Abstract
Fifty-nine monophasic Salmonella enterica serovar Typhimurium isolates, collected in Belgium during the period from 2008 to 2011, have been serotyped as 4,[5]:i:− and shown to harbor an fljB coding sequence. The genetic differences between these strains and phenotypically biphasic Salmonella Typhimurium were analyzed through PCR and DNA sequencing. Genetic alterations in the fljB promoter region affecting expression of the phase 2 flagellin were observed in 53 isolates. Other genetic events in the invertible region carrying the fljB promoter were observed in 2 isolates. For the remaining 4 isolates, no molecular differences with a reference biphasic Salmonella Typhimurium strain could be observed. Next-generation sequencing of one representative isolate affected in the fljB promoter region revealed a 26-kb IS26 composite transposon insertion along with a local genomic rearrangement. Several other IS26 element-mediated alterations of this genomic region were observed. This group of monophasic Salmonella Typhimurium isolates was genetically heterogeneous, as revealed by multilocus variable-number tandem-repeat analysis (MLVA), PCR, and sequencing. Pigs and pork represented a major source of such monophasic isolates in Belgium, as reported in other countries. Three out of 5 isolates of human origin presented genetic profiles identical to those of food isolates, demonstrating the pathogenic potential of the newly characterized variants and potential dissemination along the food chain. This study highlighted the key role played by IS26 insertions in the loss of phase 2 flagellin expression and the subsequent generation of multiple monophasic variant lineages from biphasic Salmonella Typhimurium ancestors.
INTRODUCTION
The Salmonella genus is currently divided into 2 species, Salmonella bongori and Salmonella enterica, the latter being classified into 6 subspecies, S. enterica subsp. enterica, salamae, arizonae, diarizonae, houtenae, and indica (1). Most salmonellae possess two flagellin genes, fliC and fljB, encoding the so-called phase 1 and phase 2 flagellins, respectively. Only one flagellin type is expressed at a time in a Salmonella cell. Isolates are termed biphasic, monophasic, or nonmotile if they can express both, one, or no flagellar phases, respectively. Expression of the two genes is under the control of a switch mechanism called “phase inversion.” The switch mechanism consists of the DNA inversion of a sequence segment, the so-called invertible region, by recombination of the two flanking sites, hixL and hixR, mediated by an invertase encoded by the hin gene included in this segment (2). The promoter of the fljBA operon is born on this invertible region. The fljBA operon encodes the FljB flagellin and FljA, a repressor of the fliC gene. The orientation of the invertible region determines whether the fljBA operon or the fliC gene will be expressed (2).
Characterization of Salmonella spp. below the subspecies level is traditionally obtained by serotyping based on the Kauffmann-White-Le Minor scheme (1). Serovars are determined following agglutination of entire bacteria with specific sera to identify variants of the somatic (O) and flagellar (H) antigens. Serotyping results in an antigenic formula, indicating the O, H1, and H2 antigens separated by colons. The different O and H antigens are named by numbers, alphabetical characters, or both. Some O antigens can be present or absent without interfering with serovar identification. These antigens are interesting only as epidemiological markers within a given serovar. These accessory O antigens are underlined when their presence is linked to the integration of a prophage. O or H antigens are written within square brackets if they may be present or absent without relation to phage conversion. The 1,4,[5],12:i:1,2 antigenic formula, corresponding to S. enterica serovar Typhimurium, therefore indicates that (i) the O4 and O12 antigens are always present in this serovar, (ii) the O1 antigen can be present or absent due to phage conversion, (iii) the O5 antigen can be present or absent without relation to phage conversion, and (iv) the H1 antigen “i” and the H2 antigens “1” and “2” are always present. If no H1 or H2 antigen can be detected, a minus sign (−) is written at the corresponding place in the antigenic formula and the strain is termed monophasic.
Monophasic Salmonella Typhimurium strains serotyped as 1,4,[5],12:i:− have emerged in the mid-1990s and account currently for one of the most common serotypes observed in many countries worldwide (3). Since the public health risk posed by Salmonella 1,4,[5],12:i:− strains is considered comparable to that of Salmonella Typhimurium strains (4), epidemiological surveillance and molecular characterization of this serovar should be part of the Salmonella control programs. It has been suggested that the success of Salmonella 1,4,[5],12:i:− variants might be related to the selective advantage offered by multidrug-resistant profiles associated with stable genetic elements, which also carry virulence features. Such variants might be well adapted to the animal production context and capable of causing human infections (5). However, other studies have shown that not all Salmonella 1,4,[5],12:i:− variants are systematically associated with multidrug-resistant profiles (3, 4). The explanation may therefore be more complex and certainly deserves further investigation.
