Abstract
Haemophilus parasuis, the causative agent of Glässer's disease, is one of the early colonizers of the nasal mucosa of piglets. It is prevalent in swine herds, and lesions associated with disease are fibrinous polyserositis and bronchopneumonia. Antibiotics are commonly used in disease control, and resistance to several antibiotics has been described in H. parasuis. Prediction of H. parasuis virulence is currently limited by our scarce understanding of its pathogenicity. Some genes have been associated with H. parasuis virulence, such as lsgB and group 1 vtaA, while biofilm growth has been associated with nonvirulent strains. In this study, 86 H. parasuis nasal isolates from farms that had not had a case of disease for more than 10 years were obtained by sampling piglets at weaning. Isolates were studied by enterobacterial repetitive intergenic consensus PCR and determination of the presence of lsgB and group 1 vtaA, biofilm formation, inflammatory cell response, and resistance to antibiotics. As part of the diversity encountered, a novel 2,661-bp plasmid, named pJMA-1, bearing the blaROB-1 β-lactamase was detected in eight colonizing strains. pJMA-1 was shown to share a backbone with other small plasmids described in the Pasteurellaceae, to be 100% stable, and to have a lower biological cost than the previously described plasmid pB1000. pJMA-1 was also found in nine H. parasuis nasal strains from a separate collection, but it was not detected in isolates from the lesions of animals with Glässer's disease or in nontypeable Haemophilus influenzae isolates. Altogether, we show that commensal H. parasuis isolates represent a reservoir of β-lactam resistance genes which can be transferred to pathogens or other bacteria.
INTRODUCTION
Haemophilus parasuis is an early colonizer and a member of the normal microbiota of the upper respiratory tract of piglets. H. parasuis initial acquisition occurs through direct contact with the sow after birth, and the bacterium establishes colonization in the upper respiratory tract, with a maximum level of colonization occurring at about 2 months of age (1). Under certain circumstances, some strains spread to the lungs to cause pneumonia or invade systemic sites. Systemic invasion produces fibrinous polyserositis and arthritis, which are the characteristic lesions of Glässer's disease (2, 3).
Although H. parasuis is an important swine pathogen, its host-pathogen interactions remain to be well understood. Different H. parasuis strains can be isolated from the nasal cavity of a given animal, and these nasal strains are not particularly stable, since they experience turnover during the life of the pigs (1, 4–6). Strains of H. parasuis are heterogeneous and include virulent and nonvirulent strains. It is common to find nonvirulent strains in the upper respiratory tract of healthy animals, but virulent strains can also be found (6). Different methods have been developed to differentiate H. parasuis strains. Genotyping methods include multilocus sequence typing (MLST) (7, 8), partial sequence of the 60-kDa heat shock protein-encoding gene hsp60 (9), enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) (10), or pulsed-field gel electrophoresis (11). A search for genetic markers to identify putative virulent H. parasuis isolates revealed the potential of the virulence-associated trimeric autotransporter-encoding (vtaA) genes to be a diagnostic tool (12) and suggested an association of the lsgB gene, which encodes a sialyltransferase involved in the sialylation of the lipooligosaccharide (LOS) molecule, with virulent strains (13). A number of other bacterial factors have been associated with H. parasuis virulence, including 6-phosphogluconate dehydrogenase (14), a complete LOS molecule (15), cytolethal distending toxin (16, 17), immunoglobulin A protease (18), capsule (19), and the outer membrane protein OmpP2 (20–22).
Serovar diversity and the high number of nonserotypeable isolates that have been reported have negatively affected the development of effective cross-protective vaccines, due to limited cross protection among H. parasuis strains (23). Since no definite vaccine is available, antimicrobial treatment continues to be the strategy used to control the disease. Tetracyclines are the major antimicrobials used against this bacterium, but resistance has been found in many instances (24–26), suggesting the need for more effective molecules to treat infected animals (27). Penicillins and aminopenicillins are being used as alternative treatments for infections due to H. parasuis. Of note, clinical isolates resistant to β-lactams have been found in large numbers in several countries (26, 27), and biofilm formation by H. parasuis strains may have a positive correlation with resistance to β-lactam antibiotics (28). β-Lactam resistance in H. parasuis clinical strains isolated from Glässer's disease lesions has been related to plasmid pB1000, which bears the ROB-1 β-lactamase, belongs to the ColE1 superfamily, is mobilized into Escherichia coli using the conjugation machinery of an IncP plasmid, and has also been found in Pasteurella multocida and Haemophilus influenzae clinical isolates (11, 29, 30). However, antibiotic resistance in the colonizer population of H. parasuis has not been previously examined, and its potential as a reservoir of antibiotic resistance genes has not been explored. Here, we describe the novel small plasmid pJMA-1, which was isolated from strains of H. parasuis found in the nasal cavities of healthy animals and which carries ROB-1-mediated β-lactam resistance. pJMA-1 sequence features and its relationship with previously described mobile elements, functionality, transmissibility, and stability were analyzed. The implications of antibiotic resistance carried by a plasmid in H. parasuis nasal strains are further discussed.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. parasuis strains were grown overnight at 37°C with 5% CO2 on chocolate agar plates (bioMérieux, France). The H. parasuis strains belonged to three isolate collections: (i) 86 strains isolated from nasal swabs taken from healthy pigs at weaning in Navarra, Spain (generated in this study), (ii) 20 strains isolated from nasal swab samples taken from healthy pigs at weaning in Catalonia and Mallorca, Spain (8, 9, 31), and (iii) 24 strains isolated from Glässer's disease lesions (8, 9, 31). H. parasuis Nagasaki (virulent), SW114 (nonvirulent), BB1033 (10), and BB1023 (10) were used as reference strains, when necessary.
Nontypeable (NT) Haemophilus influenzae strains were grown overnight at 37°C with 5% CO2 on chocolate agar plates or on brain heart infusion (BHI) agar plates supplemented with 10 μg/ml hemin and 10 μg/ml β-NAD, referred to as sBHI agar. We employed 20 NT H. influenzae isolates recovered from the nasopharynges of healthy children in day care centers and schools in Oviedo, Spain (32), 16 pediatric ear isolates (Hospital Universitario Germans Trias i Pujol [HUGTiP], Badalona, Spain) (this study), and 41 respiratory strains from pulmonary patients (HUGTiP and Hospital Universitario Bellvitge, Barcelona, Spain) (this study). H. influenzae Rd KW20 (33) was used as a reference strain for plasmid transmissibility and stability analysis. When necessary, sBHI agar containing ampicillin (AMP) at 25 μg/ml was used.
Isolation of H. parasuis strains from nasal swab samples from healthy piglets.
From April to September 2012, nasal swab samples were taken at weaning (3 to 4 weeks of age) from healthy pigs on six farrow-to-finish farms and two farrow-to-weaning farms that had not had a case of Glässer's disease since 2000. The farms were located in an area of 9,500 km2 in Navarra, northern Spain. The farms were included on the basis of the veterinarians' knowledge of the farm and were selected because of their different patterns regarding the respiratory health status of the pigs and because they used the most common herd health patterns, housing practices, and herd management practices encountered in the field at the time. The sampling procedure was part of a periodic veterinarian farm surveillance routine. The number of sows, production type, and weaning age for each farm are summarized in Table 1. The nares of four animals per farm were sampled. Swab (Deltalab, Spain) specimens from the nasal cavities of all piglets were placed in Amies transport medium and kept on ice until inoculation on chocolate agar plates. The plates were incubated at 37°C with 5% CO2, and suspect colonies of H. parasuis were selected and subcultured on chocolate agar and blood agar (bioMérieux) plates. Colonies growing on chocolate agar but not on blood agar were identified by conventional biochemical methods, by mass spectrometry using a matrix-assisted laser desorption ionization biotyper (version 3.0; Bruker), and by colony PCR based on species-specific amplification of the 16S RNA gene with primers HPS-F and HPS-R (Table 2), rendering an 821-bp product (34). For species-specific PCR, crude DNA templates derived directly from colonies on agar were prepared by harvesting a loopful of bacteria from a culture that had been grown overnight on a chocolate agar plate and placing it into 0.5 ml of sterile water of the purity required for DNA analysis; Tween 20 was added to a final concentration of 0.5%, and three cycles of boiling (for 5 min) and cooling on ice (for 2 min) were performed. The samples were centrifuged for 10 s at 15,000 × g and immediately placed on ice. An aliquot of 1 μl of the supernatant was used in the PCR. When a nasal swab sample rendered a positive H. parasuis identification, independent colonies from each swab (i.e., per animal) were separately tested and, if positive, stored in tryptic soy broth–20% glycerol at −80°C.
TABLE 1.
