Abstract
The human oral cavity provides the perfect portal of entry for viruses and bacteria in the environment to access new hosts. Hence, the oral cavity is one of the most densely populated habitats of the human body containing some 6 billion bacteria and potentially 35 times that many viruses. The role of these viral communities remains unclear; however, many are bacteriophage that may have active roles in shaping the ecology of oral bacterial communities. Other implications for the presence of such vast oral phage communities include accelerating the molecular diversity of their bacterial hosts as both host and phage mutate to gain evolutionary advantages. Additional roles include the acquisitions of new gene functions through lysogenic conversions that may provide selective advantages to host bacteria in response to antibiotics or other types of disturbances, and protection of the human host from invading pathogens by binding to and preventing pathogens from crossing oral mucosal barriers. Recent evidence suggests that phage may be more involved in periodontal diseases than were previously thought, as their compositions in the subgingival crevice in moderate to severe periodontitis are known to be significantly altered. However, it is unclear to what extent they contribute to dysbiosis or the transition of the microbial community into a state promoting oral disease. Bacteriophage communities are distinct in saliva compared to sub- and supragingival areas, suggesting that different oral biogeographic niches have unique phage ecology shaping their bacterial biota. In this review, we summarize what is known about phage communities in the oral cavity, the possible contributions of phage in shaping oral bacterial ecology, and the risks to public health oral phage may pose through their potential to spread antibiotic resistance gene functions to close contacts.
Keywords: bacteriophage, virus, microbiome, virome, metagenome, oral microbiome
Bacteriophage are widely distributed and have long been known to inhabit the human oral cavity (1–3). They are hypothesized to be distributed wherever their potential hosts exist. They have either lytic lifestyles where they infect their hosts and kill them rapidly to spread their progeny, or lysogenic lifestyles where they integrate into their host genomes and potentially contribute valuable gene functions. Lysogenic phage are able to lyse their hosts and spread their progeny in response to host and other external signals, but the frequency with which these phage lyse their hosts in the oral microbiome is poorly understood.
Early studies of bacteriophage in the oral cavity identified phage that parasitize oral pathogens such as Aggregatibacter actinomycetemcomitans (4, 5). In these studies, the presence of A. actinomycetemcomitans phage was positively correlated with rapidly destructive periodontitis (6, 7), which suggested a role for oral phage in bacterial virulence. However, other studies showed that these phage were not associated with periodontal disease (8, 9), so their role in the oral microbiome is still unclear. Regardless of their role in oral disease, previous studies show that phage in the oral cavity can act both as commensals (10) and pathogens (11), which suggests that they play significant roles in the ecology of the human oral microbiome.
Numerous culture-based attempts have been made to isolate novel phage from saliva and dental plaque, but overall these studies failed to consistently identify a presence of phage (10, 12, 13). Some of the studies identified phage from bacteria that were not thought to be representative of the normal oral flora or were of relatively low abundance. For example, phage capable of parasitizing Enterococcus faecalis (10) and Proteus mirabilis (13) were identified, but these bacteria were not considered to belong to the normal oral flora. The lack of plausible evidence for widespread phage in the human oral cavity led these authors to hypothesize that the interactions between phage and their hosts do not heavily influence oral microbial ecology. One caveat to these studies is that they were conducted during a time when most phage were considered to have a relatively narrow host range. More recently, there has been much more widespread acceptance of the concept of generalist strategies for phage, in which certain phage are known to have relatively broad host ranges (generalists) compared to the traditional view of phage with narrow host ranges (specialists) (14–17). These studies highlight that the concept of narrow host range arose from model systems that lacked suitable hosts and utilized lysis as the sole basis to determine whether there was a phage infection. As a result, these model systems underestimate phage host range. In reassessment of the early data suggesting the presence of E. faecalis and P. mirabilis phage in the human oral cavity, a more plausible explanation may be that the host range of these phage may have been greater than what could be tested for in vitro. While there have been some oral E. faecalis isolates shown to harbor lysogenic phage (18), whether these phage may be capable of generalist strategies are not known. Techniques such as single-cell PCR and in-situ hybridization that can detect phage within a single host cell may greatly expand the ability to characterize bacteriophage host range (19, 20), particularly in complex microbial communities.
The oral cavity is populated by communities of phage
Early studies of phage in the human oral cavity relied upon the presence of virus-like particles (VLPs) using electron microscopy to speculate that there may be many phage present in dental plaque (1). Because these types of studies could not also taxonomically characterize the phage present, it was unclear whether the presence of VLPs represented a few relatively abundant phage or many different evenly distributed phage. Using epifluorescence microscopy, studies have shown that there are approximately 108 VLPs per mL of fluid from oropharyngeal swabs (21), 108 VLPs per mL of saliva (22), and 107 VLPs per milligram of dental plaque (23). Culture- and morphology-based techniques have not been sufficient to characterize the diversity of phage in the oral cavity, but the utilization of metagenomics techniques based on shotgun sequencing approaches have proven effective in uncovering the membership and diversity of oral phage communities (22, 24).
By using next generation sequencing approaches, such as metagenomics, we now recognize that the oral cavity is home to a large population of viruses, many of which can be identified as bacteriophage (22–26). These studies also have identified some eukaryotic viruses including torque teno viruses, circoviruses, herpesviruses (HSV), and Epstein–Barr virus (EBV) among a few others, but phage appear to be more highly abundant, which may reflect the high ratio of bacterial cells to our own cells in the oral cavity. Another potential explanation for the abundance of phage compared to eukaryotic viruses are enrichment techniques such as cesium chloride (CsCl) density gradient centrifugation and sequential filtration, which could result in technical biases by removing viruses from the oral virome (27). Enveloped viruses such as HSV and EBV have been found in the oral virome, indicating that enveloped viruses may be identified, but larger viruses such as mimiviruses may be trapped by filtration (27, 28). Smaller viruses such as human papillomaviruses are readily detected after CsCl gradient enrichment (29), so the extent of virion size biases is difficult to quantify for smaller viruses. Most studies of human viromes have focused only on DNA viruses, so the constituents and potential roles of RNA phage communities lags significantly behind (30). Many phage in the oral virome may be intracellular at the time of analysis, and thus, they could also go unrecognized or their relative abundance underestimated in oral virome analysis (24, 27, 31).
