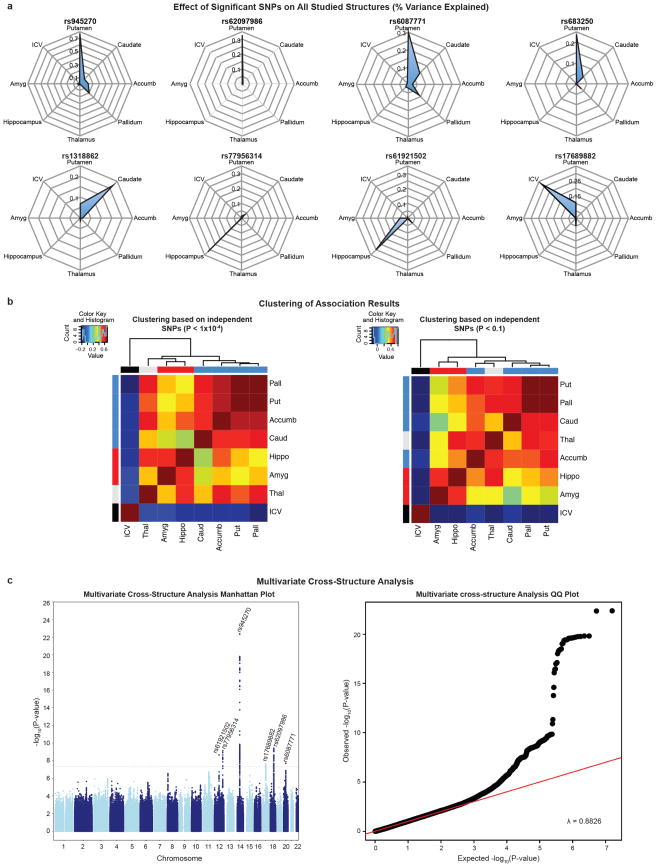
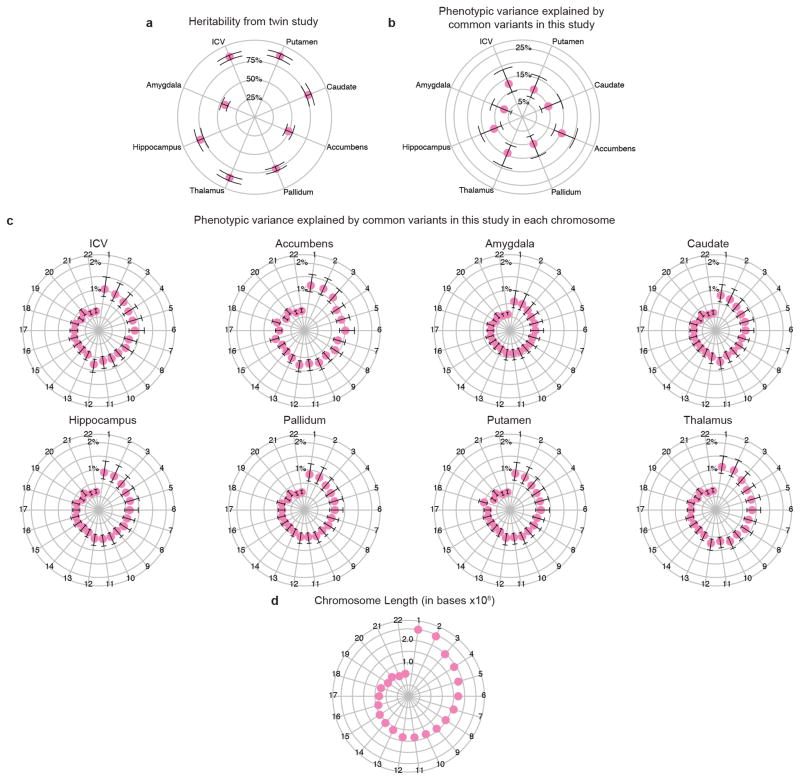
In our cohorts, significant loci were associated with 0.51–1.40% differences in volume per risk allele, explaining 0.17–0.52% of the phenotypic variance (Table 1); such effect sizes are similar to those of common variants influencing other complex quantitative traits such as height7 and bodymass index8. The full genome-wide association results explained 7–15% of phenotypic variance after controlling for the effects of covariates (Extended Data Fig. 11). Notably, the genome-wide significant variants identified here showed specific effects on single brain structures rather than pleiotropic effects across multiple structures, despite similar developmental origins as in the case of caudate and putamen (Extended Data Fig. 6a). Nevertheless, when we subjected the subcortical meta-analysis results to hierarchical clustering, genetic determinants of the subcortical structures were mostly grouped into larger circuits according to their developmental and functional subdivisions (Extended Data Fig. 6b). Genetic variants may therefore have coherent effects on functionally associated subcortical networks. Multivariate cross-structure9 analyses confirmed the univariate results, but no additional loci reached genome-wide significance (Extended Data Fig. 6c). The clustering of results into known brain circuits in the absence of individually significant genetic variants found in the cross-structure analysis suggests variants of small effect may have similar influences across structures. Most variants previously reported to be associated with brain structure and/or function showed little evidence of large-scale volumetric effects (Supplementary Table 8). We detected an intriguing association with hippocampal volume at a single nucleotide polymorphism (SNP) with a genome-wide significant association with schizophrenia10 (rs2909457; P = 2.12 × 10−6; where the A allele is associated with decreased risk for schizophrenia and decreased hippocampal volume). In general, however, we detected no genome-wide significant association with brain structure for genome-wide significant loci that contribute risk for neuropsychiatric illnesses (Supplementary Table 9).
