Abstract
Background and Objective
Reports on MRSA strains are increasing worldwide. The aim of this study was to find the prevalence of MRSA strains isolated from clinical specimens and to evaluate their resistance profile. Additionally we compared the phenotypic and genotypic methods for detection of methicillin resistance.
Materials and Methods
In this cross-sectional study, a total of 41 isolates of S. aureus were collected from clinical specimens at two teaching hospitals in Ardabil, Iran. All isolates were identified at the species level by standard biochemical tests. The methicillin resistance were evaluated using three methods: PCR for mecA gene, agar dilution for determination of oxacillin MIC and disk diffusion test to detect methicillin, oxacillin and cefoxitin resistance. Antimicrobial resistance patterns were determined by disk diffusion method.
Results
The results identified 19 (46.3%) out of 41 isolates as MRSA. Most of the MRSA strains (68.4%) were isolated from patients hospitalized in ICU. All isolates were susceptible to vancomycin, mupirocin and linezolid. Among other antibiotics co-trimoxazole was more active against MRSA isolates. Using PCR as reference method all the phenotypic tests showed 100% specificity. The sensitivity for MIC test and cefoxitin was 100% and for methicillin and oxacillin disks was 77.7% and 89.5%, respectively.
Conclusion
The prevalence of MRSA strains in our hospitals especially in ICU ward was high and disk diffusion testing using cefoxitin or oxacillin MIC test as an alternative to PCR for detection of MRSA is recommended.
Keywords: Staphylococcus aureus, Methicillin resistance, Antibiotic resistance
INTRODUCTION
Staphylococcus aureus is one of the most important and frequent cause of nosocomial infections worldwide (1). Emergence of methicillin-resistant S. aureus strain (MRSA) in 1961 made staphylococcal infections as a major therapeutic challenge (2). Initially MRSA infections were observed in hospitalized patients and those with chronic illnesses. These types of infections are caused by strains named as hospital-associated MRSA (HA-MRSA) (3). In 1990s another type of MRSA strain was emerged that primarily causes skin and soft tissue infections in healthy people. It is called community-associated MRSA (CA-MRSA) (3). MRSA strains show distinct microbiological, therapeutic and clinical features compared to their methicillin-susceptible (MSSA) counterparts. From microbiological aspect, HA-MRSA strains are resistant to multiple classes of antibiotics. This characteristic limits proper therapeutic options against staphylococcal infections (4). Clinically, infections caused by HA-MRSA strains are associated with higher mortality and morbidity (5). Some CA-MRSA strains express additional virulence factors that enable them to causes more serious diseases (6).
Currently, MRSA strains account for many of staphylococcal infections and reports of MRSA strains are increasing worldwide (7). There are also several reports from Iran showing the prevalence of methicillin resistance among clinical isolates of S. aureus (8-11). A meta-analysis study recently carried out in Iran by Askari et al., showed that the average prevalence rate of MRSA isolates among clinical specimens were 52.7% (12). Understanding the prevalence, antibiotic resistance patterns and information on accurate and reliable detection methods of MRSA strains are necessary for appropriate antibiotic treatment and effective infection control. Considering these, the current study was performed to find the prevalence and evaluate the antimicrobial resistance profile of MRSA strains isolated from clinical specimens in Ardabil, the northwest region of Iran. Additionally we compared the phenotypic and genotypic methods for detection of methicillin resistance.
MATERIALS AND METHODS
Bacterial Isolates
From July to December 2011, a total of 41 S. aureus isolates were collected from patients admitted to two teaching hospitals affiliated to Ardabil University of Medical Sciences. Isolates were examined by conventional methods such as colony morphology on blood agar, Gram stain characteristics and catalase production then identified as S. aureus by tube coagulase and DNase tests. Identified strains were stored at -80°C in Mueller-Hinton broth containing 15% glycerol.
