Abstract
To examine to what extent fresh vegetables imported into Switzerland represent carriers of extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae, 169 samples of different types of fresh vegetables imported into Switzerland from the Dominican Republic, India, Thailand, and Vietnam were analyzed. Overall, 25.4% of the vegetable samples yielded one or more ESBL-producing Enterobacteriaceae, 78.3% of which were multidrug resistant. Sixty isolates were obtained: Escherichia coli, 26; Klebsiella pneumoniae, 26; Enterobacter cloacae, 6; Enterobacter aerogenes, 1; and Cronobacter sakazakii, 1. We found 29 isolates producing CTX-M-15, 8 producing CTX-M-14, 7 producing CTX-M-55, 3 producing CTX-M-65, 1 each producing CTX-M-1, CTX-M-3, CTX-M-27, and CTX-M-63, 5 producing SHV-2, 3 producing SHV-12, and 1 producing SHV-2a. Four of the E. coli isolates belonged to epidemiologically important clones: CTX-M-15-producing B2:ST131 (1 isolate), D:ST405 (1 isolate), and D:ST38 (2 isolates). One of the D:ST38 isolates belonged to the extraintestinal enteroaggregative E. coli (EAEC) D:ST38 lineage. Two of the K. pneumoniae isolates belonged to the epidemic clones sequence type 15 (ST15) and ST147. The occurrence of antibiotic-resistant pathogenic and commensal Enterobacteriaceae in imported agricultural foodstuffs constitutes a source of ESBL genes and a concern for food safety.
INTRODUCTION
The production of extended-spectrum β-lactamases (ESBLs) is one of the most important mechanisms of antibacterial resistance in Enterobacteriaceae. Most ESBLs can be divided into 4 groups: TEM, SHV, OXA, and CTX-M types (1). Currently, CTX-Ms are the most prevalent type of ESBLs described (2, 3). The last decade has seen a rapid and massive global spread driven primarily by their carriage on resistance plasmids and by the spread of extraintestinal pathogenic Escherichia coli clones (4, 5). Important clonal lineages include E. coli strains belonging to multilocus sequence type 131 (ST131) (often associated with CTX-M-15) and enteroaggregative E. coli (EAEC) ST38 (6). In addition to these widespread ESBLs, less frequently occurring ESBLs have been detected on regional scales, e.g., GES, PER, and VEB types (7).
In recent years, it has been widely recognized that the dissemination of ESBL-producing bacteria is an issue that is no longer restricted to the medical/health care system but represents a growing problem involving food safety and environmental integrity. There is increasing evidence that antimicrobial drug use in the livestock sector plays an important role in the contamination of food with ESBL-producing bacteria (8, 9), but little is yet known about the burden of ESBL-producing Enterobacteriaceae on fresh vegetables. In the crop production sector, products can be contaminated through application of manure (animal origin) or sewage sludge (human origin) to the soil or through application of treated or untreated wastewater that is used for irrigation of crops (10).
In Switzerland, as in most industrialized countries, preharvest intervals (i.e., intervals between application of manure to the soil and the subsequent growth phase) restrict the application of manure to the soil, and wastewater is treated before reuse, with high ecological standards and levels of hygiene applied at all stages of culture and harvesting (11). Hence, the bacteriological burden of vegetable crops is low. In contrast, in many developing countries, most prominently Vietnam, China, and India, wastewater without treatment or with insufficient treatment is commonly used for agriculture, producing negative effects on human health and the environment (12, 13).
Analyses of alimentary consumption trends in Switzerland record an increase in Asian and Latin American cuisine and point to a demand for fresh produce (14). Import trade statistics show that imports to Switzerland of edible vegetables from India have doubled over the last decade, and those from the Socialist Republic of Vietnam have quadrupled. Over the last 4 years, Switzerland imported an average of 701.25 metric tons per annum of edible vegetables from the Dominican Republic, India, Thailand, and Vietnam (Swiss Federal Customs Administration [FCA] [https://www.swiss-impex.admin.ch/pages/bereiche/waren/query.xhtml]).