Farm, number of sows, type of production, piglet weaning age, animal sampled, isolates, ERIC-PCR profiles, and genetic features of H. parasuis isolates collected in this study
| Farm | No. of sows | Production type/weaning age (days) | Animal | H. parasuis isolate | ERIC-PCR profile | Presence of the following: |
|||
|---|---|---|---|---|---|---|---|---|---|
| lsgB | vtaA1 | blaROB-1 | blaROB-1-containing plasmid (plasmid size [kb]) | ||||||
| A | 250 | Farrow to finish/28 | A1 | A1.1 | NA-1 | − | − | − | − |
| A1 | A1.2 | NA-2 | − | − | − | − | |||
| A1 | A1.3 | NA-1 | − | − | − | − | |||
| A2 | A2.1 | NA-1 | − | − | − | − | |||
| A2 | A2.2 | NA-1 | − | − | − | − | |||
| A2 | A2.3 | NA-1 | − | − | − | − | |||
| A3 | A3.1 | NA-7 | − | − | + | + (2.6) | |||
| A3 | A3.2 | NA-7 | − | − | + | + (2.6) | |||
| A3 | A3.3 | NA-7 | − | − | + | + (2.6) | |||
| A4 | A4.1 | NA-10 | − | − | − | − | |||
| A4 | A4.2 | NA-10 | − | − | − | − | |||
| A4 | A4.3 | NA-10 | − | − | − | − | |||
| B | 475 | Farrow to weaning/21 | B1 | B1.1 | NA-2 | − | − | − | − |
| B1 | B1.2 | NA-2 | − | − | − | − | |||
| B1 | B1.3 | NA-15 | + | + | − | − | |||
| B2 | B2.1 | NA-16 | + | + | − | − | |||
| B2 | B2.2 | NA-17 | + | + | − | − | |||
| B2 | B2.3 | NA-18 | − | + | − | − | |||
| B3 | B3.1 | NA-17 | + | + | − | − | |||
| B3 | B3.2 | NA-21 | + | + | − | − | |||
| B3 | B3.3 | NA-21 | + | + | − | − | |||
| B4 | B4.1 | NA-21 | + | + | − | − | |||
| B4 | B4.2 | NA-21 | − | + | − | − | |||
| B4 | B4.3 | NA-21 | + | + | − | − | |||
| C | 95 | Farrow to finish/28 | C1 | C1.1 | NA-25 | − | − | − | − |
| C1 | C1.3 | NA-27 | − | − | − | − | |||
| C2 | C2.1 | NA-27 | − | − | − | − | |||
| C2 | C2.2 | NA-29 | − | − | − | − | |||
| C2 | C2.3 | NA-29 | − | − | − | − | |||
| C3 | C3.1 | NA-29 | − | − | − | − | |||
| C3 | C3.2 | NA-29 | − | − | − | − | |||
| C3 | C3.3 | NA-29 | − | − | − | − | |||
| C4 | C4.1 | NA-29 | − | − | − | − | |||
| C4 | C4.2 | NA-29 | − | − | − | − | |||
| C4 | C4.3 | NA-29 | − | − | − | − | |||
| D | 180 | Farrow to finish/28 | D1 | D1.1 | NA-37 | − | − | − | − |
| D1 | D1.2 | NA-37 | − | − | − | − | |||
| D1 | D1.3 | NA-39 | − | − | − | − | |||
| D2 | D2.1 | NA-40 | − | − | − | − | |||
| D2 | D2.2 | NA-41 | − | − | − | − | |||
| D3 | D3.1 | NA-37 | − | − | − | − | |||
| D3 | D3.2 | NA-37 | − | − | − | − | |||
| D3 | D3.3 | NA-37 | − | − | − | − | |||
| D4 | D4.1 | NA-46 | − | − | + | + (2.6) | |||
| D4 | D4.2 | NA-46 | − | − | + | + (2.6) | |||
| D4 | D4.3 | NA-48 | − | − | + | + (4.6) | |||
| E | 500 | Farrow to finish/28 | E1 | E1.1 | NA-67 | − | − | − | − |
| E1 | E1.2 | NA-67 | − | − | − | − | |||
| E1 | E1.3 | NA-67 | − | − | − | − | |||
| E2 | E2.1 | NA-52 | − | − | − | − | |||
| E2 | E2.2 | NA-52 | − | − | − | − | |||
| E2 | E2.3 | NA-54 | − | − | − | − | |||
| E3a | |||||||||
| E4a | |||||||||
| F | 260 | Farrow to finish/28 | F1 | F1.1 | NA-29 | − | − | − | − |
| F1 | F1.2 | NA-29 | − | − | − | − | |||
| F1 | F1.3 | NA-29 | − | − | − | − | |||
| F2 | F2.1 | NA-29 | − | − | − | − | |||
| F2 | F2.2 | NA-29 | − | − | − | − | |||
| F2 | F2.3 | NA-29 | − | − | − | − | |||
| F3 | F3.1 | NA-2 | − | − | − | − | |||
| F3 | F3.2 | NA-2 | − | − | − | − | |||
| F3 | F3.3 | NA-2 | − | − | − | − | |||
| F4 | F4.1 | NA-64 | + | − | + | + (2.6) | |||
| F4 | F4.2 | NA-64 | + | − | + | + (2.6) | |||
| F4 | F4.3 | NA-64 | − | − | + | + (2.6) | |||
| G | 160 | Farrow to finish/21 | G1 | G1.1 | NA-67 | − | − | − | − |
| G1 | G1.2 | NA-67 | − | − | − | − | |||
| G1 | G1.3 | NA-67 | − | − | − | − | |||
| G2 | G2.1 | NA-70 | − | − | − | − | |||
| G2 | G2.2 | NA-70 | − | − | − | − | |||
| G2 | G2.3 | NA-29 | − | − | − | − | |||
| G3 | G3.1 | NA-73 | − | − | − | − | |||
| G3 | G3.2 | NA-73 | − | − | − | − | |||
| G3 | G3.3 | NA-73 | − | − | − | − | |||
| G4 | G4.1 | NA-76 | − | + | − | − | |||
| G4 | G4.2 | NA-73 | − | − | − | − | |||
| G4 | G4.3 | NA-73 | − | − | − | − | |||
| H | 800 | Farrow to weaning/28 | H1 | H1.1 | NA-79 | − | + | − | − |
| H1 | H1.2 | NA-80 | − | − | − | − | |||
| H1 | H1.3 | NA-79 | − | + | − | − | |||
| H2 | H2.1 | NA-82 | − | − | − | − | |||
| H3 | H3.1 | NA-79 | − | + | − | − | |||
| H3 | H3.2 | NA-79 | − | + | − | − | |||
| H3 | H3.3 | NA-79 | − | + | − | − | |||
| H4 | H4.1 | NA-88 | − | − | − | − | |||
| H4 | H4.2 | NA-88 | − | − | − | − | |||
| H4 | H4.3 | NA-88 | − | − | − | − | |||
H. parasuis was not isolated from animals E3 and E4.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′) | Reference or source |
|---|---|---|
| HPS-F | GTGATGAGGAAGGGTGGTGT | 34 |
| HPS-R | GGCTTCGTCACCCTCTGT | 34 |
| ERIC1R | ATGTAAGCTCCTGGGGATTCAC | 10 |
| ERIC2 | AAGTAAGTGACTGGGGTGAGCG | 10 |
| lsgB-F1 | ATGAATTTGATTATTTGTATGACTCCATTT | 13 |
| lsgB-F2-2 | ATTCGGTCTGGAATGATAAATATCAGTATT | This study |
| lsgB-R1 | CTATTGGCATGTGTAGTCAATTACTTC | 13 |
| lsgB-R2-2 | AAGAGGAGCTCCACTATAGAACGTATAAAT | This study |
| YADAF1 | TTTAGGTAAAGATAAGCAAGGAAATCC | 12 |
| PADHR1 | CCACACAAAACCTACCCCTCCTCC | 12 |
| YADAF3 | AATGGTAGCCAGTTGTATAATGTT | 12 |
| PADHR3 | CCACTG TAATGCAATACCTGCACC | 12 |
| rob-1F | TGTTGCAATCGCTGCC | 11 |
| rob-1R | TTATCGTACACTTTCCA | 11 |
| rob-1U | ATCGTCATGCCTTTGCCAACG | 11 |
| rob-1D | AATTGGTTGGACAATAACGCA | 11 |
| pl-SEQ-rob-1F | CCCGACCGCTTTCAGCGGTCAAAA | This study |
| pl-SEQ-rob-1R | TAGGAAGTACTCATCATTTGGAAG | This study |
| pB1000-F1 | TCCATCTAAAGAATGTGAAGTATTACT | This study |
| pB1000-R2 | CTGCTAACGCCCTGCGGTTGTTAGACT | This study |
| plSeqF2 | TTAGCACTATAAGGCTAAGCGACTAGG | This study |
| plSeqR2 | AGGGAAGTGGTTATTGTGGGCGGTAGG | This study |
| plSeqF3 | AGTGATAACGTTCTTAATACG | This study |
| Pl298-SEQ-F | GGCTCACGATGTTCGCCTAAT | This study |
| Pl298-SEQ-R | ACGGCTAGGAAGGGAATAGGG | This study |
| Pl298-SEQ-rob-1F | CGAATTTCCGCGCAATGTACG | This study |
| Pl298-SEQ2-F | GGCAGGCTTGAGCTGAATTTC | This study |
| Pl157-SEQ-R | TGTCCACTGCTACACAAGGCT | This study |
| Tet(B)-F | ACGTTACTCGATGCCAT | This study |
| Tet(B)-R | AGCACTTGTCTCCTGTT | This study |
| Tet(H)-F | ATACTGCTGATCACCGT | This study |
| Tet(H)-R | TCCCAATAAGCGACGCT | This study |
| Tet(M/O/S)-F | ATAGAYACGCCAGGMCATAT | This study |
| Tet(M/O/S)-R | GAAGCCCAGAAAGGATTYGGY | This study |
| Tet(B)invF | GGTTAGTTTTCCCTGTTTTA | 29 |
| Tet(B)invR | ACCAACCGAACCACTTCACG | 29 |
| Tet(H)invF | GGGTCATCTTACCAGCATTA | 29 |
| Tet(H)invR | AGAAACCAAAATAGCGAGTT | 29 |
Molecular typing by ERIC-PCR.
Crude DNA was prepared following the protocol described above and quantified. In each case, 100 ng was used as the template for ERIC-PCR with primers ERIC1R and ERIC2 (Table 2). We followed a previously published protocol (10) and included an extra final extension step of 20 min. PCR products were analyzed by electrophoresis (70 V, 3 h) in a 2% agarose gel. Band patterns were visualized by staining with ethidium bromide, and comparison of the patterns was performed visually.
Serotyping.
Serotype determination was performed by indirect hemagglutination at the Animal Health Department of the Veterinary School at the University of León (León, Spain) following a previously published protocol (35). Soluble antigen was obtained after boiling of a bacterial suspension and subsequent centrifugation to eliminate insoluble debris.
Detection of the lsgB and group 1 vtaA genes.
Crude DNA templates derived directly from colonies on agar were prepared as described above. The lsgB gene was PCR amplified using Taq polymerase (Biotools, Spain) and the four primer pairs lsgB-F1 and lsgB-R1, lsgB-F1 and lsgB-R2-2, lsgB-F2-2 and lsgB-R1, and lsgB-F2-2 and lsgB-R2-2 (Table 2). The vtaA translocator domains from groups 1 and 3 were amplified in a multiplex format following a previously published protocol (12). Multiplex PCR tubes contained GoTaq buffer, which consisted of 2 mM MgCl2, 0.4 mM each deoxynucleoside triphosphate dNTP, 800 nM primers YADAF1 and PADHR1 (each), 400 nM primers YADAF3 and PADHR3 (each), 1 U GoTaq polymerase (Promega, USA) and 10 ng genomic DNA in a final volume of 25 μl. Cycling conditions were 5 min at 94°C, followed by 25 cycles of 45 s at 94°C, 45 s at 64°C, and 1 min at 72°C, and then a final incubation was carried out at 72°C for 7 min. In all cases, the virulent reference strain H. parasuis Nagasaki and the nonvirulent reference strain H. parasuis SW114 were used as controls. PCR for vtaA group 3 (with primers YADAF3 and PADHR3), previously shown to be present in all H. parasuis strains (12), was used as a control.
