Abstract
Leptospirosis, an emerging zoonotic disease, remains poorly understood because of a lack of genetic manipulation tools available for pathogenic leptospires. Current genetic manipulation techniques include insertion of DNA by random transposon mutagenesis and homologous recombination via suicide vectors. This study describes the construction of a shuttle vector, pMaORI, that replicates within saprophytic, intermediate, and pathogenic leptospires. The shuttle vector was constructed by the insertion of a 2.9-kb DNA segment including the parA, parB, and rep genes into pMAT, a plasmid that cannot replicate in Leptospira spp. and contains a backbone consisting of an aadA cassette, ori R6K, and oriT RK2/RP4. The inserted DNA segment was isolated from a 52-kb region within Leptospira mayottensis strain 200901116 that is not found in the closely related strain L. mayottensis 200901122. Because of the size of this region and the presence of bacteriophage-like proteins, it is possible that this region is a result of a phage-related genomic island. The stability of the pMaORI plasmid within pathogenic strains was tested by passaging cultures 10 times without selection and confirming the presence of pMaORI. Concordantly, we report the use of trans complementation in the pathogen Leptospira interrogans. Transformation of a pMaORI vector carrying a functional copy of the perR gene in a null mutant background restores the expression of PerR and susceptibility to hydrogen peroxide comparable to that of wild-type cells. In conclusion, we demonstrate the replication of a stable plasmid vector in a large panel of Leptospira strains, including pathogens. The shuttle vector described will expand our ability to perform genetic manipulation of Leptospira spp.
INTRODUCTION
Leptospirosis, which is caused by one of the 10 pathogenic Leptospira spp. described to date, is a neglected zoonotic disease that has a worldwide distribution with a high incidence in tropical countries. The virulence mechanisms and, more generally, the biology of pathogenic Leptospira spp. remain largely unknown. This hindrance is partly due to a lack of efficient genetic tools available for use in pathogenic Leptospira spp. (1, 2). While genetic modification tools allow flexible manipulation of the genome of the saprophyte Leptospira biflexa, including targeted mutagenesis and cis/trans complementation (2), genetic modification of the pathogen is limited primarily to random transposon mutagenesis.
Previously, genetic analysis of Leptospira was impeded by the absence of methods for the introduction of DNA into leptospiral cells. Currently, DNA can be introduced into Leptospira spp. by electroporation (3) or conjugation between Escherichia coli and Leptospira spp. by using RP4 derivative conjugative plasmids (4). Transformed Leptospira can be visualized on solid medium as subsurface colonies after 1 week for saprophytes and up to 4 weeks for pathogens. Markers for the selection of transformants include kanamycin, spectinomycin, and gentamicin resistance cassettes (3, 5, 6). The replication origins of L. biflexa phage LE1 (3), L. biflexa plasmid p74 (7), and a phage-related genomic island from L. interrogans (8) have previously been used to construct L. biflexa-E. coli plasmid shuttle vectors. However, no shuttle vector construct for intermediate or pathogenic Leptospira spp. has been reported.
