Abstract
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium that considered as important opportunistic human pathogen. One of the mechanisms that help bacteria to tolerate survival in adverse conditions and resistance to antibiotics is biofilm formation through quorum sensing (QS) signals and toxin-antitoxin (TA) systems. QS and TA are two systems that have important roles in biofilm formation. QS is a global regulatory mechanism that enable bacteria to communicate with each other by production of auto inducers (AI) molecules in population. Because of importance biofilm formation in P. aeruginosa infections, here, we studied frequency of QS and TA genes among clinical isolates of P. aeruginosa with ability of biofilm formation.
Materials and Methods
One hundred and forty clinical isolates of P. aeruginosa were collected from Tehran and Ilam hospitals. The isolates were identified by biochemical tests. Biofilm formation was evaluated by microplate method. After DNA extraction by boiling method, the frequency of QS genes (lasIR, rhlIR), and TA genes (mazEF, relBE, hipBA, ccdAB and mqsR) were analyzed by PCR.
Results
Our results showed that maximum resistance is related to aztreonam (72.85%) antibiotic. Most of isolates were able to produce biofilm (87.15%) and the majority of them formed strong biofilm (56.42%). PCR results showed that frequency of mazEF, relBE, hipBA, ccdAB, mqsR, lasIR and rhlIR genes were 85.71, 100, 1.42, 100, 57.14, 93.57 and 83.57 percent, respectively.
Conclusion
Clinical isolates of P. aeruginosa had high ability to form biofilm, and QS and TA system genes among these isolates were very high (except hipBA genes). There are significaut correlation between biofilm for mation and present of QS and TA system genes.
Keywords: P. aeruginosa, Quorum sensing, Toxin-antitoxin systems
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic Gram-negative non-fermenting bacillus that belongs to the family Pseudomonadaceae. It was first isolated from green pus in 1882. P. aeruginosa has minimal nutrition requirements, which contribute to its broad ecological adaptability and distribution (1). It is one the most important nosocomial pathogens that causes infections in the respiratory tract, blood, urinary tract, ear, skin, soft tissues, eye, central nervous system, heart, bone, joint and gastrointestinal tract (2). Infection with P. aeruginosa is a major health problem for immune-compromised patients and individuals with cystic fibrosis. Because it tolerates a variety of harsh physical conditions and has highly adaptable ability to survive in different environments, like hospital environments and equipments such as mechanical ventilators, intravenous lines, urinary or dialysis catheters, pacemakers, endoscopes and sinks and stability in these conditions can be potential reservoirs for contamination (3). Biofilm formation is a mechanism that helps bacteria to resist against antibiotics and immune systems and helps bacteria to survive in poor nutrient conditions (4). Several mechanisms are involved in the biofilm formation i.e. QS and TA systems (5, 6). In fact, QS is a cell signaling mechanism used by many species in response to extracellular signals (autoinducer). Genetic studies show that las (lasIR) and rhl (rhlIR) are two QS systems in P. aeruginosa and AHL or N-acylated hemoserine lactone are as messenger molecules in these systems (7). These systems have important roles in some of physiological and metabolism behaviors such as biofilm formation, virulence, antibiotic resistance and motility (8). TA systems are another systems which are effective in biofilm formation and they are present in almost all prokaryotes (9). A typical TA system consists of two genes (they are coded by the chromosomal or plasmid or both of them), one for a stable toxin and another for an unstable antitoxin. mazEF (mazF toxin and mazE anti-toxin), relBE (relB toxin and relE anti-toxin), hipBA (hipA toxin and hipB anti-toxin), ccdAB (ccdB toxin and ccdA anti-toxin) and mqsRA (mqsR toxin and mqsA anti-toxin) are TA systems that have been identified in bacteria (5, 10-13). TA systems are related to some bacterial behaviors such as biofilm formation, Plasmid maintenance, phase variation, virulence regulation, and genetic competence (14). Because of importance of biofilm formation in pathogenesis of P.aeruginosa, the correlation between lasIR and rhlIR genes and also mazEF, relBE, hipBA, ccdAB and mqsR with ability of biofilm formation in clinical isolates were determined in the present study.
