Abstract
Paddy fields represent a unique ecosystem in which regular flooding occurs, allowing for rice cultivation. However, the taxonomic identity of the microbial functional guilds that catalyze soil nitrification remains poorly understood. In this study, we provide molecular evidence for distinctly different phylotypes of nitrifying communities in a neutral paddy soil using high-throughput pyrosequencing and DNA-based stable isotope probing (SIP). Following urea addition, the levels of soil nitrate increased significantly, accompanied by an increase in the abundance of the bacterial and archaeal amoA gene in microcosms subjected to SIP (SIP microcosms) during a 56-day incubation period. High-throughput fingerprints of the total 16S rRNA genes in SIP microcosms indicated that nitrification activity positively correlated with the abundance of Nitrosospira-like ammonia-oxidizing bacteria (AOB), soil group 1.1b-like ammonia-oxidizing archaea (AOA), and Nitrospira-like nitrite-oxidizing bacteria (NOB). Pyrosequencing of 13C-labeled DNA further revealed that 13CO2 was assimilated by these functional groups to a much greater extent than by marine group 1.1a-associated AOA and Nitrobacter-like NOB. Phylogenetic analysis demonstrated that active AOB communities were closely affiliated with Nitrosospira sp. strain L115 and the Nitrosospira multiformis lineage and that the 13C-labeled AOA were related to phylogenetically distinct groups, including the moderately thermophilic “Candidatus Nitrososphaera gargensis,” uncultured fosmid 29i4, and acidophilic “Candidatus Nitrosotalea devanaterra” lineages. These results suggest that a wide variety of microorganisms were involved in soil nitrification, implying physiological diversification of soil nitrifying communities that are constantly exposed to environmental fluctuations in paddy fields.
INTRODUCTION
Rice feeds over half of the world's population and is usually considered the most important food source in Asia (1). China has approximately 29.2 million ha of rice fields, accounting for 35.8% of the grain-sowing area (2, 3). China is indeed the largest producer of rice on the planet, and the yield production of rice accounts for 43.7% of the total national grain production (4). The growth and production of rice crops depend heavily on anthropogenic management such as irrigation and fertilization. The intensified application of synthetic N fertilizers has increased significantly over the past decades to meet the demand for food productivity in China (5). The excessively high load often leads to the saturation of nitrogen nutrients in paddy soils and causes severe environmental pollution (6).
Flood management is the most dominant regime for growing semiaquatic rice plants (7). Irrigated paddy fields thus may serve as a model system for studying the microbial ecology of nitrifying communities in terrestrial environments (8). Nitrification is executed by functional microbial guilds, including ammonia-oxidizing bacteria and archaea (AOB and AOA, respectively), as well as nitrite-oxidizing bacteria (NOB). AOA and AOB perform the first and rate-limiting step of aerobic nitrification by oxidizing ammonia to nitrite, which is then rapidly converted to nitrate by NOB. During rice-growing seasons under flooding conditions, aerobic nitrification can proceed in surface and rhizosphere soils where oxygen is delivered by atmospheric diffusion and root release (9). Paddy fields during flooding seasons thus resemble freshwater ecosystems to some extent, with limited availability of dissolved oxygen. For example, Nitrosomonas-like AOB have been frequently observed in freshwater ecosystems (10, 11) and were also shown to be an important component of nitrifying communities in a Philippine rice field (12). However, unlike aquatic habitats, rice fields experience drastic changes in environmental conditions due to agricultural management. For example, paddy fields are often drained in winter for wheat cultivation and flooded in summer during the rice-growing season. Moreover, paddy fields have been subjected to strong anthropogenic disturbances with intensified fertilization regimes and other interventions that could selectively drive community diversification of nitrifying communities (9). The constantly changing environment might have served as the primary force in shaping the structure and activity of nitrifying communities in paddy fields (13).
Numerous studies have focused on rice fields in warm, humid, subtropical zones with high precipitation (8, 14, 15). However, little information is available about nitrifying community structures in cold regions, arguably the most fertile and highest-quality regions, that account for 13% of the cultivated land in northeastern China (16). Furthermore, the mere presence of the respective genes does not always indicate functional importance. The direct link between the nitrification process and taxonomic identity of active microorganisms in paddy fields of northern China remains elusive. Therefore, we employed microcosms subjected to DNA-based stable isotope probing (SIP microcosms) and high-throughput pyrosequencing to assess the autotrophic nitrification in paddy soils from northeastern China.
MATERIALS AND METHODS
Soil sampling.
The soil samples for microcosm incubation were collected from an irrigated paddy field in Wuchang County, Haerbin City of Heilongjiang Province in northeastern China (45°10′N, 127°00′E). The soil is classified as a phaeozem, according to the World Reference Base for Soil Resources 2006 (17). The paddy field is located in the center of the Mollisols in Northeastern China. The mollisol was derived from the parent materials that were sedimentary materials of loamy loess, according to U.S. Department of Agriculture soil taxonomy. The sampling site was characterized as a temperate continental monsoon climate, with a mean annual temperature of 3.5°C and precipitation of 625 mm. The field usually received urea fertilization with 200 to 400 kg N ha−1, which is equivalent to 87.2 to 174 μg of urea-N g−1(dry weight) of soil (gDWS−1), assuming an effective soil depth of 20 cm for fertilizer application. The composite soil sample was collected from 0- to 20-cm surface soils by mixing six random soil cores. The composite soil sample was passed through a 2.0-mm-pore-size sieve and stored at 4°C before construction of microcosms. The soil pH was determined using a Mettler Toledo 320-S pH meter (Mettler-Toledo Instruments Co. Ltd., Shanghai, China) with a water-to-soil ratio of 2.5. The soil organic matter content was determined using the dichromate oxidation method. Total N was determined by the Kjeldahl method. Ammonium and nitrate were extracted from soil samples with 2 M KCl, and the levels were determined using a Skalar San Plus segmented flow analyzer (Skalar Inc., Breda, The Netherlands).
DNA-SIP microcosms.