Plasmid analysis of β-lactam-resistant strains.
Plasmid DNA was extracted from bacteria grown on chocolate agar plates with a QIAprep spin miniprep kit (Qiagen, Germany). PCRs were performed using Taq polymerase (Biotools) with crude DNA (see above) or the extracted plasmid as the template. Identification of plasmids bearing blaROB-1 in β-lactam-resistant H. parasuis strains (strains for which the MICs of β-lactam antibiotics were elevated) was performed using primers rob-1F and rob-1R, rendering a 457-bp amplicon, and with divergent blaROB-1 primers rob-1D and rob-1U, which rendered a 2,238-bp or a 4,189-bp amplicon, depending on the strain (Table 2). Purification of the PCR amplicons was performed using a NucleoSpin gel and PCR cleanup kit (Macherey-Nagel, Germany). Plasmid pJMA-1 was Sanger sequenced by PCR walking with primers pl-SEQ-rob-1F, rob-1D, pB1000-F1, plSeqF2, plSeqF3, rob-1F, rob-1R, and pl-SEQ-rob-1R (Table 2). Plasmid pB1000 was Sanger sequenced by PCR walking with primers rob-1D, rob-1U, plSeqF3, Pl298-SEQ-rob-1F, Pl298-SEQ-F, Pl298-SEQ-R, Pl298-SEQ2-F, and Pl157-SEQ-R (Table 2).
Antimicrobial susceptibility testing.
In vitro antimicrobial susceptibility was determined by microdilution methods following Clinical and Laboratory Standards Institute (CLSI) guidelines (36, 37) and the methods described in previous studies (11, 29). Briefly, bacterial inocula were prepared from colonies freshly grown for 24 h on chocolate agar plates, and the inoculum was adjusted to a 0.5 McFarland standard in haemophilus test medium (HTM; Becton Dickinson, USA); 50 μl of each bacterial suspension was used per well. The antimicrobials used for microdilution were AMP and amoxicillin (AMX). Stock solutions were prepared from the pure powder form according to the CLSI standard method (36, 37), and the antimicrobials were used over a concentration range from 1 to 2,048 μg/ml. Alternatively, we used commercially available dehydrated Sensititre plate panels (EUMVS2; Trek Diagnostics Inc., USA) for cefotaxime (CTX), ceftazidime (CAZ), sulfamethoxazole (SXT), gentamicin (GEN), ciprofloxacin (CIP), tetracycline (TET), streptomycin (STR), trimethoprim (TMP), chloramphenicol (CHL), colistin (CST), florfenicol (FFN), kanamycin (KAN), and nalidixic acid (NAL). For both the microdilution and Sensititre assays, panel quality controls were performed with strains H. parasuis BB1023 (β-lactam resistant) and BB1033 (β-lactam sensitive) (11). The MIC was defined as the lowest antimicrobial concentration that inhibited bacterial growth. Given that a standard technique for determination of the antimicrobial susceptibility of H. parasuis and agreed-upon interpretation criteria do not currently exist (38), the MIC results were reviewed and the distribution of strains over the MIC range was considered.
Biofilm formation.
H. parasuis strains were grown on chocolate agar plates for 16 h at 37°C in 5% CO2. Four to five freshly grown colonies were used to inoculate 10 ml BHI with 10 μg/ml β-NAD and grown at 37°C with 5% CO2 to an optical density at 600 nm (OD600) of ∼0.7. Ten microliters of the grown culture was diluted in 100 μl sBHI on 96-well polystyrene plates (Iwaki, Japan); alternatively, 100 μl of the grown culture was diluted in 586 μl sBHI on 24-well polystyrene plates (Costar; Corning, USA). The plates were incubated statically for 36 h at 37°C in 5% CO2. The culture medium was then carefully removed, the plates were washed in distilled water, and the attached bacteria were fixed for 15 min with 100 μl (96-well plates) or 586 μl (24-well plates) methanol. The methanol was removed, and the plates were air dried for 15 min. Once the plates were dry, 100 μl (96-well plates) or 586 μl (24-well plates) of 5% crystal violet was added to each well for 5 min and the plates were incubated at room temperature. The plates were washed in distilled water and then dried for 30 min at 37°C. Stained biofilms were resolubilized with 100 or 586 μl (96-well and 24-well plates, respectively) 80% ethanol–20% acetic acid. Biofilm formation was quantified by measuring the absorbance at 595 nm. To assess the nature of the biofilm matrix, biofilm assays were performed on 24-well plates. Once biofilms formed, the wells were washed in distilled water and treated with 100 μl (96-well plates) or 586 μl (24-well plates) 10 mM sodium meta-periodate dissolved in 50 mM sodium acetate buffer (pH 4.5) or with 100 μg/ml proteinase K dissolved in a buffer containing 100 mM NaCl, 20 mM Tris (pH 7.5), as previously described (39). Controls with adequate buffers (50 mM sodium acetate buffer [pH 4.5] or 100 mM NaCl and 20 mM Tris [pH 7.5], respectively) were run in parallel. After treatment, the remaining material on the wells was fixed, and the plates were stained and monitored as described above. In all cases, assays were carried out in triplicate and on at least three independent occasions (n ≥ 9). Means and standard deviation (SDs) of the absorbance values were calculated.
Cell culture, bacterial infection, and quantification of IL-8 secretion.
PK-15 immortalized pig kidney epithelial cells (ATCC CCL-33) were maintained in RPMI 1640 medium supplemented with 10 mM HEPES, 10% fetal calf serum (FCS), 100 units/ml penicillin, and 0.1 mg/ml streptomycin (complete medium). Cells were seeded at 8 × 104 cells per well in 24-well tissue culture plates for 24 h. The cells were then serum starved for 16 h before infection by replacement of the complete medium with medium lacking FCS. A confluence of 90% was reached at the time of infection. H. parasuis cells grown to stationary phase were recovered with 1 ml phosphate-buffered saline (PBS) from a chocolate agar plate and adjusted with PBS to an OD600 of 1 (∼109 CFU/ml). Cells in 1 ml Earle's balanced salt solution (EBSS) were infected with the freshly obtained bacterial suspension at a multiplicity of infection of approximately 100:1. The plates were centrifuged at 400 × g for 5 min and incubated at 37°C in 5% CO2 for 2 h. Infected cells were washed 3 times with PBS and incubated with fresh RPMI 1640 medium containing 10 mM HEPES, 10% FCS, 100 μg/ml gentamicin, and 5 μg/ml penicillin for 6 h. Supernatants were removed from the wells, the cell debris was removed by centrifugation, and samples were frozen at −80°C. The interleukin-8 (IL-8) levels in the supernatants were measured using an enzyme-linked immunosorbent assay (ELISA) kit (Kingfisher Biotech, USA). Infection experiments were carried out in duplicate and on at least two independent occasions (n ≥ 4). Means and SDs were calculated, and comparison of means for statistically significant differences was performed using the two-tailed t test (Prism software, version 4, for the personal computer). A P value of <0.05 was considered statistically significant.
Stability of pJMA-1.
Plasmid pJMA-1 was transformed into H. influenzae Rd KW20 electrocompetent cells (40). Transformants were selected on sBHI agar plates containing 25 μg/ml ampicillin. The rate of curing of plasmid pJMA-1 in H. influenzae Rd KW20 transformants was determined as previously described (29). Briefly, 4 to 5 colonies of bacteria harboring pJMA-1 grown on chocolate agar containing 25 μg/ml ampicillin were inoculated in 10 ml sBHI and grown at 37°C in 5% CO2 for 12 h. This culture was serially passaged eight times by serial 1:100 dilution in 10 ml sBHI and growth for 12 h. In each subculture step, bacteria were plated on chocolate agar, and the proportion of resistant colonies harboring pJMA-1 was deduced by replica plating of at least 100 colonies on sBHI agar and sBHI agar plates containing 25 μg/ml ampicillin. The rate of plasmid curing was calculated as the percentage of ampicillin-resistant colonies at cycle 8/total number of colonies replicated at cycle 8.
Fitness cost determination.
The fitness cost of plasmid pJMA-1 was determined by eight independent competition experiments between H. influenzae Rd KW20 and H. influenzae Rd KW20(pJMA-1), as previously described (30, 41). Strains were grown in HTM broth for 16 h at 37°C with 5% CO2. Then, 106 CFU of H. influenzae Rd KW20 was mixed with 106 CFU of H. influenzae Rd KW20(pJMA-1) in 2 ml of antibiotic-free HTM. The mixture was grown at 37°C in 5% CO2 at 100 rpm. A total of 2 × 106 CFU was transferred to 2 ml of fresh HTM (1:1,000 dilution) every 24 h. Samples were taken every 24 h for 5 days. For each sample, aliquots were plated on nonselective chocolate agar, and the proportion of resistant colonies was deduced by replica plating of at least 50 colonies on chocolate agar plates containing ampicillin at 128 μg/ml. Relative fitness is expressed as the competition index (CI), calculated as the ratio of the mean number of CFU from eight independent competition experiments between the resistant and susceptible strains at a given time point (t1) divided by the same ratio at time zero. The selection coefficient (s) was calculated as the slope of the linear regression model: s = ln(CI)/t, where t is time, which was the number of bacterial generations calculated as the log2 value of the dilution factor. The selection coefficient estimates the difference between the relative fitness of the two competitors over the entire competition experiment (42).
Nucleotide sequence accession numbers.
The plasmid nucleotide sequences obtained in this study have been deposited in GenBank under the following accession numbers: KP164835 for pJMA-1 from strain F4.1, KP164833 for pB1000 from strain D4.3, KP164834 for pJMA-1 from strain PM5-4, and KP164832 for pB1000 from strain NU5-3.
RESULTS
Genetic features of H. parasuis isolates collected from the nasal cavities of healthy pigs at weaning.