MATERIALS AND METHODS
Collection of bacterial isolates
A total of 140 non-duplicate, clinical isolates of P. aeruginosa were collected from a nationwide distribution of several hospitals in two cities in Iran (Tehran and Ilam) between October 2012 and June 2013 and identified using conventional biochemical tests. All isolates stored at –80°C in TSB containing 20% glycerol.
MIC detection
MIC of antibiotics ceftazidime, piperacillin, ticarcillin, carbenicillin, aztreonam, meropenem, gentamicin and amikacin were done aaccording o CLSI protocol with microdilution method (15).
Polymerase chain reaction amplification
Polymerase chain reaction (PCR) was done to screen all 140 isolates for the presence of the 11 genes, lasI, lasR, rhlI, rhlR, ccdA, ccdB, relA, relB, mqsR, mazE and mazF (Table 1). PCR reaction was done by C1000TM Thermal Cycler (BIO RAD, USA). For DNA extraction, one colony of each isolate cultured in LB broth medium for overnight and the DNA was extracted by boiling method. PCR were performed in a total volume of 25 μl containing 1 μl PCR buffer, 2 mM MgCl2, 2 mM dNTPs, 10 pmol of primers, 0.25 U Taq DNA polymerase (CinnaGen Co, Iran) and 5 μl of template DNA. PCR products were analyzed by electrophoresis on 1% (w/v) agarose gel (Merck, Germany) containing DNA safe. Agarose gels visualized by gel documentation (Gel Doc™ XR+, USA) (16).
Table 1.
Primers used for detection of target genes and PCR programs for amplification
| Target gene | Primer sequences (5’ to 3’) | Amplicon size(bp) | Cycle: 33 |
|||
|---|---|---|---|---|---|---|
| Denaturation (1min) | Annealing (45sec) | Extension (1min) | Final extension (10min) | |||
| lasI | F: GTGTTCAAGGAGCGCAAAGG | 238 | 94 | 61.9 | 72 | 72 |
| R: AACGGCTGAGTTCCCAGATG | ||||||
| lasR | F: TCGAACATCCGGTCAGCAAA | 128 | 94 | 61.9 | 72 | 72 |
| R: GTTCACATTGGCTTCCGAGC | ||||||
| rhlI | F: CCGTTGCGAACGAAATAGCG | 308 | 94 | 61.9 | 72 | 72 |
| R:CAGTTCGACCATCCGCAAAC | ||||||
| rhlR | F:TCGCTCCAGACCACCATTTC | 284 | 94 | 61.9 | 72 | 72 |
| R: GACGGAGGCTTTTTGCTGTG | ||||||
| ccdA | F: GACAGTTGACAGCGACAGCT | 199 | 94 | 58.8 | 72 | 72 |
| R: TCACCAGTCCCTGTTCTCGTC | ||||||
| ccdB | F: GAGAGAGCCGTTATCGTCTGTT | 272 | 94 | 58.7 | 72 | 72 |
| R: TCCCCAGAACATCAGGTTAATG | ||||||
| relE | F: GACGAGCGGGCACTAAAGGAAT | 267 | 94 | 58.6 | 72 | 72 |
| R: TCAGAGAATGCGTTTGACCG | ||||||
| relB | F: ATGGGTAGCATTAACCTGCGT | 240 | 94 | 58.8 | 72 | 72 |
| R: TCAGAGTTCATCCAGCGT | ||||||
| mqsR | F: ACGCACACCACATACACGTT | 194 | 94 | 58.7 | 72 | 72 |
| R: GCCTGGGTCTGTAAACATCCT | ||||||
| mazE | F: ATGATCCACAGTAGCGTAAAGCGT | 249 | 94 | 58.7 | 72 | 72 |
| R: TTACCAGACTTCCTTATCTTTCGG | ||||||
| mazF | F: ATGGTAAGCCGATACGTACCC | 288 | 94 | 58.5 | 72 | 72 |
| R: TGGGGCAACTGTTCCTTT3 | ||||||
| hipA | F: CTTGTCACTTGGATGAACAACCAG | 1314 | 94 | 58.8 | 72 | 72 |
| R: TCACTTACTACCGTATTCTCGGC | ||||||
| hipB | F: AGCCCAACGCAATTGGCGAATGCA | 225 | 94 | 58.7 | 72 | 72 |
| R: CTGTTCTGTTGATTCTGGCGAGGC | ||||||
Microtiter plate biofilm assay
For biofilm formation; (1) isolates were incubated overnight at 37°C in LB broth, (2) Optical density (OD) of bacterial suspension was adjusted between 0.4-0.6 at 600 nm by spectrophotometer, (3) one hundred and ninety μl from LB broth medium was added to wells of polyvinylchloride 96-well microtiter plates and then, (4) 10 μl from bacterial suspension was added to each wells. The isolates were continuously incubated with shaking at 30 rpm at 37°C for overnight. Biofilm assay was performed as triplicate for each isolate, and LB broth medium used as negative control.