Microcosms were constructed in triplicate as described previously and treated in three different ways (18). The microcosms were incubated with 5% (vol/vol) 13CO2 plus [13C]urea for the labeled treatment, and control microcosms were incubated with 5% 12CO2 plus [12C]urea and 5% 13CO2 plus [13C]urea with 100 Pa C2H2. For each treatment, fresh soil (equivalent to 5.0 gDWS) was incubated at approximately 60% maximum water-holding capacity and at 28°C in the dark for 56 days in a 120-ml serum bottle sealed with a butyl stopper. The headspace of the bottles was aerated with synthetic air (20% O2 and 80% N2) for 1 min on a weekly basis to maintain oxic conditions and renewed with fresh gas and substrate accordingly. A total of 100 μg of urea-N gDWS−1 was pulsed into the soil once a week to establish a substrate-rich environment for nitrifying communities over the incubation period of 56 days. Urea was the main N fertilizer used during growing seasons, which was believed to be immediately hydrolyzed by extracellular ureases into ammonia and carbon dioxide, both of which allow ammonia oxidization in soils (19, 20). The 12-day preincubation was performed so that the soil-respired CO2 would remain at a consistently low level during the entire period of incubation. For acetylene-amended microcosms, an additional 7-day preincubation with 100 Pa C2H2 was executed for full inactivation of ammonia oxidizers in soils. Soil samples at day zero were collected immediately after the 12-day preincubation for subsequent analysis. The destructive sampling was performed after 56 days. Then, 4.0-g soil samples were homogenized with 20 ml of 2 M KCl for inorganic nitrogen determination of NH4+-N and NO3−-NO2−-N using a Skalar San Plus segmented flow analyzer (Skalar Inc., Breda, The Netherlands). The remaining soil samples were kept frozen at −80°C for subsequent molecular analysis.
DNA extraction and SIP gradient fractionation.
The total DNA in the soil samples was extracted and purified from 0.5 g (dry weight) of soil using a FastDNA spin kit for soil (MP Biomedicals, Cleveland, OH, USA), according to the manufacturer's instructions. The quantity and quality of DNA extracts were assayed using a NanoDrop ND-1000 UV-visible light spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For isopycnic density gradient centrifugation to resolve 13C-DNA from native 12C-DNA in the total DNA extract, approximately 3.0 μg of DNA was mixed with a CsCl stock solution to achieve a final volume of ∼5.5 ml with a CsCl buoyant density of 1.725 g ml−1 before ultracentrifugation at 177,000 × g and 20°C for 44 h. The DNA fractions for each sample were collected by displacing the gradient medium with sterile water from the top of the ultracentrifuge tube using an NE-1000 single-syringe pump (New Era Pump Systems, Inc., Farmingdale, NY, USA) precisely controlled at a flow rate of 0.38 ml min−1. A total of 15 gradient fractions were generated with equal volumes of 380 μl, and 60 μl of each fraction was used for refractive index measurement using an AR200 digital hand-held refractometer (Reichert, Inc., Buffalo, NY, USA). The fractionated DNA was purified with 70% ethanol after polyethylene glycol (PEG) 6000 precipitation and dissolved in 30 μl of Tris-EDTA (TE) buffer.
Real-time qPCR of amoA and 16S rRNA genes.
Real-time quantitative PCR (qPCR) was performed in triplicate on a CFX96 Optical Real-Time detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) to determine the copy number of amoA genes in soil DNA extracts and DNA gradient fractions to observe the changes in population sizes of AOA and AOB communities and to assess the 13CO2 labeling of amoA-carrying ammonia oxidizers. A universal 16S rRNA gene qPCR was conducted to assess the total copy numbers of bacterial and archaeal communities in soils with different treatments. The PCR primers and thermal conditions are detailed in Table S1 in the supplemental material. The real-time quantitative PCR standard was generated using plasmid DNA from one representative clone containing bacterial or archaeal amoA and 16S rRNA genes, and a dilution series of standard template over eight orders of magnitude per assay was used. Amplification efficiencies ranged from 82% to 105%, with R2 values of 0.996 to 0.999. In addition, total DNA extracts were diluted in a series to assess the possible PCR inhibition by coextraction of humic substances, and DNA extracts were diluted 10-fold for subsequent analysis. Melting curve analysis and standard agarose gel electrophoresis were always performed at the end of a PCR run to verify the amplification specificity.
Pyrosequencing of 16S rRNA and amoA genes.
Pyrosequencing was employed to investigate the community shift of nitrifying phylotypes by analyzing the V4 regions of 16S rRNA genes in soils. The total microbial communities were analyzed in soil microcosms using universal primers for 16S rRNA genes to investigate the proportional changes of nitrifying phylotypes during active nitrification in the 56-day SIP microcosms. Meanwhile, pyrosequencing of the total 16S rRNA genes was also performed in the DNA gradient fractions from the 13CO2-labeled and 12CO2 control microcosms. For each treatment, a total of 12 fractions were selected for analysis of the total microbial communities to determine the 13CO2 labeling of the 16S rRNA genes of nitrifying communities. Each DNA sample was amplified using the universal 515F-907R primer pair with adaptor, key sequence, and unique tag sequence (only in forward primer 515F). The PCR primers and conditions are described in Table S1 in the supplemental material. The resulting PCR products were gel purified and combined in equimolar ratios into a single tube in preparation for pyrosequencing analysis. The pyrosequencing was conducted on a Roche 454 GS FLX Titanium sequencer (Roche Diagnostics Corporation, Branford, CT, USA).
Bacterial and archeal amoA genes from the 13C-labeled DNA fraction with a CsCl buoyant density of 1.7399 (heavy fraction) were amplified for pyrosequencing. The primer pairs amoA-1F/2R and CrenamoA-23f/616r (see Table S1 in the supplemental material) fused with the tag sequence in the forward primers were used for the amplification of bacterial and archeal amoA genes, respectively. The resulting PCR products were prepared and pyrosequenced as described above.
All pyrosequencing data were processed using the mothur software package (21). The amoA and 16S rRNA gene reads for each sample were sorted by specific tag sequences. The resulting reads were further trimmed, and only sequences of >350 bp in length with an average quality score >30 and no ambiguous base calls were included for subsequent analysis. The high-quality reads of amoA genes were further converted into their protein sequences for quality control, and sequences with stop codons and frameshifts were removed (22). For 16S rRNA genes, the high-quality sequences were aligned and classified into different taxonomic categories based on the SILVA database embedded in mothur. The target reads of nitrifying phylotypes were further screened for phylogenetic analyses of AOB and AOA. All reads classified as belonging to the genus Nitrosospira and the phylum Crenarchaeota were selected and clustered into operational taxonomic units (OTUs) with a 97% similarity cutoff (23). A representative sequence of each OTU was used for phylogenetic analysis using the MEGA software package, version 4.0, through inference of a neighbor-joining tree using the Kimura two-parameter distance with 1,000 replicates to produce bootstrap values (24). Taxonomic assignment was accomplished by binning sequences into OTUs with a 97% similarity cutoff and analyzing a representative sequence by MEGA as described above.
Statistical analysis.
One-way analysis of variance (ANOVA) with Tukey's post hoc test was performed for multiple comparisons. All analyses were conducted using the SPSS, version 13.0, package for Windows (SPSS, Inc.). Test results with a P level of <0.05 were regarded as statistically significant.
Nucleotide sequence accession numbers.