Isolates serotyped as S. enterica subsp. enterica 1,4,[5],12:i:− express only the phase 1 flagellin. They can be monophasic variants of several S. enterica serovars: S. Typhimurium, S. Lagos, S. Agama, S. Farsta, S. Tsevie, S. Gloucester, and S. Tumodi. A PCR assay was developed by Tennant et al. (6) to determine whether an isolate serotyped as 1,4,[5],12:i:− was related to Salmonella Typhimurium or to the rarely observed S. Farsta. This PCR assay targets (i) the fliB-fliA intergenic region hosting an IS200 element found only in S. Typhimurium or S. Farsta or their variants (among serovars expressing the O:4 and H:i antigens) and (ii) fljB, the gene coding for the phase 2 flagellin. Salmonella 1,4,[5],12:i:− isolates related to Salmonella Typhimurium and lacking the fljB coding sequence, as demonstrated by this PCR assay, were recognized as Salmonella Typhimurium monophasic variants by the European Food Safety Authority (EFSA) (4). Beside these isolates, here referred to as fljB-negative Salmonella Typhimurium monophasic variants (STMV ΔfljB strains), Salmonella 1,4,[5],12:i:− strains characterized by a typical Salmonella Typhimurium IS200 signature and the presence of the fljB coding sequence (STMV fljB+ strains) were also reported (7–9). These STMV fljB+ strains are neither phenotypically biphasic nor typical Salmonella Typhimurium monophasic variants like the one recognized by EFSA (4) and the Spanish and U.S. clones described by Soyer et al. (10). Theoretically, the presence of an intact fljB coding sequence is not the unique genetic requirement for phase 2 flagellin expression. A fully operational invertible region, also called the H segment, carrying the hin gene (encoding the main invertase involved in phase inversion), a functional fljB promoter, and the two recombination sites hixL and hixR, required for the inversion mechanism, is also needed (11). In-depth molecular characterization of the entire H segment of such STMV fljB+ isolates has, to our knowledge, not been reported.
In a previous study, 59 Salmonella isolates serotyped as 1,4,[5],12:i:− and collected during the period from 2008 to 2011 at the Belgian Salmonella National Reference Laboratories for Public Health, Animal Health, and Food were reported (9). These isolates were routinely identified by classical serotyping, following at least one phase inversion assay, and were characterized by PCR analysis as STMV fljB+ strains. Such STMV fljB+ variants were also reported elsewhere (7–9). The objectives of the present study were (i) to increase awareness that not just fljB-negative strains are monophasic, (ii) to provide nucleotide sequence material for the development of improved molecular assays targeting most signatures of such strains, and (iii) to stress the need to provide a regulatory context for fljB-positive monophasic S. Typhimurium strains, given the large proportion (23.3% of Belgian monophasic strains) and the public health risk (comparable to biphasic S. Typhimurium) they represent.
MATERIALS AND METHODS
Bacterial isolates.
A collection of 59 Salmonella 4,[5]:i:− isolates, 37 from pig, 2 from poultry, 14 from pork (i.e., pig meat), 1 from unspecified meat, and 5 from human origins, was investigated. They were previously assayed with the PCR test developed by Tenant et al. (6) and identified as fljB-positive Salmonella Typhimurium (9). This STMV fljB+ strain collection includes all strains isolated during the period from 2008 to 2011 at the Belgian Salmonella National Reference Laboratories for Public Health, Animal Health, and Food. They were phenotypically confirmed after at least one phase inversion assay. Strains used for nucleotide sequencing are stored in the Belgian Co-ordinated Collections of Microorganisms (BCCM).
DNA extraction.
The genomic DNA used as a template for PCR or sequencing assays was extracted from bacteria that were grown on brilliant green agar plates (Oxoid, Aalst, Belgium) by using the DNeasy blood and tissue kit according to the manufacturer's instructions for Gram-negative bacteria (Qiagen, Valencia, CA).
PCR. (i) Invertible H segment.
PCR targeting the entire or a portion of the invertible H segment was performed with the FastStart PCR master mix (Roche Diagnostics, Mannheim, Germany) by using different primer pairs. PCRs were conducted in a 25-μl final volume consisting of FastStart PCR master mix, 0.5 μM each primer, 10 ng of template DNA, and Milli-Q water. The amplification procedure consisted of an initial denaturation at 95°C for 10 min, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C, followed by a final extension of 10 min at 72°C.
If the entire invertible H segment of a given fljB-positive STMV isolate was not PCR amplifiable (Table 1, first primer mix) or yielded a PCR amplicon whose size differed from that of the reference Salmonella Typhimurium LT2 strain, two PCRs (Table 1, second and third primer mixes) targeting different portions of the H segment (in phase 2 OFF orientation) were assayed. All PCR products were analyzed by gel electrophoresis in 1% agarose gel (Tris-acetate-EDTA [TAE] buffer) using SYBRSafe (Life Technologies Europe, Ghent, Belgium) as the intercalating dye and a 200-bp to 10-kb DNA ladder (Eurogentec, Seraing, Belgium).
TABLE 1.