The aim of this study was to evaluate the presence of ESBL-producing Enterobacteriaceae in vegetables imported from these countries and to characterize isolated strains by (i) antibiotic susceptibility testing, (ii) identification of the bla genes, (iii) multilocus sequence typing (MLST) of the E. coli and Klebsiella pneumoniae isolates, and (iv) identifying phylogenetic groups of E. coli isolates.
MATERIALS AND METHODS
Bacterial sampling.
In July and August 2014, 68 samples of raw vegetables imported via the national airport of Zürich were collected by the food control authority of the Canton Aarau, Switzerland. The vegetables consisted of cucumbers, beans, breadfruit, celery leaves, cha-om (climbing wattle; acacia), chilies, curry leaves, dill, eggplants, garlic chives, lemongrass, onions, peppermint leaves, pak-choy (Chinese cabbage), ponnangani (Asiatic pennywort), several types of squash, water mimosa, and water spinach. The countries of origin were the Dominican Republic (49 samples), India (3 samples), and Thailand (16 samples).
In addition, 101 different fresh vegetable types were purchased in the city of Zürich from 7 retail shops specializing in Asian and South American food and from 3 supermarket chains. The vegetables included basil leaves, beans, celery, Ceylon spinach, chilies, coriander, cucumbers, curry leaves, eggplant, lemon grass, moringa pods (fruits of the horseradish tree), okra (marrow), onions, shallots, dill, soy sprouts, and several types of squash. The samples had been imported from the Dominican Republic (1 sample), India (36 samples), Thailand (44 samples), and the Socialist Republic of Vietnam (20 samples).
In total, 169 vegetable samples were collected for analysis: 50 from the Dominican Republic, 39 from India, 60 from Thailand, and 20 from Vietnam.
Microbiological analysis.
Of each unwashed vegetable sample, 15 to 20 g were placed in a sterile stomacher bag. The samples were homogenized using a stomacher sample blender and incubated at a 1:10 ratio in Enterobacteriaceae enrichment (EE) broth (BD, Franklin Lakes, NJ, USA) at 37°C overnight. For the detection of ESBL producers, chromogenic Brilliance ESBL agar plates (Oxoid, Hampshire, United Kingdom) were inoculated with one loopful of each of the enrichment cultures. The plates were incubated at 37°C for 24 h under aerobic conditions. Colonies with different chromaticities and morphologies were picked from the selective plates and subcultured on sheep blood agar (Difco Columbia blood agar base EH [Becton Dickinson AG, Allschwil, Switzerland], 5% sheep blood [SB055; Oxoid AG, Pratteln, Switzerland]) at 37°C for 24 h. Identification of isolates was either outsourced and achieved by protein profiling using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Mabritec SA, Riehen, Switzerland) for the samples collected from import border control or obtained using the API ID 32 E phenotypic identification system (bioMérieux, Marcy l'Etoile, France) for the samples collected from retail stores. In cases of doubtful results, identification was verified by rpoB sequence analysis (15). The identity of Cronobacter sakazakii was confirmed by rpoB-based PCR as described previously (16). To investigate the putative enteroaggregative properties of E. coli ST38, isolates were tested by PCR for the presence of the EAEC transport regulator gene (aggR), using primers and conditions described previously (17).
ESBL confirmation and antimicrobial susceptibility testing.
ESBL production was confirmed using Etest-ESBL strips containing cefotaxime, ceftazidime, and cefepime, alone and in combination with clavulanic acid (bioMérieux, Marcy l'Etoile, France), according to the manufacturer's instructions. Additionally, the presence of β-lactamase was verified with the colorimetric β Lacta test kit (Bio-Rad, Cressier, Switzerland), as described previously (18) and according to the manufacturer's instructions.
Isolates were subjected to susceptibility testing against 13 antimicrobial agents by the disc diffusion method according to CLSI protocols and evaluated according to CLSI criteria (19). The panel included ampicillin (AM), amoxicillin-clavulanic acid (AMC), cephalothin (CF), cefotaxime (CTX), nalidixic acid (NA), ciprofloxacin (CIP), gentamicin (GM), kanamycin (K), streptomycin (S), sulfamethoxazole (SMZ), trimethoprim (TMP) tetracycline (TE), and chloramphenicol (C) (Becton, Dickinson, Heidelberg, Germany). Strains exhibiting resistance to three or more classes of antibiotics were defined as multidrug resistant (MDR).