While technical biases such as the concentration on DNA viruses and utilization of filtration techniques could lead to substantial underestimations of oral phage community diversity, there are other factors that might contribute to the overestimation of oral phage diversity. Principally among these factors include undersampling of the phage community (29) and an inability to properly assemble phage genomes from complex communities (25). One means of estimating the diversity of viral communities is a tool called Phage Communities from Contig Spectrum (PHACCS) (32, 33), which uses a rank-abundance model based on the full spectra of assembled and size-sorted contigs. Use of this tool has recently highlighted the critical role that assemblers play in estimating phage community diversity, by demonstrating that certain assemblers may provide significantly different estimations of community diversity (33). Despite the limitations imposed through the assembly process, estimates of phage diversity in human saliva suggest that there are hundreds to thousands of different phage genotypes that are relatively evenly distributed in this particular environment (22). These results indicate that the numerous VLPs in the oral cavity likely represent many different evenly distributed phage. Another study showed that phage community diversity still was substantially overestimated in oral viromes likely as a result or limitations in the phage assembly process (25). An alternative method termed the Homologous Viral Diversity Index (HVDI) uses homology amongst assembled phage contigs to identify contigs through network analysis that likely belong to the same viruses (29). A corrected contig spectra is formed from the networks and utilized to reduce the overestimation of phage genotypes. Utilization of this method indicates that phage are less evenly distributed in the human oral cavity than were originally projected (34). It also shows that phage diversity in the oral cavity is relatively homogenous between different human subjects and is far greater than is estimated in the colon.
Oral biogeography
The heterogeneity of different tissue types in the oral cavity provides a variety of surfaces available for colonization by oral bacteria and their phage (35). Studies of the oral microbiome have shown that all oral biogeographic niches are home to their own unique community members (36, 37). The supragingival areas are continuously subjected to environmental changes as we brush our teeth and ingest a variety of foods on a daily basis. This results in microbial communities in supragingival plaque that have to respond to pulses of carbohydrates and stages of either primary or secondary colonization of tooth surfaces several times during a single day. Microbial shifts during these processes can result in prolonged exposures to acidification due to the selection of acidogenic and aciduric bacteria, which can result in dental caries (38–41). Subgingival areas consist of epithelial surfaces that continually desquamate and are supplied by fluid from the gingival crevice. This particular area is primarily anaerobic and is inhabited largely by strict and facultative anaerobic bacteria. The differences in environments likely account for the observed variation in both bacteria and phage flora, which are relatively site-specific regardless of oral health status (23, 31). The general lack of oxygen and the availability of amino acids and peptides for microbial growth in the subgingival crevice (42, 43) are likely major factors responsible for the selection of a subgingival flora. A different bacterial flora that uses carbohydrates for growth inhabits the more aerobic and carbohydrate-rich supragingival areas (43). In addition, differences in pH that may be associated with distinct biogeographic sites and local periodontal health may also affect oral phage ecology; however, the effects of pH on oral phage have yet to be reported.
Few studies have demonstrated the presence of phage communities in oral plaque (23, 31). In one study, pathogen-specific phage were identified in dental plaque using metagenomic techniques (44), which could have been surmised from prior studies of phage from the oral pathogen A. actinomycetemcomitans (45). Follow-up studies now have shown that there are communities of viruses in both subgingival and supragingival plaque, with very little evidence to support that there are significant differences in phage diversity between oral aerobic and anaerobic niches (23, 31). Other studies of oral biogeography have shown that there are distinct bacterial communities on the tongue, dental plaque, buccal mucosa, keratinized gingiva, and the nasal cavity (46–53), which suggests that these surfaces also could have distinct phage communities. The relative lack of obtainable biomass from some of these sites has hampered investigation of their phage communities because there generally is not enough material available for filtration and ultracentrifugation. Other techniques such as metagenomics and metatranscriptomics, followed by filtering out discernible phage reads (54, 55) may be necessary to characterize phage communities from these oral surfaces.
Complex relationships between relative abundances of phage and their hosts
While analysis of phage evolution with their host bacteria is difficult to study in complex microbial ecosystems, there is one study that has attempted to predict putative phage hosts using their patterns of homologous sequences (22). Because many of the phage identified in the human oral cavity are predicted to have lysogenic lifestyles, their patterns of nucleotide usage and homologous sequences might be expected to resemble those of their bacterial hosts (56). Using homologous sequences to predict putative hosts of phage, one study identified putative phage of numerous different Firmicutes (includes Streptococcus, Granulicatella, and Veillonella), Bacteroidetes (includes Prevotella), Fusobacteria (includes Leptotrichia), Proteobacteria (includes Neisseria), Actinobacteria, Spirochaetes, and members of the TM7 Phylum (Table 1) (22). While BLASTX hits may provide accurate predictions of some host/phage relationships, the diversity of phage likely are much too complex to accurately predict their hosts based on homology alone. The relative abundances of bacteria do not necessarily predict the relative abundances of their phage in these studies (22, 31). In one study, the relative abundances of phage for the genera Streptococcus and Neisseria were highly concordant with the relative abundances of their hosts, while Actinomyces and Fusobacterium that primarily live in the subgingival crevice were more likely to have inverse relationships with their hosts (22). These patterns observed for anaerobic bacteria suggest that there may be ecological differences observed in host/phage relationships related to oral biogeographic sites.
Table 1.
BLASTX hits to viruses infecting specific bacterial genera from human saliva
| Phylum | Genus | Subject 1 (%) | Subject 2 (%) | Subject 3 (%) |
|---|---|---|---|---|
| Firmicutes | Streptococcus | 8.91 | 12.82 | 8.36 |
| Ruminococcus | 12.63 | 10.71 | 9.43 | |
| Granulicatella | 4.26 | 3.63 | 1.78 | |
| Lactobacillus | 11.57 | 8.35 | 3.56 | |
| Lactococcus | 4.65 | 2.70 | 5.69 | |
| Oribacterium | 0.27 | 0.67 | 1.78 | |
| Veillonella | 0.00 | 0.93 | 0.89 | |
| Total | 42.29 | 39.80 | 31.49 | |
| Bacteroidetes | Bacteroides | 17.42 | 10.88 | 13.35 |
| Prevotella | 2.66 | 6.58 | 4.09 | |
| Total | 20.08 | 17.45 | 17.44 | |
| Fusobacteria | Fusobacterium | 11.04 | 12.06 | 8.19 |
| Leptotrichia | 1.60 | 2.11 | 1.96 | |
| Total | 12.64 | 14.36 | 10.15 | |
| Proteobacteria | Neisseria | 3.86 | 4.55 | 7.12 |
| Burkholderia | 4.65 | 3.29 | 4.98 | |
| Haemophilus/Aggregatibacter | 0.40 | 1.52 | 0.18 | |
| Cardiobacterium | 0.27 | 0.34 | 0.18 | |
| Total | 9.18 | 9.70 | 12.46 | |
| Actinobacteria | Actinomyces | 3.19 | 2.19 | 2.67 |
| Spirochaetes | Treponema | 3.06 | 4.13 | 3.56 |
| Other | TM7 (Phylum level) | 3.19 | 1.01 | 0.53 |
| Other | 6.38 | 11.55 | 21.71 | |
| Total | 9.57 | 12.56 | 22.24 |
Data derived from Ref. (22).