In this study, analysis of the genomes of two genetically related strains of a recently discovered pathogenic Leptospira species (9) revealed a prophage-like region of approximately 52 kb present in only one of the strains. Further analysis of this region allowed the identification of a putative replication origin. Cloning of a DNA fragment containing this replication origin into an E. coli conjugative plasmid allowed autonomous replication in the saprophyte L. biflexa and several intermediate and pathogenic strains. Subsequent to plasmid construction and analysis, we employed this plasmid for functional trans complementation of a mutant of the pathogen L. interrogans serovar Manilae that carries an inactivation in the peroxide stress regulator-encoding gene perR. The PerR transcriptional regulator belongs to the Fur family and controls the expression of genes participating in the cell's defense against oxidants. It has been previously shown that in a Leptospira perR mutant, a catalase (katE) and a putative cytochrome c peroxidase (mauG-2) had higher expression than in the wild-type strain. Previous work has also shown that this L. interrogans perR mutant is better able to survive than wild-type cells in the presence of hydrogen peroxide (10). We report here the successful production of PerR and concomitant restoration of reduced resistance to hydrogen peroxide when PerR is expressed in trans in the perR mutant by using the plasmid described.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The following Leptospira spp. were used in this study: intermediate strains L. fainei serovar Hurstbridge strain BUT6 and L. licerasiae serovar Varillal strain VAR010; pathogenic strains L. interrogans serovar Copenhageni strain Wijnberg, L. interrogans serovar Canicola strain Hond Utrecht IV, L. interrogans serovar Icterohaemorrhagiae strain Verdun, L. interrogans serovar Copenhageni strain Fiocruz L1-130, L. interrogans serovar Manilae strain L495, L. interrogans serovar Manilae perR mutant M776 (generous gift from Gerald Murray and Ben Adler; Monash University, Melbourne, Australia) (10), L. interrogans serovar Lai strain 56601, L. mayottensis strain 200901116; and the saprophyte L. biflexa serovar Patoc strain Patoc1. Strains were grown at 30°C in Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid medium (11, 12) with or without spectinomycin (40 μg/ml; Sigma-Aldrich, St. Louis, MO). Solid EMJH medium was prepared by adding 1% (wt/vol) Noble agar to liquid EMJH medium containing spectinomycin (40 μg/ml) and incubating it at 30°C for 7 to 30 days.
E. coli strain Π1 (ΔthyA) cells were used for plasmid transformation. E. coli strain β2163 (ΔdapA) was used for transfer of vector pMaORI into Leptospira spp. via conjugation. E. coli strains were grown in Luria-Bertani broth or agar containing spectinomycin (50 μg/ml). Strain β2163 was additionally cultured with 0.3 mM diaminopimelic acid (Sigma-Aldrich), while strain Π1 was additionally cultured with 0.3 mM thymidine (Sigma-Aldrich) as previously described (13).
Plasmid construction and transformation of Leptospira spp.
The nucleotide sequence of the replication region of L. mayottensis strain 200901116 was amplified with primer pairs ori1 and ori2 (Table 1) and inserted into pCR2.1-TOPO by using the TOPO TA cloning kit in accordance with the manufacturer's instructions (Life Technologies, Waltham, MA). After BamHI-XbaI digestion and gel purification, the DNA fragment containing the replication region was inserted into the corresponding restriction sites of the conjugative spectinomycin-resistant plasmid pMAT, and the resulting construct was designated pMaORI. Plasmid constructs were introduced into Leptospira strains by conjugation as previously described (4), with E. coli β2163 containing the pMaORI plasmid construct. Transformed Leptospira strains were recovered from solid medium, grown in liquid EMJH medium with spectinomycin selection (40 μg/ml), and filtered with a 0.22-μm filter into fresh liquid EMJH medium with spectinomycin (40 μg/ml). Once these samples had grown to mid-logarithmic phase, all were visually inspected to ensure the presence of motile leptospira cells without the presence of E. coli cells or other contaminants. Once it was determined that contaminants were not present, the sample was tested for the shuttle vector pMaORI as described below.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ → 3′)a | Target |
|---|---|---|
| spcA | GGGGTGAATTTGAGAATGGA | aadA cassette |
| spcB | GTCACTGTTTGCCACATTCG | aadA cassette |
| ori1 | CGCGGATCCTAATCAGCATACTGCAATCC | ori |
| ori2 | CTAGTCTAGAAATCCGTATAGCATATTCC | ori |
| adkF | ATTCTGCTTGGCGCTCCGGG | E. coli adk |
| adkR | CCGTCAACTTTCGCGTATTT | E. coli adk |
| PerR5 | CTCCTTAGAATGGACCGAAG | perR |
| PerR3 | GGATTCTATGTAGGATCAGTAG | perR |
BamHI and XbaI restriction sites are underlined.
Amplification of aadA and adk (an E. coli specific adenylate kinase gene) was performed via PCR. The gene-specific primers used are listed in Table 1.