All pyrosequencing reads, including 16S rRNA genes and archaeal and bacterial amoA genes, have been deposited in the European Nucleotide Archive (ENA), with accession numbers ERS667669, ERS670107, and ERS670108, respectively. The 13C-labeled sequences from SIP experiments for phylogenetic tree construction have been deposited in GenBank under accession numbers KP890804 to KP890821 for the amoA and 16S rRNA genes.
RESULTS
Soil properties and nitrification activity.
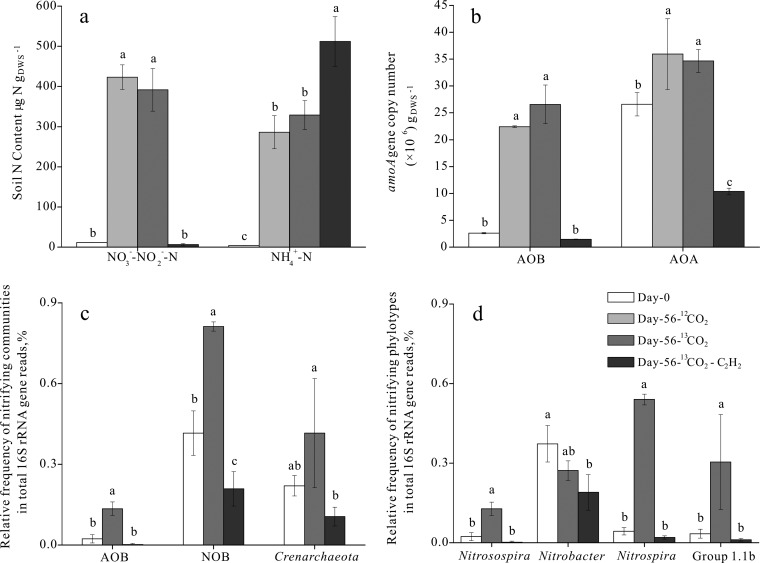
The soil used in this study has a pH value of 5.99 (H2O) and contains relatively high organic matter, with 21.9 mg per gram (dry weight) of soil (gDWS−1). Soil ammonium, nitrate, and total nitrogen levels were 2.20 μg NH4+-N gDWS−1, 6.01 μg NO3−-N g−1 gDWS−1, and 1.06 mg total N gDWS−1, respectively. Soil nitrification activity was assessed as the rate of increase in the concentrations of nitrate and nitrite in soil microcosms over the 56-day incubation period. In the absence of C2H2, significant production of soil nitrate and nitrite was observed after incubation for 56 days (Fig. 1a). There was no significant difference in soil nitrate and nitrite production between the labeled 13CO2 and control 12CO2 microcosms, in which net nitrification rates were estimated to be approximately 6.78 and 7.35 μg NO3−-NO2−-N gDWS−1 day−1, respectively. The increase in soil nitrate and nitrite content corresponded well with the decrease of NH4+-N. Moreover, C2H2 addition completely abolished production of soil nitrate and nitrite, resulting in significant accumulation of soil ammonium due to the hydrolysis of urea (Fig. 1a). C2H2 inhibition indicated that ammonium from urea hydrolysis was almost completely converted to nitrate in soil microcosms, suggesting the predominant role of nitrification in SIP microcosms. For instance, the amount of ammonium in C2H2-amended soil was largely similar to that of soil nitrate contents in soil microcosms without C2H2. Urea-derived ammonium could still be recovered after a 56-day incubation period (Fig. 1a), and acetylene completely blocked ammonia oxidation but not urea hydrolysis in the controls (Fig. 1a), indicating that the hydrolysis was independent of the nitrification process.
FIG 1.
Changes in concentrations of soil inorganic N (a), in amoA gene abundance in ammonia-oxidizing bacteria (AOB) and archaea (AOA) (b), and in relative frequencies of 16S rRNA genes of nitrifiers (c) and of specific phylotypes (d) in total pyrosequencing reads over a 56-day microcosm incubation. Soil microcosms were fertilized with 100 μg urea-N gDWS−1 on a weekly basis for 8 weeks. The relative frequency of group-specific 16S rRNA genes was defined as the proportion of target reads to the total high-quality 16S rRNA gene reads in SIP microcosms. Error bars represent standard errors of triplicate microcosms. Different letters above the columns indicate a significant difference (P < 0.05).
Abundance change of soil nitrifying communities.
Bacterial and archaeal amoA genes were quantified by real-time quantitative PCR to investigate the changes in population sizes of AOB and AOA communities over the 56-day incubation of SIP microcosms. The bacterial amoA genes increased from 2.60 × 106 ± 0.12 × 106 copies gDWS−1 at day 0 to 2.66 × 107 ± 0.36 × 107 and 2.24 × 107 ± 0.01 × 107 copies gDWS−1 at day 56, representing 10.2- and 8.62-fold increases in the 13CO2-labeled treatment and 12CO2 control microcosms, respectively (Fig. 1b). In comparison, archaeal amoA genes underwent only 1.30- and 1.35-fold increases, from 2.66 × 107 ± 0.22 × 107 copies gDWS−1 at day 0 to 3.47 × 107 ± 0.21 × 107 and 3.59 × 107 ± 0.66 × 107 copies gDWS−1 in the 13CO2-labeled treatment and 12CO2 control microcosms at day 56, respectively (Fig. 1b). In the presence of 100 Pa C2H2, soil microcosms showed no increasing trends for either bacterial or archaeal amoA genes (Fig. 1b), suggesting successful inhibition of both AOB and AOA nitrification activity by C2H2 in this study. The change in amoA gene abundance could not be unequivocally attributed to ammonia oxidation activity although the abundance of archaeal amoA genes increased at a significant level.
The high-throughput fingerprinting of 16S rRNA genes at the whole-community level provided a powerful strategy for profiling the relative abundance change in soil nitrifying communities. Pyrosequencing was performed for soil microcosms at day 0 and for SIP microcosms amended with 13CO2 in the presence and absence of C2H2 at day 56 (see Tables S2 and S4 in the supplemental material for a pyrosequencing summary). It was interesting that the incubation of SIP microcosms appeared to have resulted in no significant change in the abundance of total 16S rRNA genes and community structures at the phylum level (see Fig. S1 in the supplemental material), and the alpha diversity index of Chao1 and OTUs remained largely unchanged over the course of incubation (see Fig. S2a and b). However, the divergence of microbial composition was clearly evidenced by a multidimensional scaling (MDS) beta diversity analysis (see Fig. S2). This is likely attributed to the remarkable proportional increases of nitrifying phylotypes over the 56-day incubation. For instance, the 16S rRNA genes affiliated with AOB and NOB rose from 0.0233% and 0.416% of total reads at day 0 to 0.135% and 0.812% at day 56, representing a 5.79-fold and 1.95-fold increase over the 56-day incubation, respectively (Fig. 1c). Among the AOB and NOB communities, Nitrosospira and Nitrospira-like 16S rRNA genes responded to urea fertilization most positively, accounting for 0.128 ± 0.025% and 0.540 ± 0.020% of total 16S rRNA gene reads at day 56, respectively (Fig. 1d). Interestingly, Nitrobacter-like genes dominated NOB communities in soil microcosms at day 0 but decreased by 27.1% in frequency after the 56-day incubation with 13CO2 and urea fertilization (Fig. 1d). Crenarchaeal 16S rRNA gene reads increased from 0.220% at day 0 to 0.416% at day 56 in the 13CO2-labeled treatment (Fig. 1c). Furthermore, sequencing analysis demonstrated that members within the soil group 1.1b lineage dominated the archaeal communities in the SIP microcosms, with an 8.91-fold increase from 0.034 ± 0.017% to 0.304 ± 0.179% after the 56-day incubation (Fig. 1d). SIP microcosms treated with C2H2 showed no significant increase in relative abundances of 16S rRNA genes of nitrifiers at different taxonomic levels (Fig. 1c and d). Intriguingly, the absolute abundance of these functional guilds showed no significant changes among different treatments on the basis of pyrosequencing and real-time quantification analysis of total 16S rRNA genes (see Fig. S1a in the supplemental material).