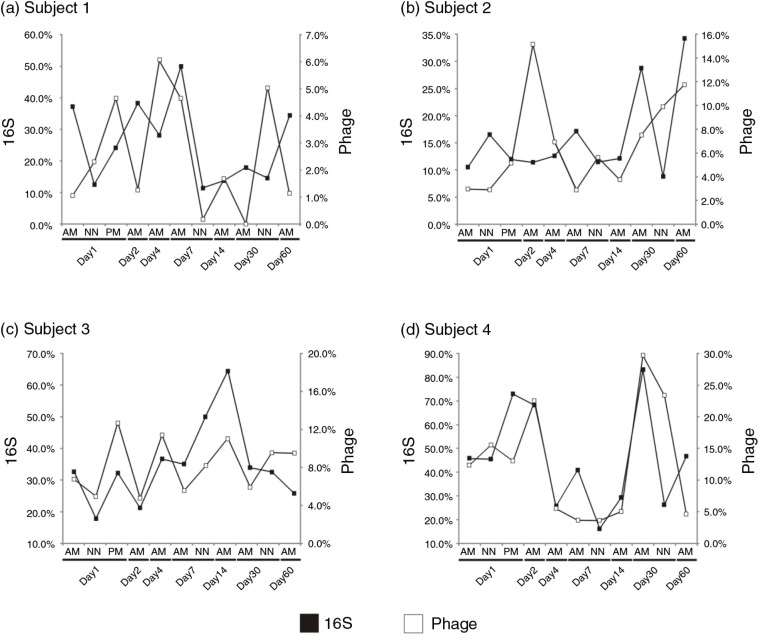
One study characterized human salivary phage and bacterial ecology at numerous time points over a 60-day period (25). Certain oral bacteria such as Streptococcus and putative streptococcal phage had relative abundances that were highly volatile over time. We plotted the relative abundances of bacteria assigned to the genus Streptococcus along with the relative abundances of putative streptococcal phage over time and observed different patterns in the different subjects studied. For example in subjects 1 and 2, at most time points there was an inverse relationship between the relative abundances of putative hosts and phage (Fig. 1, Panels a and b). This type of antagonistic relationship has been observed in Pseudomonas phage in individuals with cystic fibrosis (57) and is potentially due to the presence of predominantly lytic phage or shifts in the life cycles of temperate phage to the lytic cycle. The exact opposite relationship was found for subjects 3 and 4, where the relative abundances of putative streptococcal phage and host paralleled each other throughout the study (Fig. 1, Panels c and d), suggesting a more mutualistic relationship between host and phage where the phage in that individual did profoundly affect host relative abundance. These data suggest that there exist both antagonistic and mutualistic coevolutionary relationships between oral bacteria and their phage in the human oral cavity (58).
Fig. 1.
Relationships between putative streptococcal phage and their hosts. Relative abundances of Streptococcus species (black boxes) and putative streptococcal phage (white boxes) in the saliva of four subjects at 11 different time points over 60 days. Relative abundances of streptococci were determined based on the number of V1–V2 segment-16S rRNA reads assigned to the genus Streptococcus, and putative streptococcal phage assignments were determined based on BLASTX best hits to streptococcal phage. The percentage of all 16S rRNA reads assigned to streptococci are shown on the left y-axis, and the percentage of phage contigs assigned to streptococcal phage are shown on the right y-axis. The x-axis shows the day each subject was sampled. AM represents morning time points, NN represents noon time points, and PM represents evening time points. Panels a and b show subjects 1 and 2, and demonstrate an inverse relationship between the relative abundances of streptococcal species and their phage. Panels c and d show subjects 3 and 4 and demonstrate direct relationships.
Viral persistence in the oral cavity
Viruses and hosts may persist because their persistence could lead to an increased growth rate without any changes in phage infectivity (59). Therefore, hosts/viruses may select for each other to co-evolve and by doing so limit the ability for other phage to enter the community. Recent studies of the human gut demonstrate a high persistence of phage (60), which also was confirmed in a much larger subject group in the oral cavity (25). In the saliva samples representing eight subjects, nearly 20% of the phage that were identified during the first day of the study could also be observed 60 days later. Because many of the oral viromes were not sampled exhaustively, the 20% likely was a substantial underestimate of their persistence. More importantly, the subjects in that study were sampled at 11 different time points, and the proportion of phage that could only be identified for less than 4 days was <12%, which suggests that transience of oral phage may be the exception rather than the rule. Analysis of an individual phage over the course of the same study showed that there were only few polymorphisms, at the gene level present, suggesting that oral phage may not be under significant selective pressure to alter their nucleotide sequences. These data support that human oral phage are highly persistent and suggest that many of them likely are co-evolving with their bacterial hosts.
There are numerous different mechanisms by which oral bacteriophage may evade their host immune systems and persist. These include but are not limited to carrying their own restriction/modification enzymes (31), and avoiding cognate sequences for restriction/modification systems (56, 61, 62). In oral phage communities, there are substantial numbers of restriction/modification enzymes (25, 31), whose role may mimic those found in their host bacteria, or may potentially be utilized to broaden their host range. Restriction/Modification systems are designed to modify nucleic acids at specific residues, so that nucleic acids from parasitic entities such as viruses and plasmids that are not modified can be recognized (63). The unmodified nucleic acids are cleaved by restriction enzymes, rendering them inert for producing phage progeny or for plasmid replication. Some phage may only need modification enzymes, which allow them to evade detection when they parasitize new hosts. Having both restriction and modification enzymes, however, may provide a means by which phage can prevent competing phage from parasitizing or killing the same hosts (64–66).
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) in combination with CRISPR Associated Genes (CAS) form CRISPR/Cas systems in cellular microbiota (67–69) and are involved in acquired resistance against invading phage and plasmids. Anti-CRISPR genes (70–72) represent a diverse collection of genes that independently are capable of inhibiting CRISPR/Cas system activity. Their substantial diversity across different phage may parallel the diversity of different CRISPR/Cas systems in nature (72, 73). While relatively few anti-CRISPR genes have been identified in oral phage communities, only a minority of the phage in these communities have identifiable homologues (22, 24, 25). Given the significant number of CRISPR spacers identified in the oral cavity that match oral phage (23, 74–76), there probably is broad interplay between oral phage and CRISPR immune systems. Therefore, the use of anti-CRISPR genes may provide a means by which oral viruses persist over long time periods.
