Abstract
Iron is essential in many biological processes. However, its bioavailability is reduced in aerobic environments, such as soil. To overcome this limitation, microorganisms have developed different strategies, such as iron chelation by siderophores. Some bacteria have even gained the ability to detect and utilize xenosiderophores, i.e., siderophores produced by other organisms. We illustrate an example of such an interaction between two soil bacteria, Pseudomonas fluorescens strain BBc6R8 and Streptomyces ambofaciens ATCC 23877, which produce the siderophores pyoverdine and enantiopyochelin and the siderophores desferrioxamines B and E and coelichelin, respectively. During pairwise cultures on iron-limiting agar medium, no induction of siderophore synthesis by P. fluorescens BBc6R8 was observed in the presence of S. ambofaciens ATCC 23877. Cocultures with a Streptomyces mutant strain that produced either coelichelin or desferrioxamines, as well as culture in a medium supplemented with desferrioxamine B, resulted in the absence of pyoverdine production; however, culture with a double mutant deficient in desferrioxamines and coelichelin production did not. This strongly suggests that P. fluorescens BBbc6R8 utilizes the ferrioxamines and ferricoelichelin produced by S. ambofaciens as xenosiderophores and therefore no longer activates the production of its own siderophores. A screening of a library of P. fluorescens BBc6R8 mutants highlighted the involvement of the TonB-dependent receptor FoxA in this process: the expression of foxA and genes involved in the regulation of its biosynthesis was induced in the presence of S. ambofaciens. In a competitive environment, such as soil, siderophore piracy could well be one of the driving forces that determine the outcome of microbial competition.
INTRODUCTION
Bacteria detect, assimilate, and integrate different environmental signals in order to better adapt to their habitat and cope with changes in environmental conditions. Multiple signaling pathways allow them to communicate with each other within the same species or between different species (1). This can be achieved through the production and detection of diffusible molecules in the environment. In response to these interactions, the microorganisms have developed complex metabolic and physiological responses. One of the essential environmental factors vital for organisms is iron. It plays an essential role in many biological processes, such as DNA synthesis, respiration, and photosynthesis. Iron can adopt two different ionic forms, Fe2+ and Fe3+. This property makes it an important player in the oxidation-reduction reactions in the cell. However, while iron is an abundant element on earth, its bioavailability is reduced in aerobic environments, such as soil. Ferric iron (Fe3+) forms insoluble ferric hydroxides (with a solubility product of ∼10−39) in the presence of oxygen (2–4). Therefore, iron is a limiting factor for the growth of microorganisms.
To overcome the limitation of iron bioavailability, aerobic bacteria have developed several highly specialized strategies to acquire the metal from different sources. One of them consists of producing siderophores under conditions of iron deficiency (5). Siderophores are low-molecular-mass molecules (200 to 2,000 Da) with diverse iron affinities, and to date, more than 500 different chemical structures of siderophores have been identified. A single bacterial species can produce different siderophores, although in general, not all are produced at the same time. For example, Pseudomonas aeruginosa is able to switch between the synthesis of its siderophores pyoverdine and pyochelin depending on environmental conditions. In a severe iron depletion environment, pyoverdine, the most effective but metabolically expensive siderophore, is produced. However, in environments moderately depleted of iron, pyochelin, a less metabolically expensive siderophore, is used to take up iron (6).
In addition to acquiring iron via specific receptors for their own siderophores, many bacteria possess uptake systems for xenosiderophores, i.e., siderophores produced by other organisms. In competitive environments, like soil, this allows them to utilize exogenous siderophores in a strategy known as siderophore piracy (7, 8). A study published by Cornelis and Bodilis (9) revealed that the majority of siderophore receptors are conserved in the different representatives of a species (the core receptor), while others are acquired by horizontal gene transfer. For example, Pseudomonas fluorescens SBW25 possesses 23 other putative siderophore receptors in addition to the receptor of its own pyoverdine. This allows the bacterium to take up 19 heterologous pyoverdines from 25 different Pseudomonas isolates (10). Pseudomonas fragi, which does not produce siderophores, is able to use enterobactin, pyoverdine, and desferrioxamine B, produced by the bacterial species Escherichia coli, P. fluorescens, or P. aeruginosa and Pseudomonas stutzeri, respectively (11). Other genera of bacteria, like Yersinia (12), Erwinia (13), Vibrio (14), and Amycolatopsis (15), are also able to detect and to take up xenosiderophores, in addition to the use of their own siderophores. The wide distribution of xenosiderophore uptake genes in many bacterial species suggests that siderophore piracy is a common process in multispecies communities. However, most of these studies were performed in vitro using purified siderophores, and little is known regarding the occurrence of siderophore piracy during biotic interactions.
Actinomycetes and pseudomonads represent two of the major groups of bacteria found in soils and rhizospheres (16, 17) and are likely to simultaneously utilize similar resources, such as iron, or even to compete for them. Both groups produce siderophores to take up this essential element, but different types of molecules are produced by the two groups. Many pseudomonads produce pyoverdines, a family of high-affinity catecholate-hydroxamate siderophores, while the actinomycetes of the genus Streptomyces, for instance, secrete hydroxamate siderophores, such as desferrioxamines and coelichelin. In this study, we show, through a pairwise interaction between the two soil bacteria Streptomyces ambofaciens ATCC 23877 (18) and P. fluorescens BBc6R8 (R8) (19, 20), that the P. fluorescens strain uses the streptomycete's siderophores and does not induce the production of its own siderophores, pyoverdine and enantiopyochelin, in the presence of S. ambofaciens ATCC 23877 when grown under iron-limiting conditions. Our study reveals that P. fluorescens recognizes Streptomyces siderophores through FoxA, a TonB-dependent receptor whose biosynthesis is induced in the presence of S. ambofaciens ATCC 23877 siderophores.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
All Streptomyces, Pseudomonas, and E. coli strains used in this work are listed in Table 1. The Streptomyces strains were manipulated as described by Kieser et al. (21). Pseudomonas cell stocks were prepared by streaking each strain on Trypticase soy agar (TSA) medium (containing kanamycin at 20 μg/ml for the P18B10 and P28H6 mutants) and incubating the plates for 48 h at 26.5°C or at 37°C for P. aeruginosa PAO1. A single clone was resuspended in 50 μl water, spread on TSA medium, and then grown for 48 h. Cells were collected and centrifuged at 13,500 × g for 2 min, and the pellet was washed twice with sterile water. The pellet was then resuspended in sterile water to an optical density at 600 nm (OD600) of 0.7 (109 CFU per ml).
TABLE 1.
Bacterial strains used in this work
| Strain | Characteristics | Reference |
|---|---|---|
| S. ambofaciens ATCC 23877 | Reference strain (wild type) | 18 |
| S. coelicolor M512 | ΔredD ΔactII-ORF4 from S. coelicolor M145; deficient in undecylprodigiosin and actinorhodin production | 40 |
| S. coelicolor M512 desD | Derived from M512; deficient in desferrioxamine production; S. coelicolor M512 desD::Tn5062 | This study |
| S. coelicolor M512 cchH | Derived from M512; deficient in coelichelin production; S. coelicolor M512 cchH::Tn5062 | This study |
| S. coelicolor Δdes Δcch | Derived from M145; deficient in desferrioxamine and coelichelin production [desEFABCD:: aac(3)IV-cchABCDEFGHIJK::vph; strain W13) | 37 |
| E. coli ET12567(pUZ8002) | Donor strain in intergeneric conjugation; a methylation-defective strain | 31 |
| P. fluorescens BBc6R8 | Isolated from Laccaria bicolor | 19 |
| P. fluorescens SBW25 | Isolated from rhizosphere of sugar beet | 67 |
| P. fluorescens Pf0-1 | Isolated from agricultural soil | 68 |
| P. aeruginosa PAO1 | Human opportunistic pathogen | 69 |
| P. fluorescens P28H6 | Mutant derived from BBc6R8, Tn5 insertion in foxA | A. Deveau, unpublished data |
| P. fluorescens P18B10 | Mutant derived from BBc6R8, Tn5 insertion in foxA | Deveau, unpublished |
| Library of 60 P. fluorescens strains | Strains isolated from the bulk soil of a forest nursery, the ectomycorrhizosphere, and the ectomycorrhizas of L. bicolor/Douglas fir | 70 |
To analyze the effect of S. ambofaciens strain ATCC 23877 on the production of siderophores by P. fluorescens BBc6R8, a bioassay was set up on 26A agar medium (for 400 ml, 0.4 g glucose, 6 g tryptone, 2 g NaCl, pH 7.2). Streptomyces and Pseudomonas were streaked side by side 3 mm from each other and were incubated at 26.5°C (37°C with P. aeruginosa PAO1) for 2 days. The streaks were made up of 2 μl of a spore suspension of S. ambofaciens ATCC 23877 at 109 CFU/ml and 2 μl of Pseudomonas at 107 CFU/ml. A control experiment with Pseudomonas alone was done for each incubation condition. When indicated, the iron chelator 2,2′-bipyridyl (200 μM), desferrioxamine B mesylate (200 μM), or FeSO4 · 7H2O (12.5 μM) was added to the 26A agar medium. For fluorescence detection, a Chemidoc XRS (Bio-Rad) was used with UV transmission (302 nm).