Estimation of bacterial biofilm
After incubation, to estimate quantity of biofilm in each well: (1) micro plates were washed with distilled water. (2) The wells stained with 0.1% crystal violet and left at room temperature for 10 min and then washed with distilled water for three times (4). In final step, 200 μl of 95% ethanol was added and OD at 492 nm was measured with an ELISA reader. These OD values were considered as an index of bacteria adhering to surface and forming biofilms. For quantitative analysis of the biofilm production, the average absorbance from the control wells (Ac) was subtracted from the A492 nm of all test wells. Averages and standard deviations were calculated for all experiments. Isolates were classified as follows: A≤Ac=no biofilm producer, Ac<A≤(2×Ac)=weak biofilm producer, (2 ×Ac) <A≤(4 ×Ac) =moderate biofilm producer and (4×Ac) <A=strong biofilm producer (17).
Statistical analysis
Data expressed by percentage, mean and standard deviation (SD). To find correlation between biofilm formation and frequency QS and TA genes, X2 and Fisher’s exact test was used. Monte Carlo method with 10.000 tables with starting seed 200.000 was used when Chi-square was not value. Adjustment was done by using logistic regression. A P values <0.05 were considered to indicate statistical significance.
RESULTS
MIC results
Our findings showed that the resistance of these isolates to the antibiotic that used is high. These results are summarized in Table 2.
Table 2.
MIC results of clinical isolates of P. aeruginosa.
| Antibiotic | N (%) | ||
|---|---|---|---|
| S | I | R | |
| Ceftazidime | 34(24.28%) | 11 (7.85%) | 95(67.85%) |
| Piperacillin | 44 (31.42%) | 12 (8.57%) | 83(59.28%) |
| Ticarcillin | 42 (30%) | 12 (8.57%) | 86 (61.42%) |
| Carbenicillin | 48(34.28%) | 10(7.14%) | 82 (58.57%) |
| Aztreonam | 35 (25%) | 3 (2.14%) | 102(72.85%) |
| Meropenem | 48 (34.28%) | 8 (5.71%) | 84(60%) |
| Gentamicin | 40 (28.57%) | 9(6.42%) | 91 (65%) |
| Amikacin | 49 (35%) | 18 (12.85%) | 73(52.14%) |
| Ciprofloxacin | 47 (33.57%) | 3(2.14%) | 88 (62.85%) |
S: sensitive, I: intermediate, R: resistance.
Biofilm formation results
Microplate method showed that most isolates about 87.15% tend to form biofilm, 12.85% not producing any biofilm. Among biofilm producing strains, 56.42% formed strong biofilm (Table 3).
Table 3.
Results of biofilm formation by microplate method.
| Biofilm formation producer | no producer | Weak producer | Moderate producer | Strong producer | Total |
|---|---|---|---|---|---|
| N (%) | 18 (12.85%) | 13 (19.28%) | 30 (21.42%) | 79 (56.42%) | 140 (100%) |
PCR results
PCR results showed that frequency of mazEF, relBE, hipBA, ccdAB, mqsR, lasIR and rhlIR genes were 85.71, 100, 1.42, 100, 57.14, 93.57 and 83.57 percent, respectively. ccdAB and relBE genes had highest frequency, and in contrast, hipBA genes had the lowest frequency (Figs 1-3).
Fig 1.