Horizontal gene transfers and gene functions
The oral cavity is believed to be an excellent environment for horizontal gene transfers, and phage likely are active in that process (77). Oral bacteria may acquire new gene functions through transformations (direct uptake and incorporation of exogenous genetic material), conjugations (transfer of genetic material between cells), or transductions (the transfer of genetic material between bacteria by phage). The contribution of new gene functions by phage that result in host phenotypic changes are referred to as lysogenic conversions (78, 79). The presence of numerous integrase genes amongst the phage identified in the oral cavity indicates that many phage with lysogenic lifestyles are members of the oral virome (22). Additionally, the majority of the phage genes in the oral virome are homologous to genes identified in the Caudovirus family Siphoviridae (have long non-contractile tails). In general, siphoviruses are more likely to have temperate lifestyles, while other Caudovirus families Podoviridae (have short non-contractile tails) and Myoviridae (have long contractile tails) are more likely to have lytic lifestyles (80, 81). Siphoviruses predominate in saliva, subgingival and supragingival plaque in subjects with relative periodontal health (31), which suggests that many of the gene functions identified in the oral virome are involved in lysogenic conversions of bacteria.
There are many virulence factors that can be identified amongst the oral phage community that are putatively involved in the pathogenesis of their cellular hosts (22). The first evidence of this in the oral virome was the identification of platelet binding factors pblA and pblB in tail fiber genes of oral phage (24). Presumably, the presence of tail proteins encoding these factors contributes to the pathogenicity of their bacterial hosts. These proteins have been shown to contribute to Streptococcus mitis virulence in an animal model of endocarditis (82, 83). The prevalence of different pbl genes in the oral virome suggests that there probably are multiple different phage with different hosts that take advantage of these phage to increase their virulence potential. Other commonly identified virulence genes in oral phage are putatively involved in immune evasion, and include pspA and pspC, which are involved in complement fixation and immunoglobulin degradation, respectively (22). There also are oral virulence factors that have multiple functions, such as both the adhesion and serum resistance functions in YadA (84), which has been identified in the oral virome. Phage carrying yadA have been shown to persist in the oral cavity over relatively long time periods (22). Many different virulence genes such as pblA, pblB, pspA, and yadA have been identified in oral phage using metagenomics; however, the actual contribution of these genes to the pathogenicity of their hosts has yet to be demonstrated experimentally.
Phage attachment and traversal of mucosal surfaces
The total surface area of the average human mouth is estimated to be 215±13 cm2, with 30.5% of the surface area devoted to the teeth and hard palate, and 69.5% devoted to the tongue and other mucosa (85). This surface area may be increased in periodontal pockets in subjects with severe generalized periodontal disease (86). The oral mucosa provides a broad surface area to house phage communities, but membership of mucosal phage communities have yet to be investigated. Because saliva and dental plaque share a portion of their phage (23, 31), it is likely that some phage inhabiting mucosal surfaces also are shared with saliva.
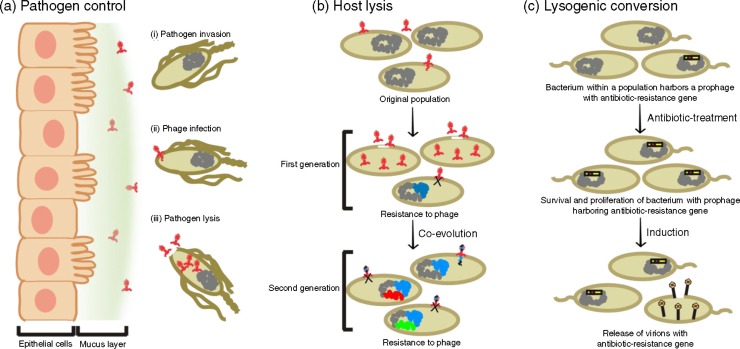
The phage to bacteria ratio in the human gums is estimated to be >35:1 compared to a <10:1 ratio for the adjacent planktonic environment (87), which indicates that the oral mucosa provides an extensive surface for phage adhesion. The high abundance of phage in mucus layers could represent an increased production of phage by bacteria in the mucus layer, a high propensity of planktonic phage to bind to mucus layers, or an ability of phage to bind to mucus layers and persist even in the absence of continual production of virions. The potential for phage virions to be highly persistent in mucus layers could have consequences for their hosts, and phage have been hypothesized to provide a protective function for the human host against invading bacterial pathogens (Fig. 2, Panel a) (87, 88).
Fig. 2.
Diagrams representing modalities by which phage may alter the diversity of the bacterial community in the human oral cavity. Panel a represents the abundance of phage capable of binding to the oral mucosa. In this example, the phage employ generalist strategies with expanded host ranges and are capable of protecting the human host against pathogens that attempt to cross mucosal barriers. Panel b represents a co-evolutionary arms race in which lytic phage drive their hosts to develop resistance phenotypes to escape predation. Panel c represents potential beneficial effects of lysogenic conversions by phage. In this example, the phage endows its host with antibiotic resistance that allows its host to survive and become more abundant in the community after antibiotic disturbances.
Phage nucleic acids have been identified in the blood of individuals who are immunosuppressed (89), which suggests mucosal barriers, and their associated immune components play a role in limiting the access of phage to the bloodstream. While many of the phage identified in the blood may gain access to the blood by way of the gastrointestinal tract, the large surface area of the oral cavity (85) suggests that breaches in the oral mucosa could also lead to translocation of phage into the bloodstream. The spread of oral phage through the bloodstream to other parts of the body could contribute to them finding additional bacterial hosts within the same individual and even their spread to other individuals (26).
Role of bacteriophage in driving oral microbial diversity
Rapid co-evolution may affect oral microbial ecology, and the persistence of many phage may result in an accelerated population dynamic where diversity of the cellular microbiota is driven by their phage. As was demonstrated with RNA phage Φ6 in an experimental system with P. syringae, when competition was introduced, the original phage rapidly evolved generalist phenotypes capable of infecting multiple different bacterial strains and species (90). These data suggest that phage in complex ecosystems compete with one another for resources, and that competition may drive the evolution of more generalist genotypes capable of expanding their host ranges (91). The consequences for oral microbial ecosystem may be that a greater proportion of the bacteria come under pressure from more generalist phage.
The different phage lifestyles in the oral cavity may help to determine the extent of selective pressure that oral bacteria are under from their phage. Those phage with primarily lytic lifestyles are capable of driving bacterial diversity through an arms race with their host, where the host develops mutations to escape phage predation and vice versa (Fig. 2, Panel b) (59). Generally less is known about diversifying selection from temperate phage, which reside primarily in the genomes of their hosts. These types of phage lyse their hosts when resources are limited as a means to spread their progeny to avoid extinction when their hosts cannot find adequate resources. In contrast, data exist suggesting that lysis occurs from some temperate phage when resources are relatively abundant as long as there are agents such as sunlight, pollutants, or other environmental factors present that might promote induction (92–96). This finding was also supported in a study that showed persistence of phage virions in the oral cavities of subjects that were sampled three times daily (25). The persistence of those virions in that study suggested that there may be a low level of lysis of their hosts even for lysogenic phage that resulted in their virions being identified at all-time points in subjects over 60 days. That temperate phage may kill their hosts even under resource abundant conditions alters the perception that they drive the diversity of their cellular hosts primarily when resources are limited.