Eighty-six H. parasuis strains were isolated in 2012 from nasal swab samples collected from healthy piglets on eight swine farms located in Navarra, Spain, that had not had a case of Glässer's disease since 2000. Table 1 summarizes the farms, the animals sampled, the isolate ERIC-PCR profiles, and the genetic features of the H. parasuis isolates. Four animals per farm were sampled (n = 32 animals); all animals were positive for the presence of H. parasuis, except for two of the animals sampled on farm E, and 86 isolates were confirmed to be H. parasuis by biochemical, mass spectrometry, and molecular methods. Isolates were genotyped by ERIC-PCR, rendering 29 profiles. Most ERIC-PCR profiles were found on a single farm (farm specific); the exceptions were 3 profiles (NA-2, -29, and -67) for isolates recovered from animals sampled on more than one farm. Nasal isolates with the same ERIC-PCR profile could be recovered from the same animal, from animals sampled on the same farm, and from animals sampled on different farms.
Given that putative virulent strains can be found in the upper respiratory tract of healthy animals (6) and that the presence of the lsgB and group 1 vtaA genes has previously been found to be associated with H. parasuis virulence (12, 13), we assessed their distribution among the collected H. parasuis nasal isolates. The lsgB gene was present in 10 isolates displaying 5 different ERIC-PCR profiles (Table 1). Most lsgB-positive strains were isolated from animals sampled on farm B. The lsgB gene was not found in isolates B4.2 and F4.3, even though they presented the same ERIC-PCR profile as other isolates rendering a positive amplification for this gene. The group 1 vtaA gene was present in 16 isolates from farms B, G, and H displaying 6 different ERIC-PCR profiles (Table 1). Isolates with the same ERIC-PCR profile did not differ regarding the distribution of the group 1 vtaA gene.
In summary, upon nasal swab sampling of 32 healthy animals on 8 farms, H. parasuis was detected in 30, and a total of 86 isolates were obtained. ERIC-PCR-based genotyping showed diversity among the isolates from the different farms. A subset of isolates contained the lsgB and group 1 vtaA genes, previously found to be associated with H. parasuis virulence potential.
Biofilm formation and epithelial inflammation caused by H. parasuis isolates collected from the nasal cavities of healthy pigs at weaning.
Given that biofilm formation has previously been related to a lack of virulence (43), we next asked if H. parasuis isolates collected from the nasal cavities of healthy piglets could form biofilms. Assessment of biofilm growth requires inoculation of comparably grown bacterial cultures. Thus, we first screened the ability of H. parasuis isolates with profiles representative of the previously established ERIC-PCR profiles to grow aerobically in BHI medium supplemented with β-NAD. We found that a significant proportion of isolates displayed aggregative growth or, alternatively, low turbidity after 16 h of growth at 37°C (data not shown). Based on reliable growth in sBHI, we selected isolates A1.2, B2.2, B3.2, F4.1, G4.1, and H1.1 and included the reference strains H. parasuis SW114 (nonvirulent) and Nagasaki (virulent). Under the conditions tested, isolates A1.2, B2.2, B3.2, F4.1, and G4.1 displayed variable biofilm growth on 96-well plates. Strains H1.1, SW114, and Nagasaki did not form biofilms (Table 3). To compare the nature of the biofilm structures, a biofilm detachment assay was performed on 24-well plates. The biofilm structures were sensitive to proteinase K and resistant to sodium meta-periodate (Table 3). These findings were independent of the amount of biofilm formed, suggesting that H. parasuis biofilm formation, although heterogeneous among isolates, may depend on the presence of proteins but not on the presence of sugar components in the extracellular matrix.
TABLE 3.
Absorbance (OD595) of bacterial biofilm detached from polystyrene wells for representative H. parasuis strains
| H. parasuis isolate | OD595 determined on: |
||||
|---|---|---|---|---|---|
| 96-well plates | 24-well plates |
||||
| Sugar dependence |
Protein dependence |
||||
| Buffer | Sodium meta-periodate | Buffer | Proteinase K | ||
| A1.2 | 0.24 ± 0.08 | NDa | ND | ND | ND |
| B2.2 | 0.43 ± 0.07 | 1.43 ± 0.2 | 1.48 ± 0.22 | 1.04 ± 0.1 | 0.11 ± 0.04 |
| B3.2 | 0.29 ± 0.05 | 0.9 ± 0.21 | 0.97 ± 0.11 | 0.57 ± 0.05 | 0.14 ± 0.07 |
| F4.1 | 0.23 ± 0.13 | ND | ND | ND | ND |
| G4.1 | 0.6 ± 0.2 | 1.77 ± 0.29 | 1.63 ± 0.25 | 1.16 ± 0.29 | 0.22 ± 0.06 |
| H1.1 | 0.02 ± 0.02 | ND | ND | ND | ND |
| Nagasaki | 0.07 ± 0.02 | ND | ND | ND | ND |
| SW114 | 0.01 ± 0.01 | ND | ND | ND | ND |
ND, not determined.
H. parasuis has also been shown to induce inflammation, which may occur to different extents depending on the strain (22, 44, 45). We analyzed the ability of the previously selected H. parasuis isolates to trigger IL-8 secretion by PK-15 porcine epithelial cells at 8 h postinfection. All strains stimulated IL-8 secretion by PK-15 cells at comparable levels, independently of their origin or virulence potential (Fig. 1).
FIG 1.
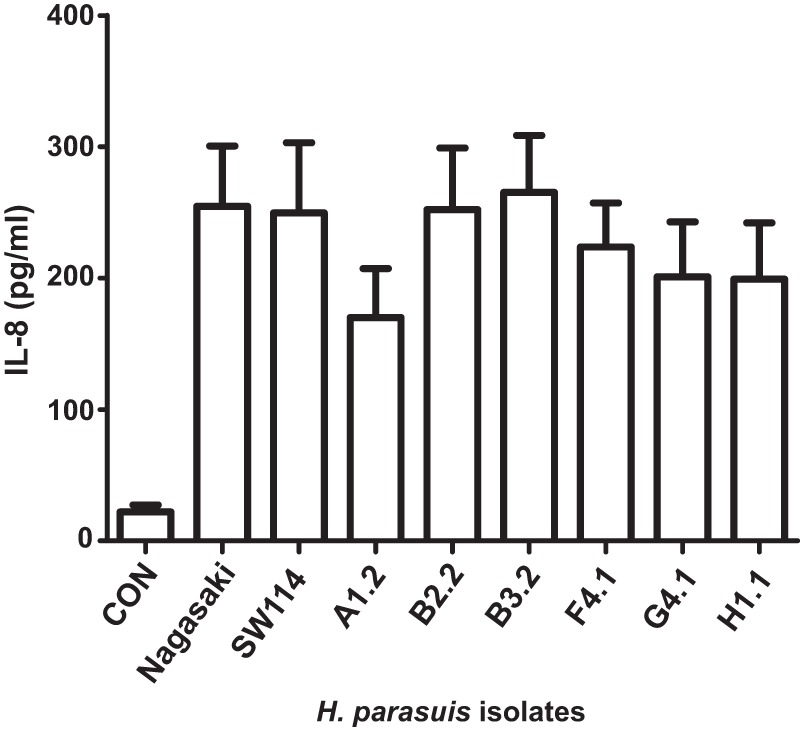
Stimulation of IL-8 secretion by H. parasuis infection of cultured porcine cells. The concentration of IL-8 secreted into the supernatant by PK-15 cells upon infection with H. parasuis strains Nagasaki, SW1174, A1.2, B2.2, B3.2, F4.1, G4.1, and H1.1 was quantified by ELISA at 8 h postinfection. CON, control (uninfected) cells.
These results show protein-dependent biofilm growth for H. parasuis nasal isolates, which, moreover, also triggered secretion of IL-8 by PK-15 cells.
Identification of a novel small plasmid bearing blaROB-1 in colonizing H. parasuis isolates.
Clinical isolates of H. parasuis and P. multocida recovered from animals in Spain have previously been reported to carry the 4,613-bp plasmid pB1000 bearing the blaROB-1 β-lactamase and conferring resistance to β-lactams (11, 29). Animals play important roles as reservoirs and sources of antimicrobial-resistant pathogens (46), due to the zoonotic circulation of resistant bacteria and the spread of antimicrobial resistance determinants between animal and human pathogens (47). In fact, pB1000 has also been found in H. influenzae clinical isolates in Spain, Italy, Australia, and North America (30, 48, 49). Moreover, biofilm formation has been associated with resistance to β-lactams in H. parasuis (28). Considering both observations, the collection of H. parasuis isolates recovered from the nasal cavities of healthy pigs at weaning generated in this study was surveyed for blaROB-1 by PCR. Nine blaROB-1-positive isolates with 4 different ERIC-PCR profiles were identified (Table 1). The β-lactam resistance of one blaROB-1-positive strain per ERIC-PCR profile was determined (Table 4). blaROB-1 is a 918-bp β-lactamase that has been found to confer resistance to ampicillin and that has been found to be carried by different plasmids in Pasteurellaceae species, such as H. influenzae (30, 48, 49), P. multocida (29), H. parasuis (11), Actinobacillus porcitonsillarum (50), and Actinobacillus pleuropneumoniae (51). To characterize the genetic platform bearing blaROB-1 in the nasal isolates under study, we performed a blaROB-1-specific inverted PCR with primers rob-1D and rob-1U, using as the template crude DNA and plasmid purified from the nine blaROB-1-positive isolates. Inverted PCR rendered an approximately 2.2-kb amplicon for isolates A3.1, A3.2, A3.3, D4.1, D4.2, F4.1, F4.2, and F4.3 and an approximately 4.2-kb amplicon for strain D4.3. Purified plasmid extracted from isolate F4.1, used as a representative strain, was completely sequenced by walking PCR using the primers listed in Table 2 (see the Materials and Methods section), rendering a novel plasmid named pJMA-1 (GenBank accession number KP164835). Purified plasmid extracted from strain D4.3 was also completely sequenced and shown to correspond to the previously described pB1000 (GenBank accession number KP164833).
TABLE 4.