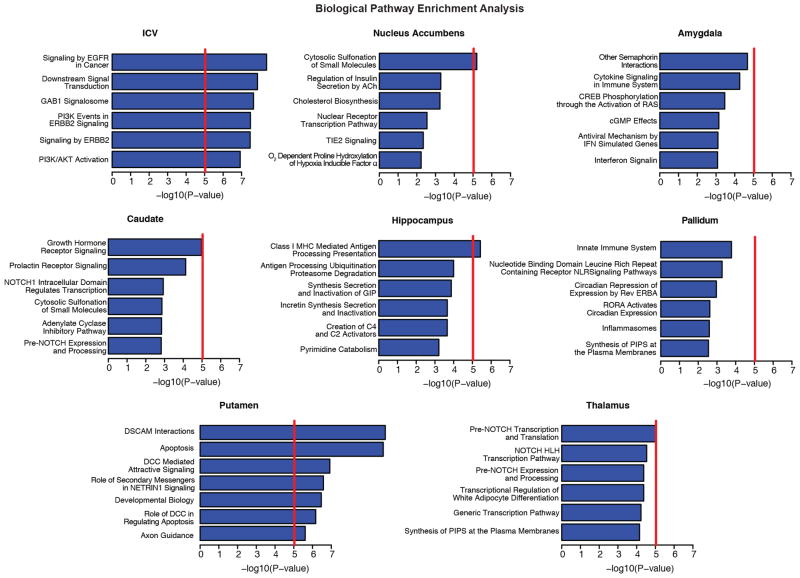
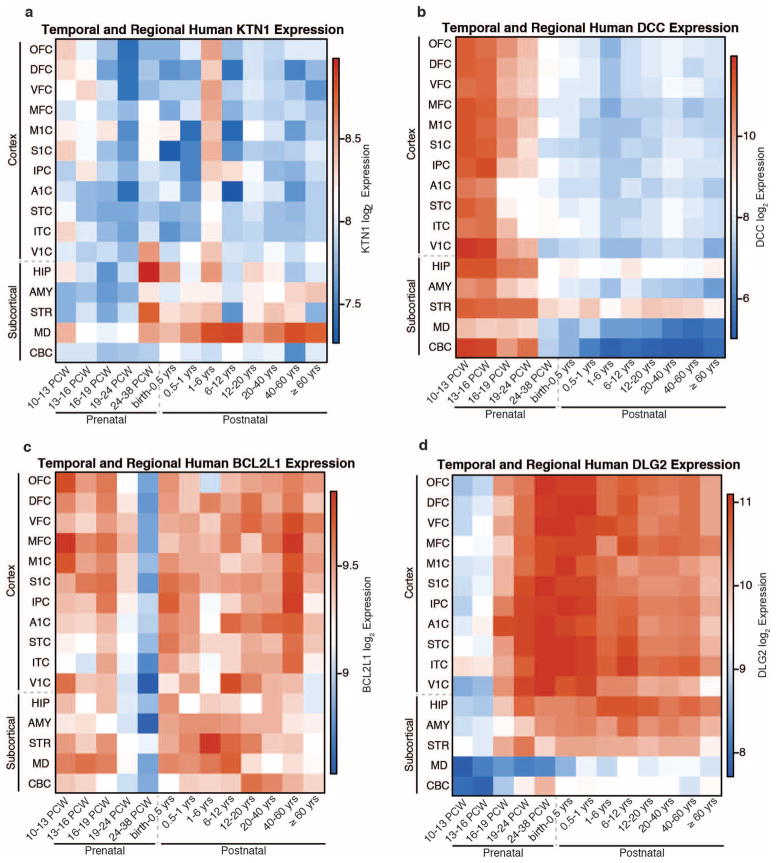
Of the four loci influencing putamen volume, we identified an inter-genic locus 50 kilobases (kb) downstream of the KTN1 gene (rs945270; 14q22.3; n = 28,275; P = 1.08 × 10−33), which encodes the protein kinectin, a receptor that allows vesicle binding to kinesin and is involved in organelle transport11. Second, we identified an intronic locus within DCC (rs62097986; 18q21.2; n = 28,036; P = 1.01 × 10−13), which encodes a netrin receptor involved in axon guidance and migration, including in the developing striatum12 (Extended Data Fig. 3b). Expression of DCC throughout the brain is highest in the first two trimesters of prenatal development13 (Extended Data Fig. 8b), suggesting that this variant may influence brain volumes early in neurodevelopment. Third, we identified an intronic locus within BCL2L1 (rs6087771; 20q11.21; n = 25,540; P = 1.28 × 10−12), which encodes an anti-apoptotic factor that inhibits programmed cell death of immature neurons throughout the brain14 (Extended Data Fig. 3c). Consistent with this, expression of BCL2L1 in the striatum strongly decreases at the end of neurogenesis (24–38 post-conception weeks (PCW); Extended Data Fig. 8c), a period marked by increased apoptosis in the putamen13,15. Fourth, we identified an intronic locus within DLG2 (rs683250; 11q14.1; n = 26,258; P = 3.94 × 10−11), which encodes the postsynaptic density 93 (PSD-93) protein (Extended Data Fig. 3d). PSD-93 is a membrane-associated guanylate kinase involved in organizing channels in the postsynaptic density16. DLG2 expression increases during early mid-fetal development in the striatum13 (Extended Data Fig. 8d). Genetic variants in DLG2 affect learning and cognitive flexibility17 and are associated with schizophrenia18. Notably, SNPs associated with variation in putamen volume showed enrichment of genes involved in apoptosis and axon guidance pathways (Extended Data Fig. 7 and Supplementary Table 7).
Hippocampal volume showed an intergenic association near the HRK gene (rs77956314; 12q24.22; n = 17,190; P = 2.82 × 10−15; Extended Data Fig. 3g) and with an intronic locus in the MSRB3gene (rs61921502; 12q14.3; n = 16,209; P = 6.87× 10−11; Extended Data Fig. 3h), supporting our previous analyses5,19 of smaller samples imputed to HapMap3 references. Caudate volume was associated with an intergenic locus 80 kb from FAT3 (rs1318862; 11q14.3; n = 15,031; P = 6.17 × 10−9; Extended Data Fig. 3e). This gene encodes a cadherin specifically expressed in the nervous system during embryonic development that influences neuronal morphology through cell–cell interactions20. The ICV was associated with an intronic locus within CRHR1 that tags the chromosome 17q21 inversion21, which has been previously found to influence ICV6 (rs17689882; 17q21.31; n = 12,822; P = 7.72 × 10−9; Extended Data Fig. 3f). Another previously identified variant with association to ICV (rs10784502)5,19 did not survive genome-wide significance in this analysis but did show a nominal effect in the same direction (P = 2.05 × 10−3; n = 11,373). None of the genome-wide significant loci in this study were in linkage disequilibrium with known functional coding variants, splice sites, or 3′/5′ untranslated regions, although several of the loci had epigenetic markings suggesting a regulatory role (Extended Data Fig. 3).