Extraction of plasmid pMaORI from Leptospira spp. and E. coli strains was performed with a QIAprep Spin Miniprep kit in accordance with the manufacturer's instructions (Qiagen, Venlo, Netherlands).
The restriction endonuclease KpnI was used to determine the restriction profile of pMaORI isolated from E. coli cells that had previously been transformed with pMaORI from Leptospira species. An 18-μl volume of the extracted plasmid was added to 2.1 μl of 10× FastDigest buffer and 1 μl (1 U) of FastDigest KpnI (Thermo Scientific, Waltham, MA). Samples were digested for 30 min at 37°C and then subjected to gel electrophoresis.
Passage experiment.
Leptospira strains were passaged 10 times to determine if the pMaORI construct could be conserved without selection. For each passage, a 100-μl aliquot was passaged into 9 ml of fresh liquid EMJH medium without selection until it reached the late logarithmic phase. Each sample was then subsequently passaged, and the remaining 8.9 ml of sample in the late logarithmic phase was plasmid extracted as described above. The plasmid extracted was tested by PCR for the aadA cassette and subsequently transformed into E. coli strain Π1, where pMaORI was verified by determination of the KpnI restriction profile.
Complementation of L. interrogans serovar Manilae perR mutant.
To complement the L. interrogans perR M776 mutant (10), the wild-type perR allele (including its own promoter region, 310 nucleotides upstream of the perR start codon) was amplified from L. interrogans serovar Manilae strain L495 with primers PerR5 and PerR3 (Table 1). This PCR product was cloned into pCR2.1-TOPO (Invitrogen). The perR DNA fragment was then released with EcoRI and inserted into the dephosphorylated EcoRI site of pMaORI to generate plasmid pMaORI-perR (pNB138). The pMaORI-perR plasmid construct was introduced by conjugation into the M776 mutant (L. interrogans perR mutant) as described above, and spectinomycin-resistant colonies were inoculated into liquid EMJH medium containing 40 μg/ml spectinomycin for further analysis.
Cellular PerR content was measured by immunoblot analysis. Exponentially growing cells were lysed by sonication in 20 mM Tris-HCl (pH 8.0)–150 mM NaCl–2 mM EDTA in the presence of protease inhibitors (Roche, Basel, Switzerland). Samples (15 μg) of total cell extracts were subjected to 15% SDS-PAGE, and proteins were transferred onto nitrocellulose. PerR protein was detected by using a rabbit anti-PerR antibody at a 1:2,000 dilution as the primary antibody, goat anti-rabbit horseradish peroxidase (HRP)-conjugated IgG at a 1:150,000 dilution (Sigma-Aldrich) as the secondary antibody, and the SuperSignal West Pico reagent (Thermo Scientific) as the HRP substrate.
Survival in the presence of H2O2 was assessed by incubating exponentially growing cells in the presence or absence of 10 mM H2O2 for 30 min at 30°C. Twenty microliters of cells was then incubated with the alamarBlue cell viability reagent (Life Technologies) according to the manufacturer's recommendations.
Bioinformatic analysis.
Bioinformatic analysis was completed with Microscope Microbial Genome Annotation & Analysis Platform web-based software (https://www.genoscope.cns.fr/agc/mage).
Accession number.
The annotated sequence of pMaORI has been deposited in GenBank under accession number KP784428.
RESULTS AND DISCUSSION
Genome analysis.
All of the members of the Leptospira genus that have been analyzed carry at least two circular replicons. The large circular (cI, >3.6 Mb) and small circular (cII, 278 to 350 kb) chromosomes carry genes involved in housekeeping and other functions (14, 15). More recently, plasmids have also been identified by whole-genome sequencing, such as p74 in the saprophyte L. biflexa (7) and pGui1 and pGui2 in the pathogen L. interrogans (16).
