Summary
Organismal development requires the precise coordination of genetic programs to regulate cell fate and function. MEF2 transcription factors (TFs) play essential roles in this process but how these broadly expressed factors contribute to the generation of specific cell types during development is poorly understood. Here we show that despite being expressed in virtually all mammalian tissues, in the retina MEF2D binds to retina-specific enhancers and controls photoreceptor cell development. MEF2D achieves specificity by cooperating with a retina-specific factor CRX, which recruits MEF2D away from canonical MEF2 binding sites, and redirects it to retina-specific enhancers that lack the consensus MEF2-binding sequence. Once bound to retina-specific enhancers, MEF2D and CRX co-activate the expression of photoreceptor-specific genes that are critical for retinal function. These findings demonstrate that broadly expressed TFs acquire specific functions through competitive recruitment to enhancers by tissue-specific TFs, and through selective activation of these enhancers to regulate tissue-specific genes.
Introduction
A remarkable feature of the development of complex multicellular organisms is that this extraordinary process is controlled by a limited number of transcription factors (TFs). TFs regulate the exquisite patterns of gene expression that determine the tissues and cell types of an organism. The diversity of TF function is especially evident in the central nervous system (CNS), where a vast array of different cell types gives rise to our ability to extract information from the external environment, process that information and respond appropriately. While a number of TFs with tissue-specific expression have been identified, most TFs function in a wide range of cell types, yet can still contribute to cell-type-specific gene expression. The best evidence that broadly expressed TFs contribute to cell-type-specific functions is that mutations in broadly expressed TFs may result in tissue-specific disease phenotypes (Amiel et al., 2007; Bienvenu et al., 2013). Nevertheless, it is not yet well understood how the function of widely expressed TFs is tailored to achieve cell type specificity. Elucidating the mechanisms that specify the function of a broadly expressed TF within a given tissue is critical for understanding how genes are differentially regulated to achieve the diversity of cell types throughout the organism.
The highly conserved myocyte enhancer factor-2 family of TFs (MEF2A–D) is expressed in virtually all cells of multi-cellular organisms yet plays specific and critical roles in the development of the brain, muscle, bone and hematopoietic lineages (reviewed in (Potthoff and Olson, 2007)). In the mammalian nervous system, MEF2 factors regulate neural progenitor differentiation, neurotrophin- and activity-dependent neuronal survival, activity-dependent restriction of excitatory synapse number as well as synaptic plasticity and behavior (Barbosa et al., 2008; Cole et al., 2012; Flavell et al., 2006; Li et al., 2008; Pulipparacharuvil et al., 2008; Shalizi et al., 2006). In humans, mutations in MEF2C can lead to severe intellectual disability, epilepsy and an absence of speech (Bienvenu et al., 2013). Despite their clear importance in the nervous system and many other tissues, relatively little is known about how these globally expressed TFs achieve cell type-specific functions in the development of the nervous system and a wide range of other tissues.
One clue as to how MEF2 family members achieve their tissue-specific functions has been provided by identification of MEF2 target genes in distinct tissues. Gain- and loss-of-function approaches and a limited number of chromatin immunoprecipitation assays have shown that MEF2 controls the transcription of synaptic regulatory proteins such as Arc and Syngap1 in neurons, Myog in myocytes, and II2 in hematopoietic cells, among other genes (Flavell et al., 2008; Pan et al., 2004; Potthoff and Olson, 2007; Sebastian et al., 2013). While to date no experiments have directly compared MEF2 target genes across different tissues, these findings suggest that MEF2 functions at least in part by regulating distinct target genes in different cell types. However, the mechanisms by which the tissue-specific targets of MEF2 are determined remain poorly understood.
The effect of MEF2 on gene expression in a given cell type must be determined in part by where MEF2 binds across the genome. MEF2 family members are known to recognize and bind to a common consensus DNA motif ((C/T)TA(A/T)4TA(A/G)) termed the MEF2-responsive element (MRE) (Potthoff and Olson 2007; Flavell et al., 2008). In vitro experiments indicate that DNA sequences that conform to this consensus site bind MEF2 with higher affinity compared to non-consensus sequences (Pollock and Treisman, 1991). These findings have led to the suggestion that MEF2 binding in a cell might be inferred from the presence of strong consensus MREs within the promoters or enhancers of genes whose expression is altered when MEF2 function is inhibited. MEF2 could therefore achieve tissue-specific function by binding to consensus MREs that are differentially accessible in each tissue due to the unique chromatin landscape of that tissue. Alternatively, MEF2 could bind to the same regulatory elements in all tissues, but function at just a subset of these sites in a given tissue. Significant innovations in high-throughput sequencing technology now make it possible to identify sites of TF binding genome-wide and to assess the activity of each bound region (Creyghton et al., 2010; Johnson et al., 2007; Kim et al., 2010; Rada-Iglesias et al., 2011); however, these approaches have not yet been used to determine whether or how MEF2 selectively regulates enhancers or promoters in a tissue-specific manner.
Once bound to an MRE that is present in an enhancer or target gene promoter, MEF2 is believed to either repress or activate nearby target genes, largely through interactions with co-factors such as histone acetyltransferases and histone deacetylases (Potthoff and Olson, 2007). In addition, it has been suggested that in myocytes MEF2 family members work together with muscle-specific bHLH factors to regulate gene expression (Molkentin et al., 1995). While several models for the possible functions of this interaction have been proposed, including cooperative binding or cooperative activation, the endogenous mechanisms by which MEF2 interacts with co-factors to regulate target genes in a tissue specific manner remain unknown.
To determine how a widely expressed TF such as MEF2 regulates tissue-specific gene expression during key steps in mammalian CNS development, we identified a cell type in the CNS, the photoreceptor cells of the mouse retina, where a single MEF2 family member, MEF2D, is predominantly expressed. A newly generated loss-of-function allele for MEF2D revealed a critical role for MEF2D in mouse retinal photoreceptor development and in the regulation of cell-type-specific gene expression, including genes that are mutated in human retinal diseases. In vivo genomic and phenotypic analyses demonstrate that MEF2D regulates cell type-specific gene expression in photoreceptors by binding to cell-type-specific enhancers together with the homeodomain TF CRX. Analysis of MEF2D binding and enhancer activation in Crx knockout retinas revealed that CRX biases a genome-wide competition among enhancers for MEF2D binding. In wild type (WT) retinas, CRX recruits MEF2D to tissue specific enhancers that lack the consensus MEF2 binding sites, thus redirecting MEF2D away from strong MREs. Once bound to retina-specific enhancers, MEF2D and CRX work cooperatively to activate the expression of photoreceptor-specific genes that are critical for retina function. Together, these findings demonstrate that broadly expressed TFs can acquire tissue-specific functions through competitive recruitment to enhancers by tissue-specific TFs and through selective activation of these enhancers to regulate tissue-specific target genes.
Results
Mef2d is required cell-autonomously for photoreceptor development and function
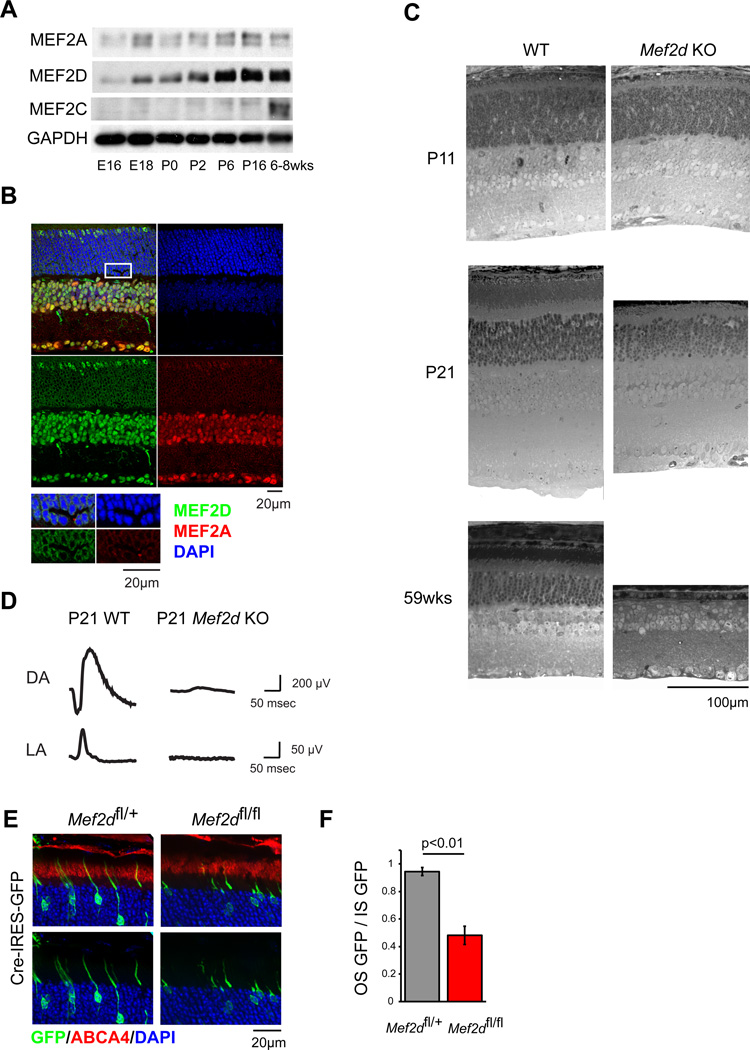
To address how the broadly expressed MEF2 TF family achieves tissue-specific function in the CNS, we focused our studies on a single family member, Mef2d, that is widely expressed and has been implicated in critical aspects of neural development (Flavell et al., 2006; Flavell et al., 2008). We first sought to identify a region of the CNS where MEF2D is the predominant family member expressed and therefore is not likely to be functionally redundant with other MEF2 family members. We reasoned that such a region could serve as an experimental system for understanding the context-dependent role of MEF2D in neuronal development. An investigation of MEF2 expression in the cortex, hippocampus and cerebellum revealed that the major cell types of these regions co-express MEF2 family members (data not shown). We therefore turned to the retina, whose well-characterized and spatially separated cell types might allow us to identify a cell type that exclusively expresses MEF2D. We found that both MEF2A and MEF2D are expressed in the developing retina (Figure 1A). In contrast, MEF2C is only expressed after retinal development is complete. While MEF2A and D are co-expressed at different levels relative to one another in retinal neurons such as horizontal, bipolar, amacrine and retinal ganglion cells (Figure 1B), the ratio of MEF2D to MEF2A expression appears to be highest in maturing photoreceptor cells in the outer nuclear layer (ONL) (Figure 1B). In these cells MEF2D is distributed in a pattern consistent with nuclear localization and binding to active euchromatin (Corbo et al., 2010). Retinal photoreceptor cells are a specialized class of primary sensory neurons that detect the incidence of photons upon the retina and transduce this event into a neural signal for processing by downstream regions of the visual system. Given the importance of MEF2 family members in neuronal development we hypothesized that MEF2D may play a critical role in the development of photoreceptor cells and that characterization of this role might yield important insights into the cell-type specific functions of MEF2 factors.