There are certain host genes that are under diversifying selection by invading phage. Receptor proteins on the host represent a primary means by which host and phage interact. The phage recognize and bind to the host cell receptors and modification or loss of these receptors as a mechanism for evading phage is an important process by which bacteria are diversified during their encounters with their phage (97, 98). Changes to these receptors often lead to fitness costs, such as the loss of the ability of oral bacteria to form viable biofilms (97, 99). While there are many studies demonstrating the co-evolution of host and phage in vitro, there also is evidence that co-evolution occurs in natural communities. P. fluorescens and phage Φ2 undergo extensive changes in soil as a result of their co-evolutionary arms race (58, 100). There also is considerable phage diversity including horizontal gene transfers in genes involved in phage attachment in other natural phage isolates, suggesting strong diversifying selective pressures among the attachment sites of these phage (58, 101, 102). Some organisms such as P. aeruginosa utilize a dense extracellular matrix to avoid exposing their receptors to phage (103); however, some phage encode their own enzymes to break down these matrices and predate upon their hosts (104). Host bacteria even encode systems that allow them to abort phage infections through cell death utilizing abortive infection systems (105).
Many oral phage persist despite host compensatory mechanisms to avoid infection (25). The presence of many siphoviruses that may have lysogenic lifestyles (31), suggests that these phage may have greater fitness in adapting to the oral environment. Oral siphoviruses may drive the diversity of oral bacteria through their contributions of pathogenic gene functions, such as mediators that allow adhesion to the oropharynx and degradation of complement and immunoglobulins (22). CRISPR/Cas systems are involved in driving reciprocal diversity between host and phage in natural environments (106), including streptococci in the human oral cavity (74). The CRISPR loci within these systems have numerous spacer sequences that target oral phage, but those matching phage persist, potentially as a result of their ability to lie dormant as prophage in their bacterial hosts.
Spread to other individuals
Oral phage must to some extent reflect the ecology of their bacterial hosts, yet the presence of both mutualistic and antagonistic relationships between phage and hosts makes it difficult to predict the dynamics of any particular host/phage relationship. Another factor that shapes oral microbial ecology is the sharing of our oral microbiota with those of our close contacts. As pertains to oral phage, many are shared between close contacts through personal contact or shared environmental reservoirs (107). A recent study showed that similar oral phage could be found between unrelated household members (26), even in the absence of such intimate behaviors such as kissing, which suggests that intimate contact is not required for the sharing of oral microbiota. However, another study recently showed that oral microbiota are shared significantly through intimate kissing (48), so it is likely that the more intimate the contact the greater is the potential to share microbiota.
There are tremendous implications for the sharing of viromes amongst close contacts, most importantly is the potential sharing of their virulence gene functions. The persistence of oral phage likely provides substantial opportunities for sharing amongst close contacts. Many oral phage carry antibiotic resistance genes, such as beta lactamases (involved in resistance to antibiotics such as penicillin) (25), which could alter the resistance of the resident microbiota to antibiotic perturbations (Fig. 2, Panel c). The fact that beta lactamases were found in oral phage of subjects that had not received antibiotics for years, suggests that antibiotic resistance in phage may not always occur as a direct result of selective antibiotic pressures. These phage identified in the oral cavity and their potential to be shared with close contacts likely contribute to the increasing trend in penicillin resistance in oral microbes (108).
Consequences of altered bacterial diversity
There have been numerous different oral pathogens implicated in the development of periodontal disease (109–111), however, there has yet to be identified any single pathogen present in every case. This has led some to hypothesize that periodontal disease may result from an altered bacterial community that creates an oral environment that is more conducive to inflammation and the subsequent development of periodontitis. Indeed there are consistent identifiable differences in the subgingival microbiota of subjects with moderate/severe periodontitis compared to relatively healthy controls (31), and it remains unclear whether some of the observed differences could be mediated by phage. A recent study has shown that there are significant differences in phage ecology in the subgingival crevice of relatively healthy subjects compared with those with moderate/severe periodontitis (31). While these differences could merely reflect variations in the bacterial community, similar changes in a number of ecosystems are not necessarily reflected in phage ecology (22, 112, 113). The phage in the subgingival crevice of subjects with moderate/severe periodontal disease were highly enriched for myoviruses rather than the siphoviruses that predominate in relative periodontal health (31), and their potentially lytic lifestyles could contribute to the altered bacterial diversity observed in the disease state. However, not all myoviruses have primarily lytic lifestyles, so the presence of myoviruses in periodontal disease does not necessarily indicate that the hosts are under added pressure from phage predation.
Another consequence of altered bacterial community composition may be an added susceptibility to pathogens. While the effects of disturbances to oral microbiota are poorly understood, disturbances that result in dysbiosis of cellular microbes on other body sites are associated with disease phenotypes (114). Some examples of dysbiosis associated with disease include vaginal candidiasis in response to antibiotic disturbances (115), pseudomembranous colitis as a result of Clostridium difficile infection after antibiotic disturbances (116), and inflammatory bowel diseases in which intestinal dysbiosis is a hallmark feature (117). In the oral cavity, dental caries occur as a result of bacterial carbohydrate fermentation leading to the acidification of the local environment and the selection of cariogenic bacteria. Periodontal disease is associated with dysbiosis of both the bacterial and phage flora in the subgingival crevice (31, 118). Other conditions such as abscesses and pharyngitis generally are caused by the presence of oral pathogens such as Group A Streptococcus (119). However, whether these conditions may also be preceded by dysbiosis that provides a selective advantage to oral pathogens is not known. Characterization of phage communities and their dynamics preceding disease development could provide insight into the role that phage play in oral health and disease.