Stable isotope probing of soil nitrifying communities.
Quantitative analysis of amoA gene abundance as a function of DNA buoyant densities of isopycnic centrifugation gradients was exploited to assess the labeling efficiency of the amoA gene carrying ammonia oxidizers in SIP microcosms. A shift toward heavy fractions was observed for bacterial amoA gene abundance in the 13CO2-labeled treatment compared to fractions in the 12CO2 controls (Fig. 2a). The highest copy number of bacterial amoA genes in the 13CO2-labeled treatment peaked in the heavy DNA fraction, with a buoyant density of 1.740 g ml−1, whereas in the 12CO2 control treatment, the highest number of amoA genes occurred in the light DNA fraction, with a buoyant density of 1.723 g ml−1 (Fig. 2a). These results indicated that a considerable amount of 13C was assimilated by AOB in the labeled soils, leading to a significant shift of amoA gene-carrying genomic DNA into the heavy DNA fractions. In comparison, only a minor shift into the heavier fractions was detected for archaeal amoA genes from the labeled soils, suggesting that AOA were labeled to a lesser extent than AOB (Fig. 2b).
FIG 2.
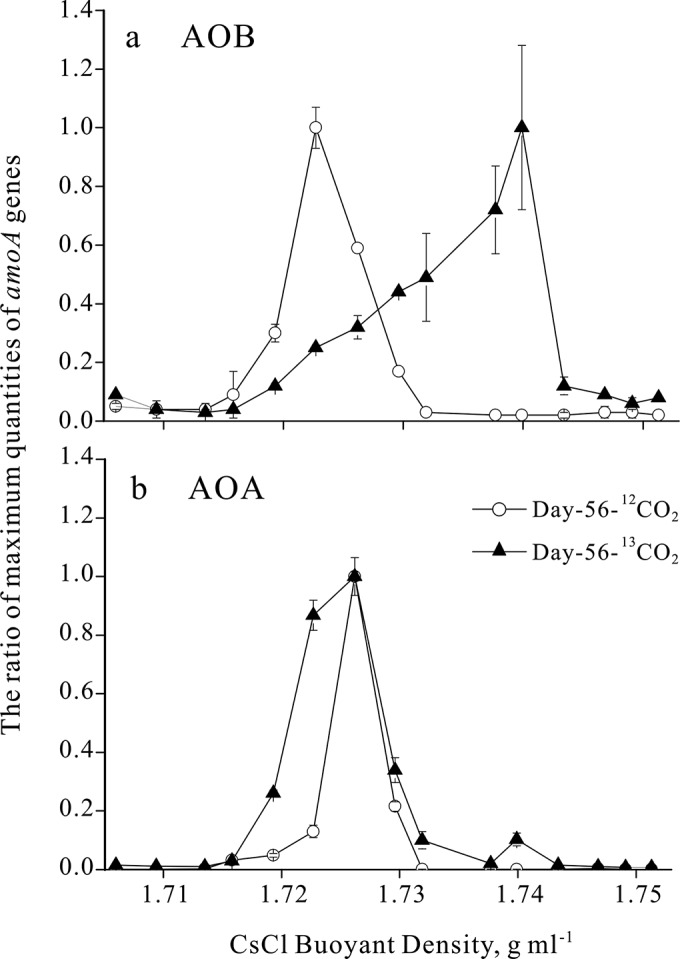
The quantitative distribution of bacterial (a) and archaeal (b) amoA genes across the entire buoyant density gradient of the DNA fractions from soil microcosms incubated with 12CO2 and 13CO2. The normalized data are the ratios of the gene copy number for each DNA gradient to the maximum quantities for each treatment. The error bars represent the standard errors of triplicate microcosms, and each contains three technical replicates.
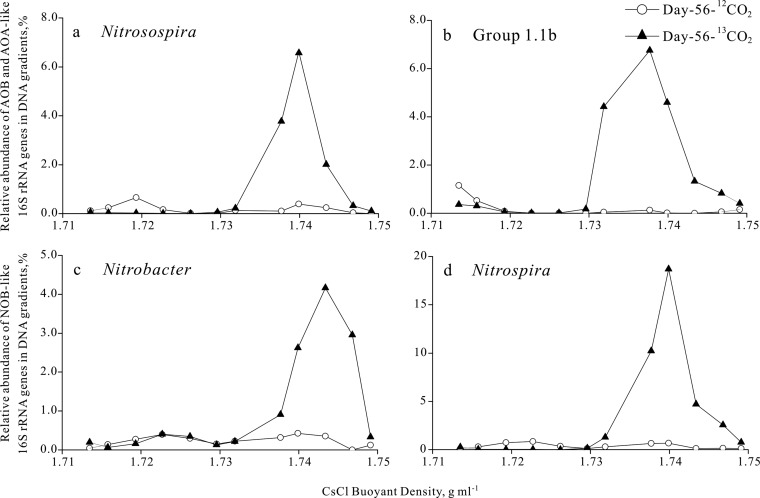
Pyrosequencing of total 16S rRNA genes in the fractionated DNA across a wide range of buoyant densities was performed to assess the enrichment of target nitrifying communities in the heavy DNA fractions from 13CO2-labeled treatment in comparison to that of those from the 12CO2 control microcosms (see Table S5 in the supplemental material). High-quality 16S rRNA gene reads were subjected to taxonomic classification, and nitrifying phylotypes were selected for further analyses (see Table S5). For 16S rRNA genes affiliated with AOB and NOB, a significant enrichment was observed in the heavy DNA fractions from 13CO2-labeled treatment but not from 12CO2 control microcosms. For instance, Nitrosospira-like AOB accounted for 6.67% of the total 16S rRNA gene reads in the heavy DNA fraction from the labeled treatment but represented only a tiny fraction, no more than 0.654%, of the total reads in the 12CO2 control microcosms (Fig. 3a). Similar results were obtained for the NOB communities. Nitrobacter- and Nitrospira-like 16S rRNA genes were enriched up to 4.16% and 18.7% in the heavy DNA fractions from 13CO2-labeled soil, respectively (Fig. 3c and d). In stark contrast, the highest relative frequencies of Nitrobacter- and Nitrospira-like 16S rRNA genes were only 0.423% and 0.844%, respectively, in the DNA fractions from 12CO2 control microcosms (Fig. 3c and d).