Figure 1. MEF2D is required cell-autonomously for photoreceptor development and function.
(A) Immunoblot of MEF2 family member expression in mouse retina over development. (B) Immunofluorescence of P25 WT retinal cross-sections for MEF2A (red) and MEF2D (green) with DAPI (blue). Inset highlights rod photoreceptor nuclei. ONL, outer nuclear layer; INL, inner nuclear layer; GCL ganglion cell layer. (C) Toluidine blue-stained cross-sections of WT and Mef2d KO littermate retinas. OS, photoreceptor outer segments; IS, photoreceptor inner segments. (D) Representative electroretinograms (ERGs) from a P21 WT mouse and a Mef2d KO littermate under dark-adapted (DA) and light-adapted (LA) conditions. (E) Representative immunofluorescence images of P21 retinas electroporated at P0 for sparse expression of Cre recombinase and GFP (green) in either Mef2dfl/+ or Mef2dfl/fl mice. ABCA4 immunostaining (red) was used to identify photoreceptor cell outer segments (OS). (F) Quantification of OS from GFP-positive photoreceptors as shown in (E). Mean GFP intensity in the ABCA4-positive region (OS GFP) was normalized to mean GFP intensity in the inner segments (IS GFP) to measure OS development while controlling for electroporation density (Mef2dfl/+, N=6; Mef2dfl/fl, N=3 retinas). Error bars represent S.E.M.
To explore the role of MEF2D in photoreceptors we generated a Mef2d knockout allele in which the first five protein-coding exons of Mef2d, including the entirety of the highly conserved MADS and MEF2 DNA-binding and dimerization domains, were removed (Figure S1A–F). This allele differs from a previously generated mutant allele of Mef2d, in which only the second coding exon of Mef2d was removed (Kim et al., 2008). This previously generated allele results in a highly expressed truncated protein product with a partial DNA binding domain. The presence of this residual protein product could potentially complicate the interpretation of phenotypes in these mice. This potential problem is eliminated in the new line of conditional MEF2D knockout mice, termed Mef2dΔ2–6 (Figure S1A–F). These mice were crossed with the germline EIIA:cre deleter strain to produce a stable constitutive knockout line. The absence of MEF2D expression in all cells of the retina was confirmed by immunofluorescence. This constitutive Mef2d knockout line, referred to below as Mef2d KO was used for all subsequent analyses, except where noted.
Mef2d KO mice are born in Mendelian ratios and are fertile but exhibit a slightly decreased body weight compared to WT littermates (p=1.35e–6; Figure S1G). While MEF2D is expressed throughout the CNS, the brains of Mef2d KO mice appear normal, most likely due to compensation by other MEF2 family members. By contrast, the retinas of Mef2d KO mice display a significant defect in the maturation of rod and cone photoreceptors. At postnatal day 11 (p11) Mef2d KO retinas are grossly normal and contain all major cell types. However, by p21, Mef2d KO photoreceptor cells differ strikingly from WT photoreceptors in that they lack the outer segment structures that are necessary for vision (Figure 1C). Photoreceptor cell outer segments are an apical organelle of stacked membranous discs in which phototransduction occurs and where the cascade of neuronal signaling that underlies the visual response to light begins. The development of these structures normally occurs between p11 and p28, which corresponds to the functional maturation of photoreceptor cells. The failure of outer segment development in Mef2d KO retinas should lead to a deficit in photoreceptor cell function. Correspondingly, in vivo electroretinograms (ERGs) performed at p21 revealed that visual responses are almost completely absent in Mef2d KO mice compared with WT littermates in both dark and light-adapted conditions (Figure 1D, Figure S1H). This indicates that both rod and cone photoreceptor cells have markedly impaired function in Mef2d KO mice. Very few apoptotic cells are present in Mef2d KO retinas at p21, however the failure of Mef2d KO photoreceptor cells to develop normally eventually leads to a slow retinal degeneration (Figure 1C). Taken together these findings indicate that MEF2D is required for photoreceptor cell development, long-term survival and vision.
Since MEF2D is the only MEF2 family member expressed in retinal photoreceptors, the developmental failure of photoreceptors to form outer segments in Mef2d KO retinas seems likely to be due to a cell-intrinsic requirement for Mef2d. However, to rule out the possibility that a MEF2D-dependent alteration in the extracellular environment during photoreceptor development is responsible for the deficits observed we selectively removed Mef2d from individual developing photoreceptors by sparsely introducing Cre recombinase into MEF2Dfl/fl photoreceptors using in vivo electroporation. We found that at p21 photoreceptors in which the expression of MEF2D is disrupted have highly abnormal outer segments when compared to photoreceptors still expressing MEF2D (Figure 1E, F & S1I). The abnormal morphology was also reproduced in photoreceptors where MEF2D shRNA was selectively introduced in an otherwise WT retina (Figure S1J–L). Finally, the disruption of photoreceptor development was reversed when the MEF2D shRNA was co-expressed with an shRNA-resistant form of MEF2D (Figure S1M & N). Taken together these findings suggest that MEF2D functions cell-autonomously to promote photoreceptor development, and that in the absence of MEF2D, photoreceptors fail to mature, are non-functional, and ultimately die.
MEF2D directly regulates critical cell-type specific targets and disease genes in the retina
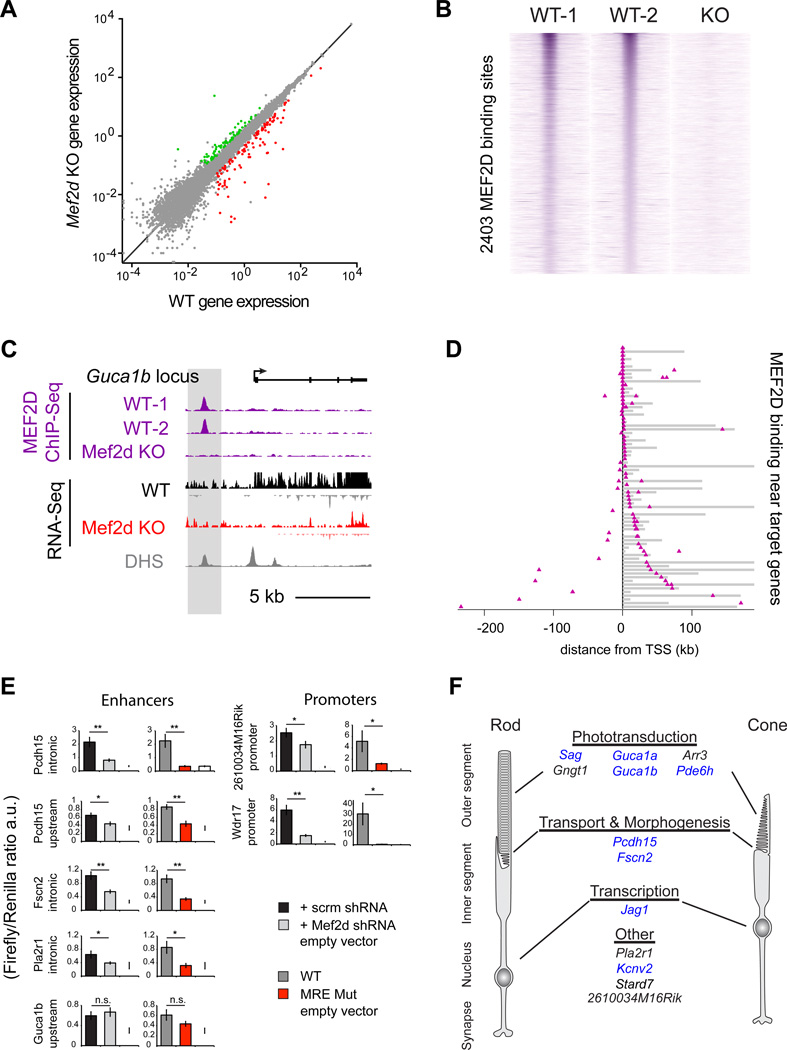
Given that MEF2D is a TF, it is likely that the gene targets of MEF2D regulate aspects of photoreceptor development and function. One possibility is that MEF2D regulates photoreceptor-specific target genes that encode proteins necessary for photoreceptor development and function. Alternatively, since MEF2 family members are expressed in a multitude of different tissues, it could be that MEF2D controls photoreceptor differentiation by regulating a core set of target genes that are shared across cell types. To distinguish between these possibilities we carried out a series of experiments to identify and characterize the direct targets of MEF2D in the retina. We reasoned that a gene must meet three criteria to be considered a direct target. First, candidate target genes must be misregulated in Mef2d KO retinas. Second, target genes must have MEF2D bound at their promoters or associated enhancers. Third, the binding of MEF2D at those promoters or enhancers must be required for proper regulation of direct target genes. Genes that met each of these three criteria were defined as bona fide targets of MEF2D in the retina.