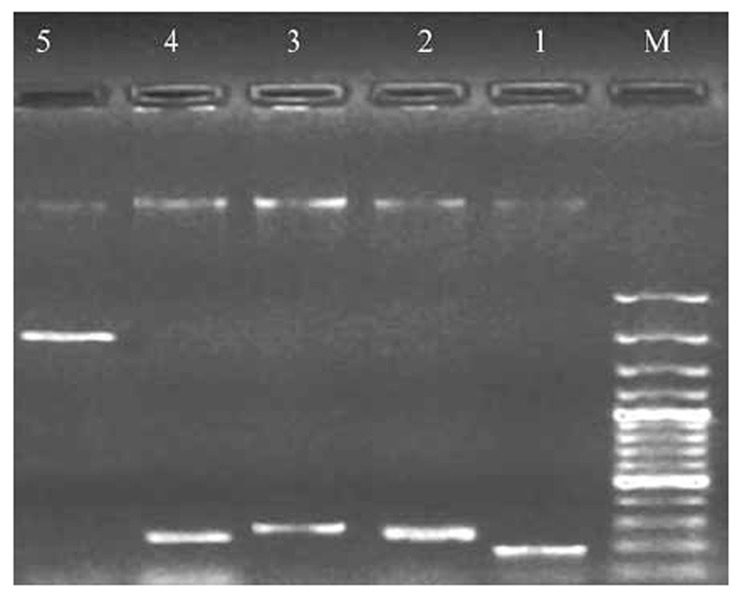
Electrophoresis of PCR product of TA systems genes on the agarose gel 1%.
M: marker 100 bp, 1: mqsR gene 194 bp, 2: ccdB gene272 bp, 3: mazF gene 249 bp, 4: relE gene267 bp, 5: hipA gene 1314 bp
Fig 3.
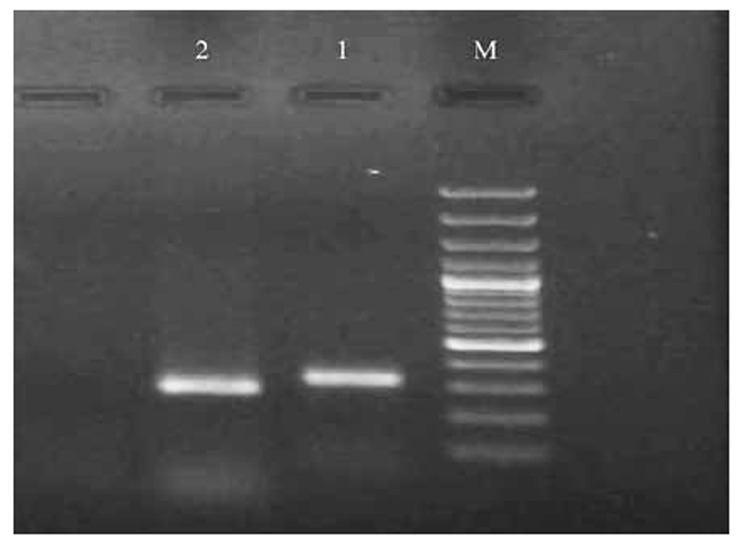
Electrophoresis of PCR products of rhlR and rhlI genes on the agarose gel 1%.
M: marker 100 bp, 1: rhlI gene 308 bp, 2: rhlR gene284 bp
Correlation between the presence of QS system genes with biofilm formation
Monte Carlo method with 10.000 tables with starting seed 200.000 was used to finding correlation between the presence of QS system genes and biofilm formation. Results showed that there are significant correlation between lasIR genes (P=0.012) and rhlIR genes (P=0.030) with biofilm formation.
Correlation between the presence of TA system genes with biofilm formation
Monte Carlo method with 10.000 tables with starting seed 200.000 was used to finding correlation between the presence of QS system genes and biofilm formation. Our finding showed that there are significant correlation between mazEF genes (P=0.002) and mqsR genes (P=0.001) with biofilm formation.
Correlation between the MIC values with biofilm formation
According to our finding of statistical analysis that mentioned above, the results showed that there are significant correlation between MIC values of ceftazidime (P=0.003), meropenem (P=0.002) and amikacin (P=0.001) with biofilm formation.
Correlation between the presence of TA system genes with MIC values
There are significant correlation between the frequency of mazEF genes with resistance to gentamicin (P=0.027), meropenem (P=0.022), piperacillin (P=0.011) and amikacin (P=0.004).