To test the ability of the S. coelicolor Δdes Δcch mutant (strain W13) to take up exogenous siderophores, two different experiments were carried out. The first experiment was as follows. One hundred microliters of a spore suspension of S. coelicolor M512 at 2 × 108 CFU/ml or 100 μl of a cell suspension of P. fluorescens BBc6R8 at 2 × 107 CFU/ml was spread on a 26A agar plate supplemented or not with 2,2′-bipyridyl at 100 or 200 μM. After 3 days of growth at 26.5°C, plugs (8 mm in diameter) were obtained from confluent regions and placed on 26A agar plates containing 200 μM 2,2′-bipyridyl with spores of strain W13 (106 or 109 CFU) evenly spread on them. The plates were incubated at 26.5°C for 3 days. The halo of growth around the plugs was then analyzed. In the second experiment, strains M512 and BBc6R8 (104 CFU) were streaked on both side of a 26A agar plate supplemented with 200 μM 2,2′-bipyridyl. The plates were incubated at 26.5°C for 3 days, and the W13 mutant (2 μl at 109 CFU/ml) was then streaked at about 3 mm alongside strain M512 or BBc6R8.
DNA manipulation and transcriptional analysis.
Isolation, cloning, and manipulation of DNA were carried out as previously described for Streptomyces (22, 23), Pseudomonas (24), and E. coli (25). Amplification of DNA fragments by PCR was performed with Dreamtaq DNA polymerase (Fermentas). All primers are described in Table 2. The transposon insertion site in P. fluorescens BBc6R8 was determined using a double-round nested-PCR-based sequencing approach. First-round PCR was performed using the primers gfptns2, nCEKG2A, nCEKG2B, and nCEKG2C at a ratio of 3:1:1:1. The PCR conditions were 94°C for 5 min, followed by cycles of 94°C for 30 s and 62°C for 30 s, lowered in successive cycles by 1°C until 54°C was reached, and 72°C for 3 min. A further 27 cycles of annealing at 54°C were performed, followed by a final extension of 72°C for 3 min. A second-round PCR was performed using the primers gfptns2 and CEKG4, followed by sequencing using the primer gfptnS2.
TABLE 2.
Primer pairs used in qPCR analyses or to determine transposon insertion sites in the P. fluorescens BBc6R8 genome
| Name | Sequence (5′–3′) | Studied gene |
|---|---|---|
| hrdB-F | CGCGGCATGCTCTTCCT | hrdB |
| hrdB-R | AGGTGGCGTACGTGGAGAAC | |
| selR-F | CGACCCGGAGCAGTACAA | selR |
| selR-R | GCAGCAGATGCAGTGGTAGA | |
| pvdD-F | TTGAGCGTGAGGACATTCTG | pvdD |
| pvdD-R | TCGAACCCTTGCAGGTAATC | |
| pvdE-F | ACGTGGGGTGATCAATGAAT | pvdE |
| pvdE-R | TTTTGCCCCACATAATTGGT | |
| pvdO-F | AAGAAAACCGGCCATCACTA | pvdO |
| pvdO-R | TGTCCACTCGTAGACGTTGC | |
| pvdS-F | ACCATCACGTCATCGTTCAA | pvdS |
| pvdS-R | TTCTCCAGCGTCGAAAAGTT | |
| pvdQ-F | TGCGTTTCTACGAGATGCAC | pvdQ |
| pvdQ-R | AAATAGCGAGTCGGGTCCTT | |
| phcf-F | ACGGCTACCAACAAATCCTG | phcF |
| phcf-R | CACCAGCAGATCCACTGAGA | |
| desC-F | ACTGACCGGGCTGTACGA | desC |
| desC-R | CTTCTCCGGCTTCTGGATCT | |
| sam0552-F | CTTCGTCCTGCAGAACTTCC | sam0552 |
| sam0552-R | AGTACGCGCAGGTAGTCGTC | |
| foxI-F | ACGTGCGTTGATTCTGGAAT | foxI |
| foxI-R | GCACATAACAGTGATAAAGC | |
| foxR-F | CGGCAGGCATTGAAGGTATT | foxR |
| foxR-R | CGCTATCGGTATTGAGCTGC | |
| foxA-F | GCAGACAACGTGATCGAGAA | foxA |
| foxA-R | CACACCCTTCAGTCCAACCT | |
| pepSY-F | GTTTCTGATGATGGCGGGAC | pepSY |
| pepSY-R | ACTTGCAGTTTGGGATGCTG | |
| fur-F | CCAGTTTGAAGCAGCAGGAC | fur |
| fur-R | TCCACCAAATCGAATCCATGC | |
| EZR1a | ATGCGCTCCATCAAGAAGAG | |
| EZL2a | TCCAGCTCGACCAGGATG | |
| gfptns2b | ATCACCTTCACCCTCTCCAC | |
| nCEKG2Ac | GGCCACGCGTCGACTAGTACNNNNNNNNNNAGAG | |
| nCEKG2Bc | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC | |
| nCEKG2Cc | GGCCACGCGTCGACTAGTACNNNNNNNNNNATAT | |
| CEKG4d | GGCCACGCGTCGACTAGTAC |
Tn5062-specific primer.
For first- and second-round PCR of transposon mutants.
For first-round PCR of transposon mutants.
For second-round PCR of transposon mutants.
For RNA extraction, cells were collected from streaks grown on 26A agar medium for 2 days at 26.5°C. Streptomyces was grown on cellophane membranes. The extraction was performed with an Aurum Total RNA minikit (Bio-Rad) according to the manufacturer's instructions, except for the addition, during cell lysis, of a sonication step (3 times for 10 s each time) at high frequency using a Bioruptor apparatus (Diagenode). RNAs were eluted in a final volume of 30 μl and quantified with a NanoDrop-1000 apparatus. Before reverse transcription, RNA samples were treated with DNase I (1 U; Fermentas) in the presence of 10 U of Ribolock RNase Inhibitor (Fermentas) per 1 μg of RNA. The DNase I was then inactivated at 65°C for 10 min in the presence of EDTA (2.3 mM). The absence of residual genomic DNA was verified by a 35-cycle PCR using RNAs as templates and the primer pairs hrdB-F/hrdB-R and selR-F/selR-R for Streptomyces and Pseudomonas, respectively. Reverse transcription was performed with an iScript Advanced cDNA synthesis kit for real-time quantitative PCR (RT-qPCR) (Bio-Rad) according to the manufacturer's instructions. The sequences of the primer pairs used to amplify cDNAs and their target genes are listed in Table 2. RT-qPCRs were carried out on a CFX96 (Bio-Rad) with microplates (Multiplate 96-well unskirted PCR plates, low profile; Bio-Rad) covered with a film (Microseal Bb adhesive seals; Bio-Rad). The reaction mixture was composed of 5 μl of SYBR green Supermix (Bio-Rad), 0.2 μM each primer pair, and 4 μl of cDNA diluted 1/10. The qPCR conditions were as follow: 30 s at 95°C and 40 cycles of 5 s at 95°C and 30 s at 60°C. To verify the absence of secondary products, melting curves were produced from 65 to 95°C with an increase of 0.5°C/cycle. Total RNA levels were normalized using transcripts from the housekeeping genes hrdB (26) for Streptomyces and selR (MHB_002629 [27]) for Pseudomonas as a control. The gene hrdB encodes the major sigma factor of Streptomyces and was used as an internal control to quantify the relative expression of target genes, as it is expressed fairly constantly throughout growth (26, 28). selR was defined, with two other genes, as a potential housekeeping gene based on microarray data (accession number GSE38243 on the Gene Expression Omnibus at NCBI [27]) and was chosen as a reference after checking the stability of its expression in the present setup. Both hrdB and selR transcript levels were experimentally confirmed to be stable under our growth conditions (data not shown).