Molecular biological analysis of β-lactamase genes.
Isolates identified as potential ESBL producers were further analyzed by PCR. DNA was extracted by a standard heat lysis protocol and analyzed by PCR for the presence of bla genes. Synthesis of primers and DNA custom sequencing were carried out by Microsynth (Balgach, Switzerland). Purification of amplicons was performed using a PCR purification kit (Qiagen, Courtaboeuf, France). Screening for blaTEM and blaSHV was carried out using primers described previously (20), and the resulting amplicons were custom sequenced. Screening for blaCTX-M alleles belonging to CTX-M groups 1, 2, 8, 9, and 25 was done as described by Woodford et al. (21). Amplicons for sequencing individual open reading frames belonging to groups 1, 2, and 9 were generated using primers described previously (8). Group 8 blaCTX-M genes were amplified using the newly designed primers gr. 8 CTX-M-fw (5′-ATG AGA CAT CGC GTT AAG CGG ATG-3′) and gr. 8 CTX-M-rev (5′-CAC GAC GAC TTT CTG CCT TCT GC-3′). The C. sakazakii isolate was additionally tested by PCR for the presence of blaVEB. (22).
Nucleotide sequences were analyzed with CLC Main Workbench 7.0.2. Database searches were performed using the BLASTN program of NCBI (http://www.ncbi.nlm.nih.gov/blast/).
Phylogenetic classification of E. coli isolates.
DNAs from E. coli isolates were subjected to triplex PCR targeting the chuA gene, the yjaA gene, and an unspecified DNA fragment termed TspE4.C2, as described previously (23). Each isolate was assigned to one of the four phylogenetic groups designated A, B1, B2, and D. Groups A and B1 typically contain commensal E. coli strains, while groups B2 and D consist of virulent extraintestinal strains (24).
Multilocus sequence typing of E. coli and K. pneumoniae.
Multilocus sequence types of E. coli isolates were determined as described by Wirth et al. (25). Sequences were imported into the E. coli MLST database website (http://mlst.ucc.ie/mlst/dbs/Ecoli) to determine multilocus sequence types.
MLST of the K. pneumoniae isolates was performed according to previously described methods (26). Sequence types were determined according to the MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
Alleles and STs that had not been previously described were submitted to the curators of the databases and were assigned new designations.
RESULTS
Prevalence of ESBL-producing Enterobacteriaceae in imported vegetables.
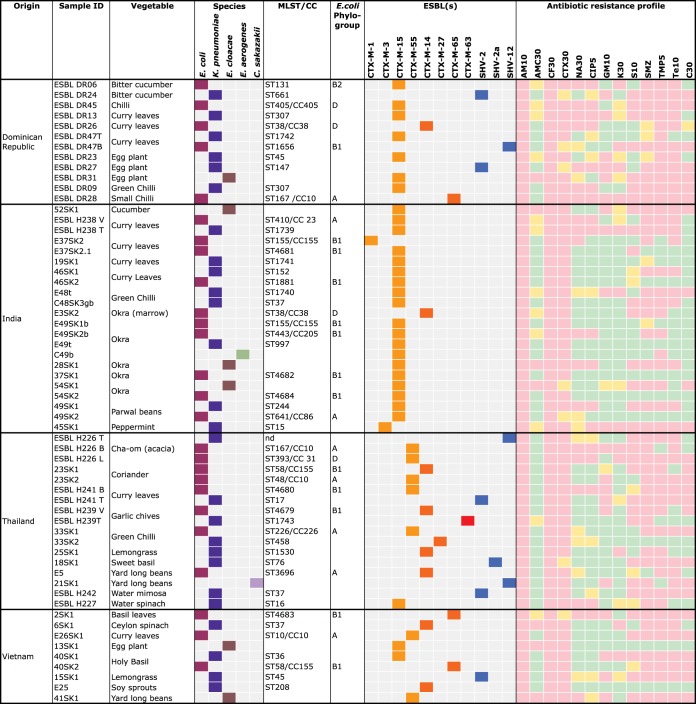
Overall, 43 (25.4%) of the 169 vegetable samples yielded ESBL-producing Enterobacteriaceae. They included 11 (22%) of the 50 samples collected from the Dominican Republic, 13 (33.3%) of the 39 samples from India, 11 (18.3%) of the 60 samples from Thailand, and 8 (40%) of the 20 samples from Vietnam. ESBL producers were detected in 25% of the samples collected at the airport and in 25.7% of the retail store samples. Of the 43 contaminated vegetables, 14 (32.6%) contained multiple isolates (two or more distinct ESBL producers). The types of contaminated vegetables, their origins, and the number of isolates per sample are shown in Fig. 1.