To begin to identify the targets genes of MEF2D we first performed high-throughput RNA sequencing (RNA-seq) of total RNA from WT and Mef2d KO retinas at p11 (Figure 2A, S2A & B). At p11, MEF2D is strongly expressed but WT and Mef2d KO retinas are morphologically indistinguishable. Thus, differences in gene expression between WT and Mef2d KO retinas at p11 should be primarily due to the disruption of MEF2D-dependent transcriptional programs rather than due to cell attrition or the secondary effects of disrupted retinal development. We find that the expression of most genes is unchanged when WT and Mef2d KO p11 retinas are compared (Spearman’s correlation coefficient ρ= 0.989) (Figure 2A). However, a subset of 185 genes is strongly misregulated in Mef2d KO retinas (Figure 2A, S2C & Table S1). This set of misregulated genes is significantly enriched for retina-specific genes (modified Fisher’s exact test; p=5e-9) according to the DAVID web tool (Huang da et al., 2009a, b). Furthermore, gene ontology (GO) analysis found that these misregulated genes are most enriched for genes involved in processes such as visual perception and sensory perception of a light stimulus (modified Fisher’s exact test; p=1.51e-4 and p=1.59e-4, respectively) (Figure S2D). This analysis is most consistent with the hypothesis that MEF2D promotes photoreceptor development by regulating a network of genes essential for photoreceptor function rather than a common set of core target genes shared across cell types and tissues.
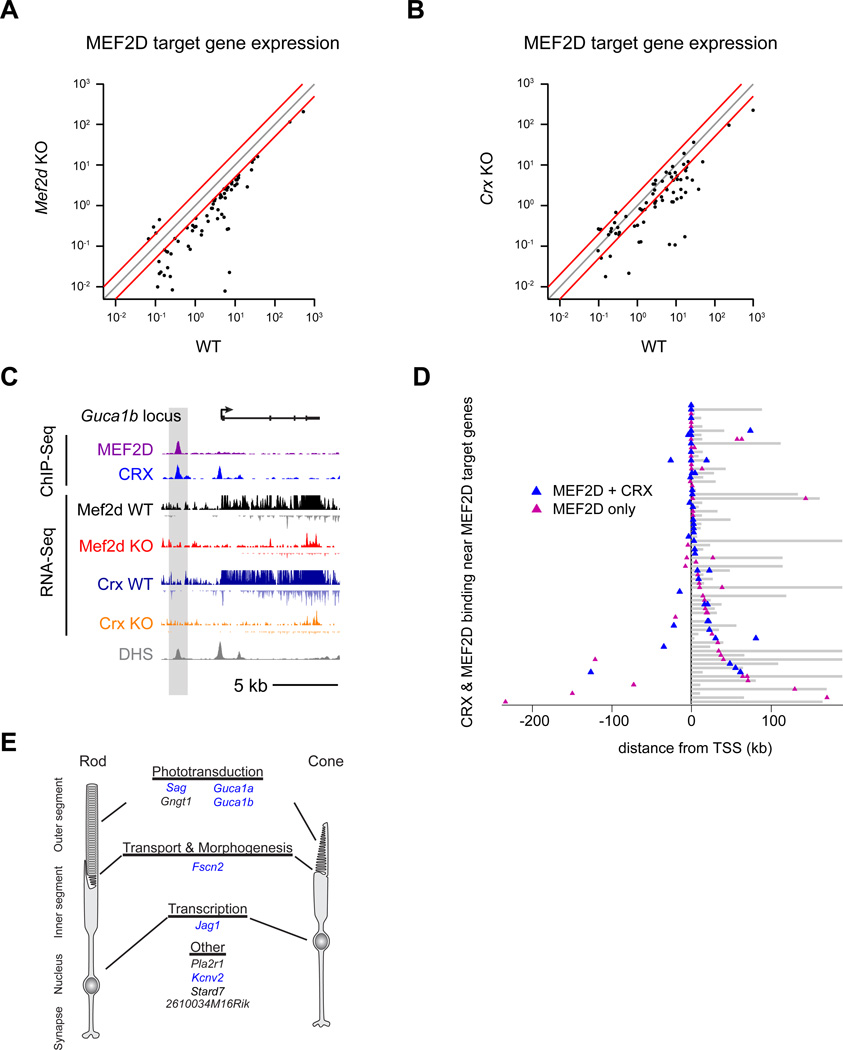
Figure 2. MEF2D directly regulates critical cell-type specific targets and disease genes in the retina.
(A) RNA-seq average exon density for individual genes in P11 WT and MEF2D KO retinas are displayed in gray (n=2 per genotype). Genes were considered upregulated (green) or downregulated (red) only if expressed at >0.1 in 2 WT or 2 KO samples, if KO/WT exon density was ≥2× or <0.5 and if the log p-value of the difference of mean log-densities between KO and WT datasets <0.05 (Student’s t test). (B) ChIP-seq signal at 2,403 high confidence MEF2D genomic binding sites for two WT and one MEF2D KO experiment from P11 retinas. Each MEF2D binding site is represented as a single horizontal line (purple) centered on the peak summit. Intensity of color correlates with peak size. MEF2D peaks are ordered according to peak size. (C) UCSC genome browser tracks for MEF2D ChIP-seq and RNA-seq experiments as well as DNAse hypersensitivity (DHS) data (ENCODE Consortium) at the Guca1b genomic locus. (D) Distribution of MEF2D binding (purple triangles) in the retina as determined by ChIP-seq with respect to 71 candidate target genes. Target gene bodies (gray) are aligned at their transcriptional start sites (TSS, black line). MEF2D binds at promoters (open triangles) and/or enhancers (filled triangles) of candidate target genes in the retina. Gene loci are ordered according to the distance of MEF2D binding to the TSS. (E) Luciferase reporter assays performed in retinal explants to determine the activity of WT MEF2D-bound target gene enhancers or promoters with scrambled (scrm) or Mef2d shRNA knockdown or with mutated MEF2D–binding sites (MRE mut) (Student’s t test; *p-value <0.05; n.s. not significant). (F) Examples of direct MEF2D target genes relevant to photoreceptor cell biology. Genes implicated in human retinal diseases are highlighted in blue.
To determine with high confidence which candidate MEF2D target genes had MEF2D bound at their promoters or nearby enhancers we performed two bioreplicates of MEF2D ChIP-Seq using p11 wild type retinas, and performed MEF2D ChIP-Seq in Mef2d KO retinas as a control for specificity. Because the majority of cells in the mouse retina are photoreceptors (73–80%), it is likely that the vast majority of identified MEF2D binding sites represent MEF2D binding in photoreceptors instead of other retinal cell types (Young RL., 1985; Jeon et al., 1998). Each MEF2D ChIP-Seq replicate alone yielded ~12,000 unique MEF2D-binding sites with an overlap of ~4,000 reproducible binding sites between the two replicates, demonstrating that a high degree of biological or technical noise is inherent to these experiments. The number of high-confidence MEF2D binding sites was further decreased to 2,403 when we considered only those peaks that are specifically reduced in the MEF2D knockout (Figure 2B & S2E). Of these sites, 400 MEF2D binding sites were identified as promoters, while 2003 were identified as genetic enhancers as they are located >1000bp away from the transcriptional start site (TSS) of the nearest gene. This suggested that MEF2D acts chiefly at enhancers in photoreceptor cells. Strikingly, 2,403 is a large number of MEF2D binding sites compared to 185 highly misregulated target genes, suggesting that only a small number of the MEF2D–bound sites actively regulate gene expression. We next asked which of these MEF2D binding sites were proximal to genes that are highly misregulated in the MEF2D KO. MEF2D was found to bind at 18 promoter regions and 75 enhancers near genes that are highly misregulated in Mef2d KO retinas, including many genes that are photoreceptor-specific and associated with retinal diseases (Monte Carlo analysis, p<1×10−5, Figure 2C & D, S2E). Thus, MEF2D appears to regulate many of its photoreceptor-specific targets by binding at proximal regulatory elements, particularly at enhancers.