Conclusions
The advent of next generation sequencing technologies has dramatically increased our knowledge of phage diversity and population dynamics in human health and disease. Yet, very little is known about the ecological and evolutionary mechanisms at the gene and molecular level that are responsible for the transition from a healthy virome to a disease-associated virome. Current research shows that the traditionally narrow phage host-range boundaries are being reevaluated by using novel molecular tools such as single-cell PCR and in-situ hybridization. Research is also expanding on the mechanisms that determine the mixed roles of phage in different oral biogeographic sites and their potential roles as drivers of bacterial diversity in oral disease. In some conditions such as periodontitis the presence of phage may be linked to more virulent bacteria, while in other oral conditions they may have more commensal roles. Oral phage research has opened new exiting areas in both basic and applied science fields, and in the near future we will be able to move beyond taxonomic classification and gene-association studies to functional meta-omics studies and controlled experimental host-microbial model systems. Such systems either used in isolation or in a combinatory experimental approach, will allow systematic investigation of the impact of viromes on the health and disease of the human host. We foresee continued progress in understanding the role of phage in oral dysbiosis and their potential impacts on indigenous oral microbiota including their potential for spread of antibiotic resistance.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Brady JM, Gray WA, Caldwell MA. The electron microscopy of bacteriophage-like particles in dental plaque. J Dent Res. 1977;56:991–3. doi: 10.1177/00220345770560082901. [DOI] [PubMed] [Google Scholar]
- 2.Halhoul N, Colvin JR. Virus-like particles in association with a microorganism from human gingival plaque. Arch Oral Biol. 1975;20:833–6. doi: 10.1016/0003-9969(75)90062-x. [DOI] [PubMed] [Google Scholar]
- 3.Shimizu Y. [Experimental studies on the bacteriophages of the Veillonella strains isolated from the oral cavity] Shigaku. 1968;55:533–41. [PubMed] [Google Scholar]
- 4.Tylenda CA, Calvert C, Kolenbrander PE, Tylenda A. Isolation of Actinomyces bacteriophage from human dental plaque. Infect Immun. 1985;49:1–6. doi: 10.1128/iai.49.1.1-6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen I, Namork E, Myhrvold V. Electron microscopy of phages in serotypes of Actinobacillus actinomycetemcomitans . Oral Microbiol Immunol. 1993;8:383–5. doi: 10.1111/j.1399-302x.1993.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 6.Preus HR, Olsen I, Gjermo P. Bacteriophage infection—a possible mechanism for increased virulence of bacteria associated with rapidly destructive periodontitis. Acta Odontol Scand. 1987;45:49–54. doi: 10.3109/00016358709094353. [DOI] [PubMed] [Google Scholar]
- 7.Preus HR, Olsen I, Namork E. Association between bacteriophage-infected Actinobacillus actinomycetemcomitans and rapid periodontal destruction. J Clin Periodontol. 1987;14:245–7. doi: 10.1111/j.1600-051x.1987.tb00974.x. [DOI] [PubMed] [Google Scholar]
- 8.Sandmeier H, van Winkelhoff AJ, Bar K, Ankli E, Maeder M, Meyer J. Temperate bacteriophages are common among Actinobacillus actinomycetemcomitans isolates from periodontal pockets. J Periodontal Res. 1995;30:418–25. doi: 10.1111/j.1600-0765.1995.tb01296.x. [DOI] [PubMed] [Google Scholar]
- 9.Willi K, Sandmeier H, Asikainen S, Saarela M, Meyer J. Occurrence of temperate bacteriophages in different Actinobacillus actinomycetemcomitans serotypes isolated from periodontally healthy individuals. Oral Microbiol Immunol. 1997;12:40–6. doi: 10.1111/j.1399-302x.1997.tb00365.x. [DOI] [PubMed] [Google Scholar]
- 10.Bachrach G, Leizerovici-Zigmond M, Zlotkin A, Naor R, Steinberg D. Bacteriophage isolation from human saliva. Lett Appl Microbiol. 2003;36:50–3. doi: 10.1046/j.1472-765x.2003.01262.x. [DOI] [PubMed] [Google Scholar]
- 11.Stevens RH, Preus HR, Dokko B, Russell DT, Furgang D, Schreiner HC, et al. Prevalence and distribution of bacteriophage phi Aa DNA in strains of Actinobacillus actinomycetemcomitans . FEMS Microbiol Lett. 1994;119:329–37. doi: 10.1111/j.1574-6968.1994.tb06909.x. [DOI] [PubMed] [Google Scholar]
- 12.Sandmeier H, Bar K, Meyer J. Search for bacteriophages of black-pigmented gram-negative anaerobes from dental plaque. FEMS Immunol Med Microbiol. 1993;6:193–4. doi: 10.1111/j.1574-695X.1993.tb00324.x. [DOI] [PubMed] [Google Scholar]
- 13.Hitch G, Pratten J, Taylor PW. Isolation of bacteriophages from the oral cavity. Lett Appl Microbiol. 2004;39:215–19. doi: 10.1111/j.1472-765X.2004.01565.x. [DOI] [PubMed] [Google Scholar]
- 14.Heilmann S, Sneppen K, Krishna S. Sustainability of virulence in a phage-bacterial ecosystem. J Virol. 2010;84:3016–22. doi: 10.1128/JVI.02326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. Statistical structure of host–phage interactions. Proc Natl Acad Sci USA. 2011;108:E288–97. doi: 10.1073/pnas.1101595108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores CO, Valverde S, Weitz JS. Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phages. ISME J. 2013;7:520–32. doi: 10.1038/ismej.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB, et al. Phage-bacteria infection networks. Trends Microbiol. 2013;21:82–91. doi: 10.1016/j.tim.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Stevens RH, Porras OD, Delisle AL. Bacteriophages induced from lysogenic root canal isolates of Enterococcus faecalis . Oral Microbiol Immunol. 2009;24:278–84. doi: 10.1111/j.1399-302X.2009.00506.x. [DOI] [PubMed] [Google Scholar]
- 19.Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allers E, Moraru C, Duhaime MB, Beneze E, Solonenko N, Barrero-Canosa J, et al. Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ Microbiol. 2013;15:2306–18. doi: 10.1111/1462-2920.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haynes M, Rohwer FL. The human virome. In: K Nelson., editor. Metagenomics of the human body. New York, NY: Springer Science + Business Media, LLC; 2011. [Google Scholar]
- 22.Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA, 3rd, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915–26. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naidu M, Robles-Sikisaka R, Abeles SR, Boehm TK, Pride DT. Characterization of bacteriophage communities and CRISPR profiles from dental plaque. BMC Microbiol. 2014;14:175. doi: 10.1186/1471-2180-14-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci USA. 2011;108:4547–53. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, et al. Human oral viruses are personal, persistent and gender-consistent. ISME J. 2014;8:1753–67. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT. Association between living environment and human oral viral ecology. ISME J. 2013;7:1710–24. doi: 10.1038/ismej.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popgeorgiev N, Temmam S, Raoult D, Desnues C. Describing the silent human virome with an emphasis on giant viruses. Intervirology. 2013;56:395–412. doi: 10.1159/000354561. [DOI] [PubMed] [Google Scholar]
- 28.Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M, et al. First isolation of Mimivirus in a patient with pneumonia. Clin Infect Dis. 2013;57:e127–34. doi: 10.1093/cid/cit354. [DOI] [PubMed] [Google Scholar]