FIG 3.
The relative frequency of Nitrosospira-like AOB (a), soil group 1.1b AOA (b), Nitrobacter-like NOB (c), and Nitrospira-like NOB (d) across the entire buoyant density gradient fractionated from DNA extraction in 13CO2-labeled and 12CO2 control soil microcosms. The relative frequency of 16S rRNA genes of each nitrifier group was determined as the proportion of target reads to all high-quality 16S rRNA gene reads in each fraction.
Pyrosequencing analysis further indicated apparent enrichment of archaeal communities in the heavy DNA fractions from 13CO2-labeled microcosms but not from the 12CO2 control microcosms (see Table S5 in the supplemental material). Crenarchaeal 16S rRNA genes were selected from the total reads and further clustered into OTUs with a 97% cutoff. Sequencing analysis indicated that the total reads of archaeal 16S rRNA genes within the soil group 1.1b lineage were enriched up to 6.75% in the heavy DNA fractions from 13CO2-labeled treatment (Fig. 3b). Furthermore, the relative frequency of members within the marine 1.1a-associated lineage was also apparently higher in the labeled treatment than in the control microcosms, even though members of this lineage were much less abundant than the soil 1.1b-associated archaea (see Table S5). These results suggested substantial assimilation of 13C label by AOA, AOB, and NOB cells in the soil tested, implying an active role in soil nitrification.
Phylogenetic analysis of active ammonia-oxidizers.
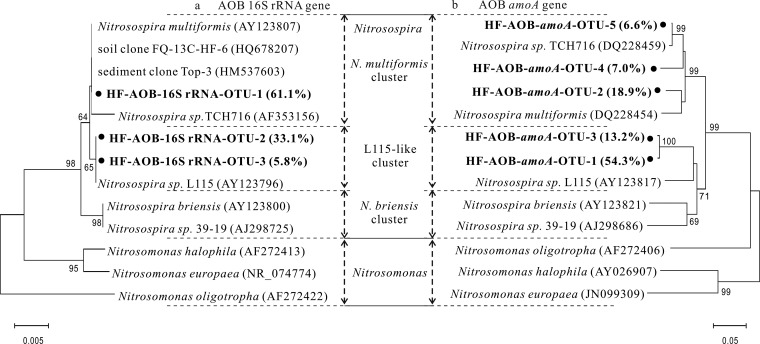
Phylogenetic analysis of all 849 of the AOB-like 16S rRNA gene reads in the 13C-DNA fraction from the 13CO2-labeled treatment revealed that the active AOB genes were predominately contributed by four Nitrosospira-like OTUs. The representative reads from these OTUs were phylogenetically most closely related to AOB strains Nitrosospira sp. strain L115 and N. multiformis within the Nitrosospira cluster 3 lineage (Fig. 4a). Bacterial amoA genes from the same fraction predominantly belonged to five OTUs, with representative reads also related to Nitrosospira sp. strain L115 and N. multiformis (Fig. 4b).
FIG 4.
Phylogenetic analysis of the 16S rRNA (a) and amoA genes (b) of ammonia-oxidizing bacteria (AOB) in 13C-labeled DNA from the 13CO2-treated microcosms of 56-day microcosms. The phylogenetic tree shows the relationship of gene sequences of AOB from the heavy fraction (HF) to those deposited in GenBank. The notation is according to the following examples: HF-AOB-16S rRNA-OTU-1 (61.1%) represents OTU-1 and contains 61.1% of the total AOB 16S rRNA gene reads in the 13C-DNA reads with >97% sequence similarity; HF-AOB-amoA-OTU-1 (54.3%) represents OTU-1 and contains 54.3% of the bacterial amoA gene reads in the 13C-DNA reads with >97% sequence similarity. Bootstrap values higher than 60% are indicated at the branch nodes. GenBank accession numbers are given in parentheses.
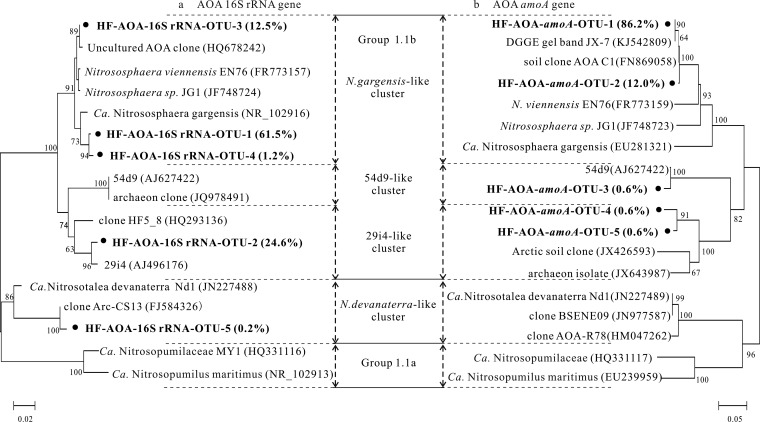
A total of 607 reads assigned to the phylum Crenarchaeota were retrieved in heavy DNA fraction 5 for phylogenetic analysis of AOA (Fig. 5a). Of the Crenarchaeota reads, 97.4% fell within AOA-like lineages, and the majority of the crenarchaeal 16S rRNA gene reads were phylogenetically closely related to “Candidatus Nitrososphaera gargensis” and fosmid clone 29i4, both within soil group 1.1b lineages (25). Archaeal amoA genes from the same fraction clustered into “Candidatus Nitrososphaera gargensis,” fosmid 29i4, and 54d9-related OTUs (Fig. 5b) although the abundance of “Ca. Nitrososphaera gargensis”-like genes was significantly higher (Fig. 5b).
FIG 5.
Phylogenetic analysis of the 16S rRNA (a) and amoA genes (b) of ammonia-oxidizing archaea (AOA) in 13C-labeled DNA from the 13CO2-treated microcosms of 56-day microcosms. The phylogenetic tree shows the relationship of gene sequences of AOA from the heavy fraction (HF) to those deposited in GenBank. The notation is according to the following examples: HF-AOA-16S rRNA-OTU-1 (61.5%) represents OTU-1 and contains 61.5% of the total AOA 16S rRNA gene reads in the 13C-DNA reads with >97% sequence similarity; HF-AOA-amoA-OTU-1 (86.2%) represents OTU-1 and contains 86.2% of the archaeal amoA gene reads in the 13C-DNA reads with >97% sequence similarity. Bootstrap values higher than 60% are indicated at the branch nodes. GenBank accession numbers are given in parentheses.