FIG 1.
Source data, identities, and distributions of sequence types, clonal complexes, bla genes, and antibiotic susceptibility patterns of ESBL-producing Enterobacteriaceae isolated from fresh vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. The colors of the squares categorizing ESBLs are as follows: light orange, CTX-M group 1; dark orange, CTX-M group 9; red, CTX-M group 8; blue, SHV enzymes. The colors of the squares categorizing antibiotic resistance profiles are as follows: pink, resistant; yellow, intermediate; green, susceptible. MLST, multilocus sequence type; CC, clonal complex; ID, identifier. The numbers after the drug abbreviations under “Antibiotic resistance profile” represent the amount of drug (in µg) on the discs.
In total, 60 ESBL producers were retrieved. Of these, 26 (43.3%) were identified as E. coli, 26 (43.3%) were classified as Klebsiella pneumoniae subsp. pneumoniae, 6 (10%) were Enterobacter cloacae, 1 (1.7%) was Enterobacter aerogenes, and 1 (1.7%) was C. sakazakii (Fig. 1).
ESBL genes.
All 60 isolates were characterized with respect to their ESBL genotypes. Overall, blaCTX-M genes were detected in 51 (85%) strains. Thirty-eight of the blaCTX-M genes belonged to CTX-M group 1 (74.5% of the blaCTX-M genes) and 12 (23.5%) belonged to CTX-M group 9. One representative of CTX-M group 8 was detected (2%). No genes from CTX-M group 2 were found.
Nine strains were identified as SHV-type ESBL producers. Five (55.6%) were SHV-2, and three (33.3%) were SHV-12. One (11.1%) SHV-2a producer was detected.
Overall, of the 26 E. coli isolates, 17 (65.8%) E. coli strains produced CTX-M group 1 ESBLs and 8 (30.8%) produced CTX-M group 9 ESBLs. Ten (38.5%) harbored blaCTX-M-15, six (23%) blaCTX-M-55, five (19.2%) blaCTX-M-14, and three (11.5%) blaCTX-M-65. One isolate (3.8%) tested positive for blaCTX-M-1, and one (3.8%) harbored SHV-12.
Of the 26 K. pneumoniae isolates, 14 (53.8%) K. pneumoniae strains produced CTX-M group 1 ESBLs and 5 produced (19.2%) CTX-M group 9 ESBLs. One isolate (3.8%) produced a CTX-M group 8 ESBL, 13 (50%) harbored blaCTX-M-15, and 3 (11.5%) carried blaCTX-M-14. One isolate (3.8%) harbored blaCTX-M-3, one blaCTX-M-27, and one blaCTX-M-63.
Five K. pneumoniae (19.2%) isolates harbored SHV-2, one (3.8%) carried SHV-2a, and one carried SHV-12. As an incidental finding, it was noted that the non-ESBL genes blaSHV-26 and blaSHV-36 and blaLEN-like and blaOKP-5a-like genes were present in several K. pneumoniae isolates (data not shown).
The E. cloacae isolates harbored the CTX-M group 1 gene blaCTX-M-15 in five cases (83.3%). One isolate carried blaCTX-M-55.