For each treatment, at least three biological replicates were performed, and the average and standard deviation of the expression relative to that of the reference gene hrdB or selR of each transcript were calculated (29). One-factor analyses of variance (ANOVA) were done with R to test for differences in transcript levels between treatments.
Construction of S. coelicolor M512 desD and cchH mutant strains.
The cosmids C105.2.E01 and F76.2.F08 from the S. coelicolor transposon insertion single-gene knockout library (30) were used to replace the wild-type alleles of desD and cchH, respectively, in S. coelicolor M512. The mutated cosmids were introduced into S. coelicolor M512 by intergenic conjugation from E. coli ET12567/pUZ8002 (31). Gene replacements were confirmed by PCR analysis using the flanking and internal primers desC-F and EZR1 for the mutant S. coelicolor M512 desD and sam0552-R and EZL2 for the mutant S. coelicolor M512 cchH.
RESULTS
P. fluorescens BBc6R8 does not produce its siderophore pyoverdine in the vicinity of S. ambofaciens ATCC 23877.
P. fluorescens BBc6R8 produced a green-yellow pigment and fluoresced under UV light when grown on iron-depleted 26A agar medium (Fig. 1a and b). When the medium was supplemented with FeSO4 · 7H2O (12.5 μM), P. fluorescens BBc6R8 no longer fluoresced (Fig. 1d). This green-yellow pigmentation reflects the presence of the pyoverdine siderophore produced by P. fluorescens BBc6R8 under conditions of iron limitation (32). During the screening of pairwise interactions between S. ambofaciens ATCC 23877 and other soil bacteria, we observed that when grown on 26A agar medium in close proximity to S. ambofaciens, P. fluorescens BBc6R8 partially lost its ability to fluoresce (Fig. 1c, e, and f). Similar results were observed in the presence of the high-affinity iron chelator 2,2′-bipyridyl (200 μM) in the agar medium (see Fig. S1 in the supplemental material). Interestingly, a stronger effect was observed when S. ambofaciens ATCC 23877 was cultivated on 26A agar for 30 h before the petri dish was inoculated with strain BBc6R8. Under these conditions, neither the green-yellow pigment nor fluorescence could be observed (Fig. 1g). Similar results were obtained when 2,2′-bipyridyl was present in the agar medium (see Fig. S1 in the supplemental material). The negative effect on pyoverdine production was proportional to the lag between the inoculations of the two bacteria on the plate. In addition, streaking the bacterial partners perpendicular to one another revealed that the effect of S. ambofaciens on pyoverdine production by P. fluorescens occurred only in the area surrounding the streak of Streptomyces (Fig. 1h and i). Altogether, these data show that in the presence of S. ambofaciens ATCC 23877, strain BBc6R8 does not produce its own siderophore.
FIG 1.
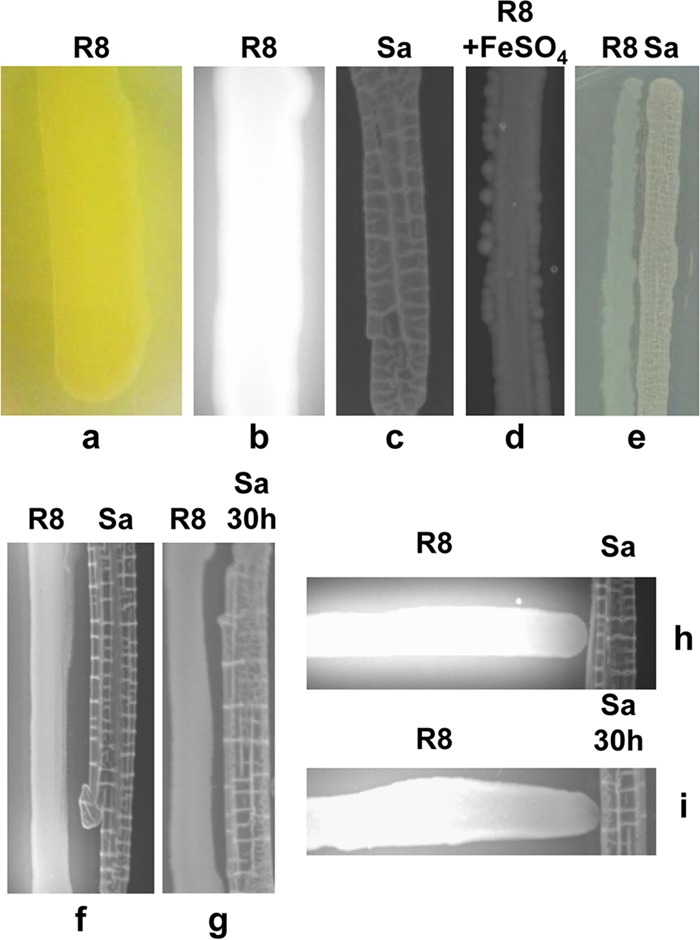
Effect of S. ambofaciens ATCC 23877 on green-yellow fluorescent pigment production by P. fluorescens BBc6R8. (a and b) P. fluorescens BBc6R8 (R8). (c) S. ambofaciens ATCC 23877 (Sa). (d) P. fluorescens BBc6R8 cultivated on medium supplemented with FeSO4. (e and f) P. fluorescens BBc6R8 plus S. ambofaciens ATCC 23877. (g) P. fluorescens BBc6R8 plus S. ambofaciens ATCC 23877 that had been streaked on a plate 30 h before strain BBc6R8. (h) P. fluorescens BBc6R8 plus S. ambofaciens ATCC 23877 cocultivated perpendicularly. (i) P. fluorescens BBc6R8 plus S. ambofaciens ATCC 23877 (streaked on a plate 30 h before strain BBc6R8) cocultivated perpendicularly. (b, c, d, f, g, h, and i) The plates were visualized under UV light. The images were taken from below the plates after a 2-day (co)culture with incubation at 26.5°C on 26A agar medium.
Transcription of the pyoverdine and enantiopyochelin synthesis genes is no longer induced in the presence of S. ambofaciens ATCC 23877.
The biosynthesis and uptake of pyoverdine, as well as their regulation, have been thoroughly studied in P. aeruginosa (33–36) and in P. fluorescens SBW25 (10). A search in the P. fluorescens BBc6R8 genome sequence (27) indicated that all the genes involved in these processes are conserved in the strain (see Table S1 in the supplemental material), suggesting similar mechanisms of biosynthesis, uptake, and regulation in strain BBc6R8 and in P. aeruginosa. Therefore, we tested the influence of S. ambofaciens on the expression of these genes by varying the duration of preincubation of Streptomyces on the plate before the addition of Pseudomonas. Messenger RNAs from cell lysates of P. fluorescens BBc6R8 grown on 26A agar medium in the absence or presence of S. ambofaciens were quantified using RT-qPCR. The duration of preincubation of Streptomyces varied between 0 and 54 h before strain BBc6R8 was streaked on the plate. The expression of genes involved in the biosynthesis of pyoverdine (pvdD) or its regulation (pvdO, pvdS, and pvdQ) and export (pvdE) and in the biosynthesis of the second type of siderophore of P. fluorescens BBc6R8, enantiopyochelin (phcF), were monitored after 2 days of growth. As expected, all the siderophore-related genes were transcribed when Pseudomonas was cultivated alone, reflecting the iron limitation in the 26A medium. In contrast, transcript levels of the pyoverdine genes were significantly reduced when P. fluorescens BBc6R8 was grown in the presence of S. ambofaciens (Fig. 2) (P < 0.01; one-factor ANOVA), except for pvdD and pvdS, when the two bacterial strains were simultaneously streaked on the agar plate. As for the production of pyoverdine, the decrease in transcript levels was proportional to the time lag between the seeding of the two bacterial strains on the plate for all genes analyzed. The expression of the enantiopyochelin gene phcF was similarly affected by the presence of S. ambofaciens ATCC 23877. Altogether, these data confirm our initial observation that P. fluorescens BBc6R8 reduces and even stops the production of its own siderophores on iron-limited medium in the presence of S. ambofaciens ATCC 23877.