Summary of PCRs and primers used to map the 26-kb transposon insertion and other genetic alterations of the invertible H segment
| PCR target | Primer mix (name, nucleotide sequence from 5′ to 3′) | Amplicon size in Salmonella Typhimurium LT2 (bp) | Amplicon size in Salmonella 4,5:i:− VAR-2009/08643/1 (bp) |
|---|---|---|---|
| Entire invertible H segment | BG1132, CAGTGCGGACTGGGATTTGTTCA | 1,200 | No amplicon |
| BG1133, AATGTTGCGAACGAACAGCTCCT | |||
| hixR-hina | BG1132, CAGTGCGGACTGGGATTTGTTCA | 600 | 600 |
| BG1137, AGCCGACTAATCTGTTCCTGT | |||
| hin-fljB promoter-hixLa | BG1136, CAGAGCGCAAGGACGA | 600 | No amplicon |
| BG1133, AATGTTGCGAACGAACAGCTCCT | |||
| Left junction of inserted transposon or intact hin-iroB regiona,b | BG1215, ATACAGGCGTGTGGCATAAATAAACCGA | 600 (with BG1215 and BG1218) | 1,000 (with BG1215 and BG1216) |
| BG1216, GCAATCTGTGCGGCCAGT | |||
| BG1218, CAAGGTAACGGGAAACGTAGGGAGT | |||
| Right junction of inserted transposon or intact hin-iroB regiona,b | BG1218, CAAGGTAACGGGAAACGTAGGGAGT | 600 (with BG1215 and BG1218) | 1,200 (with BG1232 and BG1218) |
| BG1232, TGGCGGGAAATGCGTGTTTACA | |||
| BG1215, ATACAGGCGTGTGGCATAAATAAACCGA | |||
| Left IS26 element of inserted transposona | BG1215, ATACAGGCGTGTGGCATAAATAAACCGA | No amplicon | 600 |
| BG1225, TGAATCGCGGGATCTGCCACTTCT | |||
| Right IS26 element of inserted transposon | BG1218, CAAGGTAACGGGAAACGTAGGGAGT | No amplicon | 500 |
| BG1225, TGAATCGCGGGATCTGCCACTTCT |
A PCR product will be observed with this primer mix only if the H segment is in phase 2 OFF orientation.
Three-primer PCR yielding two possible PCR products.
(ii) Junction PCRs.
As shown in Fig. 1, the insertion of a transposon was observed in the invertible region of a representative STMV fljB+ isolate. Two 3-primer PCR assays were designed to assess the occurrence of this transposon in the other STMV fljB+ strains (Table 1, fourth and fifth primer mixes). These two PCR assays targeted either the left (LJ) or right (RJ) junction of the inserted transposon or the intact fljB promoter region found in the Salmonella Typhimurium LT2 reference (biphasic) strain. The left junction PCR amplifies either the transposon left junction (primers BG1215 and BG1216) in the target fljB+ STMV isolates or the conserved hin-iroB intergenic region in phase 2 OFF orientation (control primers BG1215 and BG1218). The right junction PCR amplifies either the transposon right junction (primers BG1218 and BG1232) or the conserved hin-iroB intergenic region (control primers BG1215 and BG1218). Both PCRs were conducted in a 25-μl final volume consisting of FastStart PCR master mix, 0.5 μM each primer, 10 ng of template DNA, and Milli-Q water. The amplification procedure consisted of an initial denaturation at 95°C for 10 min followed by 15 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C, followed by another 15 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C (for the last step, with 5-s increment at each cycle), followed by a final extension of 10 min at 72°C. PCR products were analyzed by gel electrophoresis in 1% agarose gel (TAE buffer) by using SYBRSafe (Life Technologies Europe, Ghent, Belgium) as the intercalating dye and a 200-bp to 10-kb DNA ladder (Eurogentec, Seraing, Belgium). Where no amplification was observed with either the left or the right junction PCR nor with the control primers BG1215 and BG1218, PCRs targeting just the IS26 element either at the left (BG1215 and BG1225) or right (BG1218 and BG1225) junction were performed in the same conditions as for the transposon junction PCRs. Amplification of the conserved hin-iroB intergenic region was designed to occur only if the tested strain was in phase 2 OFF orientation (Fig. 1, Table 1).
FIG 1.
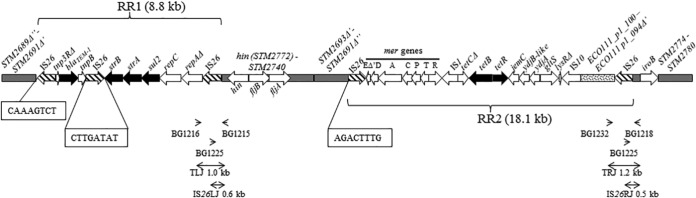
Map of the fljBA operon region of the monophasic variant strain VAR-2009/08643/1 (83,173 bp). IS26 copies are drawn with hatched arrows. Black arrows delineate antibiotic resistance genes. Gray boxes indicate regions homologous to Salmonella Typhimurium LT2 (GenBank accession number AE006468). The dotted box is homologous to a region found on the E. coli plasmid pO111_1 (GenBank accession number AP010961). Antibiotic resistance regions 1 (RR1) and 2 (RR2) are similar to a Salmonella 4,[5],12:i:− chromosomal sequence (GenBank accession number HQ331538) except for (i) a deletion covering partial merE, urf2, and tniAΔ1, (ii) an inverted IS26 element, (iii) a complete IS10 copy, and (iv) an additional region (dotted box) in RR2. Δ′ and Δ″ refer to deletions noticed in the 3′ or 5′ part of some genes, respectively. Boxed nucleotides refer to IS26-associated repeats. The insertion of a 26-kb transposon consisting of the RR1 and RR2 elements occurred at an intermediate stage and was followed by a genomic rearrangement between the first and fourth IS26 copies, leading to an inverted DNA sequence in between. This is supported by the finding of inverted repeats bordering the first and fourth IS elements (boxed nucleotides) and the orientation of the genes within the inverted segment (gray boxes). The arrowheads at the bottom point to primers used to demonstrate the presence of IS26 elements (possibly as part of the 26-kb transposon). Double-headed arrows show the corresponding amplicons with their sizes (TLJ, transposon left junction; IS26LJ, IS26 left junction; TRJ, transposon right junction; IS26RJ, IS26 right junction).