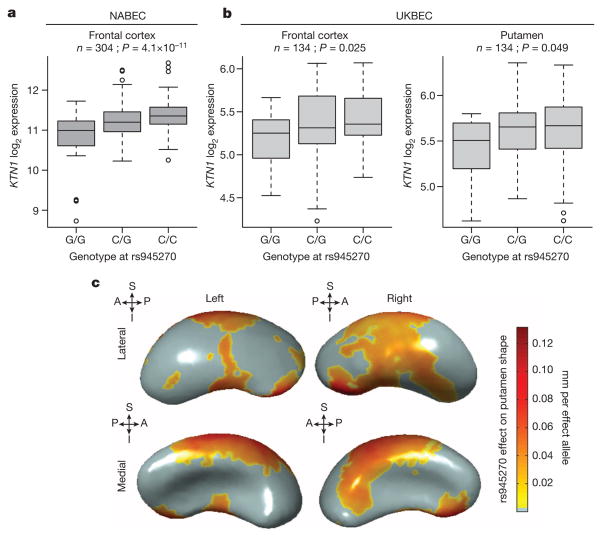
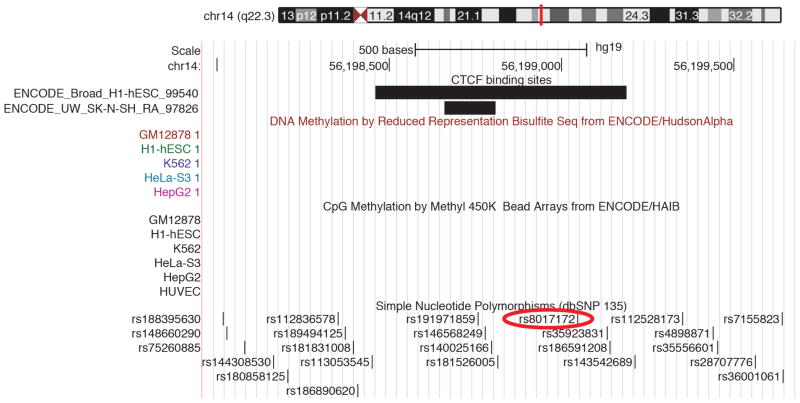
Given the strong association with putamen volume, we further examined the rs945270 locus. Epigenetic markers suggest insulator functionality near the locus as this is the lone chromatin mark in the intergenic region22 (Extended Data Fig. 3a). Chromatin immunoprecipitation followed by sequencing (ChIP-seq) indicate that a variant (rs8017172) in complete linkage disequilibrium with rs945270 (r2 = 1.0) lies within a binding site of the CTCF (CCCTC-binding factor) transcription regulator23 (Extended Data Fig. 9) in embryonic stem cells. To assess potential functionality in brain tissue, we tested for association with gene expression 1 megabase (Mb) up/downstream. We identified and replicated an effect of rs945270 on the expression of the KTN1 gene. The C allele, associated with larger putamen volume, also increased expression of KTN1 in the frontal cortex (discovery sample: 304 neuropathologically normal controls24 (P = 4.1 × 10−11); replication sample: 134 neuropathologically normal controls (P = 0.025)), and putamen (sample: 134 neuropathologically normal controls25 (P = 0.049); Fig. 2a, b). In blood, rs945270 was also strongly associated with KTN1 expression26 (P = 5.94 × 10−31; n = 5,311). After late fetal development, KTN1 is expressed in the human thalamus, striatum and hippocampus; it is more highly expressed in the striatum than the cortex13 (Extended Data Fig. 8a). KTN1 encodes the kinectin receptor facilitating vesicle binding to kinesin, and is heavily involved in organelle transport11. Kinectin is only found in the dendrites and soma of neurons, not their axons; neurons with more kinectin have larger cell bodies27, and kinectin knockdown strongly influences cell shape28. The volumetric effects identified here may therefore reflect genetic control of neuronal cell size and/or dendritic complexity. Using three-dimensional surface models of putamen segmentations in MRI scans of 1,541 healthy adolescent subjects, we further localized the allelic effects of rs945270 to regions along the superior and lateral putamen bilaterally, independent of chosen segmentation protocol (Fig. 2c and Extended Data Fig. 10). Each copy of the C allele was associated with an increase in volume along anterior superior regions receiving dense cortical projections from dorsolateral prefrontal cortex and supplementary motor areas29,30.
Figure 2. Effect of rs945270 on KTN1 expression and putamen shape.
a, b, Expression quantitative trait loci study in brain tissue demonstrates the effect of rs945270 on KTN1 gene expression in frontal cortex tissue from 304 subjects from the North American Brain Expression Cohort (NABEC25) (a) and in an independent sample of 134 subjects from the UK Brain Expression Cohort (UKBEC) (b), sampled from both frontal cortex and putamen. Boxplot dashed bars mark the twenty-fifth and seventy-fifth percentiles. c, Surface-based analysis demonstrates that rs945270 has strong effects on the shape of superior and lateral portions of the putamen in 1,541 subjects. Each copy of the rs945270-C allele was significantly associated with increased width in coloured areas (false discovery rate corrected at q = 0.05), and the degree of deformation is labelled by colour, with red indicating greater deformation. Orientation is indicated by arrows. A, anterior; I, inferior; P, posterior, S, superior.
In summary, we discovered several common genetic variants underlying variation in different structures within the human brain. Many seem to exert their effects through known developmental pathways including apoptosis, axon guidance and vesicle transport. All structure volumes showed high heritability, but individual genetic variants had diverse effects. The strongest effects were found for putamen and hippocampal volumes, whereas other structures delineated with similar reliability such as the thalamus showed no association with these or other loci (Supplementary Table 4). Discovery of common variants affecting the human brain is now feasible using collaborative analysis of MRI data, and may determine genetic mechanisms driving development and disease.
METHODS
Details of the GWAS meta-analysis are outlined in Extended Data Fig. 1. All participants in all cohorts in this study gave written informed consent and sites involved obtained approval from local research ethics committees or Institutional Review Boards. The ENIGMA consortium follows a rolling meta-analysis framework for incorporating sites into the analysis. The discovery sample comprises studies of European ancestry (Extended Data Fig. 2) that contributed GWAS summary statistics for the purpose of this analysis on or before 1 October 2013. The deadline for discovery samples to upload their data was made before inspecting the data and was not influenced by the results of the analyses. The meta-analysed results from discovery cohorts were carried forward for secondary analyses and functional validation studies. Additional samples of European ancestry were gathered to provide in silico or single genotype replication of the strongest associations as part of the replication sample. A generalization sample of sites with non-European ancestry was used to examine the effects across ethnicities. In all, data were contributed from 50 cohorts, each of which is detailed in Supplementary Tables 1–3.