FIG 2.

Effect of S. ambofaciens ATCC 23877 on the expression levels of P. fluorescens BBc6R8 genes involved in the biosynthesis of pyoverdine and enantiopyochelin as measured by RT-qPCR. The expression levels are expressed as the transcript levels of the target genes relative to the transcript level of the housekeeping gene selR, which is stable under the tested growth conditions. The data are expressed as the mean value of three biological replicates. The error bars denote standard errors. For each transcript, values with the same letter are not significantly different according to a one-factor ANOVA (P > 0.01). R8, P. fluorescens BBc6R8 grown on 26A agar medium in the absence of S. ambofaciens; 0 to 54, P. fluorescens BBc6R8 grown in the presence of S. ambofaciens ATCC 23877 with S. ambofaciens streaked on the 26A plate 0 h, 24 h, 30 h, 48 h, or 54 h before strain BBc6R8. Total RNAs were extracted 48 h after the plates were inoculated with P. fluorescens and incubated at 26.5°C.
S. ambofaciens ATCC 23877 expresses genes necessary for desferrioxamine and coelichelin production on 26A medium.
S. ambofaciens ATCC 23877 produces two types of tris-hydroxamate siderophores: desferrioxamine (B and E) and coelichelin (37). We hypothesized that S. ambofaciens produces these siderophores when grown on 26A agar medium and that the siderophores would then be accessible to strain BBc6R8. To test this hypothesis, we analyzed the expression of the genes of the biosynthesis pathways of the two siderophores by RT-qPCR. The transcription of desC (acyl-coenzyme A [CoA] acyltransferase; desferrioxamine) (37) and samR0552-cchH (nonribosomal peptide synthetase [NRPS]; coelichelin) (37) was monitored over a 2.5-day time course. Both genes were expressed under these conditions, and their transcription peaked at around 30 h (Fig. 3). This indicates that S. ambofaciens also detects iron deficiency on 26A medium and induces the expression of its siderophore-biosynthetic genes, strongly suggesting that desferrioxamine and coelichelin are produced.
FIG 3.
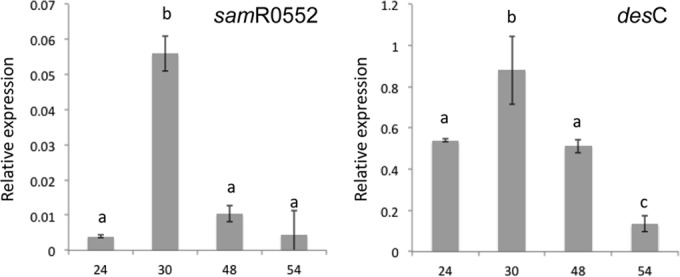
Expression levels of S. ambofaciens ATCC 23877 genes controlling the synthesis of desferrioxamine B and E and coelichelin as measured by RT-qPCR. The desC gene (right) is involved in the synthesis of desferrioxamines, and the samR0552 gene (left) is involved in coelichelin biosynthesis. Total RNAs were extracted after 24, 30, 48, and 54 h of growth on 26A agar medium at 26.5°C. The expression levels are shown as the transcript levels of the target genes relative to the transcript level of the housekeeping gene hrdB, which is stable under the tested growth conditions. The data are expressed as the mean value of three biological replicates. The error bars denote standard errors. For each transcript, values with the same letter are not significantly different according to a one-factor ANOVA (P > 0.01).
P. fluorescens BBc6R8 utilizes siderophores produced by Streptomyces.
Consequently, P. fluorescens BBc6R8 could use ferrioxamine(s) and/or ferricoelichelin as a xenosiderophore to cope with the lack of iron in the 26A medium. To test this hypothesis, P. fluorescens BBc6R8 was cultivated on a 26A agar plate supplemented with desferrioxamine B mesylate at 200 μM. After 48 h of culture, P. fluorescens BBc6R8 did not fluoresce in the presence of the exogenous purified siderophore, in contrast to the control (Fig. 4a and b). The addition of 8 μM desferrioxamine B to the medium was sufficient to inhibit pyoverdine production (Fig. 4c). This indicates that P. fluorescens BBc6R8 stops producing its own siderophore in the presence of desferrioxamine B, likely utilizing the compound as a xenosiderophore.
FIG 4.

Effects of desferrioxamine and coelichelin on the production of pyoverdine by P. fluorescens BBc6R8. (a) P. fluorescens BBc6R8 (R8). (b) P. fluorescens BBc6R8 cultivated in the presence of 200 μM desferrioxamine B mesylate. (c) P. fluorescens BBc6R8 cultivated in the presence of 8 μM desferrioxamine B mesylate. (d) P. fluorescens BBc6R8 plus S. coelicolor M512 cchH. (e) P. fluorescens BBc6R8 plus S. coelicolor M512 desD. (f) P. fluorescens BBc6R8 plus S. coelicolor Δdes Δcch. The Streptomyces strains were streaked on plates 30 h before the inoculation of P. fluorescens. The images were taken under UV light from below the plates after a 2-day (co)culture on 26A agar medium at 26.5°C.
Streptomyces coelicolor A3(2) produces the same siderophores as S. ambofaciens ATCC 23877 (37–39). Since cosmids with the biosynthetic desD (type C siderophore synthetase) (37) and cchH (NRPS) (39) genes mutated are available from the S. coelicolor transposon insertion single-gene knockout library (30), we decided to disrupt these genes in S. coelicolor M512 (a derivative of the S. coelicolor A3(2) M145 strain unable to produce the pigmented antibiotics actinorhodin and undecylprodigiosine [40]). Pairwise cultures of P. fluorescens BBc6R8 and S. coelicolor cchH and desD mutants revealed that Pseudomonas did not fluoresce in the presence of either of the two single mutants after 2 days of culture (Fig. 4d and e). However, in the presence of the S. coelicolor Δdes Δcch strain, a double mutant deficient in the biosynthesis of desferrioxamine and coelichelin (37), the production of the green-yellow pigment by P. fluorescens was not affected (Fig. 4f). Therefore, our data suggest that P. fluorescens BBc6R8 has the ability to recognize and to take up both ferrioxamine and ferricoelichelin and consequently no longer produces its own siderophores, pyoverdine and enantiopyochelin.
Streptomyces is unable to take up P. fluorescens BBc6R8 siderophores.
The question of whether Streptomyces, conversely to Pseudomonas, is able to use pyoverdine and/or enantiopyochelin as a xenosiderophore arose. It has been reported that the S. coelicolor Δdes Δcch strain (strain W13) cannot grow on agar medium supplemented with 200 μM 2,2′-bipyridyl due to an extreme iron deficiency (37). Indeed, in our experimental setup, it was unable to grow on a 26A plate containing the iron chelator (data not shown). Therefore, we examined whether P. fluorescens BBc6R8 could compensate for this deficiency by providing its own siderophores. A plug was collected from a 3-day culture of BBc6R8 on a 26A agar plate with or without 2,2′-bipyridyl (200 μM) and placed onto a 26A agar plate supplemented with the iron chelator (200 μM) with the W13 strain evenly spread on it. Alternatively, strain W13 was streaked on a 26A plate containing 2,2′-bipyridyl (200 μM) alongside the BBc6R8 strain, inoculated on the plate 3 days earlier. As a control, similar experiments were carried out with S. coelicolor M512 instead of P. fluorescens BBc6R8. While M512 efficiently promoted the growth of W13, BBc6R8 had no effect (see Fig. S2 in the supplemental material). Therefore, we conclude that S. coelicolor is unable to use pyoverdine and enantiopyochelin as xenosiderophores. By extension, we expect similar behavior for S. ambofaciens, since the two species encode the same extracellular siderophore binding proteins (41 and data not shown).