We previously reported a group of Leptospira strains that were isolated from the blood of patients with leptospirosis in Mayotte (Indian Ocean) and identified as belonging to a novel pathogenic species (9, 17). High-throughput genome sequencing of two representative strains, 200901116 and 200901122, of this new species, designated Leptospira mayottensis, was performed at the J. Craig Venter Institute (http://gsc.jcvi.org/). The draft genome sequences of strains 200901116 (4,135,276 bp, 84 contigs) and 200901122 (4,161,553 bp, 85 contigs) have been deposited in GenBank under accession numbers AKWB00000000 and AKWM00000000, respectively. The G+C content of these strains is 39.5 mol%, which is within the range of 35 to 45 mol% reported for members of the genus Leptospira (9). Comparative genome analysis was performed with the MaGe interface in the SpiroScope database (https://www.genoscope.cns.fr/agc/mage) (18).
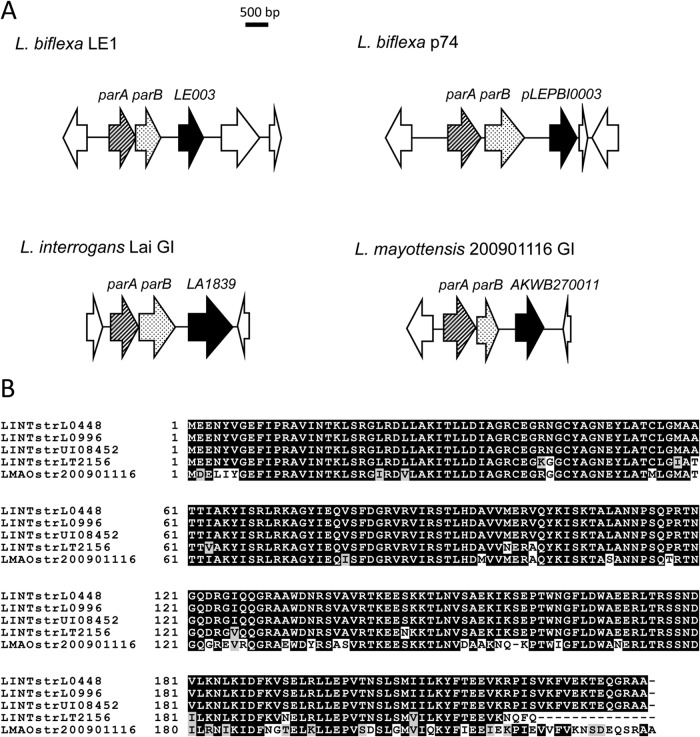
The genomes of strains 2009001116 and 200901122 are highly similar, as demonstrated by the high number of genes they have in common; they have 3,501 coding sequences (CDSs) with an average pairwise amino acid identity of >99% in common. In comparison, by the same criteria, strain 200901116 has only 34 CDSs in common with L. interrogans strain Fiocruz L1-130 and 194 CDSs in common with L. borgpetersenii strain L550, with >99% amino acid identity. By analyzing genome disparity, we found a unique region of approximately 52 kb carried by strain 200901116 but not strain 200901122 at approximate positions NT440186 to NT492268 (Fig. 1). Most of the genes in this region encode hypothetical proteins. The genes identified in this region include phage-related gene homologues coding for integrase, late control protein D, portal protein, GpA terminase, bacteriophage resistance factor (PF05565 family), and a toxin (RelE-like family)-antitoxin (Xre-like family). Another gene (AKWB270011) in this region codes for a putative protein of 234 amino acids containing an N-terminal helix-turn-helix domain (25 to 76 amino acids), which is typically associated with DNA binding. Upstream of this gene are two genes, one of which, AKWB270009, exhibited similarities to homologs of the ParA partition protein. The other gene (AKWB270010) could constitute parB of the putative partition locus. This genetic organization is similar to the replication origins of plasmid/prophage origin identified to date in L. biflexa serovar Patoc strain Patoc1 and L. interrogans serovar Lai strain 56601 (3, 7, 8) and includes genes coding for a DNA-binding protein that could constitute a Rep protein for initiation of plasmid replication and a partitioning system (Fig. 2A). Interestingly, orthologues of the L. mayottensis Rep-like protein were also found in L. interrogans pathogenic strains with a sequence identity of >75% (Fig. 2B). Together, this suggests that the 52-kb genomic island located in L. mayottensis strain 200901116 may represent a putative prophage. The occurrence of the L. mayottensis rep gene in other sequenced Leptospira strains suggests a widespread distribution of this prophage in L. interrogans strains.