Correlation between the presence of QS with TA system genes
Significant correlations were found between the frequency of mqsR genes and the frequency of lasIR genes (P=0.005) as well as rhlIR genes (P=0.032).
DISCUSSION
Pseudomonas aeuroginosa is an opportunistic pathogen that causes urinary tract infection, respiratory tract infection and burn infections. Cancer, AIDS, immunocompromised statute and patients suffering from cystic fibrosis are more susceptible to these bacteria (18). The organism persists in hospital environment because of its strong resistance to antimicrobial agents and causes nosocomial infections (19). In the environment, bacteria expose to numerous stresses, and response to stress and ensure survival in the population by different mechanisms including biofilm formation, QS and TA systems (5, 20). Bioinformatic analysis of published prokaryotic genomes has been demonstrated the position of TA and QS loci. However, little effort has been made to survey large collections of clinical bacterial isolates for the presence and functionality of these systems (21).
Here, the results of antimicrobial susceptibility showed that resistance of these isolates to antibiotics that used in this study were high. Maximum and minimum resistances were related to aztreonam (72.85%) and amikacin (52.14%). One of the reasons for this high resistance is widespread using of antibiotics in Iran. Antibiotic resistance of strains and their strong ability to form biofilm hinder the eradication of infections with this organism (22). It has been estimated that antibiotic concentration required to kill bacteria in the biofilm is 100 to 1000 fold more than their planktonic form (23).
In this study, the majority of isolates were able to form biofilm (87.15%) and strong biofilm formation was observed in 56.42% of isolates. To find correlation between the MIC values and biofilm formation, we observed significant correlation between MIC values of ceftazidime (P=0.003), meropenem (P=0.002) and amikacin (P=0.001) with biofilm formation. The more biofilm formation, the higher MIC values.
QS systems are mechanisms that regulate biofilm formation. These systems influence on the initiation of biofilm formation and also in process of biofilm maturation (21). In our study, PCR results showed that frequency of QS genes among these isolates was high. In the study by Cabrol et al (2003) frequency of lasR gene was 100% on sixty six isolates of P. aeuroginosa (24).
According to results, there was a significant correlation between the lasIR genes (P=0.012) and rhlIR genes (P=0.030) with biofilm formation. It suggests that the frequency of these genes among the biofilm producing isolates was high.
Concerning TA systems, early studies reported that these systems did not play any role in biofilm formation. For example, in Streptococcus mutans which lacking homologues of the mazF and relE toxin genes had no effect on biofilm formation to compare with parental strains (25). But, recent studies showed that TA systems are involved in biofilm formation (17).
Here, frequency of TA system genes for ccdAB, relBE, mazEF, mqsR and hipBA were 100, 100, 85.71, and 1.42, respectively. In other study; Williams and et al (2011) reported that frequency of relBE and mazEF among clinical isolates of P. aeuroginosa and MRSA were 100%; also Moritz et al (2007) reported that frequency of relBE and mazEF among clinical isolates of vancomycin-resistant enterococci (VRE) were 13% and 93%, respectively (21, 26). However in our study, hipBA genes had minimum frequency. It seems that these genes had no effect on the biofilm formation. But, hipBA genes are involved in biofilm formation in some species such as E.coli (27).
For finding correlation between the presence of TA system genes with biofilm formation, there was a significant correlation between mazEF genes (P=0.002) and mqsR genes (P=0.001) with biofilm formation. It could be concluded that among the isolates that formed biofilm, the frequency of these genes was high.
TA systems are also related to antibiotic resistance (28). In this study, we also observed a significant correlation between the MIC values of gentamicin (P=0.027), meropenem (P=0.022), piperacillin (P=0.011) and amikacin (P=0.004) with mazEF genes. It means that among the isolates that had high frequency of mazEF genes, the MIC values these antibiotics against the isolates were high.
Because both QS and TA systems are involved in biofilm formation, the relation of QS and TA systems with each other was investigated. In previous studies, the relation of MqsR with motility, biofilm formation and produce of autoinducer-2 quorum sensing system, and also the relation of MazF toxin with activity of death factor (EDF) of QS were cleared (29, 30). Here, we also observed significant correlation between mqsR genes with QS system genes and among the isolates that had high frequency of mqsR genes, the frequency of lasIR genes (P=0.005) and rhlIR genes (P=0.032) were high.