P. fluorescens BBc6R8 uses a TonB-dependent receptor to detect S. ambofaciens ATCC 23877 ferrisiderophores.
To identify the genes of P. fluorescens BBc6R8 involved in the uptake of ferrioxamines and ferricoelichelin, pairwise cultures of S. ambofaciens ATCC 23877 and 4,400 clones from a transposon mutant library of P. fluorescens BBc6R8 (P. Burlinson and A. Deveau, personal communication) were performed on solid 26A medium. S. ambofaciens was streaked on the plates 30 h before the Pseudomonas mutants to allow accumulation of ferrioxamines and ferricoelichelin in the medium. Two mutants, P28H6 and P18B10, still fluoresced under UV light under these conditions (Fig. 5a to d). Similar results were observed when the P28H6 and P18B10 clones were grown near the S. coelicolor M512 desD and cchH mutants that were unable to produce desferrioxamine and coelichelin, respectively (Fig. 5e and f). In addition, production of pyoverdine was also observed in the two Pseudomonas mutants when cultivated on 26A agar plates containing 200 μM desferrioxamine B (Fig. 5g and h). Therefore, we concluded that the mutants P28H6 and P18B10 of P. fluorescens BBc6R8 are no longer able to recognize and/or to take up the Streptomyces siderophores. Analysis of the mutants revealed that both P28H6 and P18B10 have the same gene mutated, the MHB_05767 gene (see Fig. S3 in the supplemental material). A BLASTp search showed that the product of this gene shares 88%, 84%, and 67% identity with FoxA, a TonB-dependent ferrioxamine B receptor, of P. fluorescens Pf0-1, Pseudomonas protegens Pf-5, and P. aeruginosa PAO1 (42, 43), respectively (see Fig. S4 in the supplemental material). By analogy, we named the gene of P. fluorescens BBc6R8 foxA. From this set of experiments, we conclude that P. fluorescens BBc6R8 would be able to detect and to take up the siderophores ferrioxamine B and ferricoelichelin via the outer membrane receptor FoxA.
FIG 5.

Effect of the disruption of the foxA gene on pyoverdine production by P. fluorescens BBc6R8. (a) P. fluorescens P28H6. (b) P. fluorescens P18B10. (c and d) P. fluorescens P28H6 (c) and P. fluorescens P18B10 (d) cultivated with S. ambofaciens ATCC 23877 (Sa). (e and f) P. fluorescens P28H6 cultivated with S. coelicolor M512 cchH (e) and with S. coelicolor M512 desD (f). (g and h) P. fluorescens P28H6 (g) and P. fluorescens P18B10 (h) cultivated in the presence of 200 μM desferrioxamine B mesylate. The Streptomyces strains were streaked on plates 30 h before the inoculation of P. fluorescens. The images were taken under UV light from below the plates after a 2-day (co)culture at 26.5°C on 26A agar medium.
S. ambofaciens ATCC 23877 induces expression of genes involved in TonB-dependent receptor synthesis.
In P. fluorescens Pf0-1, P. protegens Pf-5, and P. aeruginosa PAO1, foxA belongs to an operon with the genes foxI (encoding an ECF sigma factor), foxR (encoding an anti-sigma factor), and pepSY (encoding a PepSY TM helix protein), which is under the regulation of the Fur protein (43). Analysis of the genome sequence of P. fluorescens BBc6R8 indicated that the homologs of the genes foxI, foxR, and pepSY were also present in BBc6R8 (see Table S1 in the supplemental material) and were organized in a cluster similar to those in P. fluorescens Pf0-1, P. protegens Pf-5, and P. aeruginosa PAO1. A homolog of the fur gene was also retrieved in the genome sequence of the BBc6R8 strain (see Table S1 in the supplemental material). This suggests that the mechanism of regulation of the desferrioxamine-mediated iron uptake system in P. fluorescens BBc6R8 is similar to that of P. aeruginosa (33, 35). After 24 h of incubation with S. ambofaciens, the expression levels of the foxA, foxI, foxR, and pepSY genes increased compared to the control treatment (P < 0.01; one-factor [R8 alone] ANOVA) (Fig. 6). The transcription level of fur was significantly modified by the presence of S. ambofaciens, although the fold change was much smaller than for the other transcripts (Fig. 6) (P < 0.01; one-factor [R8 alone] ANOVA). These data show that in the presence of S. ambofaciens, and under conditions stimulating siderophore production, the P. fluorescens BBc6R8 foxA transcriptional-regulation cascade is induced, most likely resulting in the production of the FoxA receptor and in the uptake of the Streptomyces siderophores.
FIG 6.
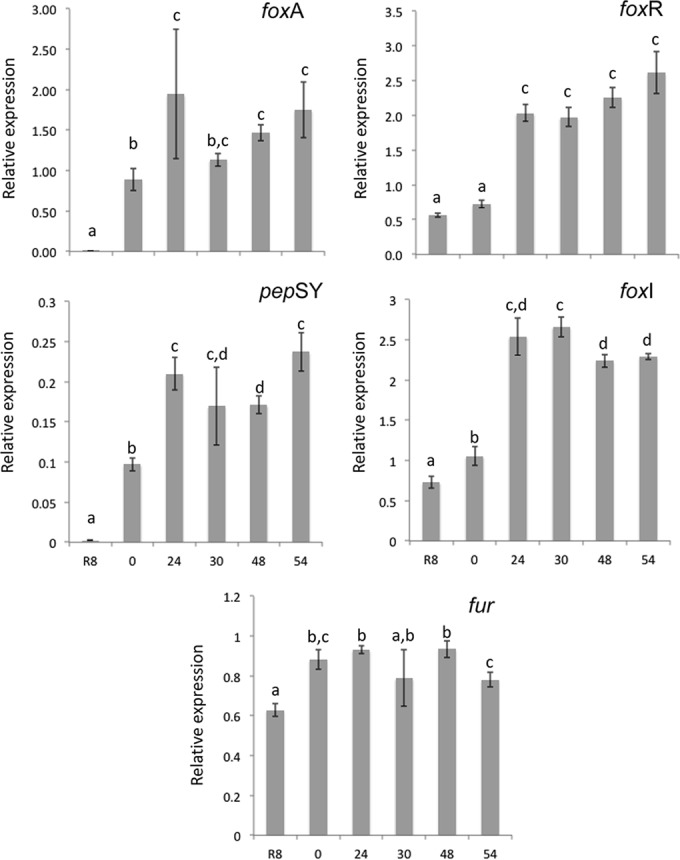
Effects of S. ambofaciens ATCC 23877 on the expression levels of P. fluorescens BBc6R8 genes controlling the synthesis of the FoxA receptor. Analysis was done by RT-qPCR. R8, P. fluorescens BBc6R8 grown on 26A agar in the absence of S. ambofaciens; 0 to 54, P. fluorescens BBc6R8 grown in the presence of S. ambofaciens ATCC 23877 that was streaked on solid 26A plates 0 h, 24 h, 30 h, 48 h, or 54 h before strain BBc6R8. The expression levels are expressed as the transcript levels of the target genes relative to the transcript level of the housekeeping gene selR, which is stable under the tested growth conditions. The data are expressed as the mean value of three biological replicates. The error bars denote standard errors. For each transcript, values with the same letter are not significantly different according to a one-factor ANOVA (P > 0.01).
Utilization of desferrioxamine is shared among P. aeruginosa and other fluorescent pseudomonads.