PCR amplicon sequencing.
Sanger sequencing was completed on PCR fragments amplified from strains displaying atypical genetic profiles (listed in Table S1 in the supplemental material) by using the primers listed in Table 1. Sequencing reaction mixtures were obtained using the BigDye Terminator v3.1 cycle sequencing kit and the BigDye XTerminator purification kit. Analyses were conducted on an ABI 3130 genetic analyzer according to the instructions of the manufacturer (Applied Biosystems, Foster City, CA).
Next-generation sequencing.
Genomic DNA of the Salmonella 4,5:i:− VAR-2009/08643/1 isolate was extracted as explained above. A sequencing library was prepared from the extracted DNA according to the manufacturer's instructions for the Titanium Series rapid library preparation method (Roche, Mannheim, Germany). Sequencing was performed with a GS Junior instrument (Roche, Mannheim, Germany) with Titanium Series reagents and run protocol (200 cycles), generating 97,082 reads with an average read length of 480 bases. Three hundred ninety-seven contigs were generated by de novo assembly by using GS De Novo Assembler software v2.7 (Roche, Mannheim, Germany) with the default parameters. This analysis also produced a contig graph indicating potential contig links. Since repeated elements were found in the genomic region of interest (surrounding the fljB promoter region), contigs were joined by PCRs to assemble an 83,173-bp sequence (GenBank accession number KJ999732) (primers are specified in Table S2 in the supplemental material). This sequence was annotated according to the published sequences of Salmonella Typhimurium LT2 (GenBank accession number AE006468) (12), the pO111_1 plasmid of Escherichia coli O111:H− strain 11128 (GenBank accession number AP010961.1) (13), and Salmonella 1,4,[5],12:i:− strain 105/7/03 (GenBank accession number HQ331538) (14). Due to the inherent limits of the technology used for whole-genome sequencing, four loci with homopolymers apparently different from the Salmonella Typhimurium LT2 and pO111_1 plasmid sequences were corrected by Sanger sequencing (nucleotides 6996 to 6999, 18916 to 18921, 19376 to 19381, and 24165 to 24171 of GenBank accession number KJ999732).
MLVA.
Multilocus variable-number tandem-repeat analyses (MLVA) were conducted as described by Lindstedt et al. (15), following the nomenclature proposed by Larsson et al. (16). Allele assignment was performed using a correction table created with 33 calibration strains provided by the European Centre of Disease Prevention and Control (17). MLVA profiles are reported as a string of 5 numbers (STTR9-STTR5-STTR6-STTR10-STTR3), representing the number of repeats at the corresponding locus, or as “NA” when no amplification product was obtained.
Nucleotide sequence accession numbers.
All sequences used in this study were sent to GenBank, and accession numbers KJ999729 to KJ999733 were assigned.
RESULTS
Fifty-nine Salmonella isolates serotyped as 4,[5]:i:− previously reported as STMV fljB+ strains (9) and isolated in Belgium during the period from 2008 to 2011 were investigated to decipher the molecular differences between phenotypically biphasic Salmonella Typhimurium and these monophasic variants. They were isolated at different stages of the food chain: pigs (37 isolates), poultry (2 isolates), pork (14 isolates), unspecified meat (1 isolate), and humans (5 isolates). Thirty-five isolates were serotyped as Salmonella 4:i:− isolates and 24 as Salmonella 4,5:i:− isolates (only the O:4 and O:5 antigens were tested).
At the first stage, the invertible H segment carrying the fljB promoter was targeted by PCR with different primer pairs, allowing the amplification of various parts of this region in the 39 STMV fljB+ animal isolates to map possible insertions or deletions compared to the Salmonella Typhimurium LT2 reference genome. Part of the H segment, carrying the fljB promoter and hixL, could not be amplified in 33/39 STMV fljB+ animal isolates (84.6%). In two isolates (5.1%), a genetic event had occurred at the other side of the invertible H segment carrying hixR and the 5′ part of hin, while in two other isolates (5.1%), PCRs indicated genetic alteration encompassing the entire invertible H segment. No sequence length alteration could be identified in the PCR profiles generated from the last two animal isolates (5.1%) compared to that of Salmonella Typhimurium LT2.
One representative isolate (VAR-2009/08643/1) of the largest variant population displaying a profound alteration in the DNA sequence around the fljB promoter was fully sequenced (GenBank accession number KJ999732). Comparison with the Salmonella Typhimurium LT2 genome (GenBank accession number AE006468) identified a 26-kb IS26 composite transposon insertion in this region, along with a 451-bp deletion encompassing the fljB promoter and hixL. This transposon consists of two resistance regions (Fig. 1) carrying several antibiotic resistance genes involved in resistance to ampicillin (blaTEM-1), streptomycin (strA, strB), sulfonamides (sul2) (region RR1), and tetracycline (tetB and tetR) (region RR2). Phenotypical resistance to these four antibiotics (known as the ASSuT profile) was confirmed using the Kirby-Bauer disc diffusion method by following recommendations of the Clinical and Laboratory Standards Institute. In addition to this IS26 composite transposon insertion, the fljB promoter-surrounding region of strain VAR-2009/08643/1 still differs from the reference genome of Salmonella Typhimurium LT2 by a 40-kb sequence inversion involving the whole fljBA operon, with only part of the invertible H segment. More precisely, after insertion of the transposon, which is supposed to have occurred without further rearrangement in an ancestor lineage not formally identified in our study, inversion of a segment bordered by the first and fourth IS26 copies subsequently occurred, as suggested by the presence of inverted repeats bordering the IS26 elements found at the edges of the inverted sequence and by the orientation of the genes born on the inverted sequence element (Fig. 1). This genomic rearrangement resulted in the dissociation of the two resistance regions. Eventually, a 451-bp deletion encompassing the fljB promoter and hixL occurred which irreversibly canceled FljB flagellin expression.