DISCUSSION
Rice paddy ecosystems receive intensive fertilization with ammonia-based fertilizers to support crop production (9). Agricultural interventions such as frequent flooding and fertilizations lead to strong fluctuations in available ammonia and oxygen that could likely shape habitat-specific communities of soil AOA and AOB. Our results provide compelling evidence for active nitrification that was catalyzed by a wide variety of phylogenetically distinct microorganisms in a complex rice soil compared to microcosms of other upland soils (26–28). AOB and NOB apparently dominated soil nitrification in the incubations, whereas active archaeal ammonia oxidizers were phylogenetically closely related to both soil group 1.1b- and marine group 1.1a-associated lineages. These results suggested that nitrification was driven by diverse nitrifying phylotypes with potentially distinct physiological versatilities, implying that the life strategy of soil nitrifying communities is more complex than previously appreciated.
DNA-based SIP strongly indicated bacterial dominance of nitrification activity in microcosms of paddy soil tested over the incubation period. The nitrification process supported greater AOB multiplication than that of AOA, as demonstrated by a much higher enrichment of bacterial amoA gene copies in the heavy fractions of 13C-labeled DNA. Quantitative PCR showed that 53.9% of AOB communities were labeled during nitrification (Fig. 2a). Assuming that one AOB cell contains 2.5 copies of amoA genes on average, ammonia oxidation by labeled AOB would reach an estimated cell-specific rate of 3.53 fmol N cell−1 h−1, which is well within the previously reported range for AOB communities (29). In fact, the 13C-labeled AOB in this study were phylogenetically closely related to Nitrosospira sp. strain L115, with a similar specific activity of 5.3 fmol N cell−1 (30). Thus, AOB alone could account for the soil nitrification activity observed in SIP microcosms over the 56-day incubation course. In stark contrast, the labeled AOA cells (approximately 4.73%) (Fig. 2b) would contribute no more than 8.13% of the total nitrate production, assuming that they all reached the maximum specific rate of archaeal ammonia oxidation previously documented (29, 31). These results agreed well with previous findings that AOB dominated ammonia oxidation in ammonia-rich soils (18, 27, 32, 33). Similarly, the paddy field in our study received relatively high nitrogen input with urea-based fertilizers during growing seasons, which might select for AOB communities with a much higher growth rate and activity than those of AOA (34). This explains the rapid growth of AOB upon urea fertilization in this study (Fig. 1b). It is worth noting that the samples were collected only once at day 56. Therefore, the cell-specific rates of ammonia oxidation in the SIP microcosm was calculated based on an assumption that a linear increase of nitrate production and cell propagation had occurred over a 56-day incubation period. Temporal sampling would likely reduce the uncertainty associated with cell-specific rate estimation and result in a better understanding of distinct nitrification potentials among nitrifiers in the soil tested.
A direct link between ammonia oxidation and AOA within soil group 1.1b in complex soil was reported in only a few studies (27, 33, 35, 36) although this group was often considered to be numerically dominant in neutral and/or alkaline soils (37–39). Our results provided evidence that archaeal ammonia oxidation in complex soil was not restricted to “Ca. Nitrososphaera gargensis”-like AOA, as previously reported (27, 33), but, rather, 29i4-like AOA catalyzed a portion of archaeal nitrification in the paddy soil tested in this study (Fig. 5; see also Table S6 in the supplemental material). Nitrification activity of “Ca. Nitrososphaera gargensis”-like AOA was often associated with upland soils, whereas 29i4-like AOA appeared to be well adapted to low-temperature arctic soil (40) and regularly oxygen-deficient niches in freshwater sediments (41). The widespread presence of the fosmid clone 29i4-related lineage has long been observed (42), but the physiological capability for ammonia oxidation has been demonstrated only very recently in arctic soils (40). We speculate that 29i4-like AOA might possess a greater capacity for stress resistance; thus, niche differentiation of archaeal communities might have been stronger in paddy fields than in upland soils. Furthermore, the acidophilic “Candidatus Nitrosotalea devanaterra”-like AOA are often found in acidic soil and oligotrophic freshwater environments. Our results demonstrated that these AOA could survive in the neutral paddy soil tested in this study (Fig. 5a; see also Table S5 in the supplemental material). Although the sequences affiliated with this marine AOA-associated lineage have been obtained from neutral or only slightly acidic environments (43–45), the evidence directly linking this group to ammonia oxidation was missing. Thus, a greater physiological versatility of this marine lineage was observed in this study, despite reduced labeling compared to that of AOA associated with soil group 1.1b.
Nitrite oxidation has received much less attention than ammonia oxidization, and the physiological diversity of NOB is poorly understood (46). Our results showed that active NOB in this study were restricted to the well-known Nitrospira- and Nitrobacter-like groups. Surprisingly, a distinct ecophysiology was observed between Nitrospira- and Nitrobacter-like NOB in the paddy soil tested. The latter predominated in the background communities of nitrite oxidizers, accounting for 89.7% of total NOB-like reads before SIP microcosm incubation (Fig. 1d). However, the former boomed in active cell division and reached 89.6% of the total NOB 16S rRNA reads in the 13C-DNA (Fig. 2d). Nitrospira and Nitrobacter were considered to be the two main players for nitrification in terrestrial ecosystems (47) but with distinct kinetic strategies for cell growth and activity. Nitrospira was hypothesized to be a K strategist, which possesses high nitrite affinity but much lower cell-specific activity than the r strategist Nitrobacter, as shown in culture studies (48–50) and soil assays (51). In fact, pyrosequencing of the total microbial communities showed a remarkable increase of Nitrospira-like but not Nitrobacter-like NOB in SIP microcosms during active nitrification (Fig. 1d). This drastic difference in the relative abundances between 13C-labeled Nitrobacter- and Nitrospira-like NOB indicated their different roles in nitrite oxidation. This difference also implies that soil nitrite was kept at a constantly low concentration in the SIP microcosms although the temporal dynamics of soil nitrite were not measured (Fig. 1a).