As similar biosynthetic and uptake systems are present in P. fluorescens BBc6R8, P. aeruginosa PAO1 (9, 42), P. protegens Pf-5, and P. fluorescens Pf0-1 (43), we expected that these bacterial strains might also be able to react to the presence of the siderophores produced by S. ambofaciens ATCC 23877. Indeed, we found that P. fluorescens Pf0-1, P. aeruginosa PAO1, and P. protegens Pf-5 no longer produced a green pigment in the presence of S. ambofaciens ATCC 23877 and did not fluoresce under UV light. Interestingly, P. fluorescens SBW25, which does not have the foxA gene, fluoresced even in the presence of S. ambofaciens (see Fig. S5 in the supplemental material). In the presence of purified desferrioxamine B, P. fluorescens Pf0-1, P. aeruginosa PAO1, and P. protegens Pf-5 no longer fluoresced, again in contrast to strain SBW25 (see Fig. S5 in the supplemental material). In cocultures with S. coelicolor M512 desD, P. fluorescens Pf0-1, P. aeruginosa PAO1, and P. protegens Pf-5 fluoresced less than in single culture, suggesting that they are also able to utilize coelichelin (see Fig. S5 in the supplemental material). Interestingly, a screening of a library of 60 fluorescent pseudomonad strains isolated from forest soil (Table 1) revealed that all the strains were also able to utilize S. ambofaciens siderophores (data not shown).
DISCUSSION
Our study reports interspecies adaptive behavior during common utilization of limited iron resources between the two soil inhabitants Pseudomonas and Streptomyces. In particular, we show that, under iron-deficient conditions, P. fluorescens BBc6R8 does not induce the production of the fluorescent siderophore pyoverdine in the presence of S. ambofaciens ATCC 23877. Instead, P. fluorescens very likely utilizes the S. ambofaciens siderophores desferrioxamine B and coelichelin as xenosiderophores, thanks to its FoxA receptor. In contrast to most studies, in which the use of xenosiderophores was revealed indirectly through the addition of purified siderophore to the growth medium (44–47), binding affinity assays (11, 48), or native PAGE and surface plasmon resonance (49) or with labeled iron (11, 50, 51), we revealed potential iron piracy through a direct interaction between a Gram-positive and a Gram-negative bacterium. This potential piracy likely occurs only in one direction. Indeed, the growth of an S. coelicolor Δdes Δcch mutant under iron-deficient conditions could not be rescued in the presence of P. fluorescens BBc6R8 (see Fig. S2 in the supplemental material). Similar behavior is expected from S. ambofaciens ATCC 23877, since the strain is extremely close phylogenetically to S. coelicolor and they encode the same extracellular siderophore binding proteins. Similar piracy involving streptomycetes has been reported previously, but only with other actinomycetes (e.g., Streptomyces and Amycolatopsis) (15, 44, 52). It is interesting that these examples also involved desferrioxamines and that competition for iron could either stimulate (44, 52) or curtail (15) the growth and/or development of Streptomyces. The effect of the siderophore piracy by P. fluorescens on S. ambofaciens remains to be identified. Indeed, we could not observe any effect on the morphological differentiation of S. ambofaciens, as it forms only vegetative mycelium on the 26A medium.
Although our model bacteria were isolated from independent ecological niches (Laccaria bicolor sporocarp for P. fluorescens BBc6R8 [53] and soil in the Picardie region, France, for S. ambofaciens [18]) and the experiments were carried out under laboratory growth conditions, this interaction for iron capture between Streptomyces and Pseudomonas likely occurs in natural environments. Indeed, several studies have reported that these bacterial genera share common ecological niches within soils, including environments such as the rhizosphere or bulk soils (54–56). Therefore, they are expected to use common pools of scarce but essential elements, such as iron. The piracy would occur through the production by P. fluorescens BBc6R8 of the TonB-dependent receptor FoxA. Interestingly, strain BBc6R8 does not produce ferrioxamine, which is the most effective compound for iron scavenging, followed by pyoverdine and enantiopyochelin, both of which it does produce (56). In a competitive environment, such as soil, it is certainly more advantageous to use efficient chelators produced by its neighbors rather than to produce its own siderophores, especially if they are less effective and metabolically costly, as demonstrated previously (6). In addition, since desferrioxamines are synthesized by many soil organisms, in this way, P. fluorescens BBc6R8 could obtain iron at a low energy cost in many competitive situations. Interestingly, it should be noted that purified desferrioxamine added to the culture medium had a positive effect on the growth of P. fluorecens BBc6R8 (data not shown). In contrast, S. ambofaciens negatively impacted the growth of BBc6R8 in cocultures, presumably because of the secretion of secondary metabolites and/or through nutrient competition (data not shown).
The production of desferrioxamines seems to be conserved throughout streptomycetes (57, 58). The desABCD genes that direct the synthesis of these chelating agents (38, 59) have been found in all Streptomyces sequenced genomes and in other, related genera, such as Salinispora (60). Furthermore, Kobayakawa and Kodani detected by high-performance liquid chromatography (HPLC) the production of desferrioxamines in 78% of their Streptomyces collection (61). Desferrioxamines are also produced by other actinomycetes and some Gram-negative bacteria (62, 63).
Our data indicate that the BBc6R8 outer membrane receptor FoxA would allow the uptake not only of desferrrioxamines of S. coelicolor, but also of the hydroxamate siderophore ferricoelichelin. The ability seems to be widespread among fluorescent pseudomonads. Indeed, the FoxA receptor is present in three different subclades of the genus Pseudomonas (64), and based on our screening of 60 P. fluorescens environmental strains, it is likely that they also possess a foxA gene and even a foxA operon. A BLASTp analysis also showed that FoxA homologs are present in other Pseudomonas species, such as P. stutzeri, P. aeruginosa, Pseudomonas fulva, Pseudomonas putida, and Pseudomonas resinovorans (data not shown), and also in members of different genera of bacteria, like Yersinia enterocolitica (12) and Erwinia herbicola (13). In these genera, FoxA is known to bind a collection of ferrioxamine derivatives with different chain lengths or bridges (65). The tris-hydroxamate siderophore coprogen is also recognized to a certain extent by FoxA (65). Desferrioxamine B, coelichelin, and coprogen are linear siderophores (37, 65). These data suggest that FoxA could bind other linear ferric-tris-hydroxamate siderophores (65). Therefore, P. fluorescens BBc6R8 may have the ability to obtain a wider variety of siderophores than were investigated in this study through the production of the FoxA receptor and thus to very efficiently compete with other bacteria. FoxA could then be considered an outer membrane receptor with a broad spectrum, at least for siderophores of the tris-hydroxamate family, and as a widespread receptor. Other bacteria, such as Streptomyces, also possess broad-spectrum tris-hydroxamate receptors. Indeed, in S. ambofaciens ATCC 23877 and S. coelicolor A3(2), CdtB, a siderophore binding protein involved in iron-siderophore transport, is able to bind ferrioxamines and ferricoelichelin with high affinity (37, 41). Moreover, DesE, a second siderophore binding protein, binds different ferric-tris-hydroxamates, with the exception of ferricoelichelin (41). Possessing some broad-spectrum siderophore receptors may be a frequent strategy among microorganisms selected during evolution to be more competitive in soil.
The capability to take up xenosiderophores can be amplified if bacteria possess multiple xenosiderophore receptors. This property is particularly well developed in the genus Pseudomonas. Indeed, several Pseudomonas species possess numerous TonB-dependent receptors, usually more than 20, with P. protegens Pf-5 encoding the highest number (45) of TonB-dependent receptors (9). Twenty-nine have been identified so far in the draft genome sequence of the BBc6R8 strain (27). Several bacteria possessing multiple siderophore binding proteins, like P. fragi, have even lost the ability to produce siderophores and rely exclusively on siderophore piracy (11). Whether siderophore piracy results from the loss of the ability to synthesize the cognate siderophores during evolution or the acquisition of receptor genes through horizontal transfer remains an open question. Nevertheless, our data and those of others strongly support the idea that the capacity for siderophore piracy is widespread among bacteria and that piracy by “cheaters” does happen in vivo (66). What is unclear is to what extent this piracy really occurs in natural ecosystems such as soils and how it impacts community dynamics. Metagenomics combined with metatranscriptomics may help to answer this question in the future by providing a more complete picture of who is producing what and who is “cheating” in natural complex microbial communities.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the French National Research Agency through the Laboratory of Excellence ARBRE (ANR-11-LABX-0002-01), by the French National Institute for Agricultural Research (INRA), and by Région Lorraine. J.G. was supported by a CJS (Contrat Jeune Scientifique) grant from INRA.