Two PCR assays were designed to assess the occurrence of this “new” genetic profile in STMV fljB+ strains isolated from animals, food, and humans. These PCRs targeted the left or right transposon junctions, i.e., segments encompassing distal parts of the composite transposon, and the fljB promoter region from the original (intact) genome. A control amplification primer was added to each PCR assay to amplify the intact fljB promoter region in strains where no mobile element had inserted at that position. Amplification of both left and right transposon junctions (PCR profile “TLJ, TRJ” in Table S1 in the supplemental material) was observed for 31 isolates, 1 of human, 21 of pig, 1 of poultry, 7 of pork, and 1 of unspecified meat origin (Table 2; see also Table S1). In 12 isolates (9 from pigs and 3 from pork), an IS26 element, alone or as part of the composite transposon, was detected at the left junction along with different profiles at the right junction (Table 2; see also Table S1). One IS26 copy, combined with either a 482-bp or 536-bp deletion encompassing the fljB promoter and hixL, was observed in 1 isolate of pig origin and 2 isolates of human and pork origins (GenBank accession numbers KJ999731, KJ999730, and KJ999729) (Fig. 2 and Table 2; see also Table S1). Thus, in 78.0% (46/59) of the isolates, an IS26 element either found alone or as part of a composite transposon was inserted between the hin gene and the fljB promoter in phase 2 OFF orientation, affecting expression of the phase 2 flagellin gene. In 7 other isolates, a genetic alteration in the fljB promoter region was observed by PCRs targeting the invertible H segment, but no IS26 or transposon was identified at this place with the junction PCRs (Table 2). In total, genetic alterations in the fljB promoter region affecting the expression of the phase 2 flagellin were observed in 53 isolates. For 2 isolates, a genetic alteration was observed in the other part of the H segment (Table 2). For the remaining 4 isolates, no molecular differences with Salmonella Typhimurium LT2 could be observed in the invertible H segment. Overall, 17 different genetic profiles were identified by different PCR and sequencing reactions among the 59 STMV fljB+ isolates (see Table S1).
TABLE 2.
Summary of genetic alterations supporting the monophasic phenotype found in the invertible H segment of the 59 fljB-positive Salmonella 1,4,[5],12:i:− isolates collected during the period from 2008 to 2011 in Belgium
| Genetic alteration | Transposable element found in the invertible H segment | PCR profilea associated with the genotype (amplicon size[s] in kb) | No. of isolates |
|---|---|---|---|
| 451-bp deletion encompassing the fljB promoter and hixL | 26-kb IS26 composite transposonb inserted between hin and iroB | Amplification of both left and right transposon junctions (1.0 and 1.2) | 31 |
| Disruption of the invertible H segment between hin and the fljB promoter | IS26 element associated or not with a transposon inserted between hin and the fljB promoter | Amplification of the left IS26 element of the inserted transposon (0.6) | 12 |
| 536-bp deletion encompassing the fljB promoter and hixL | One single IS26 copy inserted between hin and iroB | Atypical fragment observed with the two 3-primer junction PCR assays (0.9) | 2 |
| 482-bp deletion encompassing the fljB promoter and hixL | One single IS26 copy inserted between hin and iroB | Atypical fragment observed with the two 3-primer junction PCR assays (1.0) | 1 |
| Other genetic alteration in the fljB promoter region | Not found | No part of the invertible H segment amplified | 6 |
| Only the hixR-hin portion of the invertible H segment amplified (0.6) | 1 | ||
| Genetic alteration in the H segment part containing hixR and a portion of hin | Not found | Only the hin-hixL portion of the invertible H segment amplified (0.6) | 2 |
| Not found | Not found | Entire invertible H segment amplified (1.2) | 4 |
Profiles obtained when performing the invertible H segment and junction PCRs described in Materials and Methods.
This is the 26-kb composite transposon identified in this study and to which GenBank entry KJ999732 refers.
FIG 2.