- 29.Santiago-Rodriguez TM, Ly M, Bonilla N, Pride DT. The human urine virome in association with urinary tract infections. Front Microbiol. 2015;6:14. doi: 10.3389/fmicb.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delwart EL. Viral metagenomics. Rev Med Virol. 2007;17:115–31. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ly M, Abeles SR, Boehm TK, Robles-Sikisaka R, Naidu M, Santiago-Rodriguez T, et al. Altered oral viral ecology in association with periodontal disease. MBio. 2014;5:e01133–14. doi: 10.1128/mBio.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, et al. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics. 2005;6:41. doi: 10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguirre de Carcer D, Angly FE, Alcami A. Evaluation of viral genome assembly and diversity estimation in deep metagenomes. BMC Genomics. 2014;15:989. doi: 10.1186/1471-2164-15-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shira R, Abeles ML, Santiago-Rodriguez TM, Pride DT. Effects of long term antibiotic therapy on human oral and fecal viromes. 2015 doi: 10.1371/journal.pone.0134941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi N. Microbial ecosystem in the oral cavity: metabolic diversity in an ecological niche and its relationship to oral diseases. Int Congr Ser. 2005;1284:103–12. [Google Scholar]
- 36.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2014;9:968–79. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinberg I, Jenkins GN, Chatterjee R, Wijeyeweera L. The antimony pH electrode and its role in the assessment and interpretation of dental plaque Ph. J Dent Res. 1982;61:1139–47. doi: 10.1177/00220345820610100601. [DOI] [PubMed] [Google Scholar]
- 39.Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 2000;193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 40.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–71. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 41.Hodgson RJ, Lynch RJ, Watson GK, Labarbe R, Treloar R, Allison C. A continuous culture biofilm model of cariogenic responses. J Appl Microbiol. 2001;90:440–8. doi: 10.1046/j.1365-2672.2001.01263.x. [DOI] [PubMed] [Google Scholar]
- 42.Kadowaki T, Nakayama K, Okamoto K, Abe N, Baba A, Shi Y, et al. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J Biochem. 2000;128:153–9. doi: 10.1093/oxfordjournals.jbchem.a022735. [DOI] [PubMed] [Google Scholar]
- 43.Nishihara T, Koseki T. Microbial etiology of periodontitis. Periodontol 2000. 2004;36:14–26. doi: 10.1111/j.1600-0757.2004.03671.x. [DOI] [PubMed] [Google Scholar]
- 44.Al-Jarbou AN. Genomic library screening for viruses from the human dental plaque revealed pathogen-specific lytic phage sequences. Curr Microbiol. 2012;64:1–6. doi: 10.1007/s00284-011-0025-z. [DOI] [PubMed] [Google Scholar]
- 45.Castillo-Ruiz M, Vines ED, Montt C, Fernandez J, Delgado JM, Hormazabal JC, et al. Isolation of a novel Aggregatibacter actinomycetemcomitans serotype b bacteriophage capable of lysing bacteria within a biofilm. Appl Environ Microbiol. 2011;77:3157–9. doi: 10.1128/AEM.02115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bassis CM, Tang AL, Young VB, Pynnonen MA. The nasal cavity microbiota of healthy adults. Microbiome. 2014;2:27. doi: 10.1186/2049-2618-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013;14:631–40. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kort R, Caspers M, van de Graaf A, van Egmond W, Keijser B, Roeselers G. Shaping the oral microbiota through intimate kissing. Microbiome. 2014;2:41. doi: 10.1186/2049-2618-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faveri M, Feres M, Shibli JA, Hayacibara RF, Hayacibara MM, de Figueiredo LC. Microbiota of the dorsum of the tongue after plaque accumulation: an experimental study in humans. J Periodontol. 2006;77:1539–46. doi: 10.1902/jop.2006.050366. [DOI] [PubMed] [Google Scholar]
- 50.Tanner AC, Paster BJ, Lu SC, Kanasi E, Kent R, Jr., Van Dyke T, et al. Subgingival and tongue microbiota during early periodontitis. J Dent Res. 2006;85:318–23. doi: 10.1177/154405910608500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 2014;111:E2875–84. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papaioannou W, Gizani S, Haffajee AD, Quirynen M, Mamai-Homata E, Papagiannoulis L. The microbiota on different oral surfaces in healthy children. Oral Microbiol Immunol. 2009;24:183–9. doi: 10.1111/j.1399-302X.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- 54.Dereure O, Cheval J, Du Thanh A, Pariente K, Sauvage V, Manuguerra JC, et al. No evidence for viral sequences in mycosis fungoides and Sezary syndrome skin lesions: a high-throughput sequencing approach. J Invest Dermatol. 2013;133:853–5. doi: 10.1038/jid.2012.371. [DOI] [PubMed] [Google Scholar]
- 55.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pride DT, Wassenaar TM, Ghose C, Blaser MJ. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics. 2006;7:8. doi: 10.1186/1471-2164-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James CE, Davies EV, Fothergill JL, Walshaw MJ, Beale CM, Brockhurst MA, et al. Lytic activity by temperate phages of Pseudomonas aeruginosa in long-term cystic fibrosis chronic lung infections. ISME J. 2014 doi: 10.1038/ismej.2014.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brockhurst MA. Bacteria–virus coevolution. In: OS Soyer., editor. Evolutionary systems biology. New York, NY: Springer Science + Business Media, LLC; 2012. [Google Scholar]
- 59.Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution. 2008;62:1–11. doi: 10.1111/j.1558-5646.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 60.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450–5. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S, Ward TJ, Siletzky RM, Kathariou S. Two novel type II restriction-modification systems occupying genomically equivalent locations on the chromosomes of Listeria monocytogenes strains. Appl Environ Microbiol. 2012;78:2623–30. doi: 10.1128/AEM.07203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res. 2003;13:145–58. doi: 10.1101/gr.335003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr Opin Microbiol. 2005;8:466–72. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 64.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci USA. 2012;109:17621–6. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furuse K, Osawa S, Kawashiro J, Tanaka R, Ozawa A, Sawamura S, et al. Bacteriophage distribution in human faeces: continuous survey of healthy subjects and patients with internal and leukaemic diseases. J Gen Virol. 1983;64:2039–43. doi: 10.1099/0022-1317-64-9-2039. [DOI] [PubMed] [Google Scholar]