Studies showed that different agents are involved in biofilm formation among P. aeuroginosa isolates and QS and TA system are important systems. Thus, knock out essential genes (such as QS and TA system genes) in biofilm formation in the planktonic state can be considered as potential targets to prevention of biofilm formation (31). Understanding the molecular basis of biofilm formation and disturbing these mechanisms can provide a novel approach to prevent biofilm formation and eradication of infections.
finally, our finding showed that these isolates have high ability to biofilm formation (especially strong biofilm) and QS and TA system genes that influence biofilm formation and antibiotic resistance had high frequency in these isolates (except hipBA genes). Thus, because of important physiologic roles of these systems in bacterial, they can candidutes as novel targets for treating infections of multidrug-resistant bacteria specially by using drugs that activate silent toxins of TA systems or drugs that disturbing QS systems (18, 32-35).
Fig 2.
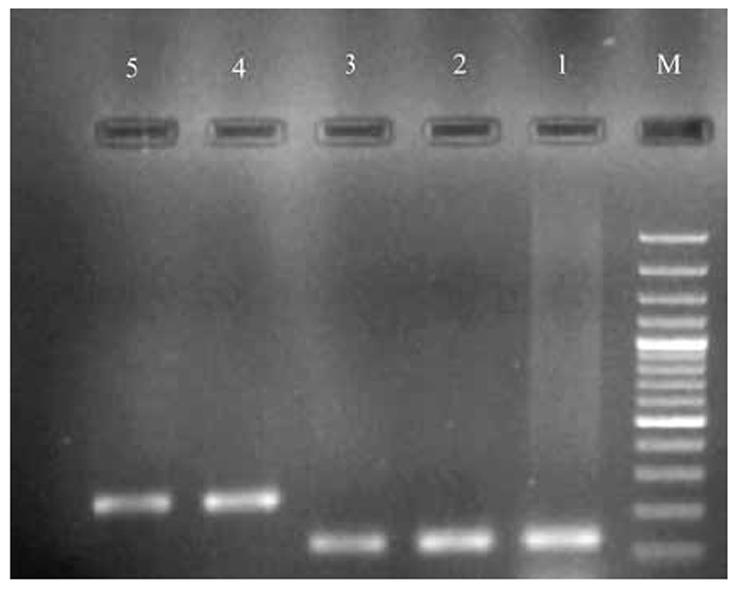
Electrophoresis of PCR products of lasR and lasI genes on the agarose gel 1%.
M: marker 100 bp, 1: lasR gene 128 bp, 2: lasI gene238 bp
Acknowledgments
This research was supported by a grant from Ilam University of Medical Sciences. Authors gratefully acknowledge all the staff in Clinical Microbiology Research Center especially Hassan Valadbeigi.
References
- 1.El Solh AA, Alhajhusain A. Update on the treatment of Pseudomonas aeruginosa pneumonia. J. Antimicrob Chemother. 2009 doi: 10.1093/jac/dkp201. dkp201. [DOI] [PubMed] [Google Scholar]
- 2.Neidig A, Yeung AT, Rosay T, Tettmann B, Strempel N, Rueger M, et al. Typ A is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC microbiology. 2013;13:77. doi: 10.1186/1471-2180-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshe RB, Edwards KM, Vesikari T, Black SV, Walker RE, Hultquist M, et al. Live attenuated versus inactivated influenza vaccine in infants and young children. N Engl J Med. 2007;356:685–696. doi: 10.1056/NEJMoa065368. [DOI] [PubMed] [Google Scholar]
- 4.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. PNAS. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartmann A, Schikora A. Quorum sensing of bacteria and trans-kingdom interactions of N-acyl homoserine lactones with eukaryotes. J Chem Ecol. 2012;38:704–713. doi: 10.1007/s10886-012-0141-7. [DOI] [PubMed] [Google Scholar]
- 8.Szabó MÁ, Varga GZ, Hohmann J, Schelz Z, Szegedi E, Amaral L, et al. Inhibition of quorum-sensing signals by essential oils. Phytother Res. 2010;24:782–786. doi: 10.1002/ptr.3010. [DOI] [PubMed] [Google Scholar]
- 9.Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biology. 2011;9:e1001033. doi: 10.1371/journal.pbio.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett. 2013;340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi Y, Park J-H, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 12.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Drèze P, Van Melderen L. Diversity of bacterial type II toxin–antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr131. gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SJ, Son WS, Lee B-J. Structural overview of toxin–antitoxin systems in infectious bacteria: A target for developing antimicrobial agents. Biochim Biophys Acta. 2013;1834:1155–1167. doi: 10.1016/j.bbapap.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 14.Ghafourian S, Raftari M, Sadeghifard N, Sekawi Z. Toxin-antitoxin Systems: Classifcation, Biological Function and Application in Biotechnology. CIMB,Curr Iss Mol Biol. 2013;16:9–14. [PubMed] [Google Scholar]