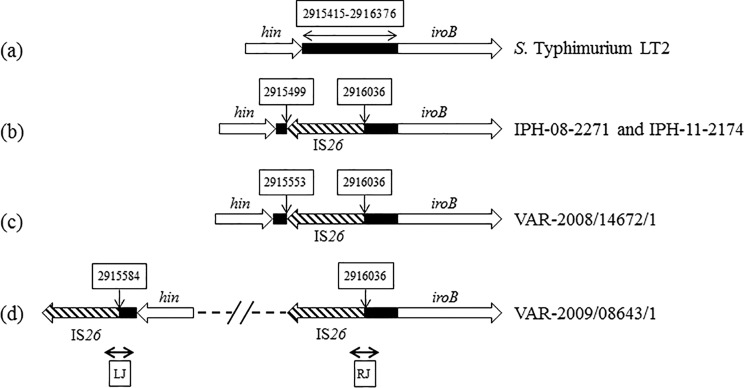
Map of the hin-iroB intergenic sequence of various monophasic variants. The intergenic sequence lying between the hin and iroB genes and observed in the Salmonella Typhimurium LT2 reference strain as well as in some representative isolates of the monophasic variants described in this work is shown as a black box. IS26 elements are indicated by hatched arrows. Nucleotide positions in the intergenic sequence bordering IS26 elements are given in white boxes above vertical arrows and follow the nucleotide sequence numbering of Salmonella Typhimurium LT2 (AE006468). Compared to S. Typhimurium LT2 (panel a), the hin-iroB intergenic sequence was deleted by 536 (panel b), 482 (panel c), or 451 (panel d) bp. LJ and RJ indicate the left and right junctions, respectively, amplified by the junction PCRs described in Materials and Methods. The dotted line in panel d delineates the large (56.6-kb) intervening region detailed in Fig. 1.
MLVA was then performed to complete the molecular characterization of these isolates. Twenty-one different MLVA subtypes were observed among the 59 isolates. The STTR10 locus could not be detected in any of the strains under study, and the STTR9 locus showed no variability (see Table S1 in the supplemental material). A Simpson diversity index of 0.900 was associated with STMV fljB+ isolates and was slightly lower than the one associated with STMV ΔfljB isolates (0.942) recovered during the same period in Belgium (9). Thirteen out of all STMV fljB+ isolates (22.0%), 12 of pig and 1 of pork origin, displayed the same MLVA profile (3-13-10-NA-0211), junction PCR profile (“TLJ, TRJ”), and antigenic formula (4:i:−) (see Table S1). Other small clusters of strains, including up to 4 isolates and displaying the same antigenic and molecular typing characteristics, were also observed.
DISCUSSION
This study revealed several genotypes supporting the monophasic phenotype of STMV fljB+ isolates. In 78.0% (46/59) of the isolates, the insertion of an IS26 element, either alone or as part of a composite transposon, between the hin gene and the fljB promoter affected expression of the phase 2 flagellin gene. Nonexpression of fljB in 9 isolates could be explained by other genetic alterations in the fljB promoter or in the other part of the invertible region. Future whole-genome sequencing studies could help to decipher the nature of the genetic events associated with the lack of phase 2 flagellin expression in the 4 remaining isolates.
The insertion of one IS26 element at nucleotide position 2916036 (Salmonella Typhimurium LT2 reference genome) in the hin-iroB intergenic region illustrated in Fig. 2b, c, and d was observed in 47 isolates (PCR profiles detailed in Table S1 in the supplemental material) and was reminiscent of previously characterized Salmonella Typhimurium monophasic strains (14) (C. Lucarelli, personal communication) (GenBank accession number GU226187). However, the genomic sequence at the other side of the inserted IS26 element in these 47 isolates was characterized by various deletions, yielding genetically distinct groups within the above-described general category. This suggests the existence of a common ancestor harboring a single IS26 copy at position 2916036 with no further genomic deletion or rearrangement event. This ancestor would be a phenotypically biphasic strain which would have generated different STMV lineages after the insertion of another IS26 copy (possibly as part of a composite transposon) at several positions, followed by genetic recombination between both IS26 copies and deletion of the sequence in between. These deletions suppressed the invertible character of the fljB promoter region and rendered the strains phenotypically monophasic. The diverse STMV fljB+ population investigated in this study and characterized by numerous PCRs and MLVA profiles would therefore not represent a single lineage but rather a diversity of lineages originating from a few common ancestors.
Our study thus highlighted the key role played by IS26 insertions in the loss of genetic functionality required for phase 2 flagellin expression. In addition, IS26 composite transposon insertion in some monophasic variants caused acquisition of antibiotic resistance genes in the bacterial chromosome, illustrating again the importance of IS26 in the dissemination of antibiotic resistance genes, as reported by Tremblay (18). The two resistance regions, RR1 and RR2 (Fig. 1), born on an IS26 composite transposon, are 100% identical at the nucleotide level to two regions of the Escherichia coli pO111_1 plasmid (GenBank accession number AP010961.1). This perfect conservation suggests that the IS26 composite transposon is easily transferable and may not be restricted to the Salmonella genus, as proposed earlier (19). This is supported by the occurrence of the RR1 resistance element on the E. coli pO111_1 plasmid and on Tn6029, a transposon inserted in an IncHI2 plasmid isolated from Salmonella Typhimurium. (GenBank accession number GQ150541) (20, 21). The IS26 composite transposon could have been inserted through homologous recombination with IS26 elements already present in the Salmonella strain, as suggested by the absence of paired direct repeats flanking the third and fifth IS26 elements (22) (Fig. 1). Chromosomal integration of closely related RR1 and RR2 resistance elements was also reported at other positions in Salmonella 1,4,[5],12:i:− strain 105/7/03 (GenBank accession number HQ331538) (14) (Fig. 1). The ASSuT resistance profile associated with these resistance genes cannot therefore be used as a phenotypical discrimination marker, since it is frequently observed in both monophasic and biphasic Salmonella Typhimurium isolates (4, 23).