- 66.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 67.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 68.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 69.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–7. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 70.Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa . MBio. 2014;5:e00896. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seed KD, Lazinski DW, Calderwood SB, Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–91. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–32. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pride DT, Salzman J, Relman DA. Comparisons of clustered regularly interspaced short palindromic repeats and viromes in human saliva reveal bacterial adaptations to salivary viruses. Environ Microbiol. 2012;14:2564–76. doi: 10.1111/j.1462-2920.2012.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pride DT, Sun CL, Salzman J, Rao N, Loomer P, Armitage GC, et al. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res. 2011;21:126–36. doi: 10.1101/gr.111732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robles-Sikisaka R, Naidu M, Ly M, Salzman J, Abeles SR, Boehm TK, et al. Conservation of streptococcal CRISPRs on human skin and saliva. BMC Microbiol. 2014;14:146. doi: 10.1186/1471-2180-14-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roberts AP, Mullany P. Genetic basis of horizontal gene transfer among oral bacteria. Periodontol 2000. 2006;42:36–46. doi: 10.1111/j.1600-0757.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- 78.Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 80.Wichels A, Biel SS, Gelderblom HR, Brinkhoff T, Muyzer G, Schutt C. Bacteriophage diversity in the North Sea. Appl Environ Microbiol. 1998;64:4128–33. doi: 10.1128/aem.64.11.4128-4133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus . Nature. 2003;424:1047–51. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 82.Mitchell J, Siboo IR, Takamatsu D, Chambers HF, Sullam PM. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol Microbiol. 2007;64:844–57. doi: 10.1111/j.1365-2958.2007.05703.x. [DOI] [PubMed] [Google Scholar]
- 83.Bensing BA, Siboo IR, Sullam PM. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect Immun. 2001;69:6186–92. doi: 10.1128/IAI.69.10.6186-6192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roggenkamp A, Ackermann N, Jacobi CA, Truelzsch K, Hoffmann H, Heesemann J. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J Bacteriol. 2003;185:3735–44. doi: 10.1128/JB.185.13.3735-3744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Collins LM, Dawes C. The surface area of the adult human mouth and thickness of the salivary film covering the teeth and oral mucosa. J Dent Res. 1987;66:1300–2. doi: 10.1177/00220345870660080201. [DOI] [PubMed] [Google Scholar]
- 86.Nesse W, Abbas F, van der Ploeg I, Spijkervet FK, Dijkstra PU, Vissink A. Periodontal inflamed surface area: quantifying inflammatory burden. J Clin Periodontol. 2008;35:668–73. doi: 10.1111/j.1600-051X.2008.01249.x. [DOI] [PubMed] [Google Scholar]
- 87.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gorski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci. 2005;62:511–19. doi: 10.1007/s00018-004-4403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–87. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bono LM, Gensel CL, Pfennig DW, Burch CL. Competition and the origins of novelty: experimental evolution of niche-width expansion in a virus. Biol Lett. 2013;9:20120616. doi: 10.1098/rsbl.2012.0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Duplessis M, Levesque CM, Moineau S. Characterization of Streptococcus thermophilus host range phage mutants. Appl Environ Microbiol. 2006;72:3036–41. doi: 10.1128/AEM.72.4.3036-3041.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cochran PK, Paul JH. Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl Environ Microbiol. 1998;64:2308–12. doi: 10.1128/aem.64.6.2308-2312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang SC, Paul JH. Gene transfer by transduction in the marine environment. Appl Environ Microbiol. 1998;64:2780–7. doi: 10.1128/aem.64.8.2780-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ortmann AC, Lawrence JE, Suttle CA. Lysogeny and lytic viral production during a bloom of the cyanobacterium Synechococcus spp. Microb Ecol. 2002;43:225–31. doi: 10.1007/s00248-001-1058-9. [DOI] [PubMed] [Google Scholar]
- 95.Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008;2:579–89. doi: 10.1038/ismej.2008.35. [DOI] [PubMed] [Google Scholar]
- 96.Wommack KE, Hill RT, Muller TA, Colwell RR. Effects of sunlight on bacteriophage viability and structure. Appl Environ Microbiol. 1996;62:1336–41. doi: 10.1128/aem.62.4.1336-1341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 98.Icho T, Iino T. Isolation and characterization of motile Escherichia coli mutants resistant to bacteriophage chi. J Bacteriol. 1978;134:854–60. doi: 10.1128/jb.134.3.854-860.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Samuel AD, Pitta TP, Ryu WS, Danese PN, Leung EC, Berg HC. Flagellar determinants of bacterial sensitivity to chi-phage. Proc Natl Acad Sci USA. 1999;96:9863–6. doi: 10.1073/pnas.96.17.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gomez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–9. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 101.Ceyssens PJ, Glonti T, Kropinski NM, Lavigne R, Chanishvili N, Kulakov L, et al. Phenotypic and genotypic variations within a single bacteriophage species. Virol J. 2011;8:134. doi: 10.1186/1743-422X-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature. 2011;474:604–8. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- 103.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 104.Glonti T, Chanishvili N, Taylor PW. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa . J Appl Microbiol. 2010;108:695–702. doi: 10.1111/j.1365-2672.2009.04469.x. [DOI] [PubMed] [Google Scholar]
- 105.Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–9. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 106.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–50. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 107.Rusin P, Maxwell S, Gerba C. Comparative surface-to-hand and fingertip-to-mouth transfer efficiency of gram-positive bacteria, gram-negative bacteria, and phage. J Appl Microbiol. 2002;93:585–92. doi: 10.1046/j.1365-2672.2002.01734.x. [DOI] [PubMed] [Google Scholar]
- 108.Sweeney LC, Dave J, Chambers PA, Heritage J. Antibiotic resistance in general dental practice—a cause for concern? J Antimicrob Chemother. 2004;53:567–76. doi: 10.1093/jac/dkh137. [DOI] [PubMed] [Google Scholar]
- 109.Sela MN. Role of Treponema denticola in periodontal diseases. Crit Rev Oral Biol Med. 2001;12:399–413. doi: 10.1177/10454411010120050301. [DOI] [PubMed] [Google Scholar]
- 110.Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 111.Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 112.Bratbak G, Heldal M, Thingstad TF, Tuomi P. Dynamics of virus abundance in coastal seawater. FEMS Microbiol Ecol. 1996;19:263–69. [Google Scholar]
- 113.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. Phage–host interaction: an ecological perspective. J Bacteriol. 2004;186:3677–86. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kurowski K, Ghosh R, Singh SK, Beaman KD. Clarithromycin-induced alterations in vaginal flora. Am J Ther. 2000;7:291–5. doi: 10.1097/00045391-200007050-00004. [DOI] [PubMed] [Google Scholar]
- 116.Schubert AM, Rogers MA, Ring C, Mogle J, Petrosino JP, Young VB, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;5:e01021–14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 118.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 119.Shulman ST, Bisno AL, Clegg HW, Gerber MA, Kaplan EL, Lee G, et al. Clinical practice guideline for the diagnosis and management of group A streptococcal pharyngitis: 2012 update by the Infectious Diseases Society of America. Clin Infect Dis. 2012;55:e86–102. doi: 10.1093/cid/cis629. [DOI] [PMC free article] [PubMed] [Google Scholar]