FIG 1.
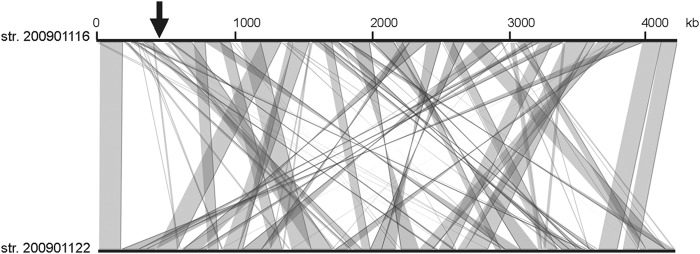
Comparative genome analysis of the draft genomes of strains (str.) 200901116 and 200901122. Comparative genome analysis was performed with the MaGe interface (18) in the SpiroScope database (https://www.genoscope.cns.fr/agc/mage). The arrow indicates the phage-like region present in strain 200901116 but not in strain 200901122.
FIG 2.
Genetic organization of replication origins of prophages and plasmids identified in Leptospira spp. (A) Comparison of the organization of parA and parB in saprophytic and pathogenic Leptospira spp. Sequence analysis of L. mayottensis strain 200901116 reveals a putative replication origin constituted by genes encoding a DNA-binding protein that could constitute a Rep protein for initiation of plasmid replication and a ParA/ParB partitioning system. This genetic organization is similar to previously identified replication origins in L. biflexa (3, 7) and L. interrogans (8). (B) Sequence alignment of putative Rep proteins from L. mayottensis serovar Mini strain 200901116, L. interrogans strain UI08452, L. interrogans serovar Medanensis strain L0448, L. interrogans strain L0996, and L. interrogans serovar Zanoni strain LT2156. Identical residues are shaded in black, and conserved residues are shaded in gray.
Design of a replicative vector.
To construct the pMaORI replicative vector, a 2.9-kb autonomously replicating sequence consisting of the parA locus (AKWB270009), the parB locus (AKWB270010), and rep (AKWB270011) located within the sequence of L. mayottensis strain 200901116 was amplified and inserted into 2.2-kb conjugative plasmid pMAT, which was derived from pSW29T (4). The pMAT vector includes the aadA spectinomycin resistance cassette, ori R6K for replication, and oriT RK2/RP4 for conjugation. The engineered vector was 5,035 bp long and was named pMaORI (Fig. 3).
FIG 3.
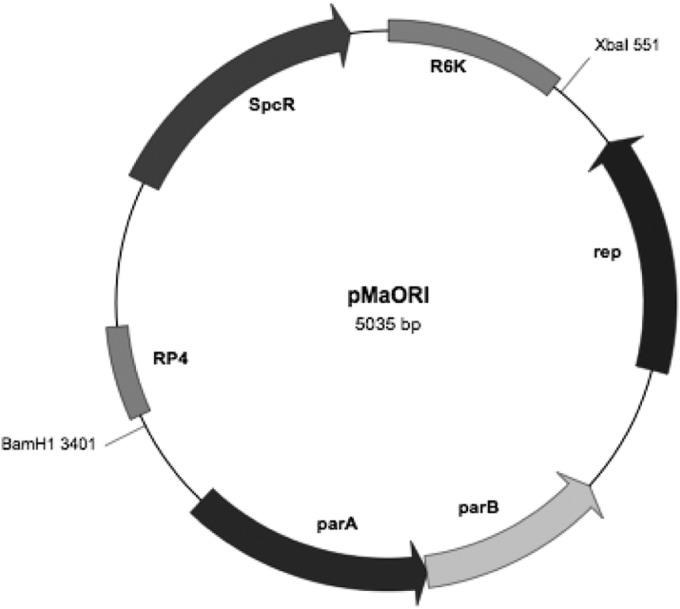
Schematic representation of plasmid vector pMaORI. Plasmid pMaORI (5,035 bp) was generated by cloning the replication origin of L. mayottensis strain 200901116, which carries the rep, parA, and parB genes, into the XbaI-BamHI restriction sites of conjugative plasmid pMAT, which contains the spectinomycin resistance cassette (aadA/Spcr), ori R6K, and oriT RK2/RP4. The aadA gene is expressed from the Borrelia burgdorferi flgB promoter.