This study demonstrated the existence of diverse phylotypes of active nitrifiers with potential physiological distinctions. This outcome suggested the strong adaptation of nitrifiers to constantly changing environmental conditions in the paddy soil tested. Rice cultivation necessitates regular flooding and intensive fertilization, leading to drastic changes in soil oxygen and ammonia concentrations in the field. Nitrifying communities could thus have experienced stronger stresses in paddy fields than in upland soils, which constantly receive oxygen diffusion from the atmosphere without blockage from the water layer. We speculated that the long-term environmental fluctuation in paddy fields could create certain scenarios where distinctly different nitrifiers were selectively favored with different competitive statuses under specific conditions. The generally high availability of ammonia and oxygen in our microcosms apparently favored growth of AOB over that of AOA groups. Nitrification leading to possible decreases in pH and ammonia concentration might have stimulated AOA growth occasionally. Furthermore, agricultural management allows the creation of soil microsites with highly heterogeneous environmental conditions in the paddy soil; therefore, nitrification might be performed by different nitrifier combinations. These hypotheses might partially explain the existence of distinct active phylotypes usually found in different niches. For instance, active AOA in this study were phylogenetically most closely affiliated with three environmentally distinct cultures and enrichments. “Ca. Nitrososphaera gargensis”-and 29i4-like AOA were enriched from a hot spring and cold arctic soil, respectively, while “Ca. Nitrosotalea”-like sequences were isolated only from oligotrophic freshwater or acid soils. It is very likely that NOB communities have evolved in close association with ammonia oxidizers (52). However, it remains unclear how these functional groups interact with one another and generate distinct nitrifying communities in the paddy soil tested.
It is worth noting that artificial incubation of SIP microcosms in this study could not represent what is naturally occurring under field conditions. Paddy soils during rice growing season would have rarely experienced high concentrations of oxygen and ammonium simultaneously, as they did in this study. The enrichment of nitrifying communities might have resulted from the succession of microbial communities over the course of incubation of SIP microcosms (see Fig. S2 in the supplemental material). For instance, numerically less abundant Nitrospira-like NOB under in situ conditions significantly outnumbered Nitrobacter-like NOB after laboratory incubation for 56 days (see Table S3 in the supplemental material), suggesting preferential growth of the former under artificial conditions. In addition, the population dynamics during the 56-day laboratory incubation remain unclear due to the lack of temporal sampling. However, one recent study has shown that nitrogen fertilization intensity and the sampling time point had only small effects on the abundance, composition, and activity of AOA, AOB, and NOB populations (47). Our recent study also demonstrated that the labeled AOA showed high degrees of similarity in SIP microcosms at day 28 and day 56 (26). Furthermore, the labeled nitrifiers are largely similar to those found under in situ conditions, implying that these functional groups might be selected for and acclimated to the paddy soil tested. We speculate that long-term physiochemical conditions formed seed banks of active nitrifiers and that incubation of SIP microcosms could provide some clues as to which organisms might have been actively involved in nitrification under field conditions. The actual nitrification dynamics of different phylotypes under field conditions should be further studied, in addition to cultivation studies and comparative ecophysiological analysis of functional guilds across physiochemically different environments.
Taken together, the results of this study clearly demonstrated autotrophic nitrification by AOA, AOB, and NOB communities in a neutral paddy soil that is exposed to periodic flooding and fertilization regimes. Stable isotope probing suggested that the genomes of phylogenetically distinct nitrifiers were labeled to different extents. The active AOB were phylogenetically closely related to the strains isolated from acidic soil, while the labeled AOA showed high sequence similarity to those from hot springs and arctic soils. These results imply that phylogenetic relatedness might not necessitate physiological similarity and that the activity of phylogenetically distinct nitrifiers might vary with environmental fluctuations under field conditions in a paddy field.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Science Foundation of China (41090281), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB15040000), and the Distinguished Young Scholar Programme of Jiangsu Province (BK2012048).
We thank Guanghua Wang and his colleague for sampling assistance and our lab colleagues for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00426-15.
REFERENCES
- 1.Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. 2010. Food security: the challenge of feeding 9 billion people. Science 327:812–818. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- 2.Luo YF, Fu HL, Traore S. 2014. Biodiversity conservation in rice paddies in China: toward ecological sustainability. Sustainability 6:6107–6124. doi: 10.3390/su6096107. [DOI] [Google Scholar]
- 3.Liu SL, Huang DY, Chen AL, Wei WX, Brookes PC, Li Y, Wu JS. 2014. Differential responses of crop yields and soil organic carbon stock to fertilization and rice straw incorporation in three cropping systems in the subtropics. Agric Ecosyst Environ 184:51–58. doi: 10.1016/j.agee.2013.11.019. [DOI] [Google Scholar]
- 4.Zhang XX, Yin S, Li YS, Zhuang HL, Li CS, Liu CJ. 2014. Comparison of greenhouse gas emissions from rice paddy fields under different nitrogen fertilization loads in Chongming Island, Eastern China. Sci Total Environ 472:381–388. doi: 10.1016/j.scitotenv.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Ju XT, Xing GX, Chen XP, Zhang SL, Zhang LJ, Liu XJ, Cui ZL, Yin B, Christie P, Zhu ZL, Zhang FS. 2009. Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc Natl Acad Sci U S A 106:3041–3046. doi: 10.1073/pnas.0813417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebilo M, Mayer B, Nicolardot B, Pinay G, Mariotti A. 2013. Long-term fate of nitrate fertilizer in agricultural soils. Proc Natl Acad Sci U S A 110:18185–18189. doi: 10.1073/pnas.1305372110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kögel-Knabner I, Amelung W, Cao Z, Fiedler S, Frenzel P, Jahn R, Kalbitz K, Kölbl A, Schloter M. 2010. Biogeochemistry of paddy soils. Geoderma 157:1–14. doi: 10.1016/j.geoderma.2010.03.009. [DOI] [Google Scholar]
- 8.Wu YC, Lu L, Wang BZ, Lin XG, Zhu JG, Cai ZC, Yan XY, Jia ZJ. 2011. Long-term field fertilization significantly alters community structure of ammonia-oxidizing bacteria rather than archaea in a paddy soil. Soil Sci Soc Am J 75:1431–1439. doi: 10.2136/sssaj2010.0434. [DOI] [Google Scholar]
- 9.Liesack W, Schnell S, Revsbech NP. 2000. Microbiology of flooded rice paddies. FEMS Microbiol Rev 24:625–645. doi: 10.1111/j.1574-6976.2000.tb00563.x. [DOI] [PubMed] [Google Scholar]