The E. aerogenes isolate harbored blaCTX-M-15, and the C. sakazakii isolate carried blaSHV-2.
Regarding the geographical distribution of the ESBLs, CTX-M group 1 enzymes were detected in 7 of 12 isolates (58.3%) from the Dominican Republic and in 19 of 22 isolates (86.3%) from India. In both countries, CTX-M-15 was the predominant enzyme. In contrast, CTX-M group 9 enzymes were detected more frequently from isolates from Thailand and from Vietnam (5 of 17 isolates [29.4%] and 4 of 9 isolates [44.4%], respectively).
Notably, none of the isolates originating from India contained any SHV ESBLs.
Antimicrobial susceptibility patterns.
Disc diffusion tests showed that all 60 isolates were resistant to ampicillin and to the narrow-spectrum cephalosporin cephalothin. Resistance to cefotaxime was noted for 53 (88.3%) of the isolates.
Disc diffusion tests performed for other categories of antibiotics revealed that 19 (31.7%) isolates were resistant to the quinolone antibiotic nalidixic acid and 18 (30%) were resistant to the fluoroquinolone ciprofloxacin. Resistance to aminoglycosides was detected in 20 (33.3%) isolates resistant to gentamicin, 12 (20%) resistant to kanamycin, and 31(51.7%) resistant to streptomycin. Resistance to the folate pathway inhibitors sulfamethoxazole and trimethoprim was noted in 44 (73.3%) isolates and 45 (75%) isolates, respectively. Tetracycline resistance was found in 39 (65%) and chloramphenicol resistance in 28 (46.7%) isolates, respectively.
Multidrug resistance was detected in 47 (78.3%) of the isolates: 11 isolates (91.6%) from the Dominican Republic, 13 (59%) of the 22 isolates from India, 16 (94%) of the 17 isolates originating from Thailand, and 7 (77.8%) of the 9 strains from Vietnam.
Epidemiological characteristics of E. coli and K. pneumoniae isolates. (i) Phylogenetic groups and MLST of E. coli.
Phylogenetic typing allocated 21 (80.8%) of the E. coli isolates to group A or B1, which typically contain commensal E. coli strains. Five isolates (19.2%) belonged to extraintestinal pathogenic phylogroups B2 and D (one and four isolates, respectively).
Multilocus sequence typing of the 26 E. coli isolates identified 22 different sequence types (Fig. 1). There were four new allelic combinations (isolates E37SK2.1, 37SK1, ESBL H241 B, and ESBL H239 V). Two isolates contained new allelic variants of the fumC and recA genes: isolate 54SK2 with ST4684 (fumC, allele number 604) and isolate 2SK1 ST4683 (recA, allele number 326).
Among the pathogenic groups B2 and D, four isolates belonged to the epidemiologically important sequence types ST131, ST405, and ST38: isolate ESBL DR06 was assigned to the internationally disseminated CTX-M-15-producing B2:ST131 clone; isolate ESBL DR45 belonged to D:ST405, which belongs to the clonal complex CC405; and two isolates, ESBL DR26 and E3SK2, were identified as D:ST38, which belongs to the clonal complex CC38. Since this clone may include enteroaggregative E. coli strains, these two isolates were tested by PCR for the EAEC-specific marker gene aggR. The results revealed that one isolate (E3SK2) belonged to the extraintestinal EAEC D:ST38 lineage. One further isolate (ESBL H226 L) was classified as D:ST393.
(ii) MLST of K. pneumoniae.
Multilocus sequence typing revealed high diversity among the 26 K. pneumoniae isolates. Five isolates exhibited new sequence types (isolates ESBL H238T, E48T, 19SK1, ESBL DR47T, and ESBL H239T). Two isolates, 45SK1 and ESBL DR27, belonged to epidemic clones ST15 and ST147, respectively. For isolate ESBL H226 T, the ST could not be determined, because the mdh gene could not be amplified.
DISCUSSION
Recent studies indicate that fresh vegetables constitute a source of ESBL producers and represent a possible route for the dissemination of resistance genes via the consumer in the community (27–29).