We thank Gregory Challis (University of Warwick, United Kingdom) for the S. coelicolor Δdes Δcch double mutant and Paul Dyson (University of Swansea, United Kingdom) for the cosmids C105.2.E01 and F76.2.F08. We also thank Joyce Loper (Oregon State University), Gail Preston (University of Oxford), and Stuart Levy (Tufts University) for the Pseudomonas Pf5, SBW25, and Pf0-1 strains, respectively. We acknowledge Jean-Selim Medot for his help in this work. We are grateful to Rachel Gregor (Ben-Gurion University of the Negev) for proofreading the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03520-14.
REFERENCES
- 1.Konovalova A, Søgaard-Andersen L. 2011. Close encounters: contact-dependent interactions in bacteria. Mol Microbiol 81:297–301. doi: 10.1111/j.1365-2958.2011.07711.x. [DOI] [PubMed] [Google Scholar]
- 2.Barry SM, Challis GL. 2009. Recent advances in siderophore biosynthesis. Curr Opin Chem Biol 13:205–215. doi: 10.1016/j.cbpa.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Hider RC, Kong X. 2010. Chemistry and biology of siderophores. Nat Prod Rep 27:637. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 4.Chu BC, Garcia-Herrero A, Johanson TH, Krewulak KD, Lau CK, Peacock RS, Slavinskaya Z, Vogel HJ. 2010. Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view. BioMetals 23:601–611. doi: 10.1007/s10534-010-9361-x. [DOI] [PubMed] [Google Scholar]
- 5.Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumas Z, Ross-Gillespie A, Kummerli R. 2013. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc Biol Sci 280:20131055. doi: 10.1098/rspb.2013.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luckey M, Pollack JR, Wayne R, Ames BN, Neilands JB. 1972. Iron uptake in Salmonella typhimurium: utilization of exogenous siderochromes as iron carriers. J Bacteriol 111:731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubert S, Fischer D, Heesemann J. 1999. Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J Bacteriol 181:6387–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis P, Bodilis J. 2009. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ Microbiol Rep 1:256–262. doi: 10.1111/j.1758-2229.2009.00041.x. [DOI] [PubMed] [Google Scholar]
- 10.Moon CD, Zhang X-X, Matthijs S, Schäfer M, Budzikiewicz H, Rainey PB. 2008. Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol 8:7. doi: 10.1186/1471-2180-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champomier-Vergès M-C, Stintzi A, Meyer J-M. 1996. Acquisition of iron by the non-siderophore-producing Pseudomonas fragi. Microbiology 142:1191–1199. doi: 10.1099/13500872-142-5-1191. [DOI] [PubMed] [Google Scholar]
- 12.Bäumler AJ, Hantke K. 1992. Ferrioxamine uptake in Yersinia enterocolitica: characterization of the receptor protein FoxA. Mol Microbiol 6:1309–1321. doi: 10.1111/j.1365-2958.1992.tb00852.x. [DOI] [PubMed] [Google Scholar]
- 13.Berner I, Winkelmann G. 1990. Ferrioxamine transport mutants and the identification of the ferrioxamine receptor protein (FoxA) in Erwinia herbicola (Enterobacter agglomerans). Biol Met 2:197–202. doi: 10.1007/BF01141359. [DOI] [PubMed] [Google Scholar]
- 14.Tanabe T, Funahashi T, Miyamoto K, Tsujibo H, Yamamoto S. 2011. Identification of genes, desR and desA, required for utilization of desferrioxamine B as a xenosiderophore in Vibrio furnissii. Biol Pharm Bull 34:570–574. doi: 10.1248/bpb.34.570. [DOI] [PubMed] [Google Scholar]
- 15.Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. 2012. Interspecies modulation of bacterial development through iron competition and siderophore piracy: xenosiderophores alter development in actinomycetes. Mol Microbiol 86:628–644. doi: 10.1111/mmi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shakya M, Gottel N, Castro H, Yang ZK, Gunter L, Labbé J, Muchero W, Bonito G, Vilgalys R, Tuskan G, Podar M, Schadt CW. 2013. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS One 8:e76382. doi: 10.1371/journal.pone.0076382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinnert-Sindico S. 1954. Une nouvelle espece de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n sp caracteres culturaux. Ann Inst Pasteur (Paris) 87:702–707. [PubMed] [Google Scholar]
- 19.Frey-Klett P, Pierrat JC, Garbaye J. 1997. Location and survival of mycorrhiza helper Pseudomonas fluorescens during establishment of ectomycorrhizal symbiosis between Laccaria bicolor and Douglas fir. Appl Environ Microbiol 63:139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deveau A, Brulé C, Palin B, Champmartin D, Rubini P, Garbaye J, Sarniguet A, Frey-Klett P. 2010. Role of fungal trehalose and bacterial thiamine in the improved survival and growth of the ectomycorrhizal fungus Laccaria bicolor S238N and the helper bacterium Pseudomonas fluorescens BBc6R8: role of trehalose and thiamine in mutualistic interaction. Environ Microbiol Rep 2:560–568. doi: 10.1111/j.1758-2229.2010.00145.x. [DOI] [PubMed] [Google Scholar]
- 21.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes, Norwich, United Kingdom. [Google Scholar]
- 22.Leblond P, Fischer G, Francou F-X, Berger F, Guérineau M, Decaris B. 1996. The unstable region of Streptomyces ambofaciens includes 210 kb terminal inverted repeats flanking the extremities of the linear chromosomal DNA. Mol Microbiol 19:261–271. doi: 10.1046/j.1365-2958.1996.366894.x. [DOI] [PubMed] [Google Scholar]
- 23.Pang X, Aigle B, Girardet J-M, Mangenot S, Pernodet J-L, Decaris B, Leblond P. 2004. Functional angucycline-like antibiotic gene cluster in the terminal inverted repeats of the Streptomyces ambofaciens linear chromosome. Antimicrob Agents Chemother 48:575–588. doi: 10.1128/AAC.48.2.575-588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pospiech A, Neumann B. 1995. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet 11:217–218. doi: 10.1016/S0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 26.Buttner MJ, Chater KF, Bibb MJ. 1990. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J Bacteriol 172:3367–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deveau A, Gross H, Morin E, Karpinets T, Utturkar S, Mehnaz S, Martin F, Frey-Klett P, Labbé J. 2014. Genome sequence of the mycorrhizal helper bacterium Pseudomonas fluorescens BBc6R8. Genome Announc 2:e01152-13. doi: 10.1128/genomeA.01152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth V, Aigle B, Bunet R, Wenner T, Fourrier C, Decaris B, Leblond P. 2004. Differential and cross-transcriptional control of duplicated genes encoding alternative sigma factors in Streptomyces ambofaciens. J Bacteriol 186:5355–5365. doi: 10.1128/JB.186.16.5355-5365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández-Martínez LT, Sol RD, Evans MC, Fielding S, Herron PR, Chandra G, Dyson PJ. 2011. A transposon insertion single-gene knockout library and new ordered cosmid library for the model organism Streptomyces coelicolor A3(2). Antonie Van Leeuwenhoek 99:515–522. doi: 10.1007/s10482-010-9518-1. [DOI] [PubMed] [Google Scholar]
- 31.Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor ςE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J Bacteriol 181:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamalero E, Fracchia L, Cavaletto M, Garbaye J, Frey-Klett P, Varese GC, Martinotti MG. 2003. Characterization of functional traits of two fluorescent pseudomonads isolated from basidiomes of ectomycorrhizal fungi. Soil Biol Biochem 35:55–65. doi: 10.1016/S0038-0717(02)00236-5. [DOI] [Google Scholar]