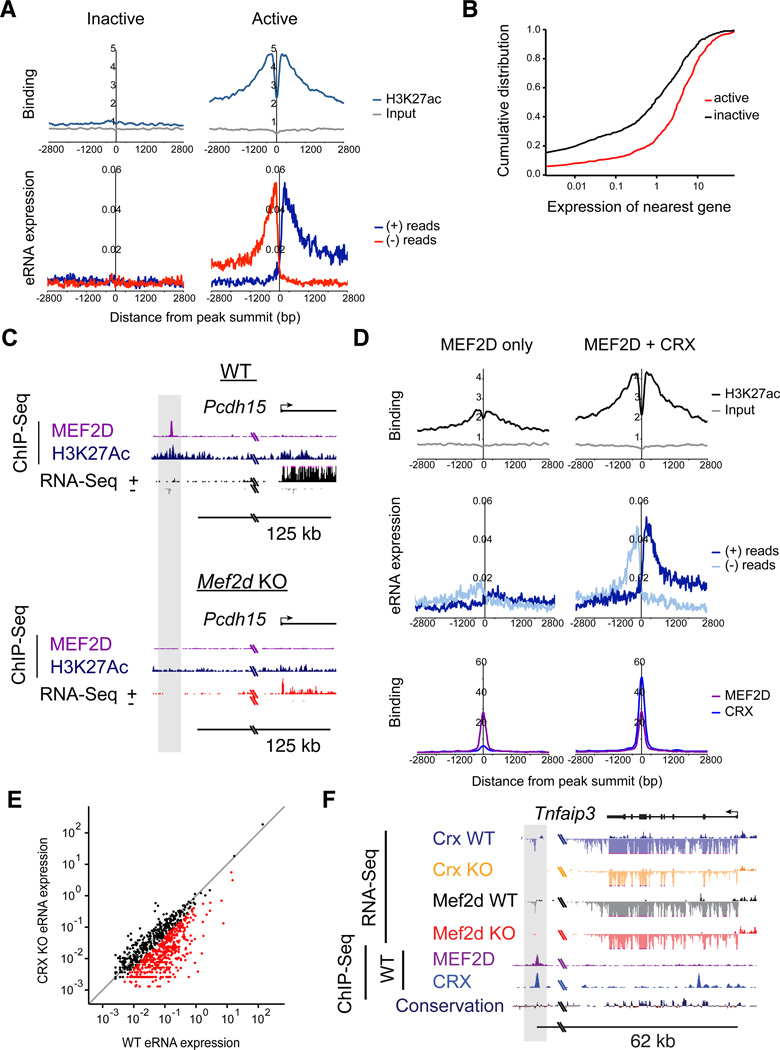
To test whether direct binding of MEF2D to the promoters or enhancers of candidate target genes is required for proper gene expression, we isolated a subset of these elements that contained consensus MREs and tested the ability of these enhancers to regulate reporter gene expression in the intact retina in a MEF2D-dependent manner. All seven of the regulatory elements that were tested are sufficient to drive reporter gene expression in photoreceptor cells and 6/7 are expressed exclusively in photoreceptor cells but not in other retinal cells as determined by electroporation of GFP expressing reporters into the developing retina (data not shown). Additionally, the activity of 6/7 of these reporters was significantly reduced in the presence of MEF2D shRNA or when the MRE was mutated as determined by electroporation of luciferase reporters into the developing retina, demonstrating that direct MEF2D binding to these elements is required to drive expression of the reporter gene (Figure 2E). The observation that expression of one of the reporters is not decreased in the presence of MEF2D shRNA, or when the MRE is mutated, suggests that MEF2D may not directly regulate gene expression through this candidate enhancer region or that this isolated sequence does not fully recapitulate the endogenous regulation of this enhancer by MEF2D. However, the analyses of the remaining reporter constructs support the hypothesis that candidate target genes identified by RNA-Seq and ChIP-Seq are indeed direct targets of MEF2D. This hypothesis was later confirmed at endogenous MEF2D–bound enhancers in vivo, by demonstrating that these enhancers lose markers of active enhancers, H3K27Ac and eRNAs, in MEF2D KO retinas (Figure 6C & S6A).
Figure 6. CRX determines the selective activation of MEF2D-bound retinal enhancers.
(A) Aggregate plots of H3K27Ac ChIP-seq signal (top) and eRNA expression (bottom) at active (N=660) or inactive (N=584) MEF2D-bound enhancers (reads per bp per peak). Plots are centered on summits of MEF2D-bound regions that are >1kb away from the nearest gene TSS. (B) Cumulative distribution of WT average exon density (from RNA-Seq data, n=2) for genes nearest active enhancers (red) versus genes nearest inactive enhancers (black) (KS test; p=9.73e-17). (C) The Pcdh15 genomic locus in WT and Mef2d KO retinas with ChIP-seq tracks for MEF2D and H3K27Ac and RNA-Seq tracks. The upstream MEF2D-bound enhancer is highlighted in gray. (D) Top, aggregate plots of H3K27Ac ChIP-seq signal at enhancers bound by MEF2D alone (left) versus those bound by MEF2D and CRX (right) in WT retinas (Student’s t test; p=4.79e-65). Middle, aggregate plots of eRNAs at enhancers bound by MEF2D alone (left) versus those bound by MEF2D and CRX (right) in WT retinas. Bottom, aggregate plots of MEF2D ChIP signal (purple) or CRX ChIP signal (blue) for same regions, demonstrating normalization of data analysis to MEF2D peak size and differential peak size of CRX. (E) Average eRNA expression at individual MEF2D-bound enhancers in WT versus Crx KO retinas. N=2 for each genotype. Read density was calculated for the 400bp window centered on the summit of each MEF2D-bound region. Gray line indicates unity. Expression of eRNAs where Crx KO/WT expression was equal or less than 0.5 is indicated with red points. (F) Expression of eRNAs at a putative enhancer bound by MEF2D and CRX upstream of the Tnfaip3 genomic locus in WT, Mef2d KO, and Crx KO retinas with RNA-seq tracks and ChIP-seq tracks for MEF2D and CRX in WT retinas. eRNA expression and MEF2D and CRX binding in the upstream enhancer is highlighted in gray.
Intriguingly, the most highly misregulated MEF2D target genes identified by these analyses have critical roles in photoreceptor function (Figure 2F & S2C; Table S2). For example, Sag, Gngt1, Arr3, Pde6h, Guca1a and Guca1b are key components of the phototransduction cascade. Misregulation of these transcripts in combination would be expected to severely disrupt phototransduction and is likely to be the primary cause of the abnormal photoresponses in Mef2d KO mice. Indeed human mutations in Sag, Pde6h, Guca1a and Guca1b are all associated with visual disorders (Fuchs et al., 1995; Kohl et al., 2012; Payne et al., 1998; Sato et al., 2005). Other photoreceptor-specific target genes which are not directly part of the phototransduction cascade may contribute to the structural defects in outer segment formation observed in Mef2d KO retinas. For example, Fscn2, a photoreceptor-specific actin-bundling protein, is mutated in retinitis pigmentosa and has been demonstrated to be necessary for outer segment elongation (Wada et al., 2001; Yokokura et al., 2005). Similarly, Pcdh15, a cadherin superfamily member, has been implicated in vesicular trafficking between the inner and outer segments and is mutated in a form of Usher Syndrome (USH1F) characterized by visual impairment and hearing loss (Cosgrove and Zallocchi, 2014). In contrast, MEF2 targets that have been identified in other neuronal cell types such as Nur77, Arc and Syngap1 (Flavell et al., 2008) are not strongly expressed in the developing retina under normal conditions. These analyses suggest that the primary function of MEF2D in the retina is to regulate the expression of genes that are critical for specific photoreceptor functions rather than genes with common functions across cell types.
MEF2D binds and regulates tissue-specific enhancers with the retina co-factor CRX
We next sought to understand the mechanisms by which MEF2D binds to and regulates its targets in photoreceptor cells. Elucidating this mechanism is of special interest because many MEF2D targets are essential for photoreceptor function and are mutated in human diseases of the retina. The simplest and most prevalent model of MEF2 function is that MEF2 binds to MEF2 consensus binding sites (MREs) in the promoters or enhancers of its target genes and thereby controls their expression (Edmondson et al., 1992; Sandmann et al., 2006). Given the photoreceptor-specific expression of many MEF2D target genes, in this model the binding of MEF2D would be expected at MREs that are accessible in photoreceptors but not other neuronal cell types. The ability of MEF2D to recognize and bind to these photoreceptor-specific MREs would somehow be specified during CNS development, for example through changes in DNA accessibility or through interaction with tissue-specific co-factors. A second possibility is that MEF2D binds to a common set of MREs accessible in all tissues and that these bound elements are selectively activated in a tissue-specific manner. To begin to distinguish between these possibilities we compared MEF2D binding across the retinal genome to that observed in two distinct cell types where MEF2 factors are known to play important roles.
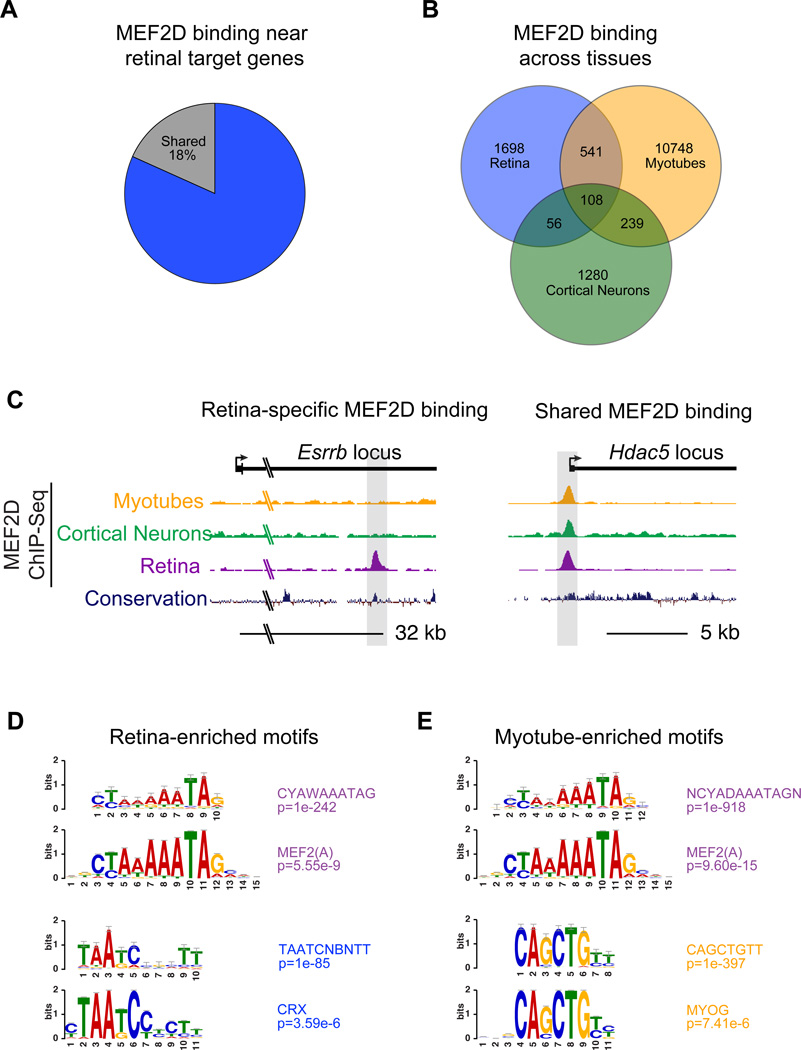
To explore the possibility that MEF2 regulates cell-type specific gene transcription by binding the genome in a cell-type specific manner, we performed ChIP-Seq for MEF2D in DIV7 cultured cortical neurons and analyzed MEF2D ChIP-Seq data in C2C12 myocytes (Sebastian et al., 2013) and compared these results to MEF2D ChIP-Seq from the retina. A comparison of MEF2D binding in each of the three tissues showed that MEF2D binding to the enhancers and promoters that are necessary for retinal-specific gene expression occurs in a highly tissue-specific manner (~82%, 76/93) (Figure 3A). Furthermore, the majority of MEF2D binding genome-wide in each tissue is tissue-specific, although some instances of shared binding did occur (Figure 3B, C & S3). These findings suggest that tissue-specific binding of MEF2D is an important mechanism governing the specific function of MEF2D in the retina.