MICs of four β-lactams for 16 H. parasuis and 2 H. influenzae strains used in this study
| Strain | MICa (μg/ml) |
|||
|---|---|---|---|---|
| Ampicillin | Amoxicillin | Cefotaxime | Ceftazidime | |
| H. parasuis A3.1 | 64 | 2,048 | 0.12 | ≤0.25 |
| H. parasuis D4.1 | 256 | 512 | ≤0.06 | ≤0.25 |
| H. parasuis D4.3 | 16 | 2,048 | ≤0.06 | ≤0.25 |
| H. parasuis F4.1 | >32 | >2,048 | ≤0.06 | ≤0.25 |
| H. parasuis F9 | 128 | >2,048 | ≤0.06 | ≤0.25 |
| H. parasuis MU21-2 | 32 | >2,048 | ≤0.06 | ≤0.25 |
| H. parasuis V56-2 | 32 | >2,048 | ≤0.06 | 2 |
| H. parasuis SC14-1 | >32 | 1,024 | ≤0.06 | ≤0.25 |
| H. parasuis 416-1 | 128 | >2,048 | ≤0.06 | ≤0.25 |
| H. parasuis MU25-5 | 32 | 1,024 | ≤0.06 | ≤0.25 |
| H. parasuis NU5-3 | >32 | >2,048 | ≤0.06 | ≤0.25 |
| H. parasuis CD7-3 | 256 | >2,048 | ≤0.06 | 1 |
| H. parasuis CD9-1 | 256 | >2,048 | ≤0.06 | ≤0.25 |
| H. parasuis PM5-4 | >32 | 512 | ≤0.06 | ≤0.25 |
| H. influenzae Rd KW20 | <2 | <4 | ND | ND |
| H. influenzae Rd KW20(pJMA-1) | 512 | 512 | ND | ND |
| H. parasuis BB1033 (Amps) | ≤2 | ≤4 | 0.25 | 2 |
| H. parasuis BB1023 (Ampr) | >1,024 | 1,024 | ≤0.06 | ≤0.25 |
Ampicillin and amoxicillin MICs were determined by microdilution. Cefotaxime and ceftazidime MICs were determined by the Sensititre assay. ND, not determined.
The 2,661-bp plasmid pJMA-1 bears blaROB-1 and shares a common backbone with other small plasmids described in the Pasteurellaceae; this common backbone is virtually identical in pJMA-1, pB1000 (30), and pB1002 (29). Furthermore, pJMA-1 is highly similar to pHS-Tet (52), which bears tet(B) instead of blaROB-1. The pJMA-1 sequence, excluding the ca. 90 nucleotides near the origin of replication, is 99% identical to the sequence of pB1002 (Fig. 2), a homologue of pB1000 which bears the transposase ISApl1 (29). ISApl1 is a 1,072-bp insertion sequence (IS) belonging to the IS30 family found in the Pasteurellaceae family (53, 54). pJMA-1 contains the first 119 nucleotides of ISApl1, including the left inverted repeat downstream of the blaROB-1 gene. In pB1000 and pB1002, ISApl1 has been suggested to mobilize blaROB-1 through integration in and duplication of 5 nucleotides (CTGAA/GACTT), caused by the insertion via ISApl1 (29). The same 5 nucleotides are present in pJMA-1 downstream of the ISApl1 deletion site. Interestingly, blaROB-1 from pJMA-1 is inserted in the 5-bp repeat (nucleotides 2126 to 2131 in the pB1000 sequence), whereas blaROB-1 from pB1000 is inserted between nucleotides 2528 and 2532 (Fig. 2), showing that the target sequence of ISApl1 is present in several locations in these plasmids. These observations suggest that the integration/excision of resistance genes through this IS is likely to be a plastic process and may elicit different combinations. pJMA-1 belongs to the ColE1 superfamily. Replication is likely to be mediated by two antisense RNAs, which occurs in all ColE1 plasmids (55). Putative DnaA boxes are encoded between positions 2248 and 2257 and positions 2421 and 2430 (TGATGGATAA and TTTATATATCA, respectively). The putative origin of replication (oriV) is encoded between nucleotides 2156 and 2174 (TTAGTAAAAAGCGTTCTTT). Additionally, plasmid pJMA-1 carries 49 nucleotides of the 5′ end of the mobC relaxase, as well as the putative origin of transfer (oriT; CAAAAAGGGTACTCTTCGAGTTTCCTTTTTGACCTTG), required for plasmid mobilization.
FIG 2.
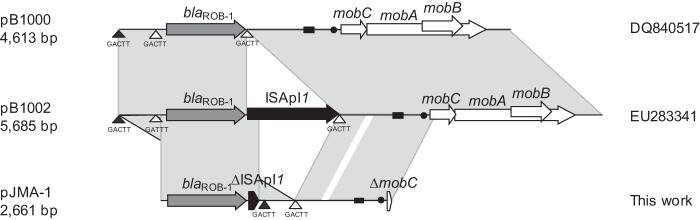
Comparison of the genetic structures of pB1000, pB1002, and pJMA-1. The reading frames for genes are shown as arrows, and the direction of transcription is indicated by arrowheads. Regions with more than 99% identity are shaded gray. Triangles, positions of the CTGAA/GACTT sequence; black triangles, sites used by ISApl1 to integrate blaROB-1 in pJMA-1; white triangles, probable insertion sites used to generate pB1000 and pB1002; black rectangles, putative origins of replication (oriV); black dots, putative transfer origin (oriT). The designations DQ840517 and EU283341 are the GenBank accession numbers for pB1000 and pB1002, respectively.
The presence of pJMA-1 in H. parasuis strains A3.1, A3.2, A3.3, D4.1, D4.2, F4.2, and F4.3 was further confirmed by PCR mapping, using as a template purified plasmid and six PCR combinations with nine primers, as follows: (i) rob-1F and rob-1R, rendering a 457-bp product; (ii) rob-1D and rob-1U, rendering a 2,394-bp product; (iii) plSeqF2 and plSeqR2, rendering a 954-bp; (iv) pB1000-F1 and pB1000-R2, rendering a 494-bp product; (v) rob-1F and pB1000-R2, rendering a 1,280-bp product; and (vi) pl-SEQ-rob-1F and rob-1U, rendering a 522-bp product.
To confirm the functionality of the plasmid and the β-lactamase, the pJMA-1 plasmids purified from the eight H. parasuis strains shown to carry this plasmid were separately transformed into H. influenzae Rd KW20 electrocompetent cells, and transformants were selected on sBHI agar plates containing 25 μg/ml ampicillin. Both H. influenzae Rd KW20(pJMA-1) crude DNA and purified plasmids were used for PCR mapping using the six primer pairs described above, rendering in all cases products compatible with pJMA-1. In conclusion, these results identify the presence of a novel plasmid bearing the blaROB-1 gene and conferring resistance to β-lactams in eight H. parasuis isolates recovered from the nasal cavities of healthy piglets.
Association of pJMA-1 with H. parasuis nasal isolates from healthy pigs at weaning.
Whereas pB1000 was described in β-lactam-resistant H. parasuis isolates recovered from characteristic lesions of Glässer's disease (11), pJMA-1 was found in β-lactam-resistant H. parasuis isolates recovered from healthy animals. To further assess the association of pJMA-1 with H. parasuis isolates from the nasal cavities of healthy pigs, we sampled 19 strains isolated from the nasal cavities of animals on farms located in Catalonia and 1 nasal strain from Mallorca, eastern Spain. Interestingly, 10 isolates were blaROB-1 positive (Table 5). A blaROB-1-specific inverted PCR with primers rob-1D and rob-1U rendered a single amplicon of approximately 2.2 kb in nine isolates and a single amplicon of approximately 4.2 kb in one isolate. The first amplicon was completely sequenced by walking PCR for one representative strain, PM5-4, and its sequences was shown to be identical to that of pJMA-1 (GenBank accession number KP164834). PCR mapping of the remaining eight blaROB-1-positive strains using the six primer pairs described above rendered PCR products with sizes compatible to those shown by pJMA-1, suggesting that all eight strains bore this plasmid. Differently, walking PCR sequencing of purified plasmid from strain NU5-3 rendered pB1000 (GenBank accession number KP164832). The β-lactam resistance profiles of the 10 blaROB-1-positive strains were determined (Table 4). Accordingly, we tested the remaining 10 strains belonging to this set of isolates from healthy piglets for β-lactam resistance and found that they presented low antimicrobial MICs, with all isolates being considered susceptible to ampicillin (data not shown). Serovar information for a subset of these nasal strains (Table 5) showed that pJMA-1-bearing strains belonged to serovars 12 and 14 (virulent), 15 (moderately virulent), and 6 and 7 (nonvirulent), which limits the ability to make a clear-cut association between a particular serovar(s) and the presence of pJMA-1. The pJMA-1 plasmids purified from these nine nasal H. parasuis strains were next separately transformed into H. influenzae Rd KW20, and transformants were selected on sBHI agar plates with 25 μg/ml ampicillin. H. influenzae Rd KW20(pJMA-1) crude DNA and purified plasmids were used for PCR mapping, as described above, rendering in all cases products compatible with pJMA-1. As expected, H. influenzae Rd KW20 transformed with pJMA-1 purified from H. parasuis PM5-4 (chosen as a representative strain) was shown to be resistant to (to have elevated MICs for) ampicillin and amoxicillin (Table 4).
TABLE 5.
Distribution of the blaROB-1 gene and blaROB-1-containing plasmid in H. parasuis nasal isolates from pigs in Catalonia and Mallorca, Spain, sampled at weaning
| H. parasuis nasal isolate | blaROB-1 presence | blaROB-1-containing plasmid size (kb) | β-Lactamase plasmid | Serovar |
|---|---|---|---|---|
| CA38-4 | − | 12 | ||
| SL8-2 | − | NDa | ||
| F9 | + | 2.6 | pJMA-1 | 6 |
| MU21-2 | + | 2.6 | pJMA-1 | 7 |
| ND14-1 | − | 7 | ||
| FL1-3 | − | 10 | ||
| VS6-2 | + | 2.6 | pJMA-1 | 15 |
| SC14-1 | + | 2.6 | pJMA-1 | 15 |
| 416N-1 | + | 2.6 | pJMA-1 | 15 |
| LH9N-4 | − | 10 | ||
| MU25-5 | + | 2.6 | pJMA-1 | 12 |
| NU5-3 | + | 4.6 | pB1000 | ND |
| VB4-1 | − | 7 | ||
| ND19-4 | − | 5 | ||
| SR2-2 | − | 12 | ||
| PO125 | − | ND | ||
| CD7-3 | + | 2.6 | pJMA-1 | 14 |
| CD9-1 | + | 2.6 | pJMA-1 | 15 |
| SN10-1 | − | ND | ||
| PM5-4 | + | 2.6 | pJMA-1 | 6 |
ND, not determined.