The brain measures examined in this study were obtained from structural MRI data collected at participating sites around the world. Brain scans were processed and examined at each site locally, following a standardized protocol procedure to harmonize the analysis across sites. The standardized protocols for image analysis and quality assurance are openly available online (http://enigma.ini.usc.edu/protocols/imaging-protocols/). The subcortical brain measures (nucleus accumbens, amyg-dala, caudate nucleus, hippocampus, pallidum, putamen and thalamus) were delineated in the brain using well-validated, freely available brain segmentation software packages: FIRST31, part of the FMRIB Software Library (FSL), or FreeSurfer32. The agreement between the two software packages has been well documented in the literature5,33 and was further detailed here (Supplementary Table 4). Participating sites used the software package most suitable for their data set (the software used at each site is given in Supplementary Table 2) without selection based on genotype or the associations present in this study. In addition to the subcortical structures of the brain, we examined the genetic effects of a measure of global head size, the ICV. The ICV was calculated as: 1/(determinant of a rotation-translation matrix obtained after affine registration to a common study template and multiplied by the template volume (1,948,105 mm3)). After image processing, each image was inspected individually to identify poorly segmented structures. Each site contributed histograms of the distribution of volumes for the left and right hemisphere structures (and a measure of asymmetry) of each subcortical region used in the analysis. Scans marked as outliers (> 3 standard deviations from the mean) based on the histogram plots were re-checked at each site to locate any errors. If a scan had an outlier for a given structure, but was segmented properly, it was retained in the analysis. Site-specific phenotype histograms, Manhattan plots and quantile–quantile plots from each participating site are available on the ENIGMA website (http://enigma.ini.usc.edu/publications/enigma-2/).
Each study in the discovery sample was genotyped using commercially available platforms. Before imputation, genetic homogeneity was assessed in each sample using multi-dimensional scaling (MDS) analysis (Extended Data Fig. 2). Ancestry outliers were excluded through visual inspection of the first two components. Quality control filtering was applied to remove genotyped SNPs with low minor allele frequency (< 0.01), poor genotype call rate (< 95%), and deviations from Hardy–Weinberg equilibrium (P < 1 × 10−6) before imputation. The imputation protocols used MaCH34 for haplotype phasing and minimac35 for imputation and are freely available online (http://enigma.ini.usc.edu/protocols/genetics-protocols/). Full details of quality control procedures and any deviations from the imputation protocol are given in Supplementary Table 3.
Genome-wide association scans were conducted at each site for all eight traits of interest including the ICV and bilateral volumes of the nucleus accumbens, amyg-dala, caudate nucleus, hippocampus, pallidum, putamen and thalamus. For each SNP in the genome, the additive dosage value was regressed against the trait of interest separately using a multiple linear regression framework controlling for age, age2, sex, 4 MDS components, ICV (for non-ICV phenotypes) and diagnosis (when applicable). For studies with data collected from several centres or scanners, dummy-coded covariates were also included in the model. Sites with family data (NTR-Adults, BrainSCALE, QTIM, SYS, GOBS, ASPSFam, ERF, GeneSTAR, NeuroIMAGE and OATS) used mixed-effects models to control for familial relationships in addition to covariates stated previously. The primary analyses for this paper focused on the full set of subjects including data sets with patients to maximize the power to detect effects. We re-analysed the data excluding patients to verify that detected effects were not due to disease alone (Extended Data Fig. 5a). The protocols used for testing association with mach2qtl (ref. 34) for studies with unrelated subjects and merlin-offline36 for family-based designs are freely available online (http://enigma.ini.usc.edu/protocols/genetics-protocols/). Full details for the software used at each site are given in Supplementary Table 3.
The GWAS results from each site were uploaded to a centralized server for quality checking and processing. Results files from each cohort were free from genomic inflation in quantile–quantile plots and Manhattan plots (http://enigma.ini.usc.edu/publications/enigma-2/). Poorly imputed SNPs (with R2 < 0.5) and low minor allele count (< 10) were removed from the GWAS result files from each site. The resulting files were combined meta-analytically using a fixed-effect, inverse-variance-weighted model as implemented in the software package METAL37. The discovery cohorts were meta-analysed first, controlling for genomic inflation. The combined discovery data set (comprised of all meta-analysed SNPs with data from at least 5,000 subjects) was carried forward for the additional analyses detailed below.
To account appropriately for multiple comparisons over the eight traits in our analysis, we first examined the degree of independence between each trait. We generated an 8 × 8 correlation matrix based on the Pearson’s correlation between all pair-wise combinations of the mean volumes of each structure in the QTIM study. Using the matSpD software38 we found that the effective number of independent traits in our analysis was 7. We therefore set a significance criteria threshold of P < (5 × 10−8/7) = 7.1 × 10−9.
Heritability estimates for mean volumes of each of the eight structures in this study were calculated using structural equation modelling in OpenMx39. Twin modelling was performed controlling for age and sex differences on a large sample (n = 1,030) of healthy adolescent and young adult twins (148 monozygotic and 202 dizygotic pairs) and their siblings from the Queensland Twin Imaging (QTIM) study. Subsequently, a multivariate analysis showed that common environmental factors (C) could be dropped from the model without a significant reduction in the goodness-of-fit (Δχ236 = 29.81; P = 0.76). Heritability (h2) was significantly different from zero for all eight brain measures: putamen (h2 = 0.89; 95% confidence interval 0.85–0.92), thalamus (h2 = 0.88; 0.85–0.92), ICV (h2 = 0.88; 0.84–0.90), hippocampus (h2 = 0.79; 0.74–0.83), caudate nucleus (h2 = 0.78; 0.75–0.82), pallidum (h2 = 0.75; 0.72–0.78), nucleus accumbens (h2 = 0.49; 0.45–0.55), amygdala (h2 = 0.43; 0.39, 0.48) (Extended Data Fig. 11a).