Figure 3. MEF2D binds at tissue-specific enhancers in the retina.
(A) Percentage of MEF2D-bound near target genes that are retina-specific or are also sites of MEF2D binding in DIV7 cultured cortical neurons or myotubes. (B) Overlap of MEF2D-bound genomic regions from P11 retina, DIV7 cultured cortical neurons, and cultured myotubes (from Sebastian et al., 2013) as determined by ChIP-Seq. (C) UCSC genome browser tracks for MEF2D ChIP-seq from cultured myotubes, cortical neurons or retina as well as genomic conservation at Esrrb and Hdac5 genomic loci (D-E) Position weight matrices (PWMs) of enriched TF binding motifs within MEF2D-bound regions specific to the retina or myotubes. Below each is a high-ranking JASPAR (http://jaspar.genereg.net) matrix corresponding to the PWM (Hypergeometric test).
We next investigated the mechanism by which MEF2D achieves tissue-specific binding to promoters and enhancers in the retina. Previous studies have suggested that interactions between MEF2 family members and particular co-factors can influence the binding of MEF2D to a given regulatory element (Black et al., 1996; Molkentin et al., 1995), however, it is unknown if these interactions mediate tissue-specific MEF2 binding and the functional importance of these interactions has not been examined in vivo in an endogenous or genome-wide context. We hypothesized that MEF2D interacts with a retina-specific TF that recruits MEF2D to tissue-specific regulatory sites. To begin to identify such co-factors in an unbiased manner, we searched MEF2 binding sites in the retina for common DNA sequence features using a de novo DNA motif search program (Heinz et al., 2010). This analysis revealed that after the MRE, the most abundant motif sequence present within retina-specific MEF2D–bound regions, is the sequence TAATCNBNTT (p=1e-85) (Figure 3D). This sequence motif matches the binding site for the homeodomain TF CRX (p=3.59e-6). In contrast, when we performed a de novo motif search of MEF2D–bound regions identified in myocytes the most prevalent sequence, besides the MRE, was CAGCTGTT (p=1e-397). This is the consensus binding motif for myogenic basic helix-loop-helix (bHLH) proteins, such as MYOD (p=7.41e-6; Figure 3E), which have been suggested to be MEF2 co-factors in this cell type (Molkentin et al., 1995). In cortical neurons, however, no single recognizable motif was significantly enriched at MEF2D binding sites apart from the MRE, likely due to the extreme heterogeneity of these cells. These findings raise the possibility that the specificity of MEF2 binding in different tissues (e.g. photoreceptors versus muscle) is determined by interactions with tissue-specific co-factors. In particular, the enrichment of the CRX consensus motif at retina-specific MEF2D-binding sites suggests that in photoreceptors CRX could influence the binding of MEF2D to the promoters and enhancers of photoreceptor-specific genes.
CRX is a retina-specific TF that is mutated in several forms of human congenital blindness. Much like MEF2D, CRX is necessary for photoreceptor outer segment development and photoreceptor function (Chen et al., 1997; Freund et al., 1997; Furukawa et al., 1997; Furukawa et al., 1999; Swain et al., 1997) and the loss of function phenotypes of these two TFs in the mouse retina closely resemble one another (Figure 1C & Furukawa et al., 1999). These shared phenotypes and the enrichment of the CRX motif at MEF2D–bound sites suggested that CRX protein might bind and function with MEF2D at MEF2D–bound regulatory elements to regulate common target genes and similar biological processes.
To determine if CRX acts with MEF2D to co-regulate target genes we first performed RNA-Seq from WT and Crx KO retinas at p11 (Figure S4). As previously observed, many genes are misregulated in absence of CRX in the retina including genes that are direct targets of MEF2D (Figure 4A, B, E, S4C & Table S2) (Hsiau et al., 2007; Tran et al., 2014). Next, we analyzed CRX ChIP-Seq data (Corbo et al., 2010) for overlap with MEF2D ChIP-Seq to determine if CRX co-regulates MEF2D target genes by co-binding to shared target gene enhancers and promoters. Strikingly, we found that ~70% of target gene-associated MEF2D regulatory elements are co-bound by CRX in the retina (Figure 4C & D). These findings suggested that MEF2D may functionally interact with CRX to achieve tissue-specific binding or regulation of the promoters and enhancers of photoreceptor genes.
Figure 4. MEF2D and CRX directly co-regulate critical shared target genes.
Average gene expression levels as quantified by exon density of MEF2D direct target genes in WT versus Mef2d KO retinas (A) and WT versus Crx KO retinas (B). N=2 per genotype. Gray line indicates unity. Red lines indicate a two-fold change from unity. (C) UCSC genome browser tracks for MEF2D ChIP-seq and CRX ChIP-seq (Corbo et al., 2010) from WT retinas, as well as for RNA-seq from the retinas of p11 littermate WT and MEF2D KO or WT and CRX KO mice, as well as DNAse hypersensitivity data (ENCODE Consortium) at the Guca1b genomic locus. (D) Distribution of MEF2D binding (magenta triangles) or MEF2D and CRX co-binding (blue triangles) as determined by ChIP-seq with respect to 71 MEF2D target genes. Target gene bodies (gray) are aligned at their transcriptional start sites (TSS, black line). Gene loci are ordered according to the proximity of the nearest MEF2D binding to the TSS. (E) Examples of shared direct MEF2D and CRX target genes relevant to photoreceptor cell biology. Genes implicated in human retinal diseases are in blue.
CRX biases a genome-wide competition for MEF2D binding toward retina-specific sites
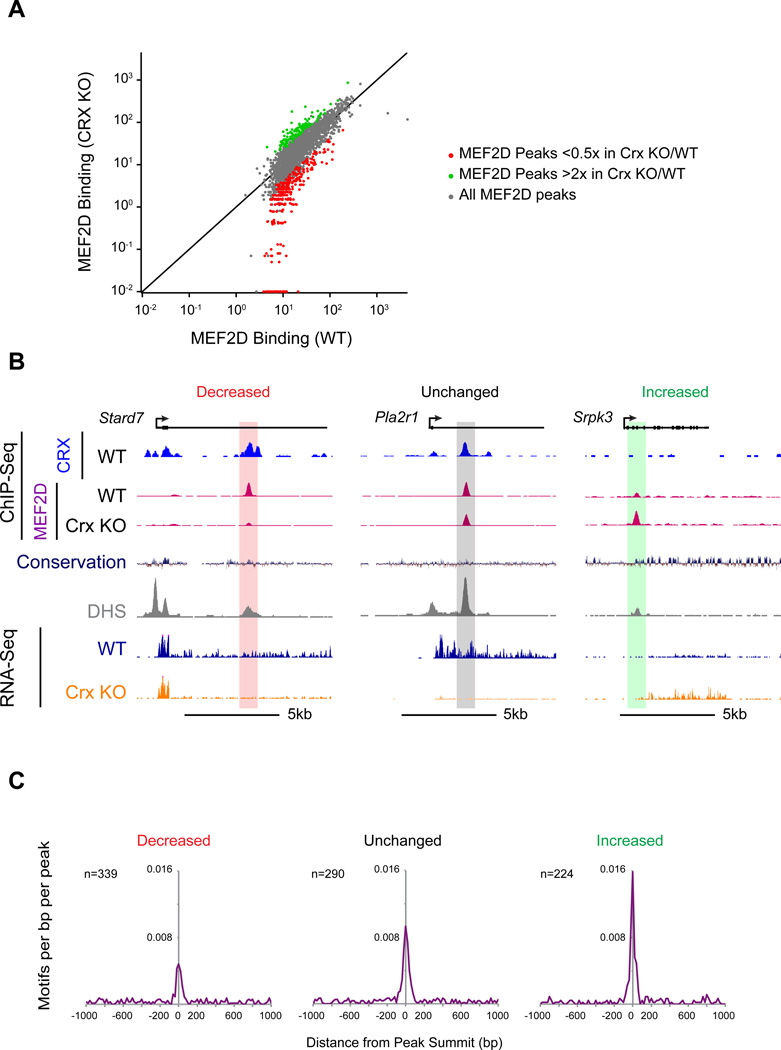
To test the possibility that CRX is required for retina-specific MEF2D binding we performed MEF2D ChIP-Seq in Crx KO retinas and compared the pattern of MEF2D binding to that of WT retinas. By ChIP-Seq we found that MEF2D binding is reduced more than twofold in Crx KO retinas at 339 MEF2D binding sites including many MEF2D regulatory elements near MEF2D target genes (Figure 5A, B & S5A, red). For example, retina-specific MEF2D binding is particularly dependent on CRX near the MEF2D target genes Gngt1, a key component of the phototransduction cascade, and Stard7, which has been shown to be required for normal photoreceptor development (Hao et al., 2012) (Figure 5B and data not shown). Mef2d is not, however, a transcriptional target of CRX and therefore the decrease in MEF2D binding at these sites is not due to decreased expression of Mef2d in Crx KO retinas (Figure S5B).