We next asked if pJMA-1 could also be found in virulent strains isolated from characteristic lesions of Glässer's disease. We sampled 24 strains isolated from polyserositis or pneumonia lesions. All strains were blaROB-1 and blaROB-1 inverted PCR negative (see Table S1 in the supplemental material). In agreement with that finding, when we tested the β-lactam resistance of this set of strains isolated from lesions, we found that all of them were susceptible to ampicillin (data not shown). Finally, we assessed nontypeable H. influenzae isolates for the presence of pJMA-1. We screened 20 nasopharyngeal isolates from healthy children, 16 ear isolates from children with otitis media, 6 respiratory isolates from children, 33 respiratory isolates from samples from adult patients suffering from bronchiectasis or chronic obstructive pulmonary disease, and 2 isolates from adult patients with pneumonia. All strains were blaROB-1 and blaROB-1 inverted PCR negative (data not shown).
In sum, our strain survey suggests an association between pJMA-1 and H. parasuis strains isolated from the nasal cavities of healthy piglets.
Antibiotic resistance profiles and coexistence with other small plasmids.
To get further insight into the antibiotic resistance of the isolates, 1 representative of each of the 14 genetically different H. parasuis nasal isolates carrying pJMA-1 or pB1000 was further explored. These isolates displayed a whole range of MIC values for sulfamethoxazole, gentamicin, streptomycin, trimethoprim, chloramphenicol, colistin, kanamycin, and nalidixic acid (see Table S2 in the supplemental material). All isolates were sensitive to ciprofloxacin and florfenicol. Of note, isolates F9, SC14-1, and PM5-4 were resistant (they showed elevated MICs) to tetracycline, displaying tetracycline MICs of 16, 16, and 8 μg/ml, respectively. To assess the genetic determinant(s) linked to tetracycline resistance, we used as a template purified plasmid from strains F9, SC14-1, and PM5-4 for PCR detection of different resistance genes (see the primers listed in Table 2). Strain F9 rendered a positive tet(B) amplification, and strain PM5-4 was tet(H) positive. Strain SC14-1 was negative for the tet genes tested [primer pairs Tet(B)-F and Tet(B)-R, Tet(H)-F and Tet(H)-R, and Tet(M/O/S)-F and Tet(M/O/S)-R]. Moreover, inverse PCR with primers Tet(B)invF and Tet(B)invR, annealing in the tet(B) gene, rendered a ca. 4-kb product for strain F9, and inverse PCR with primers Tet(H)invF and Tet(H)invR, annealing in the tet(H) gene, rendered a ca. 6-kb product for strain PM5-4.
Stability and fitness cost of pJMA-1.
We next assessed the stability of plasmid pJMA-1. To do so, cultures of H. influenzae Rd KW20(pJMA-1) were inoculated and propagated every 12 h for eight serial passages in triplicate in medium without antibiotic. One hundred percent of the colonies remained resistant to 25 μg/ml ampicillin after the eight subcultures and maintained pJMA-1, as confirmed by plasmid extraction and PCR mapping as described above.
We also determined the biological cost of pJMA-1. Competition experiments offer discriminative and precise measurements of fitness, since they reflect the competitive disadvantage during all phases of the growth cycle and in several consecutive cycles (56). As in a previous study (29), we performed competition experiments with H. influenzae Rd KW20. Briefly, 2 ml of HTM was inoculated with H. influenzae Rd KW20 and H. influenzae Rd KW20(pJMA-1), and 2 × 106 CFU from this mixture was transferred to 2 ml fresh HTM every 24 h for 5 days. The proportion of bacteria carrying pJMA-1 was measured at the 0-h time point and then every 24 h until day 5. The proportion of pJMA-1-carrying bacteria was fairly well maintained over time. The selection coefficient (s) was established to estimate the difference in relative fitness between the strains in competition over the entire experiment. H. influenzae Rd KW20 bearing pJMA-1 presented a competitive disadvantage of ca. 1.76% per 10 generations relative to the growth of H. influenzae Rd KW20 (Fig. 3).
FIG 3.
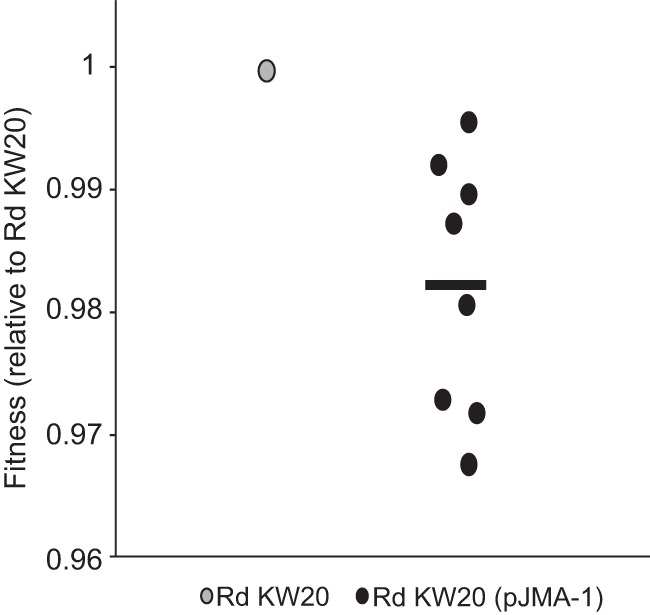
Fitness of Rd KW20(pJMA-1) relative to that of the plasmid-free ancestor strain Rd KW20. Fitness (W) was calculated as W = 1 + s, where s is the selection coefficient (see Materials and Methods). Black dots, fitness of Rd KW20(pJMA-1) relative to that of Rd KW20 (gray dot) obtained in eight independent competition assays; black line, average relative fitness of the eight measurements.
Altogether, these results indicate that pJMA-1 is stably maintained in the H. influenzae Rd KW20 heterologous host and has a weak impact on bacterial fitness.
DISCUSSION
In this study, we explored the genetic diversity of H. parasuis strains isolated from the nasal cavities of healthy piglets at weaning to examine two relevant issues associated with colonization and infection by this opportunistic pathogen: (i) the presence of genetic and phenotypic traits related to the virulence potential of colonizing strains and (ii) the presence of mobile genetic elements linked to resistance to antibiotics in colonizing strains. We first focused on H. parasuis strains isolated from the nasal cavities of healthy animals from eight farms that had not had a case of Glässer's disease since 2000 and found isolates with the same ERIC-PCR profile recovered from the same animal, from animals sampled on the same farm, and from animals sampled on different farms, suggesting the existence of circulating H. parasuis genotypes likely to be commonly found in healthy animals. The group 1 vtaA and lsgB genes, previously associated with the H. parasuis virulence potential (12, 13), were also observed in a subset of the colonizers recovered in this study, in agreement with previous evidence of the presence of strains with virulence potential in the nasal cavities of healthy pigs (57–59). Although most lsgB-positive strains were isolated from animals sampled on farm B, to our knowledge, this farm does not present any feature differentiating it from the other seven farms sampled. The absence of disease caused by the potentially virulent strains detected could be explained by the protection acquired by the piglets through lactation, i.e., the protection conferred by maternal immunity. Moreover, the lsgB gene was found in four out of five isolates presenting ERIC-PCR profile NA-21 and in two out of three isolates presenting ERIC-PCR profile NA-64. We acknowledge that isolates B4.2 (ERIC-PCR profile NA-21) and F4.3 (ERIC-PCR profile NA-64) could lack the lsgB gene or, alternatively, could present a significant number of polymorphisms in this gene, which would make them negative for PCR products after PCR using four different primer pair combinations.
Isolate heterogeneity was found not only at the genomic level but also in terms of the ability of the isolates to reproducibly grow in BHI liquid medium supplemented with β-NAD. This limitation prompted us to select for further analysis a panel of isolates with comparable growth levels. All strains tested triggered the secretion of the proinflammatory cytokine IL-8 by cultured epithelial cells to comparable levels, independently of the presence or absence of the lsgB and group 1 vtaA genes. Following this notion, H. parasuis field strains belonging to serotypes 4 and 5 have been shown to induce the release of IL-8 and IL-6 by porcine brain microvascular endothelial cells to a similar extent (60). A different finding was that H. parasuis has previously been shown to stimulate IL-8 and IL-6 release by newborn pig tracheal cells, and serotype 4 field isolates induced higher levels of these mediators than did serotype 5 isolates (44). The previously observed induction of proinflammatory IL-8 by virulent H. parasuis strains seems to be linked to the capacity of these strains to evade the immune response and multiply inside the host more than to their intrinsic capacity to induce higher levels of inflammation than nonvirulent strains (61).
In previous studies of biofilm formation by H. parasuis, a relationship between colonizers and biofilm growth was suggested (43). In this study, biofilm formation was shown to be heterogeneous; however, the biofilm was sensitive to proteinase K treatment, suggesting the relevance of currently unknown H. parasuis proteins in the formation of the biofilm extracellular matrix. An association between biofilm formation and resistance to β-lactam antibiotics has recently been established for H. parasuis (28), and a relationship between resistance to β-lactams and the presence of the pB1000 plasmid, bearing blaROB-1, has been established for H. parasuis clinical isolates (11). In this study, as part of the diversity encountered among nasal isolates, we found a novel small plasmid named pJMA-1. pJMA-1 was shown to bear blaROB-1 and to be responsible for resistance to β-lactam antibiotics. Of note, pJMA-1 could be found in strains isolated from the nasal cavities of healthy animals sampled in different geographic regions in Spain but not in H. parasuis strains isolated from clinical lesions. pJMA-1 belongs to the ColE1 superfamily and shares a backbone common to other small plasmids described in the Pasteurellaceae, such as pB1000 (30), pB1002 (29), and pHS-Tet (52). Comparative analysis of the pJMA-1, pB1000, and pB1002 nucleotide sequences revealed that β-lactamase acquisition by plasmids pB1000/pB1002 and pJMA-1 seems to have occurred via two different transposition and integration events mediated by ISApl1 within a conserved ColE1 backbone. Thus, the presence of ColE1 plasmids within a population could act both as a reservoir and as a mechanism for the dissemination of antibiotic resistance. Nucleotide analysis of pJMA-1 also showed a putative transfer origin (homologous to that described in pB1000), as well as a region of the mobC relaxase, which could mediate this dissemination. Interestingly, previous analysis of the blaROB-1 gene distribution has shown the existence of a 2.6-kb plasmid in A. pleuropneumoniae and P. multocida swine isolates (62). We currently lack information to evaluate a potential relationship between pJMA-1 and this 2.6-kb plasmid found previously.