- 15.Percival SL, Hill KE, Malic S, Thomas DW, Williams DW. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair Regen. 2011;19:1–9. doi: 10.1111/j.1524-475X.2010.00651.x. [DOI] [PubMed] [Google Scholar]
- 16.Queipo-Ortuño MI, Colmenero JDD, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008;15:293–296. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nature Rev Microbiol. 2011;9:779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M, et al. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 2009;53:2432–43. doi: 10.1128/AAC.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crespo M, Woodford N, Sinclair A, Kaufmann M, Turton J, Glover J, et al. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-β-lactamase, in a tertiary care center in Cali, Colombia. J CLIN MICROBIOL. 2004;42:5094–101. doi: 10.1128/JCM.42.11.5094-5101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mah T-FC OTG. Mechanisms of biofilm resistance to antimicrobial agents. TRENDS MICROBIOL. 2001;9:34–9. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 21.Williams JJ, Halvorsen EM, Dwyer EM, DiFazio RM, Hergenrother PJ. Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2011;322:41–50. doi: 10.1111/j.1574-6968.2011.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann U, Mansouri E, Von Specht B-U. Recombinant OprF-OprI as a vaccine against Pseudomonas aeruginosa infections. VACCINE. 2004;22:840–7. doi: 10.1016/j.vaccine.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 23.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, et al. Gene expression in Pseudomonas aeruginosa biofilms. NATURE. 2001;413:860–4. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 24.Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J BACTERIOL. 2003;185:7222–30. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemos JA, Brown TA, Abranches J, Burne RA. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin–antitoxin modules. FEMS Microbiol Lett. 2005;253:251–7. doi: 10.1016/j.femsle.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 26.Moritz EM, Hergenrother PJ. Toxin–antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc. Natl. Acad. Sci. U.S.A. 2007;104:311–6. doi: 10.1073/pnas.0601168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J MED MICROBIOL. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 28.Anderson G, O’toole G. Bacterial biofilms. 1. Springer. Verlag Berlin Heidelberg; 2008. [Google Scholar]
- 29.Belitsky M, Avshalom H, Erental A, Yelin I, Kumar S, London N, et al. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol Cell. 2011;4:625–35. doi: 10.1016/j.molcel.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 30.Barrios AFG, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–16. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, Fang X, Wood TK, Huang ZJ. A systems-level approach for investigating Pseudomonas aeruginosa biofilm formation. PloS one. 2013;8:e57050. doi: 10.1371/journal.pone.0057050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balaban N, Giacometti A, Cirioni O, Gov Y, Ghiselli R, Mocchegiani F, et al. Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J Infect Dis. 2003;187:625–30. doi: 10.1086/345879. [DOI] [PubMed] [Google Scholar]
- 33.Lioy VS, Rey O, Balsa D, Pellicer T, Alonso JC. A toxin–antitoxin module as a target for antimicrobial development. Plasmid. 2010;63:31–9. doi: 10.1016/j.plasmid.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Williams JJ, Hergenrother PJ. Artificial activation of toxin–antitoxin systems as an antibacterial strategy. Trends Microbiol. 2012;20:291–8. doi: 10.1016/j.tim.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol. 2011;37:121–40. doi: 10.3109/1040841X.2010.532479. [DOI] [PubMed] [Google Scholar]