Figure 5. CRX mediates a genome-wide competition for MEF2D binding to retina-specific sites.
(A) MEF2D ChIP-Seq signal in WT versus CRX KO retinas at individual MEF2D-bound regions. Read density was calculated for the 400bp window around the summit of each MEF2D-bound region. Data from CRX KO retinas was compared to the data from 3 different WT retinas experiments. Peaks highlighted are at least 0.5× reduced (red) or 2× increased (green) in CRX KO retinas as compared to the average ChIP-Seq read value of the 3 WT samples. To be considered changed, CRX KO versus the 3 WT values for MEF2D ChIP-seq density had to have p<0.01 significance (Student’s t test). Black line indicates unity. (B) Stard7 (left), Pla2r1 (center), Srpk3 (right) genomic loci with CRX ChIP-Seq tracks from WT retinas, MEF2D ChIP-Seq tracks from CRX WT and CRX KO retinas, genomic conservation, DNAse hypersensitivity tracks from retina (DHS, ENCODE) and RNA-Seq tracks from WT and Crx KO retinas, MEF2D- bound enhancers are highlighted according to the fold change of MEF2D binding in Crx KO versus WT retinas. (C) Aggregate plots of MEF2 consensus binding motif occurrence in a 2kb window centered on summits of MEF2D-bound sites. Motif enrichments at MEF2D peaks that are increased, unchanged, or decreased in Crx KO retinas are shown. The MRE is significantly de-enriched within a 400bp window of MEF2D peak summits where MEF2D binding is decreased in Crx KO retinas compared to sites where MEF2D binding is unchanged (Chi-squared test; p<0.002). The MRE is significantly enriched at sites where MEF2D binding increases in Crx KO retinas compared to sites where MEF2D binding is unchanged (Chi-squared test: p<0.05).
Unexpectedly, we also found that MEF2D binding is increased more than twofold in Crx KO retinas at a distinct set of 224 sites genome-wide, demonstrating that MEF2D binding in photoreceptors is not merely lost at a subset of sites in Crx KO retinas, but instead that the binding of MEF2D is redistributed to other sites across the genome (Figure 5A, B & S5A, green). Notably, many of the regions where MEF2D binding increases in Crx KO retinas are not retina-specific enhancers, but instead are sites where MEF2D binds in other tissues (Figure 3B, 5B & S5C). These sites included an upstream enhancer of Npas4, which is expressed in a stimulus-dependent manner in cortical neurons, but is not expressed in the retina, and the promoter of Srpk3, a gene that is an important target of MEF2 in muscle (Figure 5B and data not shown) (Kim et al., 2010; Nakagawa et al., 2010). In contrast, MEF2D binding sites where MEF2D binding decreases in Crx KO retinas are highly tissue-specific (Figure S5C).
This redistribution of MEF2D binding in Crx KO retinas occurs predominately at regions that are already DNase hypersensitive in WT retinas (Figure 5B & S5D), suggesting that loss of CRX does not make these sites newly accessible, but instead that without CRX the distribution of MEF2D binding shifts to other available sites. To determine if this redistribution of MEF2D binding has functional consequences for gene expression we examined the expression of the nearest genes to sites where MEF2D is lost or gained in the Crx KO. We observed decreased gene expression near certain sites where MEF2D binding is lost in the Crx KO (Figure 5B & S5E). This may be directly due to the loss of MEF2D at these sites. Alternatively, loss of CRX itself may contribute to this change. We also observed increases in gene expression near some sites where MEF2D binding increases in the Crx KO (Figure 5B & S5E). This suggests that the redistribution of MEF2D in the Crx KO has functional consequences on gene expression in photoreceptors.
While MEF2D requires CRX for binding to many important enhancers, not all MEF2D binding is CRX-dependent (Figure 5A & B). We therefore hypothesized that MEF2D may only require CRX to stabilize binding at regulatory elements with weak consensus MREs. In support of this idea, we find that the consensus MEF2D binding sequence is substantially de-enriched in regions where MEF2D binding is CRX-dependent compared to sites where MEF2D binding is unchanged (Figure 5C & S5F). In contrast, regions where MEF2D binding increases in the Crx KO are enriched for consensus MREs relative to MEF2D binding sites where the level of MEF2D binding decreases in the absence of CRX (KS test, p= 4.76e-5) (Figure 5C; Figure S5F). Taken together, these data strongly suggest that in photoreceptor cells CRX recruits MEF2D away from non-retinal consensus binding sites in favor of retina-specific sites that do not require a consensus MRE. This appears to be an important mechanism for achieving tissue-specific binding of MEF2D so that MEF2D can regulate photoreceptor-specific target genes.
MEF2D and CRX co-regulate gene expression by selective activation of retinal enhancers
As described above, tissue-specific binding of MEF2D in the retina together with CRX plays a critical role in specifying the function of MEF2D in photoreceptor cells. However, of the several thousand MEF2D–bound sites that we detect in the retina fewer than 4% are located near genes whose expression is significantly altered in the absence of MEF2D. This suggests that additional mechanisms must exist to determine which of the many MEF2D–bound sites are functionally important in the retina, possibly through the selective activation of MEF2D-bound regulatory elements. This hypothesis is consistent with previous genome-wide analyses which demonstrate that only a small fraction of TF occupancy affects the expression of nearby genes (Spitz and Furlong, 2012). We therefore sought to determine if MEF2D regulates the expression of its target genes through selective activation of enhancers and to identify the mechanisms beyond binding to DNA that regulate the action of MEF2D in photoreceptor cells.
To determine the subset of MEF2D-bound regulatory sites that are active, we identified the subset of MEF2D binding sites that display features of active enhancers, acetylation of histone 3 at lysine 27 (H3K27ac) and transcription of bidirectional enhancer RNAs (eRNAs) (Creyghton et al., 2010; Kim et al., 2010; Rada-Iglesias et al., 2011). We found that at approximately 33% (660/2003) of MEF2D-bound sequences H3K27 is acetylated and eRNAs are transcribed, suggesting that only a subset of MEF2D-bound sequences are active enhancers (Figure 6A). Consistent with these findings, genes near enhancers marked by H3K27ac and eRNA transcription are more highly expressed than genes near MEF2D binding sites with no H3K27ac or eRNA expression (Figure 6B). Furthermore, we find that MEF2D binding is necessary for full activation of MEF2D target gene enhancers, as H3K27ac levels and eRNA transcription are significantly reduced at target gene enhancers in MEF2D KO compared to WT retinas (Figure 6C & S6A). We observed that this reduction in H3K27ac and eRNA levels in MEF2D KO versus WT retinas is more significant at enhancers that are near target genes than at enhancers not associated with target genes (p<3.79e-10 for eRNAs; p<3.12e-9 for H3K27Ac, t-test). Together these results show that selective activation of MEF2D binding sites is an important feature of MEF2D target gene regulation and that MEF2D is necessary, but not necessarily sufficient for activation of enhancers near MEF2D target genes.
To determine the mechanism of selective MEF2D enhancer activation, we considered the possibility that CRX might serve as a co-activator at MEF2D-bound enhancers, not only at sites where CRX is required for MEF2D binding, but also at sites where CRX and MEF2D are bound, but where MEF2D binding is not dependent upon CRX. To test this hypothesis, we first asked if CRX binding correlated with enhancer activity at MEF2D-bound sites. When assessed across the genome, the MEF2D-bound enhancers that are co-bound by CRX were found to be significantly more active (enriched for H3K27ac and eRNA expression) than the MEF2D enhancers where CRX was not bound, even when the amount of MEF2D binding is the same (Figure 6D). This strongly suggests that the presence of CRX together with MEF2D is required for the maximal activation of MEF2D-bound enhancers in photoreceptors. However, MEF2D does not appear to be a required co-factor for the activation of CRX-bound enhancers as the majority of active CRX-bound sites do not bind MEF2D and because H3K27ac levels and eRNA expression are similar at sites where CRX and MEF2D are both bound and where CRX is bound alone (data not shown).
To determine if CRX co-binding is required for the selective activation of MEF2D-bound regulatory elements, we performed RNA-seq in WT and Crx KO retinas at p11 and quantified the levels of eRNAs at MEF2D-bound enhancers genome-wide (Figure 6E & F). We found that CRX is required for the majority of eRNA expression at MEF2D-bound enhancers and that this is observed both at the population level and at individual enhancers (Figure 6E & F). These sites include nearly all the active sites where MEF2D binding is dependent upon CRX (97%) as well as the majority of active, CRX and MEF2D co-bound sites where MEF2D is not dependent on CRX for binding (73%) (Figure S6B). These latter sites include enhancers of clinically relevant MEF2D target genes such as Pcdh15, Guca1a and Guca1b (Figure 6C and data not shown). Furthermore, the level of eRNA transcription at MEF2D binding sites shared across tissues (Figure 3B) is low and not significantly affected by deletion of CRX, consistent with our observation that there is a low level of CRX binding at these sites. Taken together these results indicate that CRX is required for the selective activation of MEF2D-bound regulatory elements not only because CRX recruits MEF2D to tissue-specific enhancers, but also because CRX directly co-activates MEF2D-bound enhancers.