Percentage variance explained by each genome-wide significant SNP was determined based on the final combined discovery data set (Extended Data Fig. 6a) or the discovery combined with the replication samples (Table 1) after correction for covariates using the following equation:
where the t-statistic is calculated as the beta coefficient for a given SNP from the regression model (controlling for covariates) divided by the standard error of the beta estimate, and where n is the total number of subjects and k is the total number of covariates included in the model (k = 10) (ref. 40). R2g|c is the variance explained by the variant controlling for covariates and R2c is the variance explained by the covariates alone. R2g|c/(1 − R2c) gives the variance explained by the genetic variant after accounting for covariate effects. The total variance explained by the GWAS (Extended Data Fig. 11b, c) was calculated by first linkage disequilibrium pruning the results without regard to significance (pruning parameters in PLINK:– –indep-pairwise 1000kb 25 0.1). The t-statistics of the regression coefficients from the pruned results are then corrected for the effects of ‘winner’s curse’ and the variance explained by each SNP after accounting for covariate effects is summed across SNPs using freely available code (http://sites.google.com/site/honcheongso/software/total-vg)40,41. As the correction for winners curse may be influenced by asymmetry in the distribution of t (arising from the choice of reference allele) we bootstrapped the choice of reference allele (5,000 iterations) to derive the median value and 95% confidence intervals of the estimates of variance explained (Extended Data Fig. 11b, c). The correction for winner’s curse corrected for upward biases when estimating the percentage variance explained by each SNP across the genome via simulation40, but this correction could still allow some bias. Future large studies will be able to evaluate independently the percentage variance explained.
We performed multivariate GWAS using the Trait-based Association Test that uses Extended Simes procedure (TATES)9. For the TATES analysis we used GWAS summary statistics from the discovery data set and the correlation matrix created from the eight phenotypes using the QTIM data set (Extended Data Fig. 6c).
We examined the moderating effects of mean age and proportion of females on the effect sizes estimated for the top loci influencing brain volumes (Extended Data Fig. 5b, c) using a mixed-effect meta-regression model such that:
In this model, the effect and variance at each site are treated as random effects and the moderator Xmod (either mean age or proportion of females) is treated as a fixed effect. Meta-regression tests were performed using the meta for package (version 1.9-1) in R.
Hierarchical clustering was performed on the GWAS t-statistics from the discovery data set results using independent SNPs clumped from the TATES results (clumping parameters: significance threshold for index SNP = 0.01, significance threshold for clumped SNPs = 0.01, r2 = 0.25, physical distance = 1 Mb; Extended Data Fig. 6b). Regions with the strongest genetic similarity were grouped together based on the strength of their pairwise correlations. The results were represented visually using hierarchical clustering with default settings from the gplots package (version 2.12.1) in R.
Gene annotation, gene-based test statistics and pathway analysis were performed using the KGG2.5 software package42 (Supplementary Table 7 and Extended Data Fig. 7). Linkage disequilibrium was calculated based on RSID numbers using the 1000 Genomes Project European samples as a reference (http://enigma.ini.usc.edu/protocols/genetics-protocols/). For the annotation, SNPs were considered ‘within’ a gene if they fell within 5 kb of the 3′/5′ untranslated regions based on human genome (hg19) coordinates. Gene-based tests were performed using the GATES test42 without weighting P values by predicted functional relevance. Pathway analysis was performed using the hybrid set-based test (HYST) of association43. For all gene-based tests and pathway analyses, results were considered significant if they exceeded a Bonferroni correction threshold accounting for the number of pathways and traits tested such that Pthresh = 0.05/(671 pathways × 7 independent traits) = 1.06 × 10−5.
Expression quantitative loci were examined in two independent data sets: the NABEC (GSE36192)24 and UKBEC (GSE46706)44,45. Detailed processing and exclusion criteria for both data sets are described elsewhere24,45. In brief, the UKBEC consists of 134 neuropathologically normal donors from the MRC Sudden Death Brain Bank in Edinburgh and Sun Health Research Institute; expression was profiled on the Affymetrix Exon 1.0 ST array. The NABEC is comprised of 304 neurologically normal donors from the National Institute of Ageing and expression profiled on the Illumina HT12v3 array. The expression values were corrected for gender and batch effects and probes that contained polymorphisms (seen > 1% in European 1000G) were excluded from analyses44. Blood expression quantitative trait loci (eQTL) data were queried using the Blood eQTL Browser (http://genenetwork.nl/bloodeqtlbrowser/)26. Brain expression over the lifespan was measured from a spatio-temporal atlas of human gene expression and graphed using custom R scripts (GSE25219; details given in13).
Fine-grained three-dimensional surface mappings of the putamen were generated using a medial surface modelling method46,47 in 1,541 healthy subjects from the IMAGEN study48 (Fig. 2c and Extended Data Fig. 10a, b). Putamen volume segmentations from either FSL (Fig. 2c and Extended Data Fig. 10a) or FreeSurfer (Extended Data Fig. 10b) were first converted to three-dimensional meshes and then co-registered to an average template for statistical analysis. The medial core distance was used as a measure of shape and was calculated as the distance from each point on the surface to the centre of the putamen. At each point along the surface of the putamen, an association test was performed using multiple linear regression in which the medial core distance at a given point on the surface was the outcome measure and the additive dosage value of the top SNP was the predictor of interest while including the same covariates that were used for volume including age, sex, age2, 4 MDS, ICV and site.
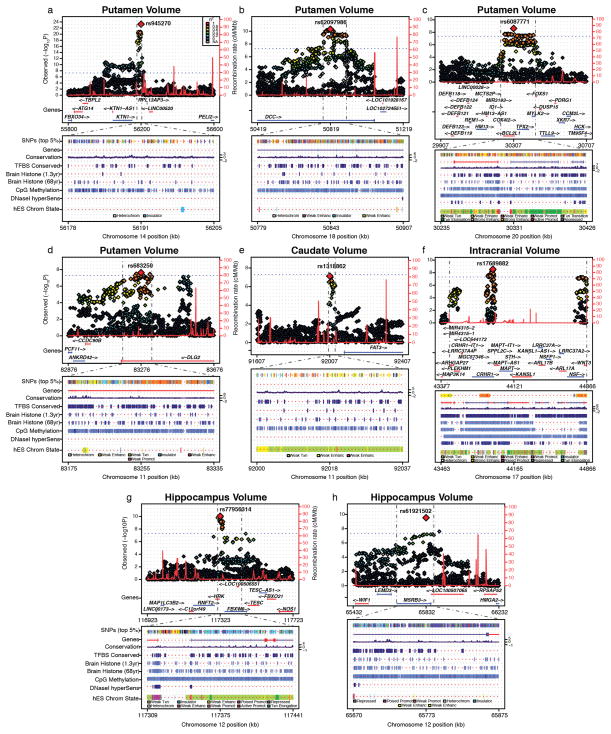
In Extended Data Fig. 3, all tracks were taken from the UCSC Genome Browser Human hg19 assembly. SNPs (top 5%) shows the top 5% associated SNPs within the locus and are coloured by their correlation to the top SNP. Genes shows the gene models from GENCODE version 19. Conservation was defined at each base through the phyloP algorithm which assigns scores as −log10 P values under a null hypothesis of neutral evolution calculated from pre-computed genomic alignment of 100 vertebrate species49. Conserved sites are assigned positive scores, while faster-than-neutral evolving sites are given negative scores. TFBS conserved shows computationally predicted transcription factor binding sites using the Transfac Matrix Database (v.7.0) found in human, mouse and rat. Brain histone (1.3 year) and brain histone (68 year) show maps of histone trimethylation at histoneH3 Lys 4 (H3K4me3), an epigenetic mark for transcriptional activation, measured by ChIP-seq. These measurements were made in neuronal nuclei (NeuN+) collected from prefrontal cortex of post-mortem human brain50. CpG methylation was generated using meth-ylated DNA immunoprecipitation and sequencing from postmortem human frontal cortex of a 57-year-old male51. DNaseI hypersens displays DNaseI hypersensitivity, evidence of open chromatin, which was evaluated in postmortem human frontal cerebrum from three donors (age 22–35), through the ENCODE consortium52. Finally, hES Chrom State gives the predicted chromatin states based on computational integration of ChIP-seq data for nine chromatin marks in H1 human embryonic stem cell lines derived in the ENCODE consortium53.