Determination of pMaORI replication in various Leptospira spp.
The pMaORI construct was confirmed to be a viable shuttle vector in saprophytic, intermediate, and pathogenic Leptospira strains (Table 2). Compared with the original conjugative vector carrying the Himar1 transposon (4), the number of transconjugants increased 2- to 5-fold when our replication vector pMaORI was inserted into L. biflexa serovar Patoc strain Patoc1, L. interrogans serovar Copenhageni strain Fiocruz LA-130, and L. interrogans serovar Manilae strain L495. It was possible to transform the pMaORI construct into E. coli β2163, conjugate pMaORI into leptospires, recover the plasmid via plasmid extraction, and then retransform pMaORI into E. coli Π1 (Table 2). Transformed Leptospira strains were initially confirmed to be positive for the pMaORI construct via aadA amplification. To ensure that this confirmation was not a result of E. coli contamination, positive Leptospira strains were tested for adk, an E. coli gene. Further, to ensure that aadA confirmation was not a result of naked-DNA contamination within the sample, pMaORI, which was extracted from Leptospira strains, was transformed into E. coli Π1, plasmid DNA was extracted from E. coli, and pMaORI was confirmed by KpnI restriction profile.
TABLE 2.
Leptospira strains are capable of replication and conservation of pMaORI
| Type | Species | Serovar | Strain | Spcr coloniesb | pMaORI conservationd |
|---|---|---|---|---|---|
| Saprophyte | L. biflexa | Patoc | Patoc1 | +++ | ND |
| Intermediate | L. fainei | Hurstbridge | BUT6 | ++ | ND |
| Intermediate | L. licerasiae | Varillal | VAR010 | + | ND |
| Pathogen | L. interrogans | Canicola | Hond Utrecht IV | +++ | Positivec |
| Pathogen | L. interrogans | Copenhageni | Fiocruz L1-130 | ++ | Positive |
| Pathogen | L. interrogans | Copenhageni | Wijnberg | + | Positive |
| Pathogen | L. interrogans | Icterohaemorrhagiae | Verdun | +++ | Positive |
| Pathogen | L. interrogans | Lai | 56601 | +++ | Positive |
| Pathogen | L. interrogans | Manilae | L495 | ++ | Positive |
| Pathogen | L. mayottensis | Minia | 200901116 | − | NA |
Serogroup.
−, no colonies; +, 1 to 40 colonies; ++, 41 to 100 colonies; +++, >100 colonies.
The strain verified until passage 4.
pMaORI conserved after 10 passages without spectinomycin selection. Results verified by KpnI restriction profile of plasmid preparations, and PCR of the aadA cassette and the E. coli adk gene. NA, not applicable. ND, not determined.
The pMaORI vector was introduced (via conjugation) into L. mayottensis strain 200901116 to determine if the pMaORI plasmid could replicate in its parental strain. Analysis revealed that this strain was incapable of maintaining the pMaORI vector (Table 2).
Following confirmation that pathogenic Leptospira strains were able to replicate and maintain the pMaORI plasmid, strains were passaged 10 times in EMJH medium without spectinomycin to determine if the pMaORI plasmid could be conserved in the absence of selection. It was confirmed that pathogenic strains were indeed capable of maintaining this shuttle vector through 10 passages without selection (Table 2). This was confirmed via extraction of plasmid pMaORI from Leptospira cultures, followed by PCR amplification of the aadA cassette, confirmation of the ability to retransform into E. coli Π1, and subsequent plasmid restriction profile analysis. A limitation of the method used to detect plasmid stability is that it does not quantify the percentage of the population that retains pMaORI. Future studies should focus on quantifying the percentage of the bacterial population that retains pMaORI and determining the number of copies per cell.