- 10.Geets J, Boon N, Verstraete W. 2006. Strategies of aerobic ammonia-oxidizing bacteria for coping with nutrient and oxygen fluctuations. FEMS Microbiol Ecol 58:1–13. doi: 10.1111/j.1574-6941.2006.00170.x. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann M, Scheibe A, Avrahami S, Kusel K. 2011. Ammonium availability affects the ratio of ammonia-oxidizing bacteria to ammonia-oxidizing archaea in simulated creek ecosystems. Appl Environ Microbiol 77:1896–1899. doi: 10.1128/AEM.02879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolaisen MH, Risgaard-Petersen N, Revsbech NP, Reichardt W, Ramsing NB. 2004. Nitrification-denitrification dynamics and community structure of ammonia oxidizing bacteria in a high yield irrigated Philippine rice field. FEMS Microbiol Ecol 49:359–369. doi: 10.1016/j.femsec.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Alam MS, Ren GD, Lu L, Zheng Y, Peng XH, Jia ZJ. 2013. Conversion of upland to paddy field specifically alters the community structure of archaeal ammonia oxidizers in an acid soil. Biogeosciences 10:5739–5753. doi: 10.5194/bg-10-5739-2013. [DOI] [Google Scholar]
- 14.Chen XP, Zhu YG, Xia Y, Shen JP, He JZ. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ Microbiol 10:1978–1987. doi: 10.1111/j.1462-2920.2008.01613.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang YA, Ke XB, Wu LQ, Lu YH. 2009. Community composition of ammonia-oxidizing bacteria and archaea in rice field soil as affected by nitrogen fertilization. Syst Appl Microbiol 32:27–36. doi: 10.1016/j.syapm.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Yue J, Shi Y, Liang W, Wu J, Wang CR, Huang GH. 2005. Methane and nitrous oxide emissions from rice field and related microorganism in black soil, northeastern China. Nutr Cycl Agroecosys 73:293–301. doi: 10.1007/s10705-005-3815-5. [DOI] [Google Scholar]
- 17.IUSS Working Group WRB. 2006. World reference base for soil resources 2006. World soil resources report no. 103. Food and Agriculture Organization of the United Nations, Rome, Italy: ftp://ftp.fao.org/agl/agll/docs/wsrr103e.pdf. [Google Scholar]
- 18.Jia ZJ, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 19.Serrano-Silva N, Luna-Guido M, Fernández-Luqueño F, Marsch R, Dendooven L. 2011. Emission of greenhouse gases from an agricultural soil amended with urea: a laboratory study. Appl Soil Ecol 47:92–97. doi: 10.1016/j.apsoil.2010.11.012. [DOI] [Google Scholar]
- 20.Hamer U, Potthast K, Makeschin F. 2009. Urea fertilisation affected soil organic matter dynamics and microbial community structure in pasture soils of southern Ecuador. Appl Soil Ecol 43:226–233. doi: 10.1016/j.apsoil.2009.08.001. [DOI] [Google Scholar]
- 21.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett 132:203–208. doi: 10.1111/j.1574-6968.1995.tb07834.x. [DOI] [PubMed] [Google Scholar]
- 23.Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. 2012. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 25.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L, Jia ZJ. 2013. Urease gene-containing Archaea dominate autotrophic ammonia oxidation in two acid soils. Environ Microbiol 15:1795–1809. doi: 10.1111/1462-2920.12071. [DOI] [PubMed] [Google Scholar]
- 27.Zhang LM, Hu HW, Shen JP, He JZ. 2012. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045. doi: 10.1038/ismej.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia WW, Zhang CX, Zeng XW, Feng YZ, Weng JH, Lin XG, Zhu JG, Xiong ZQ, Xu J, Cai ZC, Jia ZJ. 2011. Autotrophic growth of nitrifying community in an agricultural soil. ISME J 5:1226–1236. doi: 10.1038/ismej.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prosser JI, Nicol GW. 2012. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Jiang QQ, Bakken LR. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol Ecol 30:171–186. doi: 10.1111/j.1574-6941.1999.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 31.Hatzenpichler R. 2012. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol 78:7501–7510. doi: 10.1128/AEM.01960-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ. 2009. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624. doi: 10.1038/ngeo613. [DOI] [Google Scholar]
- 33.Pratscher J, Dumont MG, Conrad R. 2011. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci U S A 108:4170–4175. doi: 10.1073/pnas.1010981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.French E, Kozlowski JA, Mukherjee M, Bullerjahn G, Bollmann A. 2012. Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl Environ Microbiol 78:5773–5780. doi: 10.1128/AEM.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schleper C. 2010. Ammonia oxidation: different niches for bacteria and archaea? ISME J 4:1092–1094. doi: 10.1038/ismej.2010.111. [DOI] [PubMed] [Google Scholar]
- 37.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu HW, Zhang LM, Dai Y, Di HJ, He JZ. 2013. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J Soils Sediments 13:1439–1449. doi: 10.1007/s11368-013-0726-y. [DOI] [Google Scholar]
- 39.Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW. 2011. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci U S A 108:21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T. 2013. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7:1620–1631. doi: 10.1038/ismej.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 42.Quaiser A, Ochsenreiter T, Klenk HP, Kletzin A, Treusch AH, Meurer G, Eck J, Sensen CW, Schleper C. 2002. First insight into the genome of an uncultivated crenarchaeote from soil. Environ Microbiol 4:603–611. doi: 10.1046/j.1462-2920.2002.00345.x. [DOI] [PubMed] [Google Scholar]
- 43.Auguet JC, Casamayor EO. 2013. Partitioning of Thaumarchaeota populations along environmental gradients in high mountain lakes. FEMS Microbiol Ecol 84:154–164. doi: 10.1111/1574-6941.12047. [DOI] [PubMed] [Google Scholar]
- 44.Bollmann A, Bullerjahn GS, Mckay RM. 2014. Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior. PLoS One 9:e97068. doi: 10.1371/journal.pone.0097068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun W, Xia CY, Xu MY, Guo J, Wang AJ, Sun GP. 2013. Distribution and abundance of archaeal and bacterial ammonia oxidizers in the sediments of the Dongjiang River, a drinking water supply for Hong Kong. Microbes Environ 28:457–465. doi: 10.1264/jsme2.ME13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorokin DY, Lucker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WIC, Damste JSS, Le Paslier D, Muyzer G, Wagner M, van Loosdrecht MCM, Daims H. 2012. Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J 6:2245–2256. doi: 10.1038/ismej.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke XB, Angel R, Lu YH, Conrad R. 2013. Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ Microbiol 15:2275–2292. doi: 10.1111/1462-2920.12098. [DOI] [PubMed] [Google Scholar]
- 48.Kim DJ, Kim SH. 2006. Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res 40:887–894. doi: 10.1016/j.watres.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 49.Blackburne R, Vadivelu VM, Yuan ZG, Keller J. 2007. Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res 41:3033–3042. doi: 10.1016/j.watres.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 50.Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol 65:3690–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Attard E, Poly F, Commeaux C, Laurent F, Terada A, Smets BF, Recous S, Le Roux X. 2010. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ Microbiol 12:315–326. doi: 10.1111/j.1462-2920.2009.02070.x. [DOI] [PubMed] [Google Scholar]
- 52.Prosser JI. 2012. Ecosystem processes and interactions in a morass of diversity. FEMS Microbiol Ecol 81:507–519. doi: 10.1111/j.1574-6941.2012.01435.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.