Extended Data
Extended Data Figure 1. Outline of the genome-wide association meta-analysis.
Structural T1-weighted brain MRI and biological specimens for DNA extraction were acquired from each individual at each site. Imaging protocols were distributed to and completed by each site for standardized automated segmentation of brain structures and calculation of the ICV. Volumetric phenotypes were calculated from the segmentations. Genome-wide genotyping was completed at each site using commercially available chips. Standard imputation protocols to the 1000 Genomes reference panel (phase 1, version 3) were also distributed and completed at each site. Each site completed genome-wide association for each of the eight volumetric brain phenotypes with the listed covariates. Statistical results from GWAS files were uploaded to a central site for quality checking and fixed effects meta-analysis.
Extended Data Figure 2. Ancestry inference via multi-dimensional scaling plots.
Multi-dimensional scaling (MDS) plots of the discovery cohorts to HapMap III reference panels of known ancestry are displayed. Ancestry is generally homogeneous within each group. In all discovery samples any individuals with non-European ancestry were excluded before association. The axes have been flipped to the same orientation for each sample for ease of comparison. ASW, African ancestry in southwest USA; CEU, Utah residents with northern and western European ancestry from the CEPH collection; CHD, Chinese in metropolitan Denver, Colorado; GIH, Gujarati Indians in Houston, Texas; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, California; MKK, Maasai in Kinyawa, Kenya; TSI, Tuscans in Italy; YRI, Yoruba in Ibadan, Nigeria.
Extended Data Figure 3. Genomic function is annotated near novel genome-wide significant loci.
a–h, For each panel, zoomed-in Manhattan plots (±400 kb from top SNP) are shown with gene models below (GENCODE version 19). Plots below are zoomed to highlight the genomic region that probably contains the causal variant(s) (r2 > 0.8 from the top SNP). Genomic annotations from the UCSC browser and ENCODE are displayed to indicate potential functionality (see Methods for detailed track information). SNP coverage is low in f owing to a common genetic inversion in the region. Each plot was made using the Locus Track software (http://gump.qimr.edu.au/general/gabrieC/LocusTrack/).
Extended Data Figure 4. Quantile–quantile and forest plots from meta-analysis of discovery cohorts.
a, Quantile–quantile plots show that the observed P values only deviate from the expected null distribution at the most significant values, indicating that population stratification or cryptic relatedness are not unduly inflating the results. This is quantified through the genomic control parameter (λ; which evaluates whether the median test statistic deviates from expected)54. λ values near 1 indicate that the median test statistic is similar to those derived from a null distribution. Corresponding meta-analysis Manhattan plots can be found in Fig. 1. b, Forest plots show the effect at each of the contributing sites to the meta-analysis. The size of the dot is proportional to the sample size, the effect is shown by the position on the x axis, and the standard error is shown by the line. Sites with an asterisk indicate the genotyping of a proxy SNP (in perfect linkage disequilibrium calculated from 1000 Genomes) for replication.
Extended Data Figure 5. Influence of patients with neuropsychiatric disease, age and gender on association results.
a, Scatterplot of effect sizes including and excluding patients with neuropsychiatric disorders for nominally significant SNPs. For each of the eight volumetric phenotypes, SNPs with P < 1 × 10−5 in the full discovery set meta-analysis were also evaluated excluding the patients. The beta values from regression, a measure of effect size, are plotted (blue dots) along with a line of equivalence between the two conditions (red line). The correlation between effect sizes with and without patients was very high (r > 0.99), showing that the SNPs with significant effects on brain structure are unlikely to be driven by the diseased individuals. b, Meta-regression comparison of effect size with mean age at each site. Each site has a corresponding number and coloured dot in each graph. The size of each dot is based on the standard error such that bigger sites with more definitive estimates have larger dots (and more influence on the meta-regression). The age range of participants covered most of the lifespan (9–97 years), but only one of these eight loci showed a significant relationship with the mean age of each cohort (rs608771 affecting putamen volume). c, Meta-regression comparison of effect size with the proportion of females at each site. No loci showed evidence of moderation by the proportion of females in a given sample. However, the proportion of females at each site has a very restricted range, so results should be interpreted with caution. Plotted information follows the same convention as described in b. The sites are numbered in the following order: (1) AddNeuroMed, (2) ADNI, (3) ADNI2GO, (4) BETULA, (5) BFS, (6) BIG, (7) BIG-Rep, (8) BrainSCALE, (9) BRCDECC, (10) CHARGE, (11) EPIGEN, (12) GIG, (13) GSP, (14) HUBIN, (15) IMAGEN, (16) IMpACT, (17) LBC1936, (18) Lieber, (19) MAS, (20) MCIC, (21) MooDS, (22) MPIP, (23) NCNG, (24) NESDA, (25) neuroIMAGE, (26) neuroIMAGE-Rep, (27) NIMH, (28) NTR-Adults, (29) OATS, (30) PAFIP, (31) QTIM, (32) SHIP, (33) SHIP-TREND, (34) SYS, (35) TCD-NUIG, (36) TOP, (37) UCLA-BP-NL and (38) UMCU.