Vegetable crops originating from most European and North American countries are farmed according to regulations for applying manure/slurry to protect vegetables from contamination with pathogenic microorganisms, in accordance with the recommendations of the World Health Organization (WHO) (30). Consequently, carriers of blaESBL and multidrug resistance genes associated with vegetables have been described as predominantly saprophytic and opportunistic bacteria, which are thought to constitute a background reservoir of antibiotic resistance genes (31) and not a threat per se to human health.
In this study, we examine the presence of ESBL-producing Enterobacteriaceae in fresh vegetables imported into Switzerland from countries with very different farming standards and where the food production industry is to a certain extent underdeveloped.
The high rate of contamination (average, 25.4%) of the samples with ESBL producers and the very high rate (78.3%) of MDR Enterobacteriaceae detected in this study give rise to concern. These results contrast strongly with results from similar studies that reported lower prevalences (6% to 12%), of ESBL producers in raw vegetables (27, 28, 32).
We found national variations among the CTX-M types identified in the samples. The predominance of blaCTX-M-15 genes in isolates from India is in accordance with previous studies involving clinical isolates originating from Delhi and south India, and the frequency of group 9 CTX-M types in isolates from Thailand and Vietnam is reflective of reports from China and the Far East (3, 33). In the isolates from Thailand analyzed in this study, CTX-M-55 outnumbered CTX-M-14. Originally detected in clinical isolates of E. coli and K. pneumoniae from Thailand in 2005 (34), this particular ESBL type has been found widely in food-producing animals and humans in China and appears to be displacing CTX-M-14 as the most common CTX-M variant (35). Our data indicate that this epidemiological characteristic may hold true for Thailand and also for Vietnam. In comparison, the CTX-M type distribution of ESBL producers isolated from healthy humans in Switzerland is dominated by CTX-M-15 and CTX-M-1 (36).
The predominance of phylogenetic groups A and B1 among the E. coli isolates and the wide diversity of multilocus sequence types among the E. coli and K. pneumoniae isolates indicate that blaESBL and MDR genes are well established in commensal strains. It is already recognized that commensal bacteria constitute an important reservoir of antibiotic resistance genes in food animals (37). Our results suggest that vegetables of the types and origins analyzed in this study represent another potent and hitherto underappreciated source of antibiotic resistance genes.
The occurrence of pathogenic bacteria in food is a threat to public health. In this study, we found a CTX-M-15-producing isolate belonging to the highly virulent pandemic E. coli strain B2:ST131, which is associated with severe infections in humans (38). Furthermore, one E. coli D:ST405, two E. coli D:ST393, and two E. coli D:ST38 strains were found in this study. These strains also belong to lineages that cause extraintestinal diseases, mainly urinary tract infections, in humans and contribute to the global dissemination of ESBLs and MDR genes (6, 39). The detection of enteroaggregative properties in one of the E. coli D:ST38 strains is of particular concern. EAEC is associated with acute or persistent diarrhea in outbreak and nonoutbreak settings worldwide. Its association with CTX-M-14 has been described recently in Europe (40, 41), as well as in Asia (42), and its detection in vegetables destined for human consumption raises questions concerning food safety.
C. sakazakii is an opportunistic foodborne pathogen that can cause fatal necrotizing enterocolitis, bacteremia, and meningitis in infants and immunocompromised adults (43), and its detection, for the first time to our knowledge, as an SHV-12 producer in a vegetable sample from Thailand merits attention. Previously, a clinical isolate of C. sakazakii harboring blaVEB-1, a blaESBL gene found increasingly in Thailand, was reported (22). However, the isolate in this study tested negative for this particular gene.
Among the K. pneumoniae isolates detected in this study, two (45SK1 and ESBL DR27) belonged to epidemic clones associated with nosocomial infections in humans (44, 45), giving rise to further concern for consumer health.
In conclusion, the results of this study suggest that the international production of and trade in fresh vegetables constitute a possible route for the spread of ESBLs and pathogenic Enterobacteriaceae. Appropriate measures, such as the improvement of agricultural practices and water quality, need to be taken, and globally mandatory guidelines should be established in order to ensure consumer and public health worldwide.