Plasmid complementation of perR in L. interrogans.
A perR mutant was previously obtained by random transposon mutagenesis of L. interrogans serovar Manilae strain L495 (10). This perR mutant was shown to survive better than wild-type cells in the presence of hydrogen peroxide. However, repeated attempts to complement the mutation were unsuccessful (10).
In this study, complementation of the perR mutant was performed by cloning the perR wild-type allele with its native promoter into the pMaORI plasmid vector. Hundreds of transconjugants were obtained, and complemented cells showed expression of PerR (Fig. 4A), indicating that the perR open reading frame was expressed in trans from the pMaORI expression vector. Next, the ability of the complemented perR mutant cells to resist lethal concentrations of H2O2 were tested and compared to those of the wild type and the perR mutant. Cell viability was assessed via the alamarBlue assay, which measures the ability of cells to carry out redox reactions; reducing blue resazurin to pink resorufin. As shown in Fig. 4B, in the presence of H2O2, perR mutant cells displayed better viability than wild-type cells, as shown by their ability to reduce resazurin, indicating, as expected, that the perR mutant survived better than the wild type in the presence of peroxide. perR mutant cells that had been transformed with the pMaORI vector containing the perR allele were no longer able to reduce resazurin, which indicated that they had decreased viability in the presence of H2O2 (Fig. 4B). This demonstrated that expression of the perR CDS from the pMaORI vector could restore an H2O2-associated phenotype comparable to that of wild-type cells.
FIG 4.
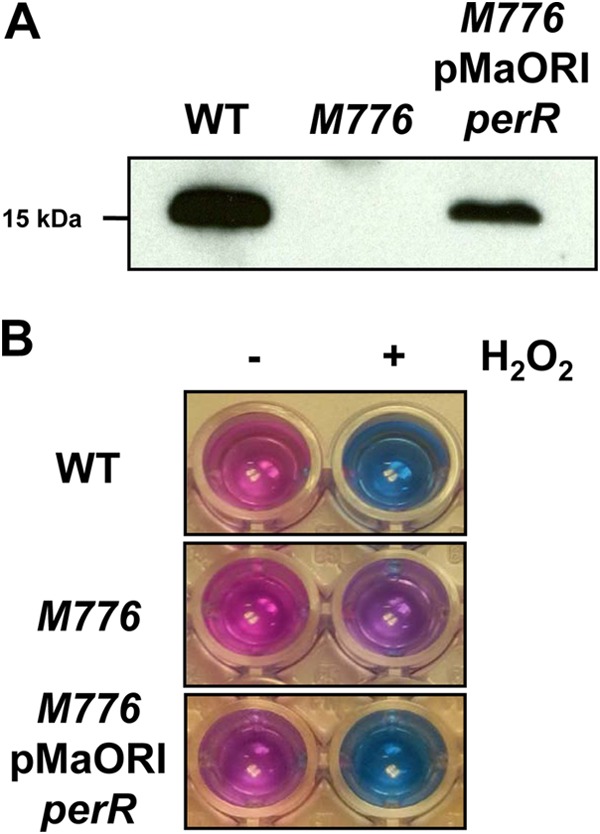
Complementation of an L. interrogans perR mutant. (A) PerR production in wild-type (WT), perR mutant (M776), and complemented perR mutant (M776/pMaORI-perR) cells. Western blot analysis shows recovery of PerR expression in the perR mutant carrying replicative plasmid pMaORI-perR. (B) Viability of wild-type (WT), perR mutant (M776), and complemented perR mutant (M776/pMaORI-perR) cells during peroxide stress. Exponentially growing wild-type, perR mutant, and complemented perR mutant cells were exposed to H2O2, and their viability was assessed in the presence of resazurin. Cell viability was measured by the capacity of the cells to carry out the redox reaction of reducing blue resazurin into bright pink resorufin.