- 33.Visca P, Leoni L, Wilson MJ, Lamont IL. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol Microbiol 45:1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 34.Ravel J, Cornelis P. 2003. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol 11:195–200. doi: 10.1016/S0966-842X(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 35.Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 36.Schalk IJ, Guillon L. 2013. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ Microbiol 15:1661–1673. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 37.Barona-Gomez F, Lautru S, Francou F-X, Leblond P, Pernodet J-L, Challis GL. 2006. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology 152:3355–3366. doi: 10.1099/mic.0.29161-0. [DOI] [PubMed] [Google Scholar]
- 38.Barona-Gómez F, Wong U, Giannakopulos AE, Derrick PJ, Challis GL. 2004. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J Am Chem Soc 126:16282–16283. doi: 10.1021/ja045774k. [DOI] [PubMed] [Google Scholar]
- 39.Lautru S, Deeth RJ, Bailey LM, Challis GL. 2005. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol 1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 40.Floriano B, Bibb M. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- 41.Patel P, Song L, Challis GL. 2010. Distinct extracytoplasmic siderophore binding proteins recognize ferrioxamines and ferricoelichelin in Streptomyces coelicolor A3(2). Biochemistry 49:8033–8042. doi: 10.1021/bi100451k. [DOI] [PubMed] [Google Scholar]
- 42.Llamas MA, Sparrius M, Kloet R, Jimenez CR, Vandenbroucke-Grauls C, Bitter W. 2006. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J Bacteriol 188:1882–1891. doi: 10.1128/JB.188.5.1882-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartney SL, Mazurier S, Kidarsa TA, Quecine MC, Lemanceau P, Loper JE. 2011. TonB-dependent outer-membrane proteins and siderophore utilization in Pseudomonas fluorescens Pf-5. BioMetals 24:193–213. doi: 10.1007/s10534-010-9385-2. [DOI] [PubMed] [Google Scholar]
- 44.Yamanaka K. 2005. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 151:2899–2905. doi: 10.1099/mic.0.28139-0. [DOI] [PubMed] [Google Scholar]
- 45.Joshi F, Archana G, Desai A. 2006. Siderophore cross-utilization amongst rhizospheric bacteria and the role of their differential affinities for Fe3+ on growth stimulation under iron-limited conditions. Curr Microbiol 53:141–147. doi: 10.1007/s00284-005-0400-8. [DOI] [PubMed] [Google Scholar]
- 46.Funahashi T, Tanabe T, Miyamoto K, Tsujibo H, Maki J, Yamamoto S. 2013. Characterization of a gene encoding the outer membrane receptor for ferric enterobactin in Aeromonas hydrophila ATCC 7966(T). Biosci Biotechnol Biochem 77:353–360. doi: 10.1271/bbb.120774. [DOI] [PubMed] [Google Scholar]
- 47.Tomaras AP, Crandon JL, McPherson CJ, Banevicius MA, Finegan SM, Irvine RL, Brown MF, O'Donnell JP, Nicolau DP. 2013. Adaptation-based resistance to siderophore-conjugated antibacterial agents by Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4197–4207. doi: 10.1128/AAC.00629-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miethke M, Kraushaar T, Marahiel MA. 2013. Uptake of xenosiderophores in Bacillus subtilis occurs with high affinity and enhances the folding stabilities of substrate binding proteins. FEBS Lett 587:206–213. doi: 10.1016/j.febslet.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 49.Veggi D, Gentile MA, Cantini F, Lo Surdo P, Nardi-Dei V, Seib KL, Pizza M, Rappuoli R, Banci L, Savino S, Scarselli M. 2012. The factor H binding protein of Neisseria meningitidis interacts with xenosiderophores in vitro. Biochemistry 51:9384–9393. doi: 10.1021/bi301161w. [DOI] [PubMed] [Google Scholar]
- 50.Weber M, Taraz K, Budzikiewicz H, Geoffroy V, Meyer J-M. 2000. The structure of a pyoverdine from Pseudomonas sp. CFML 96.188 and its relation to other pyoverdines with a cyclic C-terminus. BioMetals 13:301–309. doi: 10.1023/A:1009235421503. [DOI] [PubMed] [Google Scholar]
- 51.Barelmann I, Fernández DU, Budzikiewicz H, Meyer J-M. 2003. The pyoverdine from Pseudomonas chlororaphis D-TR133 showing mutual acceptance with the pyoverdine of Pseudomonas fluorescens CHA0. BioMetals 16:263–270. doi: 10.1023/A:1020615830765. [DOI] [PubMed] [Google Scholar]
- 52.Ueda K, Kawai S, Ogawa H, Kiyama A, Kubota T, Kawanobe H, Beppu T. 2000. Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J Antibiot 53:979–982. doi: 10.7164/antibiotics.53.979. [DOI] [PubMed] [Google Scholar]
- 53.Garbaye J, Duponnois R, Wahl JL. 1990. The bacteria associated with Laccaria laccata ectomycorrhizas or sporocarps: effect of symbiosis establishment on Douglas fir. Symbiosis Rehovot 9:267–273. [Google Scholar]
- 54.Mengoni A, Barzanti R, Gonnelli C, Gabbrielli R, Bazzicalupo M. 2001. Characterization of nickel-resistant bacteria isolated from serpentine soil. Environ Microbiol 3:691–698. doi: 10.1046/j.1462-2920.2001.00243.x. [DOI] [PubMed] [Google Scholar]
- 55.Nazaret S, Brothier E, Ranjard L. 2003. Shifts in diversity and microscale distribution of the adapted bacterial phenotypes due to Hg(II) spiking in soil. Microb Ecol 45:259–269. doi: 10.1007/s00248-002-2035-7. [DOI] [PubMed] [Google Scholar]
- 56.Srifuengfung S, Assanasen S, Tuntawiroon M, Meejanpetch S. 2010. Comparison between Pseudomonas aeruginosa siderophores and desferrioxamine for iron acquisition from ferritin. Asian Biomed 4:631–635. [Google Scholar]
- 57.Müller G, Raymond KN. 1984. Specificity and mechanism of ferrioxamine-mediated iron transport in Streptomyces pilosus. J Bacteriol 160:304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meiwes J, Fiedler H-P, Zähner H, Konetschny-Rapp S, Jung G. 1990. Production of desferrioxamine E and new analogues by directed fermentation and feeding fermentation. Appl Microbiol Biotechnol 32:505–510. [DOI] [PubMed] [Google Scholar]
- 59.Kadi N, Oves-Costales D, Barona-Gomez F, Challis GL. 2007. A new family of ATP-dependent oligomerization-macrocyclization biocatalysts. Nat Chem Biol 3:652–656. doi: 10.1038/nchembio.2007.23. [DOI] [PubMed] [Google Scholar]
- 60.Nett M, Ikeda H, Moore BS. 2009. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayakawa F, Kodani S. 2012. Screening of streptomycetes for production of desferrioxamines. J Pure Appl Microbiol 6:1553–1558. [Google Scholar]
- 62.Martinez JS, Haygood MG, Butler A. 2001. Identification of a natural desferrioxamine siderophore produced by a marine bacterium. Limnol Oceanogr 46:420–424. doi: 10.4319/lo.2001.46.2.0420. [DOI] [Google Scholar]
- 63.Essén SA, Johnsson A, Bylund D, Pedersen K, Lundström US. 2007. Siderophore production by Pseudomonas stutzeri under aerobic and anaerobic conditions. Appl Environ Microbiol 73:5857–5864. doi: 10.1128/AEM.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silby MW, Winstanley C, Godfrey SAC, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 65.Deiss K, Hantke K, Winkelmann G. 1998. Molecular recognition of siderophores: a study with cloned ferrioxamine receptors (FoxA) from Erwinia herbicola and Yersinia enterocolitica. Biometals 11:131–137. doi: 10.1023/A:1009230012577. [DOI] [PubMed] [Google Scholar]
- 66.Cordero OX, Ventouras L-A, DeLong EF, Polz MF. 2012. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci U S A 109:20059–20064. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey MJ, Lilley AK, Thompson IP, Rainey PB, Ellis RJ. 1995. Site directed chromosomal marking of a fluorescent pseudomonad isolated from the phytosphere of sugar beet; stability and potential for marker gene transfer. Mol Ecol 4:755–763. doi: 10.1111/j.1365-294X.1995.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 68.Compeau G, Al-Achi BJ, Platsouka E, Levy SB. 1988. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol 54:2432–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 70.Frey-Klett P, Chavatte M, Clausse M-L, Courrier S, Roux CL, Raaijmakers J, Martinotti MG, Pierrat J-C, Garbaye J. 2005. Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New Phytol 165:317–328. doi: 10.1111/j.1469-8137.2004.01212.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