ACKNOWLEDGMENTS
This work was partly supported by the Swiss Federal Office of Public Health, Division of Communicable Diseases.
We thank the team of curators of the Institut Pasteur MLST (Klebsiella) and the MLST database at the University of Warwick (E. coli) for importing novel alleles and Beatriz Winkenbach for technical assistance.
REFERENCES
- 1.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist 17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 4.Cantón R, Coque TM. 2006. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol 9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitout JD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:9. doi: 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naas T, Poirel L, Nordmann P. 2008. Minor extended-spectrum beta-lactamases. Clin Microbiol Infect 14:42–52. doi: 10.1111/j.1469-0691.2007.01861.x. [DOI] [PubMed] [Google Scholar]
- 8.Geser N, Stephan R, Hächler H. 2012. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet Res 8:21. doi: 10.1186/1746-6148-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ, National ESBL Surveillance Group . 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann A, Locatelli A, Amoureux L, Depret G, Jolivet C, Gueneau E, Neuwirth C. 2012. Occurrence of CTX-M producing Escherichia coli in soils, cattle, and farm environment in France (Burgundy Region). Front Microbiol 3:83. doi: 10.3389/fmicb.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Althaus D, Hofer E, Corti S, Julmi A, Stephan R. 2012. Bacteriological survey of ready-to-eat lettuce, fresh-cut fruit, and sprouts collected from the Swiss market. J Food Prot 75:1338–1341. doi: 10.4315/0362-028X.JFP-12-022. [DOI] [PubMed] [Google Scholar]
- 12.Jiménez B. 2006. Irrigation in developing countries using wastewater. Int Rev Environ Strateg 6:229–250. [Google Scholar]
- 13.Raschid-Sally L, Jayakody P. 2009. Drivers and characteristics of wastewater agriculture in developing countries: results from a global assessment. International Water Management Institute, Colombo, Sri Lanka. [Google Scholar]
- 14.Chevalley M. The fruit and vegetable market in Switzerland. Overview of the market and access information for international trading companies. http://www.swisscofel.ch. [Google Scholar]
- 15.Mollet C, Drancourt M, Raoult D. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol 26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 16.Stoop B, Lehner A, Iversen C, Fanning S, Stephan R. 2009. Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter. Int J Food Microbiol 136:165–168. doi: 10.1016/j.ijfoodmicro.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Boisen N, Scheutz F, Rasko DA, Redman JC, Persson S, Simon J, Kotloff KL, Levine MM, Sow S, Tamboura B, Toure A, Malle D, Panchalingam S, Krogfelt KA, Nataro JP. 2012. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis 205:431–444. doi: 10.1093/infdis/jir757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renvoisé A, Decré D, Amarsy-Guerle R, Huang TD, Jost C, Podglajen I, Raskine L, Genel N, Bogaerts P, Jarlier V, Arlet G. 2013. Evaluation of the βLacta test, a rapid test detecting resistance to third-generation cephalosporins in clinical strains of Enterobacteriaceae. J Clin Microbiol 51:4012–4017. doi: 10.1128/JCM.01936-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement, CLSI document M100-S23. CLSI, Wayne, PA. [Google Scholar]
- 20.Pitout JD, Thomson KS, Hanson ND, Ehrhardt AF, Moland ES, Sanders CC. 1998. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob Agents Chemother 42:1350–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother 57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 22.Girlich D, Poirel L, Leelaporn A, Karim A, Tribuddharat C, Fennewald M, Nordmann P. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum beta-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J Clin Microbiol 39:175–182. doi: 10.1128/JCM.39.1.175-182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JR, Delavari P, Kuskowski M, Stell AL. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 25.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raphael E, Wong LK, Riley LW. 2011. Extended-spectrum beta-lactamase gene sequences in gram-negative saprophytes on retail organic and nonorganic spinach. Appl Environ Microbiol 77:1601–1607. doi: 10.1128/AEM.02506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuland EA, Al Naiemi N, Raadsen SA, Savelkoul PH, Kluytmans JA, Vandenbroucke-Grauls CM. 2014. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur J Clin Microbiol Infect Dis 33:1843–1846. doi: 10.1007/s10096-014-2142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwaiger K, Helmke K, Hölzel CS, Bauer J. 2011. Antibiotic resistance in bacteria isolated from vegetables with regards to the marketing stage (farm vs. supermarket). Int J Food Microbiol 148:191–196. doi: 10.1016/j.ijfoodmicro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. 2006. WHO guidelines for the safe use of wastewater, excreta and greywater. vol 4 Excreta and greywater use in agriculture. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 31.Marti R, Scott A, Tien YC, Murray R, Sabourin L, Zhang Y, Topp E. 2013. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl Environ Microbiol 79:5701–5709. doi: 10.1128/AEM.01682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mesa RJ, Blanc V, Blanch AR, Cortés P, González JJ, Lavilla S, Miró E, Muniesa M, Saco M, Tórtola MT, Mirelis B, Coll P, Llagostera M, Prats G, Navarro F. 2006. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage). J Antimicrob Chemother 58:211–215. doi: 10.1093/jac/dkl211. [DOI] [PubMed] [Google Scholar]
- 33.Hawkey P. 2008. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect 14:159–165. doi: 10.1111/j.1469-0691.2007.01855.x. [DOI] [PubMed] [Google Scholar]
- 34.Kiratisin P, Apisarnthanarak A, Saifon P, Laesripa C, Kitphati R, Mundy LM. 2007. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn Microbiol Infect Dis 58:349–355. doi: 10.1016/j.diagmicrobio.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Zheng B, Zhao L, Wei Z, Ji J, Li L, Xiao Y. 2014. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis 14:659. doi: 10.1186/s12879-014-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geser N, Stephan R, Korczak BM, Beutin L, Hächler H. 2012. Molecular identification of extended-spectrum-β-lactamase genes from Enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob Agents Chemother 56:1609–1612. doi: 10.1128/AAC.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Food Safety Authority. 2012. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2010. EFSA J 10:2598. doi: 10.2903/j.efsa.2012.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 39.Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Blanco JE, Alonso MP, García-Garrote F, Chaves F, Orellana MÁ, Martínez-Martínez L, Calvo J, Prats G, Larrosa MN, González-López JJ, López-Cerero L, Rodríguez-Baño J, Pascual A. 2011. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother 66:2011–2021. doi: 10.1093/jac/dkr235. [DOI] [PubMed] [Google Scholar]
- 40.Chattaway MA, Jenkins C, Ciesielczuk H, Day M, DoNascimento V, Rodríguez I, van Essen-Zandbergen A, Schink AK, Wu G. 2014. Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg Infect Dis 20:1935–1937. doi: 10.3201/eid2011.131845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nüesch-Inderbinen MT, Hofer E, Hächler H, Beutin L, Stephan R. 2013. Characteristics of enteroaggregative Escherichia coli isolated from healthy carriers and from patients with diarrhoea. J Med Microbiol 62:1828–1834. doi: 10.1099/jmm.0.065177-0. [DOI] [PubMed] [Google Scholar]
- 42.Kim JS, Kim SJ, Oh KH, Kang YH, Chung GT. 2014. Characterization of CTX-M-type extended-spectrum beta-lactamase-producing diarrheagenic Escherichia coli isolates in the Republic of Korea during 2008-2011. J Microbiol Biotechnol 24:421–426. doi: 10.4014/jmb.1401.01023. [DOI] [PubMed] [Google Scholar]
- 43.Healy B, Cooney S, O'Brien S, Iversen C, Whyte P, Nally J, Callanan JJ, Fanning S. 2010. Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog Dis 7:339–350. doi: 10.1089/fpd.2009.0379. [DOI] [PubMed] [Google Scholar]
- 44.Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. 2011. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents 38:160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigues C, Machado E, Ramos H, Peixe L, Novais A. 2014. Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIK). Int J Med Microbiol 304:1100–1108. doi: 10.1016/j.ijmm.2014.08.003. [DOI] [PubMed] [Google Scholar]