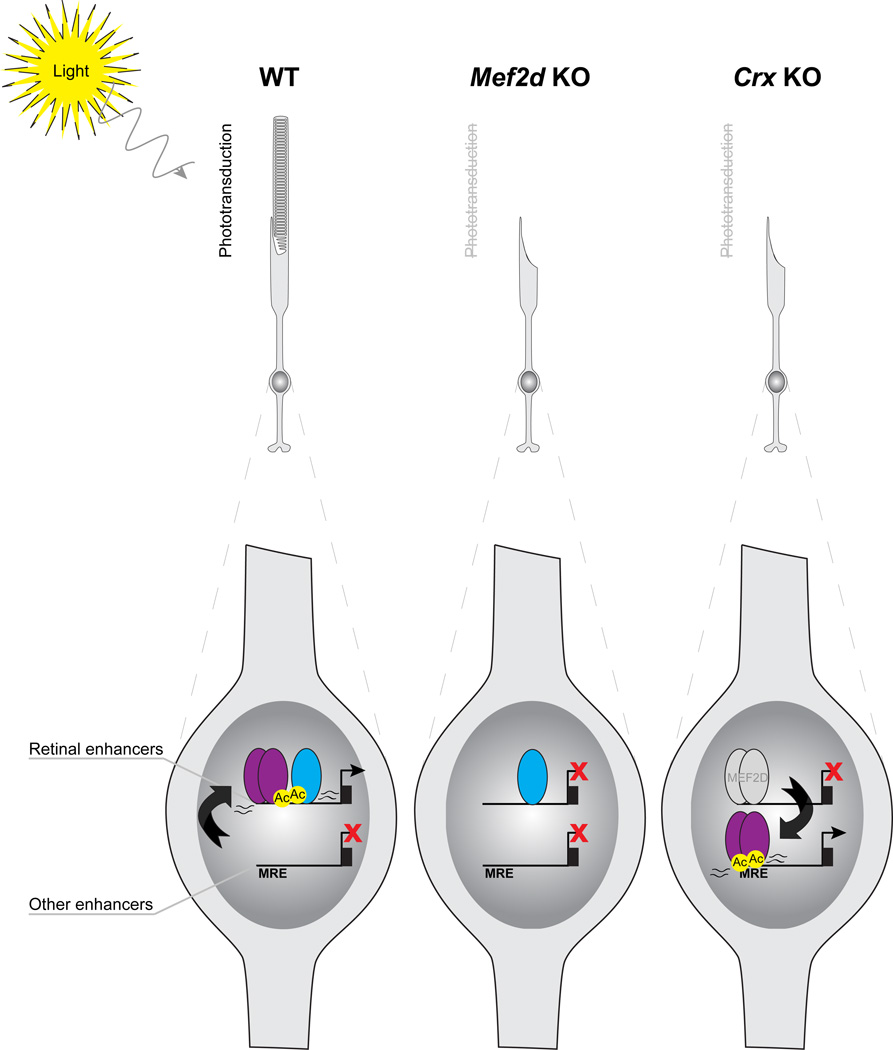
While other mechanisms undoubtedly contribute to the function of MEF2D in the retina, the data presented in this study highlight two distinct mechanisms by which MEF2D, a broadly expressed TF, achieves cell-type specific function in retinal photoreceptor development (Figure 7). First, MEF2D is recruited away from non-retinal consensus binding sites toward retina-specific enhancers and CRX stabilizes MEF2D binding at those enhancers where the MRE is particularly weak. Second, MEF2D cooperates with CRX to co-activate retina-specific enhancers as determined by increased H3K27Ac levels and eRNA production. Through these mechanisms MEF2D achieves tissue-specific function in the development of the mouse retina. Disruption of these mechanisms leads to misregulated expression of critical cell-type-specific genetic programs, a failure of photoreceptor cell development and ultimately to a loss of visual function.
Figure 7. MEF2D and CRX coordinately regulate gene expression necessary for photoreceptor development and function through tissue-specific enhancer co-binding and co-activation.
In WT photoreceptors, CRX recruits MEF2D toward retina-specific enhancers without MEF2D consensus binding sites at the expense of non-retinal enhancers with high-affinity consensus binding sites (MREs). Once bound, MEF2D and CRX selectively activate retina-specific enhancers, as determined by enriched H3K27ac (Ac) and eRNA expression, to drive shared target genes that are required for photoreceptor outer segment development and phototransduction. MEF2D is required for retinal enhancer activation and photoreceptor gene expression, leading to a failure of photoreceptor development and phototransduction in Mef2d KO photoreceptors. In the CRX KO, MEF2D binding is lost at retina-specific enhancers without consensus MREs and recruited back to non-retinal enhancers with consensus MREs resulting in a loss of gene expression necessary for photoreceptor development and function and an increase in expression of non-retinal genes.
Discussion
In this study, we identify retinal photoreceptors as a neuronal cell type in the CNS that predominantly expresses a single MEF2 family member, MEF2D, during development and that requires MEF2D cell-autonomously for functional differentiation in vivo. MEF2 TFs have been previously proposed to play a role in photoreceptors (Escher et al., 2011; Hao et al., 2011), but evidence for this has proved elusive until now. Our analyses demonstrate that MEF2D regulates photoreceptor cell development by binding to and activating photoreceptor-specific enhancers and thereby regulating critical photoreceptor-specific genes including genes that are mutated in human retinal disorders. Within the CNS, retinal photoreceptors are notable because they are a homogeneous and functionally well-characterized cell type that makes up the majority of cells in the retina. This made it possible to perform genomic and epigenetic analyses in vivo to functionally dissect the mechanisms by which MEF2D regulates photoreceptor development.
Although it is broadly expressed across many tissues, we find that MEF2D binds to and selectively activates retina-specific enhancers with the retina-specific homeodomain TF CRX, and that cooperation with CRX is critical for the tissue-specific function of MEF2D through two distinct mechanisms. First, CRX recruits MEF2D to photoreceptor-specific enhancers that lack a consensus MRE. This suggests that CRX actively stabilizes MEF2D binding rather than functioning solely as a pioneer factor that opens up chromatin to reveal MREs. It remains to be determined if MEF2D recruits CRX in a similar manner, although preliminary motif analyses suggest that this may not occur, as the majority of both CRX enhancers co-bound with MEF2D and those without MEF2D contain a consensus CRX DNA binding motif (83.17% vs 84.16%). Second, CRX works cooperatively with MEF2D to selectively co-activate CRX/MEF2D co-bound enhancers as determined by increased H3K27Ac levels and eRNA production at enhancers when both MEF2D and CRX are bound. Importantly, the cooperative action of MEF2D and CRX determines the subset of MEF2D-bound sites that are active enhancers among the thousands of MEF2D binding sites in photoreceptors.
This type of cooperative interaction may also be important for other cell types that express MEF2 factors, as CRX is closely related to two other homeobox factors OTX1 and OTX2 that are critical for the development of the CNS and are expressed in many non-neuronal cell types. Alternatively MEF2 family members may interact with distinct TF families in different cell types. We find that myocyte-specific MEF2D-binding sites are highly enriched for a tissue-specific cofactor motif, the myogenic bHLH recognition element. Indeed previous in vitro reporter studies have suggested that MEF2 may interact with myogenic bHLH heterodimers during muscle differentiation (Molkentin et al., 1995).
It will be of particular interest to determine the generality of our finding that a tissue-specific factor (CRX) can bias the binding of a widely expressed TF (MEF2D) so that the two TFs function together at tissue-specific enhancers. Previous work has demonstrated a competition between functional DNA binding sites and non-functional binding sites in satellite regions and repetitive DNA, which are thought to limit free TF concentration in the nucleus (Liu et al., 2007). Our results suggest that CRX biases the genome-wide competition for MEF2D binding toward regulatory sites that are relevant to photoreceptor gene expression and that in the absence of CRX, MEF2 binding increases at MREs that are not normally bound by MEF2 in the retina, thereby inducing expression of nearby genes. However, these newly bound sites are not merely sponges that limit available MEF2D, but are highly conserved and appear to regulate MEF2 target gene expression in other cell types (Nakagawa et al., 2010).
Once bound to the genome, we find that additional mechanisms must regulate the action of MEF2 function because only a small subset of the several thousand MEF2D binding sites are required for expression of nearby genes. We found that selective activation of a subset of MEF2D-bound enhancers plays a significant role in determining which genes require MEF2D for their expression, and that CRX contributes to this selective activation. In non-neural tissues, MEF2 co-factors have been suggested to help recruit co-activators such as histone acetyltransferases (HATs) (Youn et al., 2000). As CRX binds the HAT P300 (Yanagi et al., 2000), this raises the possibility that in photoreceptors MEF2D and CRX cooperatively recruit HATs to form a MEF2D-HAT-CRX complex. Such a tripartite complex may also stabilize MEF2D binding, in which case a single mechanism could account for the contribution of CRX to MEF2D binding and selective activation of MEF2D-bound regulatory elements. The majority of MEF2D binding in the retinal genome, however, occurs at sites that appear to be non-functional, because they lack marks of enhancer activity (e.g. eRNAs and H3K27ac). Nonetheless, this binding may be functionally significant through other mechanisms of action. Notably, MEF2 TFs are known to function in a stimulus-dependent manner elsewhere in the CNS (Flavell et al., 2006; Shalizi et al., 2006) and it is possible that MEF2D in photoreceptors could regulate enhancers in a dynamic manner. While future studies are required to test this hypothesis, it is interesting that many of the direct MEF2D target genes that have been identified in this study have been shown to be strongly regulated in photoreceptors by light and/or by the circadian clock (Storch et al., 2007).
The findings presented in this study contribute to work that has been done previously to elucidate the network of TFs that are critical for photoreceptor development. While TFs such as CRX, NRL and NR2E3 are known to be highly tissue-specific, MEF2D is broadly expressed but achieves photoreceptor-specific function through interactions with CRX. Thus, the role of tissue-specific TFs in photoreceptor development is not simply to activate target gene expression, but also to recruit potent transcriptional regulators such as MEF2 toward photoreceptor functions by directly competing MEF2D away from non-retinal enhancers and by selectively activating photoreceptor enhancers with MEF2D. Furthermore, the loss of Crx does not simply result in the loss of CRX target gene expression, but also results in the redistribution of MEF2D toward non-retinal specific enhancers to activate other programs of gene expression.