Besides its initial identification in H. parasuis, pB1000 has also been detected in P. multocida and nontypeable H. influenzae clinical strains (29, 30). Conversely, we could not detect pJMA-1 in H. influenzae strains isolated from healthy carriers or from clinical samples. In H. influenzae, high-level resistance to β-lactam antibiotics is mediated by the production of the β-lactamases TEM-1 and ROB-1, with TEM-1 being more prevalent than ROB-1 (63). Given its potentially low frequency, we cannot rule out the possibility that pJMA-1 is present in H. influenzae. Wider collections of H. influenzae β-lactamase-positive strains should be screened in future work to increase the probability of identifying pJMA-1 in clinical strains of human origin.
Furthermore, while plasmid pB1000 has a high biological cost (30) and has been found only in infected animals, plasmid pJMA-1 seems to have a weak impact on bacterial fitness (we acknowledge that fitness data for both pJMA-1 and pB1000 have been obtained in the heterologous strain H. influenzae Rd KW20) and has been found only in commensal populations. This means that in the absence of antibiotic pressure, the dynamics of plasmid loss in the population will be slow, while in the presence of antibiotic, bacteria bearing the plasmid will be selected, making the eradication of antibiotic resistance in the animals difficult. In this study, pB1000 was also found in some of the available colonizing strains (2 out of 106) but to a significantly lower extent than pJMA-1 (17 out of 106). pJMA-1 is smaller than pB1000, is stable, can be transferred to other bacterial species, and has a weak impact on bacterial fitness. We speculate that these features could be advantageous for pJMA-1 maintenance in colonizing bacteria as part of the respiratory microbiota of healthy animals and for its potential transfer to other pathogenic bacteria. In addition, we also identified the presence of two plasmids carrying the tetracycline resistance genes tet(B) and tet(H) in two nasal colonizers, and these will be the subject of future work.
In conclusion, this study emphasizes that the importance of commensal bacteria of the respiratory microbiota as a reservoir of antibiotic resistance in animals is largely unknown and remains to be further explored. These aspects have been tackled for the swine intestinal microbiome and were found to be relevant enough to be considered in agricultural cost-benefit analyses (64, 65). It becomes a critical issue when the genes providing resistance are borne on mobile elements, since commensal bacteria can interact with other bacteria, including zoonotic pathogens, and transfer resistance determinants. Altogether, the cautious and controlled use of antibiotics is needed, as it is the only effective action that can be taken to counteract the acquisition and dissemination of antibiotic resistance and the subsequent inefficacy of antibiotic therapies in animal and human health.
Supplementary Material
ACKNOWLEDGMENTS
We thank Saioa Burgui (Instituto de Agrobiotecnología) and Nuria Galofré-Milà (CReSA) for their technical help.
J.M. is funded by Ph.D. studentship BES-2013-062644 from the Ministerio Economía y Competitividad-MINECO, Spain. This work has been funded by grants from MINECO (SAF2012-31166) and the Departamento Industria Gobierno Navarra (IlQ14064.R12) to J.G., MINECO (AGL2013-45662) to V.A., and EU projects EvoTAR 282004-FP7 and EFFORT 613754-FP7 to B.G.-Z.
CIBERES is an initiative from ISCIII, Spain.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03865-14.
REFERENCES
- 1.Cerda-Cuellar M, Naranjo JF, Verge A, Nofrarias M, Cortey M, Olvera A, Segalés J, Aragón V. 2010. Sow vaccination modulates the colonization of piglets by Haemophilus parasuis. Vet Microbiol 145:315–320. doi: 10.1016/j.vetmic.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Costa-Hurtado M, Aragón V. 2013. Advances in the quest for virulence factors of Haemophilus parasuis. Vet J 198:571–576. doi: 10.1016/j.tvjl.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Tang C, Liao M, Yue H. 2014. Update on the pathogenesis of Haemophilus parasuis infection and virulence factors. Vet Microbiol 168:1–7. doi: 10.1016/j.vetmic.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira S, Pijoan C. 2004. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet Microbiol 99:1–12. doi: 10.1016/j.vetmic.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Olvera A, Cerda-Cuellar M, Nofrarias M, Revilla E, Segalés J, Aragón V. 2007. Dynamics of Haemophilus parasuis genotypes in a farm recovered from an outbreak of Glässer's disease. Vet Microbiol 123:230–237. doi: 10.1016/j.vetmic.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Turni C, Blackall PJ. 2010. Serovar profiling of Haemophilus parasuis on Australian farms by sampling live pigs. Aust Vet J 88:255–259. doi: 10.1111/j.1751-0813.2010.00592.x. [DOI] [PubMed] [Google Scholar]
- 7.Mullins MA, Register KB, Brunelle BW, Aragón V, Galofre-Mila N, Bayles DO, Jolley KA. 2013. A curated public database for multilocus sequence typing (MLST) and analysis of Haemophilus parasuis based on an optimized typing scheme. Vet Microbiol 162:899–906. doi: 10.1016/j.vetmic.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Olvera A, Cerda-Cuellar M, Aragón V. 2006. Study of the population structure of Haemophilus parasuis by multilocus sequence typing. Microbiology 152:3683–3690. doi: 10.1099/mic.0.29254-0. [DOI] [PubMed] [Google Scholar]
- 9.Olvera A, Calsamiglia M, Aragón V. 2006. Genotypic diversity of Haemophilus parasuis field strains. Appl Environ Microbiol 72:3984–3992. doi: 10.1128/AEM.02834-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rafiee M, Bara M, Stephens CP, Blackall PJ. 2000. Application of ERIC-PCR for the comparison of isolates of Haemophilus parasuis. Aust Vet J 78:846–849. doi: 10.1111/j.1751-0813.2000.tb10507.x. [DOI] [PubMed] [Google Scholar]
- 11.San Millán A, Escudero JA, Catalán A, Nieto S, Farelo F, Gibert M, Moreno MA, Domínguez L, González-Zorn B. 2007. β-Lactam resistance in Haemophilus parasuis is mediated by plasmid pB1000 bearing blaROB-1. Antimicrob Agents Chemother 51:2260–2264. doi: 10.1128/AAC.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olvera A, Pina S, Macedo N, Oliveira S, Aragón V, Bensaid A. 2012. Identification of potentially virulent strains of Haemophilus parasuis using a multiplex PCR for virulence-associated autotransporters (vtaA). Vet J 191:213–218. doi: 10.1016/j.tvjl.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Martínez-Moliner V, Soler-Llorens P, Moleres J, Garmendia J, Aragón V. 2012. Distribution of genes involved in sialic acid utilization in strains of Haemophilus parasuis. Microbiology 158:2117–2124. doi: 10.1099/mic.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 14.Fu S, Yuan F, Zhang M, Tan C, Chen H, Bei W. 2012. Cloning, expression and characterization of a cell wall surface protein, 6-phosphogluconate dehydrogenase, of Haemophilus parasuis. Res Vet Sci 93:57–62. doi: 10.1016/j.rvsc.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Xu C, Zhang L, Zhang B, Feng S, Zhou S, Li J, Zou Y, Liao M. 2013. Involvement of lipooligosaccharide heptose residues of Haemophilus parasuis SC096 strain in serum resistance, adhesion and invasion. Vet J 195:200–204. doi: 10.1016/j.tvjl.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, He Y, Xu C, Xu L, Feng S, Liao M, Ren T. 2012. Cytolethal distending toxin (CDT) of the Haemophilus parasuis SC096 strain contributes to serum resistance and adherence to and invasion of PK-15 and PUVEC cells. Vet Microbiol 157:237–242. doi: 10.1016/j.vetmic.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M, Zhang Q, Zhao J, Jin M. 2012. Haemophilus parasuis encodes two functional cytolethal distending toxins: CdtC contains an atypical cholesterol recognition/interaction region. PLoS One 7:e32580. doi: 10.1371/journal.pone.0032580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins MA, Register KB, Bayles DO, Butler JE. 2011. Haemophilus parasuis exhibits IgA protease activity but lacks homologs of the IgA protease genes of Haemophilus influenzae. Vet Microbiol 153:407–412. doi: 10.1016/j.vetmic.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Xu X, Wu Y, Li L, Cao R, Cai X, Chen H. 2013. Polysaccharide biosynthesis protein CapD is a novel pathogenicity-associated determinant of Haemophilus parasuis involved in serum-resistance ability. Vet Microbiol 164:184–189. doi: 10.1016/j.vetmic.2013.01.037. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Feng S, Xu C, Zhou S, He Y, Zhang L, Zhang J, Guo L, Liao M. 2012. Serum resistance in Haemophilus parasuis SC096 strain requires outer membrane protein P2 expression. FEMS Microbiol Lett 326:109–115. doi: 10.1111/j.1574-6968.2011.02433.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B, Xu C, Zhang L, Zhou S, Feng S, He Y, Liao M. 2013. Enhanced adherence to and invasion of PUVEC and PK-15 cells due to the overexpression of RfaD, ThyA and Mip in the ΔompP2 mutant of Haemophilus parasuis SC096 strain. Vet Microbiol 162:713–723. doi: 10.1016/j.vetmic.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Zhou S, He X, Xu C, Zhang B, Feng S, Zou Y, Li J, Liao M. 2014. The outer membrane protein P2 (OmpP2) of Haemophilus parasuis induces proinflammatory cytokine mRNA expression in porcine alveolar macrophages. Vet J 199:461–464. doi: 10.1016/j.tvjl.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Miniats OP, Smart NL, Rosendal S. 1991. Cross protection among Haemophilus parasuis strains in immunized gnotobiotic pigs. Can J Vet Res 55:37–41. [PMC free article] [PubMed] [Google Scholar]
- 24.Eaves LE, Blackall PJ, Fegan M. 1989. Characterisation and antimicrobial sensitivity of haemophili isolated from pigs. Aust Vet J 66:1–4. doi: 10.1111/j.1751-0813.1989.tb09701.x. [DOI] [PubMed] [Google Scholar]
- 25.Kofer J, Hinterdorfer F, Awad-Masalmeh M. 1992. Occurrence and drug resistance of bacteria pathogenic to the lungs from autopsy material of swine. Tierarztl Prax 20:600–604. [PubMed] [Google Scholar]