Extended Data Figure 6. Cross-structure analyses.
a, Radial plots of effect sizes from the discovery sample for all genome-wide significant SNPs identified in this study. Plots indicate the effect of each genetic variant, quantified as percentage variance explained, on the eight volumetric phenotypes studied. As expected, the SNPs identified with influence on a phenotype show the highest effect size for that phenotype: putamen volume (rs945270, rs62097986, rs608771 and rs683250), hippocampal volume (rs77956314 and rs61921502), caudate volume (rs1318862) and ICV (rs17689882). In general much smaller effects are observed on other structures. b, Correlation heat map of GWAS test statistics (t-values) and hierarchical clustering55. Independent SNPs were chosen within an linkage disequilibrium block based on the highest association in the multivariate cross-structure analysis described in Extended Data Fig. 6c. Two heat maps are shown taking only independent SNPs with either P < 1 × 10−4 (left) or P < 0.01 (right) in the multivariate cross-structure analysis. Different structures are labelled in developmentally similar regions by the colour bar on the top and side of the heat map including basal ganglia (putamen, pallidum, caudate and accumbens; blue), amygdalo–hippocampal complex (hippocampus and amygdala; red), thalamus (turquoise) and ICV (black). Hierarchical clustering showed that developmentally similar regions have mostly similar genetic influences across the entire genome. The low correlation with the ICV is owing to it being used as a covariate in the subcortical structure GWAS associations. c, A multivariate cross-structure analysis of all volumetric brain traits. A Manhattan plot (left) and corresponding quantile–quantile plot (right) of multivariate GWAS analysis of all traits (volumes of the accumbens, amygdala, caudate, hippocampus, pallidum, putamen, thalamus, and ICV) in the discovery data set using the TATES method9 is shown. Multivariate cross-structure analysis confirmed the univariate analyses (see Table 1), but did not reveal any additional loci achieving cross-structure levels of significance.
Extended Data Figure 7. Pathway analysis of GWAS results for each brain structure.
A pathway analysis was performed on each brain volume GWAS using KGG42 to conduct gene-based tests and the Reactome database for pathway definition43. Pathway-wide significance was calculated using a Bonferroni correction threshold accounting for the number of pathways and traits tested such that Pthresh = 0.05/(671 pathways × 7 independent traits) = 1.06 × 10−5 and is shown here as a red line. The number of independent traits was calculated by accounting for the non-independence of each of the eight traits examined (described in the Methods). Variants that influence the putamen were clustered near genes known to be involved in DSCAM interactions, neuronal arborization and axon guidance56. Variants that influence intracranial volume are clustered near genes involved in EGFR and phosphatidylinositol-3-OH kinase (PI(3)K)/AKT signalling pathways, known to be involved in neuronal survival57. All of these represent potential mechanisms by which genetic variants influence brain structure. It is important to note that the hybrid set-based test (HYST) method for pathway analysis used here can be strongly influenced by a few highly significant genes, as was the case for putamen hits in which DCC and BCL2L1 were driving the pathway results.
Extended Data Figure 8. Spatio-temporal maps showing expression of genes near the four significant putamen loci over time and throughout regions of the brain.
Spatio-temporal gene expression13 was plotted as normalized log2 expression. Different areas of the neocortex (A1C, primary auditory cortex; DFC, dorsolateral prefrontal cortex; IPC, posterior inferior parietal cortex; ITC, inferior temporal cortex; MFC, medial prefrontal cortex; M1C, primary motor cortex; OFC, orbital prefrontal cortex; STC, superior temporal cortex; S1C, primary somatosensory cortex; VFC, ventrolateral prefrontal cortex; V1C, primary visual cortex) as well as subcortical areas (AMY, amygdala; CBC, cerebellar cortex; HIP, hippocampus; MD, mediodorsal nucleus of the thalamus; STR, striatum) are plotted from 10 post-conception weeks (PCW) to more than 60 years old. Genes that probably influence putamen volume are expressed in the striatum at some point during the lifespan. After late fetal development, KTN1 is expressed in the human thalamus, striatum and hippocampus and is more highly expressed in the striatum than the cortex. Most genes seem to have strong gradients of expression across time, with DCC most highly expressed during early prenatal life, and DLG2 most highly expressed at mid-fetal periods and throughout adulthood. BCL2L1, which inhibits programmed cell death, has decreased striatal expression at the end of neurogenesis (24–38 PCW), a period marked by increased apoptosis in the putamen15.
Extended Data Figure 9. CTCF-binding sites in the vicinity of the putamen locus marked by rs945270.
CTCF-binding sites from the ENCODE project are displayed from the database CTCFBSDB 2.0 (ref. 23) from two different cell types: embryonic stem cells (track ENCODE_Broad_H1-hESC_99540) and a neuroblastoma cell line differentiated with retinoic acid (ENCODE_UW_SK-N-SH_RA_97826). A proxy SNP to the top hit within the locus, rs8017172 (r2 = 1.0 to rs945270), lies within a CTCF-binding site called based on ChIP-seq data in the embryonic stem cells and near the binding site in neural SK-N-SH cells. As this is the lone chromatin mark in the intergenic region (see Extended Data Fig. 3), it suggests that the variant may disrupt a CTCF-binding site and thereby influence transcription of surrounding genes.
Extended Data Figure 10. Shape analysis in 1,541 young healthy subjects shows consistent deformations of the putamen regardless of segmentation protocol.
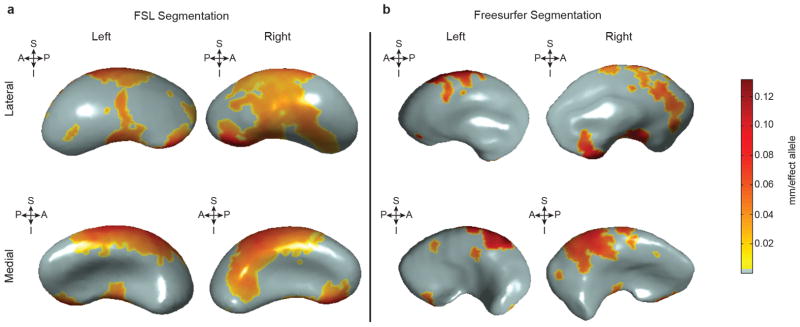
a, b, The distance from a medial core to surfaces derived from FSL FIRST (a; identical to Fig. 2c) or FreeSurfer (b) segmentations was derived in the same 1,541 subjects. Each copy of the rs945270-C allele was significantly associated with an increased width in coloured areas (false discovery rate corrected at q = 0.05) and the degree of deformation is labelled by colour. The orientation is indicated by arrows. A, anterior; I, inferior; P, posterior; S, superior. Shape analysis in both software suites gives statistically significant associations in the same direction. Although the effects are more widespread in the FSL segmentations, FreeSurfer segmentations also show overlapping regions of effect, which appears strongest in anterior and superior sections.