Finally, the present study suggests that MEF2D has the potential to be important in human retinal disease. MEF2D co-regulates critical retinal disease genes with CRX, which itself is mutated in several retinal diseases characterized by photoreceptor degeneration. Importantly, the identification of active photoreceptor enhancers allowed us to identify critical MEF2D-bound functional regulatory elements, which can be as much as 100kb away from the transcriptional start site of retinal disease genes. This is significant as these regulatory elements may correspond to sites of genetic variation in humans and may ultimately be found to harbor disease-causing mutations. For example, SNPs in these regulatory elements that affect the binding of MEF2D or CRX might disrupt enhancer activity and nearby gene expression, leading to retinal disease. Such mutations would join a growing cohort of enhanceropathies that contribute to human disease (Smith and Shilatifard, 2014). Thus these genome-wide analyses provide a rich resource for considering how non-coding regulatory regions function in normal development of the retina and potentially in human disease.
Experimental Procedures
A full description of the experimental procedures is included in the Supplemental Information.
Generation of MEF2D knockout and conditional knockout mice
Targeting of the Mef2d locus was carried out by homologous recombination in ES cells (Figure S1). Successfully targeted mice were then crossed to EIIA-Cre expressing mice (stock number 003724; The Jackson Laboratory) and offspring were analyzed for expression of the presence of Cre and the state of the targeted MEF2D allele. Mice with the neomycin cassette deleted, and with transmission of either the floxed allele or the null allele were crossed into a C57Bl/6 background and used to establish the knockout and conditional knockout lines used in this study. The Institutional Animal Care and Use Committee at Harvard University approved all of the experiments in this study.
RNA-seq
Total RNA was extracted from two p11 mouse retinas per sample. RNA was submitted to BGI (Shenzhen, China) for library construction and sequencing. Libraries were sequenced to a depth of at least 8×107 clean reads per sample on the Illumina Hiseq 2000 platform.
ChIP-seq
ChIP antibodies used were anti-MEF2D (Flavell et al., 2008) and anti-H3K27Ac (Abcam AB4729). MEF2D ChIP from mouse cortical cultures was performed as described (Kim et al., 2010). MEF2D and H3K27Ac ChIP from mouse retinas was performed as described for brain tissue (Hong et al., 2008) with modifications (Supplemental Information). Immunoprecipitated chromatin was submitted to BGI for library construction and sequencing on the Illumina Hiseq 2000 platform. For each sample, >20 million clean reads were obtained. Sequencing reads were aligned to the mouse genome (NCBI 37, mm9) using the Burrows-Wheeler Aligner.
RNA-seq and ChIP-seq datasets
Raw and analyzed datasets have been submitted to GEO (GSE61392).
Supplementary Material
Acknowledgements
We thank members of the Greenberg laboratory for helpful discussions, J. Zieg for help with figures, the MRDDRC Gene Manipulation Core (M. Thompson, Y. Zhou, and H. Ye), the EM Core (E. Benecchi), and the Rodent Pathophysiology Core (R. Bronson) for experimental assistance and advice and P. Zhang for help with mice. This work was supported by a Stuart H.Q. & Victoria Quan Fellowship (M.M.A.), the Ruth L. Kirschstein NIH Training Grant T32GM007753 (M.M.A.), the Ruth L. Kirschstein Individual Postdoctoral Award F32NS070544 (T.J.C.), and NIH grant NS028829 (M.E.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Extended Experimental Procedures, six figures with legends, and two tables.
Author Contributions
Experiments were designed by M.M.A., T.J.C. and M.E.G. Experiments were conducted and analyzed by M.M.A., T.J.C., D.A.H., A.C.B., C.L., M.H., B.P., A.N.M., S.W.F., M.A.S. and E.R. The manuscript was written by T.J.C., M.M.A. and M.E.G.
References
- Amiel J, Rio M, de Pontual L, Redon R, Malan V, Boddaert N, Plouin P, Carter NP, Lyonnet S, Munnich A, et al. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet. 2007;80:988–993. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenu T, Diebold B, Chelly J, Isidor B. Refining the phenotype associated with MEF2C point mutations. Neurogenetics. 2013;14:71–75. doi: 10.1007/s10048-012-0344-7. [DOI] [PubMed] [Google Scholar]
- Black BL, Ligon KL, Zhang Y, Olson EN. Cooperative transcriptional activation by the neurogenic basic helix-loop-helix protein MASH1 and members of the myocyte enhancer factor-2 (MEF2) family. J Biol Chem. 1996;271:26659–26663. doi: 10.1074/jbc.271.43.26659. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, Vetere G, Pekar T, Ross PJ, Neve RL, et al. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat Neurosci. 2012;15:1255–1264. doi: 10.1038/nn.3189. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Lawrence KA, Karlstetter M, Myers CA, Abdelaziz M, Dirkes W, Weigelt K, Seifert M, Benes V, Fritsche LG, et al. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome research. 2010;20:1512–1525. doi: 10.1101/gr.109405.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D, Zallocchi M. Usher protein functions in hair cells and photoreceptors. Int J Biochem Cell Biol. 2014;46:80–89. doi: 10.1016/j.biocel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol. 1992;12:3665–3677. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher P, Schorderet DF, Cottet S. Altered expression of the transcription factor Mef2c during retinal degeneration in Rpe65−/− mice. Invest Ophthalmol Vis Sci. 2011;52:5933–5940. doi: 10.1167/iovs.10-6978. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet. 1995;10:360–362. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Hao H, Kim DS, Klocke B, Johnson KR, Cui K, Gotoh N, Zang C, Gregorski J, Gieser L, Peng W, et al. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012;8:e1002649. doi: 10.1371/journal.pgen.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Tummala P, Guzman E, Mali RS, Gregorski J, Swaroop A, Mitton KP. The transcription factor neural retina leucine zipper (NRL) controls photoreceptor-specific expression of myocyte enhancer factor Mef2c from an alternative promoter. J Biol Chem. 2011;286:34893–34902. doi: 10.1074/jbc.M111.271072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Phan D, van Rooij E, Wang DZ, McAnally J, Qi X, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest. 2008;118:124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl S, Coppieters F, Meire F, Schaich S, Roosing S, Brennenstuhl C, Bolz S, van Genderen MM, Riemslag FC, European Retinal Disease C, et al. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am J Hum Genet. 2012;91:527–532. doi: 10.1016/j.ajhg.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu B, Szary J, Kofoed EM, Schaufele F. Functional sequestration of transcription factor activity by repetitive DNA. J Biol Chem. 2007;282:20868–20876. doi: 10.1074/jbc.M702547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Nakagawa O, Arnold M, Nakagawa M, Hamada H, Shelton JM, Kusano H, Harris TM, Childs G, Campbell KP, Richardson JA, Nishino I, Olson EN. Centronuclear myopathy in mice lacking a novel muscle-specific protein kinase transcriptionally regulated by MEF2. Genes Dev. 2010;19:2066–2077. doi: 10.1101/gad.1338705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Ye Z, Cheng L, Liu JO. Myocyte enhancer factor 2 mediates calcium-dependent transcription of the interleukin-2 gene in T lymphocytes: a calcium signaling module that is distinct from but collaborates with the nuclear factor of activated T cells (NFAT) J Biol Chem. 2004;279:14477–14480. doi: 10.1074/jbc.C300487200. [DOI] [PubMed] [Google Scholar]
- Payne AM, Downes SM, Bessant DA, Taylor R, Holder GE, Warren MJ, Bird AC, Bhattacharya SS. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum Mol Genet. 1998;7:273–277. doi: 10.1093/hmg/7.2.273. [DOI] [PubMed] [Google Scholar]
- Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Sato M, Nakazawa M, Usui T, Tanimoto N, Abe H, Ohguro H. Mutations in the gene coding for guanylate cyclase-activating protein 2 (GUCA1B gene) in patients with autosomal dominant retinal dystrophies. Graefes Arch Clin Exp Ophthalmol. 2005;243:235–242. doi: 10.1007/s00417-004-1015-7. [DOI] [PubMed] [Google Scholar]
- Sebastian S, Faralli H, Yao Z, Rakopoulos P, Palii C, Cao Y, Singh K, Liu QC, Chu A, Aziz A, et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev. 2013;27:1247–1259. doi: 10.1101/gad.215400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nature reviews Genetics. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;24:730–41. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Tran NM, Zhang A, Zhang X, Huecker JB, Hennig AK, Chen S. Mechanistically distinct mouse models for CRX-associated retinopathy. PLoS Genet. 2014;10:e1004111. doi: 10.1371/journal.pgen.1004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Abe T, Takeshita T, Sato H, Yanashima K, Tamai M. Mutation of human retinal fascin gene (FSCN2) causes autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2001;42:2395–2400. [PubMed] [Google Scholar]
- Yanagi Y, Masuhiro Y, Mori M, Yanagisawa J, Kato S. p300/CBP acts as a coactivator of the cone-rod homeobox transcription factor. Biochem Biophys Res Commun. 2000;269:410–414. doi: 10.1006/bbrc.2000.2304. [DOI] [PubMed] [Google Scholar]
- Yokokura S, Wada Y, Nakai S, Sato H, Yao R, Yamanaka H, Ito S, Sagara Y, Takahashi M, Nakamura Y, et al. Targeted disruption of FSCN2 gene induces retinopathy in mice. Invest Ophthalmol Vis Sci. 2005;46:2905–2915. doi: 10.1167/iovs.04-0856. [DOI] [PubMed] [Google Scholar]
- Youn HD, Chatila TA, Liu JO. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 2000;19:4323–4331. doi: 10.1093/emboj/19.16.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RL. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212(2):199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.