This study also broadens our knowledge on the genetic variability associated with the monophasic character of Salmonella 1,4,[5],12:i:− strains and the genomic plasticity of the region carrying the genetic elements needed for expression of the phase 2 flagellin. Interestingly, the hin-fljB region would have been acquired itself by horizontal gene transfer, as indicated by its low G+C content compared to that of the Salmonella core genome (24). These findings highlight the diversity of STMV isolates and the challenging identification of these isolates in routine diagnosis. Although important for epidemiological traceability, the distinction between phenotypically biphasic and monophasic Salmonella Typhimurium may not be that critical from a public health perspective. Indeed, similar pathogenicity gene repertoires have been reported in monophasic and biphasic Salmonella Typhimurium isolates (25). Our results show that PCR assays targeting the fljB coding sequence are not sufficient to rule in the monophasic character of Salmonella isolates displaying the 1,4,[5],12:i:− antigenic formula. Molecular identification of the genetic alterations in STMV fljB+ isolates supporting/explaining the monophasic phenotype is needed to confirm the classification of these isolates as S. Typhimurium variants and thereby fit to sanitary regulations targeting S. Typhimurium. PCR assays developed in this study targeting portions of the invertible H segment are now available for the identification of such variants. A recommended laboratory scenario may be to first confirm the phylogenetic relatedness of monophasic 1,4,[5],12:i:− isolates to Salmonella Typhimurium (6) and to pinpoint the fljB-positive or -negative genotype of the STMV isolates (4). Eventually, the monophasic character of ΔfljB STMV strains can be validated with the PCR assays reported in the present article, which target the genetic signatures associated with the impaired functionality of the fljB promoter. Given the large number of genetic modifications associated with the monophasic character of Salmonella 1,4,[5],12:i:− strains, multiplexed molecular tests may rapidly become a strong requirement for routine or reference laboratories. Future studies with similar PCRs and sequencing approaches could help to describe currently uncharacterized variants.
Pigs and pork represent major sources of STMV fljB+ isolates collected in Belgium, as also reported in other countries (4, 25). The proportion of isolates originating from pigs and pork among the nonhuman isolates was even higher in STMV fljB+ isolates than in STMV ΔfljB isolates (94.4% [this study] compared to 66.0% [9], respectively). Among the 5 characterized human isolates, 3 displayed PCR and MLVA types identical to those of food isolates (pork or unspecified meat), demonstrating the pathogenic potential of the new variants and their possible dissemination along the food chain. Thus, losing a flagellar phase would not be detrimental for the success of Salmonella Typhimurium as a mammalian pathogen, as discussed elsewhere (26).
In conclusion, this study revealed new Salmonella 1,4,[5],12:i:− genotypes, explaining the lack of phase 2 flagellin expression in isolates harboring an fljB coding sequence. IS26 insertions played a key role in the loss of genetic elements needed for phase 2 flagellin expression, which would have generated several STMV lineages from Salmonella Typhimurium biphasic strains. Genetic diversity observed by PCR highlighted the nonclonality of STMV isolates circulating in Belgium, as observed in other countries (8, 27, 28). New STMV will probably be observed in the future due to the genomic plasticity of the region carrying elements needed for expression of phase 2 flagellin, especially IS26 element-mediated genomic rearrangements. As these new variants were also observed in humans, molecular epidemiological follow-up is essential for tracing such contaminations throughout the food chain, particularly in the pig and pork sector.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed in part by a grant from Belgian Scientific Policy (E1122-P1).
We thank C. Ingabire, M. Van Hessche, H. Vander Veken, D. Vandergheynst, O. Ozhelvaci, M. Thirionet, D. Baeyens, and F. De Cooman for technical assistance. We are also very grateful to C. Lucarelli for sending us unpublished sequences.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00270-15.
REFERENCES
- 1.Grimont PAD, Weill FX. 2007. Antigenic formulae of the Salmonella serovars, 9th ed. Institut Pasteur & WHO Collaborating Centre for Reference and Research on Salmonella, Paris, France. [Google Scholar]
- 2.Bonifield HR, Hughes KT. 2003. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J Bacteriol 185:3567–3574. doi: 10.1128/JB.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Switt AI, Soyer Y, Warnick LD, Wiedmann M. 2009. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:−. Foodborne Pathog Dis 6:407–415. doi: 10.1089/fpd.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EFSA. 2010. Scientific opinion on monitoring and assessment of the public health risk of “Salmonella Typhimurium-like” strains. EFSA J 8(10):1826. doi: 10.2903/j.efsa.2010.1826. [DOI] [Google Scholar]
- 5.Mourão J, Machado J, Novais C, Antunes P, Peixe L. 2014. Characterization of the emerging clinically-relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:− (monophasic variant of S. Typhimurium) clones. Eur J Clin Microbiol Infect Dis 33:2249–2257. doi: 10.1007/s10096-014-2180-1. [DOI] [PubMed] [Google Scholar]