Extended Data Figure 11. The phenotypic variance explained by all common variants in this study.
a, Twin-based heritability (with 95% confidence intervals), measuring additive genetic influences from both common and rare variation, is shown for comparison with common variant based heritability (see Methods). b, The median estimated percentage of phenotypic variance explained by all SNPs (and 95% confidence interval) is given for each brain structure studied41. The full genome-wide association results from common variants explain approximately 7–15% of variance depending on the phenotype. c, The median estimated variance explained by each chromosome is shown for each phenotype. d, Some chromosomes explain more variance than would be expected by their length, for example chromosome 18 in the case of the putamen, which contains the DCC gene.
Supplementary Material
Acknowledgments
Funding sources for contributing sites and acknowledgments of contributing consortia authors can be found in Supplementary Note 3.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions Individual author contributions are listed in Supplementary Note 4.
Summary statistics from GWAS results are available online using the ENIGMA-Vis web tool: http://enigma.ini.usc.edu/enigma-vis/.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Blokland GA, de Zubicaray GI, McMahon KL, Wright MJ. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res Hum Genet. 2012;15:351–371. doi: 10.1017/thg.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kravitz AV, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poldrack RA, et al. Interactive memory systems in the human brain. Nature. 2001;414:546–550. doi: 10.1038/35107080. [DOI] [PubMed] [Google Scholar]
- 4.Pessiglione M, Seymour B, Flandin G, Dolan RJ, Frith CD. Dopamine-dependent prediction errors underpin reward-seeking behaviour in humans. Nature. 2006;442:1042–1045. doi: 10.1038/nature05051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein JL, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikram MA, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nature Genet. 2012;44:539–544. doi: 10.1038/ng.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Sluis S, Posthuma D, Dolan CV. TATES: efficient multivariate genotype-phenotype analysis for genome-wide association studies. PLoS Genet. 2013;9:e1003235. doi: 10.1371/journal.pgen.1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar J, Yu H, Sheetz MP. Kinectin, an Essential Anchor for Kinesin-Driven Vesicle Motility. Science. 1995;267:1834–1837. doi: 10.1126/science.7892610. [DOI] [PubMed] [Google Scholar]
- 12.Hamasaki T, Goto S, Nishikawa S, Ushio Y. A role of netrin-1 in the formation of the subcortical structure striatum: repulsive action on the migration of late-born striatal neurons. J Neurosci. 2001;21:4272–4280. doi: 10.1523/JNEUROSCI.21-12-04272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motoyama N, et al. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 15.Itoh K, et al. Apoptosis in the basal ganglia of the developing human nervous system. Acta Neuropathol. 2001;101:92–100. doi: 10.1007/s004010000252. [DOI] [PubMed] [Google Scholar]
- 16.Scannevin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nature Rev Neurosci. 2000;1:133–141. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- 17.Nithianantharajah J, et al. Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nature Neurosci. 2013;16:16–24. doi: 10.1038/nn.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bis JC, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nature Genet. 2012;44:545–551. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deans MR, et al. Control of neuronal morphology by the atypical cadherin Fat3. Neuron. 2011;71:820–832. doi: 10.1016/j.neuron.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefansson H, et al. A common inversion under selection in Europeans. Nature Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 22.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziebarth JD, Bhattacharya A, Cui Y. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res. 2013;41:D188–D194. doi: 10.1093/nar/gks1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez DG, et al. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis. 2012;47:20–28. doi: 10.1016/j.nbd.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramasamy A, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nature Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nature Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyoshima I, Sheetz MP. Kinectin distribution in chicken nervous system. Neurosci Lett. 1996;211:171–174. doi: 10.1016/0304-3940(96)12752-x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Kinectin-mediated endoplasmic reticulum dynamics supports focal adhesion growth in the cellular lamella. J Cell Sci. 2010;123:3901–3912. doi: 10.1242/jcs.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen MX, Schoene-Bake JC, Elger CE, Weber B. Connectivity-based segregation of the human striatum predicts personality characteristics. Nature Neurosci. 2009;12:32–34. doi: 10.1038/nn.2228. [DOI] [PubMed] [Google Scholar]
- 30.Parent A, Hazrati LN. Functional anatomy of the basal ganglia. I. The cortico-basal ganglia-thalamo-cortical loop. Brain Res Brain Res Rev. 1995;20:91–127. doi: 10.1016/0165-0173(94)00007-c. [DOI] [PubMed] [Google Scholar]
- 31.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischl B, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 33.Morey RA, et al. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum Brain Mapp. 2010;31:1751–1762. doi: 10.1002/hbm.20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 37.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boker S, et al. OpenMx: an open source extended structural equation modeling framework. Psychometrika. 2011;76:306–317. doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walters R, Bartels M, Lubke G. Estimating variance explained by all variants in meta-analysis with heterogeneity. Behav Genet. 2013;43:543. [Google Scholar]
- 41.So HC, Li M, Sham PC. Uncovering the total heritability explained by all true susceptibility variants in a genome-wide association study. Genet Epidemiol. 2011;35:447–456. doi: 10.1002/gepi.20593. [DOI] [PubMed] [Google Scholar]
- 42.Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li MX, Kwan JS, Sham PC. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012;91:478–488. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramasamy A, et al. Resolving the polymorphism-in-probe problem is critical for correct interpretation of expression QTL studies. Nucleic Acids Res. 2013;41:e88. doi: 10.1093/nar/gkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trabzuni D, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutman BA, et al. Maximizing power to track Alzheimer’s disease and MCI progression by LDA-based weighting of longitudinal ventricular surface features. Neuroimage. 2013;70:386–401. doi: 10.1016/j.neuroimage.2012.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutman BA, Wang YL, Rajagopalan P, Toga AW, Thompson PM. Shape matching with medial curves and 1-d group-wise registration. 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI); 2012. pp. 716–719. [Google Scholar]
- 48.Schumann G, et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15:1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 49.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheung I, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci USA. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 55.Hager R, Lu L, Rosen GD, Williams RW. Genetic architecture supports mosaic brain evolution and independent brain-body size regulation. Nat Commun. 2012;3:1079. doi: 10.1038/ncomms2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23:147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- 57.Brunet A, Datta SR, Greenberg ME. Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol. 2001;11:297–305. doi: 10.1016/s0959-4388(00)00211-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.