While replication of plasmid vectors in pathogens has not been previously described, these results demonstrate that plasmid complementation is possible in pathogenic Leptospira strains. The pMaORI vector will therefore be useful for the purpose of genetic complementation of mutants obtained by random transposon mutagenesis or in the generation of conditional mutants.
ACKNOWLEDGMENTS
We thank J. Vinetz and D. Fouts for their permission to use the draft genomes that are part of a project funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract HHSN272200900007C. This work was supported in part by National Science Foundation grant IIA-1159099 (C.J.P.) and the Institut Pasteur (M.P.).
REFERENCES
- 1.Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picardeau M. 2015. Genomics, proteomics, and genetics of Leptospira. Springer-Verlag, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 3.Girons IS, Bourhy P, Ottone C, Picardeau M, Yelton D, Hendrix RW, Glaser P, Charon N. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J Bacteriol 182:5700–5705. doi: 10.1128/JB.182.20.5700-5705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picardeau M. 2008. Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl Environ Microbiol 74:319–322. doi: 10.1128/AEM.02172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauby H, Saint Girons I, Picardeau M. 2003. Construction and complementation of the first auxotrophic mutant in the spirochaete Leptospira meyeri. Microbiology 149:689–693. doi: 10.1099/mic.0.26065-0. [DOI] [PubMed] [Google Scholar]
- 6.Poggi D, Oliveira de Giuseppe P, Picardeau M. 2010. Antibiotic resistance markers for genetic manipulations of Leptospira spp. Appl Environ Microbiol 76:4882–4885. doi: 10.1128/AEM.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, Creno S, Kuczek ES, Bommezzadri S, Davis JC, McGrath A, Johnson MJ, Boursaux-Eude C, Seemann T, Rouy Z, Coppel RL, Rood JI, Lajus A, Davies JK, Médigue C, Adler B. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. doi: 10.1371/journal.pone.0001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourhy P, Salaün L, Lajus A, Médigue C, Boursaux-Eude C, Picardeau M. 2007. A genomic island of the pathogen Leptospira interrogans serovar Lai can excise from its chromosome. Infect Immun 75:677–683. doi: 10.1128/IAI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourhy P, Collet L, Brisse S, Picardeau M. 2014. Leptospira mayottensis sp. nov., a pathogenic Leptospira species isolated from humans. Int J Syst Evol Microbiol 64:4061–4067. doi: 10.1099/ijs.0.066597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo M, Murray GL, Khoo CA, Haake DA, Zuerner RL, Adler B. 2010. Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect Immun 78:4850–4859. doi: 10.1128/IAI.00435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellinghausen HC, McCullough WG. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am J Vet Res 26:45–51. [PubMed] [Google Scholar]
- 12.Johnson RC, Harris VG. 1967. Differentiation of pathogenic and saprophytic leptospires. J Bacteriol 94:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Zuerner RL. 1991. Physical mapping of chromosomal and plasmid DNA comprising the genome of Leptospira interrogans. Nucleic Acids Res 19:4857–4860. doi: 10.1093/nar/19.18.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuerner RL, Herrmann JL, Saint Girons I. 1993. Comparison of genetic maps for two Leptospira interrogans serovars provides evidence for two chromosomes and intraspecies heterogeneity. J Bacteriol 175:5445–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu WN, Huang LL, Zeng LB, Zhuang XR, Chen CY, Wang YZ, Qin JH, Zhu YZ, Guo XK. 2014. Isolation and characterization of two novel plasmids from pathogenic Leptospira interrogans serogroup Canicola serovar Canicola strain Gui44. PLoS Negl Trop Dis 8:e3103. doi: 10.1371/journal.pntd.0003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourhy P, Collet L, Lernout T, Zinini F, Hartskeerl RA, van der Linden H, Thiberge JM, Diancourt L, Brisse S, Giry C, Pettinelli FPM. 2012. Human Leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J Clin Microbiol 50:307–311. doi: 10.1128/JCM.05931-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Le Fèvre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli C, Thil Smith AA, Weiman M, Médigue C. 2013. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]