- 6.Tennant SM, Diallo S, Levy H, Livio S, Sow SO, Tapia M, Fields PI, Mikoleit M, Tamboura B, Kotloff KL, Nataro JP, Galen JE, Levine MM. 2010. Identification by PCR of non-typhoidal Salmonella enterica serovars associated with invasive infections among febrile patients in Mali. PLoS Negl Trop Dis 4:e621. doi: 10.1371/journal.pntd.0000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arguello H, Sorensen G, Carvajal A, Baggesen DL, Rubio P, Pedersen K. 2014. Characterization of the emerging Salmonella 4,[5],12:i:− in Danish animal production. Foodborne Pathog Dis 11:366–372. doi: 10.1089/fpd.2013.1672. [DOI] [PubMed] [Google Scholar]
- 8.Barco L, Longo A, Lettini AA, Cortini E, Saccardin C, Minorello C, Olsen JE, Ricci A. 2014. Molecular characterization of “inconsistent” variants of Salmonella Typhimurium isolated in Italy. Foodborne Pathog Dis 11:497–499. doi: 10.1089/fpd.2013.1714. [DOI] [PubMed] [Google Scholar]
- 9.Boland C, Bertrand S, Mattheus W, Dierick K, Wattiau P. 2014. Molecular typing of monophasic Salmonella 4,[5]:i:− strains isolated in Belgium (2008–2011). Vet Microbiol 168:447–450. doi: 10.1016/j.vetmic.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 10.Soyer Y, Moreno Switt A, Davis MA, Maurer J, McDonough PL, Schoonmaker-Bopp DJ, Dumas NB, Root T, Warnick LD, Grohn YT, Wiedmann M. 2009. Salmonella enterica serotype 4,5,12:i:−, an emerging Salmonella serotype that represents multiple distinct clones. J Clin Microbiol 47:3546–3556. doi: 10.1128/JCM.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kutsukake K, Nakashima H, Tominaga A, Abo T. 2006. Two DNA invertases contribute to flagellar phase variation in Salmonella enterica serovar Typhimurium strain LT2. J Bacteriol 188:950–957. doi: 10.1128/JB.188.3.950-957.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 13.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucarelli C, Dionisi AM, Filetici E, Owczarek S, Luzzi I, Villa L. 2012. Nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in Salmonella Typhimurium and monophasic Salmonella Typhimurium strains. J Antimicrob Chemother 67:111–114. doi: 10.1093/jac/dkr391. [DOI] [PubMed] [Google Scholar]
- 15.Lindstedt BA, Vardund T, Aas L, Kapperud G. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J Microbiol Methods 59:163–172. doi: 10.1016/j.mimet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Larsson JT, Torpdahl M, Petersen RF, Sorensen G, Lindstedt BA, Nielsen EM. 2009. Development of a new nomenclature for Salmonella Typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill 1 4:pii=19174 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19174. [PubMed] [Google Scholar]
- 17.European Centre of Disease Prevention and Control (ECDC). 2011. Laboratory standard operating procedure for MLVA of Salmonella enterica serotype Typhimurium. ECDC, Solna, Sweden: http://ecdc.europa.eu/en/publications/publications/1109_SOP_Salmonella_Typhimurium_MLVA.pdf. [Google Scholar]
- 18.Tremblay S. 2007. Etude moléculaire du recrutement des gènes de résistance aux antibiotiques. M.Sc. thesis Université Laval, Québec, Canada. [Google Scholar]
- 19.Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM. 2012. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog 8:e1002837. doi: 10.1371/journal.ppat.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cain AK, Liu X, Djordjevic SP, Hall RM. 2010. Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb Drug Resist 16:197–202. doi: 10.1089/mdr.2010.0042. [DOI] [PubMed] [Google Scholar]
- 21.Hall RM, Cain AK. 2012. Comment on: nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in Salmonella Typhimurium and monophasic Salmonella Typhimurium strains. J Antimicrob Chemother 67:785. doi: 10.1093/jac/dkr525. [DOI] [PubMed] [Google Scholar]
- 22.Doublet B, Praud K, Bertrand S, Collard JM, Weill FX, Cloeckaert A. 2008. Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob Agents Chemother 52:3745–3754. doi: 10.1128/AAC.00525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuyts V, Mattheus W, De Laminne de Bex G, Wildemauwe C, Roosens NH, Marchal K, De Keersmaecker SC, Bertrand S. 2013. MLVA as a tool for public health surveillance of human Salmonella Typhimurium: prospective study in Belgium and evaluation of MLVA loci stability. PLoS One 8:e84055. doi: 10.1371/journal.pone.0084055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumler AJ, Heffron F. 1998. Mosaic structure of the smpB-nrdE intergenic region of Salmonella enterica. J Bacteriol 180:2220–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser E, Tietze E, Helmuth R, Junker E, Blank K, Prager R, Rabsch W, Appel B, Fruth A, Malorny B. 2010. Pork contaminated with Salmonella enterica serovar 4,[5],12:i:−, an emerging health risk for humans. Appl Environ Microbiol 76:4601–4610. doi: 10.1128/AEM.02991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McQuiston JR, Fields PI, Tauxe RV, Logsdon JM Jr. 2008. Do Salmonella carry spare tyres? Trends Microbiol 16:142–148. doi: 10.1016/j.tim.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Bugarel M, Vignaud ML, Moury F, Fach P, Brisabois A. 2012. Molecular identification in monophasic and nonmotile variants of Salmonella enterica serovar Typhimurium. Microbiologyopen 1:481–489. doi: 10.1002/mbo3.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasyl D, Hoszowski A. 2012. Occurrence and characterization of monophasic Salmonella enterica serovar Typhimurium (1,4,[5],12:i:−) of non-human origin in Poland. Foodborne Pathog Dis 9:1037–1043. doi: 10.1089/fpd.2012.1154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