- 26.Wissing A, Nicolet J, Boerlin P. 2001. The current antimicrobial resistance situation in Swiss veterinary medicine. Schweiz Arch Tierheilkd 143:503–510. [PubMed] [Google Scholar]
- 27.de la Fuente AJ, Tucker AW, Navas J, Blanco M, Morris SJ, Gutiérrez-Martín CB. 2007. Antimicrobial susceptibility patterns of Haemophilus parasuis from pigs in the United Kingdom and Spain. Vet Microbiol 120:184–191. doi: 10.1016/j.vetmic.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Xu C, Shen H, Li J, Guo L, Cao G, Feng S, Liao M. 2014. Biofilm formation in Haemophilus parasuis: relationship with antibiotic resistance, serotype and genetic typing. Res Vet Sci 97:171–175. doi: 10.1016/j.rvsc.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 29.San Millán A, Escudero JA, Gutiérrez B, Hidalgo L, García N, Llagostera M, Domínguez L, González-Zorn B. 2009. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob Agents Chemother 53:3399–3404. doi: 10.1128/AAC.01522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.San Millán A, García-Cobos S, Escudero JA, Hidalgo L, Gutiérrez B, Carrilero L, Campos J, González-Zorn B. 2010. Haemophilus influenzae clinical isolates with plasmid pB1000 bearing blaROB-1: fitness cost and interspecies dissemination. Antimicrob Agents Chemother 54:1506–1511. doi: 10.1128/AAC.01489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aragón V, Cerda-Cuellar M, Fraile L, Mombarg M, Nofrarias M, Olvera A, Sibila M, Solanes D, Segalés J. 2010. Correlation between clinico-pathological outcome and typing of Haemophilus parasuis field strains. Vet Microbiol 142:387–393. doi: 10.1016/j.vetmic.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 32.Puig C, Martí S, Fleites A, Trabazo R, Calatayud L, Linares J, Ardanuy C. 2014. Oropharyngeal colonization by nontypeable Haemophilus influenzae among healthy children attending day care centers. Microb Drug Resist 20:450–455. doi: 10.1089/mdr.2013.0186. [DOI] [PubMed] [Google Scholar]
- 33.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, McKenney K, Sutton G, FitzHugh W, Fields C, Gocyne JD, Scott J, Shirley R, Liu L-I, Glodek A, Kelley JM, Weidman JF, Phillips CA, Spriggs T, Hedblom E, Cotton MD, Utterback TR, Hanna MC, Nguyen DT, Saudek DM, Brandon RC, Fine LD, Fritchman JL, Fuhrmann JL, Geoghagen NSM, Gnehm CL, McDonald LA, Small KV, Fraser CM, Smith HO, Venter JC. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira S, Galina L, Pijoan C. 2001. Development of a PCR test to diagnose Haemophilus parasuis infections. J Vet Diagn Invest 13:495–501. doi: 10.1177/104063870101300607. [DOI] [PubMed] [Google Scholar]
- 35.Del Río ML, Gutiérrez CB, Rodríguez-Ferri EF. 2003. Value of indirect hemagglutination and coagglutination tests for serotyping Haemophilus parasuis. J Clin Microbiol 41:880–882. doi: 10.1128/JCM.41.2.880-882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; second informational supplement. CLSI document VET01-S2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard, 4th ed CLSI document VET01-A4 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Dayao DA, Kienzle M, Gibson JS, Blackall PJ, Turni C. 2014. Use of a proposed antimicrobial susceptibility testing method for Haemophilus parasuis. Vet Microbiol 172:586–589. doi: 10.1016/j.vetmic.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, Ramasubbu N. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol 186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason KM, Munson RS Jr, Bakaletz LO. 2003. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect Immun 71:3454–3462. doi: 10.1128/IAI.71.6.3454-3462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutiérrez B, Escudero JA, San Millán A, Hidalgo L, Carrilero L, Ovejero CM, Santos-López A, Thomas-López D, González-Zorn B. 2012. Fitness cost and interference of Arm/Rmt aminoglycoside resistance with the RsmF housekeeping methyltransferases. Antimicrob Agents Chemother 56:2335–2341. doi: 10.1128/AAC.06066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder L, Champness W. 2007. Plasmids, p 208–209. In Molecular genetics of bacteria, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 43.Jin H, Zhou R, Kang M, Luo R, Cai X, Chen H. 2006. Biofilm formation by field isolates and reference strains of Haemophilus parasuis. Vet Microbiol 118:117–123. doi: 10.1016/j.vetmic.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Bouchet B, Vanier G, Jacques M, Auger E, Gottschalk M. 2009. Studies on the interactions of Haemophilus parasuis with porcine epithelial tracheal cells: limited role of LOS in apoptosis and pro-inflammatory cytokine release. Microb Pathog 46:108–113. doi: 10.1016/j.micpath.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Jin H, Chen P, Li Z, Meng X, Liu M, Li S, Shi D, Xiao Y, Wang X, Zhou Z, Bi D, Zhou R. 2012. Haemophilus parasuis infection activates the NF-κB pathway in PK-15 cells through IκB degradation. Vet Microbiol 160:259–263. doi: 10.1016/j.vetmic.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Lloyd DH. 2007. Reservoirs of antimicrobial resistance in pet animals. Clin Infect Dis 45(Suppl 2):S148–S152. doi: 10.1086/519254. [DOI] [PubMed] [Google Scholar]
- 47.Kruse H, Sorum H. 1994. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl Environ Microbiol 60:4015–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.San Millán A, Giufre M, Escudero JA, Hidalgo L, Gutiérrez B, Cerquetti M, González-Zorn B. 2011. Contribution of ROB-1 and PBP3 mutations to the resistance phenotype of a β-lactamase-positive amoxicillin/clavulanic acid-resistant Haemophilus influenzae carrying plasmid pB1000 in Italy. J Antimicrob Chemother 66:96–99. doi: 10.1093/jac/dkq392. [DOI] [PubMed] [Google Scholar]
- 49.Tristram SG, Littlejohn R, Bradbury RS. 2010. blaROB-1 presence on pB1000 in Haemophilus influenzae is widespread, and variable cefaclor resistance is associated with altered penicillin-binding proteins. Antimicrob Agents Chemother 54:4945–4947. doi: 10.1128/AAC.00263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matter D, Rossano A, Sieber S, Perreten V. 2008. Small multidrug resistance plasmids in Actinobacillus porcitonsillarum. Plasmid 59:144–152. doi: 10.1016/j.plasmid.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Chang CF, Yeh TM, Chou CC, Chang YF, Chiang TS. 2002. Antimicrobial susceptibility and plasmid analysis of Actinobacillus pleuropneumoniae isolated in Taiwan. Vet Microbiol 84:169–177. doi: 10.1016/S0378-1135(01)00459-X. [DOI] [PubMed] [Google Scholar]
- 52.Lancashire JF, Terry TD, Blackall PJ, Jennings MP. 2005. Plasmid-encoded Tet B tetracycline resistance in Haemophilus parasuis. Antimicrob Agents Chemother 49:1927–1931. doi: 10.1128/AAC.49.5.1927-1931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, Tan C, Li J, Chen H, Xu P, He Q, Bei W, Chen H. 2008. Characterization of ISApl1, an insertion element identified from Actinobacillus pleuropneumoniae field isolate in China. Vet Microbiol 132:348–354. doi: 10.1016/j.vetmic.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 54.Tegetmeyer HE, Jones SC, Langford PR, Baltes N. 2008. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet Microbiol 128:342–353. doi: 10.1016/j.vetmic.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 55.Tomizawa JI, Ohmori H, Bird RE. 1977. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A 74:1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foucault ML, Courvalin P, Grillot-Courvalin C. 2009. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:2354–2359. doi: 10.1128/AAC.01702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amano H, Shibata M, Kajio N, Morozumi T. 1994. Pathologic observations of pigs intranasally inoculated with serovar 1, 4 and 5 of Haemophilus parasuis using immunoperoxidase method. J Vet Med Sci 56:639–644. doi: 10.1292/jvms.56.639. [DOI] [PubMed] [Google Scholar]
- 58.Amano H, Shibata M, Kajio N, Morozumi T. 1996. Pathogenicity of Haemophilus parasuis serovars 4 and 5 in contact-exposed pigs. J Vet Med Sci 58:559–561. doi: 10.1292/jvms.58.559. [DOI] [PubMed] [Google Scholar]
- 59.Brockmeier SL, Loving CL, Mullins MA, Register KB, Nicholson TL, Wiseman BS, Baker RB, Kehrli ME Jr. 2013. Virulence, transmission, and heterologous protection of four isolates of Haemophilus parasuis. Clin Vaccine Immunol 20:1466–1472. doi: 10.1128/CVI.00168-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouchet B, Vanier G, Jacques M, Gottschalk M. 2008. Interactions of Haemophilus parasuis and its LOS with porcine brain microvascular endothelial cells. Vet Res 39:42. doi: 10.1051/vetres:2008019. [DOI] [PubMed] [Google Scholar]
- 61.Costa-Hurtado M, Olvera A, Martínez-Moliner V, Galofre-Mila N, Martínez P, Domínguez J, Aragón V. 2013. Changes in macrophage phenotype after infection of pigs with Haemophilus parasuis strains with different levels of virulence. Infect Immun 81:2327–2333. doi: 10.1128/IAI.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Juteau JM, Sirois M, Medeiros AA, Levesque RC. 1991. Molecular distribution of ROB-1 β-lactamase in Actinobacillus pleuropneumoniae. Antimicrob Agents Chemother 35:1397–1402. doi: 10.1128/AAC.35.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farrell DJ, Morrissey I, Bakker S, Buckridge S, Felmingham D. 2005. Global distribution of TEM-1 and ROB-1 β-lactamases in Haemophilus influenzae. J Antimicrob Chemother 56:773–776. doi: 10.1093/jac/dki281. [DOI] [PubMed] [Google Scholar]
- 64.Hansen KH, Damborg P, Andreasen M, Nielsen SS, Guardabassi L. 2013. Carriage and fecal counts of cefotaxime M-producing Escherichia coli in pigs: a longitudinal study. Appl Environ Microbiol 79:794–798. doi: 10.1128/AEM.02399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai B, Cole JR, Hashsham SA, Tiedje JM, Stanton TB. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A 109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