Determination of methicillin resistance
Methicillin resistance was evaluated using three methods: 1) Disk diffusion test using 30 μg cefoxitin disk (≤ 21 mm indicated MRSA), 1 μg oxacillin disk (≤ 10 mm indicated MRSA), and 5 μg methicillin disk (≤ 9 mm indicated MRSA); 2) Oxacillin MIC (Minimum Inhibition Concentration) test (≥ 4 μg/ml indicated MRSA); and 3) Polymerase chain reaction (PCR) for the detection of mecA gene (positive indicated MRSA) (13,14). Antibiotic disks were obtained from Himedia (Himedia Laboratories, Pvt. Ltd, Mumbai, India) and oxacilllin powder for MIC determination was purchased from Sigma-Aldrich (St. Louis, MO, USA). All tests were compared for sensitivity and specificity with PCR for mecA gene as reference method. Sensitivity was calculated by dividing the number of mecA-positive isolates detected as resistant using phenotypic methods by the total number of mecA-positive strains (ether susceptible or resistant). Specificity was calculated through dividing the number of mecA-negative isolates classified as sensitive based on phenotypic criteria by the total number of mecA-negative isolates (15).
Antimicrobial susceptibility testing
The isolates were tested for antibiotic sensitivity using the Kirby Bauer disk diffusion method by employing the following disks (disk); penicillin (10) co-amoxiclav (30), chloramphenicol (30), tetracycline (30), ciprofloxacin (10), ceftriaxone (100), cefazolin (30), clindamycin (2), imipenem (10), co-trimoxazole (25), rifampicin (30), gentamicin (10) pristinamycin (15), linezolid (30) and mupirocin (5). MICs of oxacillin and vancomycin to the both MRSA and MSSA isolates were determined by agar dilution method. All procedures were carried out and interpreted according to CLSI guideline (13). S. aureus ATCC 25923, ATCC 29213 and ATCC 33591 were used as control strains in disk diffusion and agar dilution methods.
PCR amplification of mecA gene
Total bacterial DNA was extracted from S. aureus using DNP™ Genomic DNA Extraction Kit (Cinagen, Tehran, Iran). Oligonucleotide primers (14): 5’- AAAATCGATGGTAAAGGTTGGC-3’ (forward) and 5’- AGTTCTGCAGTACCGGATTTGC-3’ (revers) were synthesized by Bioneer company (Daejon, South Korea). PCR was performed in a 20 × μL AccuPower™ PCR PreMix (Bioneer) with 10 pmol of each primers under the following conditions: initial denaturation at 95°C for 5 min, followed by 34 cycles of 95°C for 1 min, 55°C for 1 min and 72°C for 1min, and a final incubation at 72°C for 5 min. The amplified DNA fragments (PCR product: 533 bp) were separated on 1% (w/v) agarose gel, stained with ethidium bromide and visualized under ultraviolet light. S. aureus ATCC 29213 and ATCC 33591were used as mecA negative and positive controls respectively.
Statistical analysis
Chi-square test was used to compare the prevalence of MRSA and MSSA strains between specimen type and hospital wards.
RESULTS
A total of 41 non duplicate S. aureus isolates including 22 (53.6%) MSSA and 19 (46.3%) MRSA were isolated from different clinical specimens that have been sent to the Microbiology Laboratory. The prevalence of MRSA was significantly higher (P = 0.0001) in sputum (n = 11, 57.8%) than other specimens, respectively, (Table 1). The majority of isolates with MSSA phenotype were cultured from urine specimens (n = 9, 40.9%).
Table 1.
Prevalence of S. aureus among clinical specimens in relationship with specimen type.
| Specimen type |
MSSA n (%) |
MRSA n (%) |
Total |
|---|---|---|---|
| Sputum | 6 (27.3) | 11 (57.8) | 17 |
| Blood | 6 (27.3) | 3 (15.7) | 9 |
| Urine | 9 (40.9) | 1(5.2) | 10 |
| Wound | 1 (4.5) | 3 (15.7) | 4 |
| Cerebral spinal fluid | - | 1(5.2) | 1 |
| Total | 22 (100) | 19 (100) | 41 |
Table 2 shows the distribution of isolate in relationship with hospital wards. The prevalence of MRSA isolates (68.4%) were significantly higher (P = 0.0001) in patients from intensive care unit (ICU). MRSA accounted for about 81.25% of S. aureus strains from patients at ICU.
Table 2.
Prevalence of S. aureus among clinical specimens in relationship with hospital wards.
| Ward |
MSSA n (%) |
MRSA n (%) |
|---|---|---|
| Emergency | 2 (13.3) | 2 (11.1) |
| Surgery | 1 (6.6) | 3 (16.6) |
| Infectious | 4 (26.6) | - |
| Intensive care unit | 3 (20) | 13 (68.4) |
| Outpatient (Clinic) | 4 (26.6) | - |
| Total | 22 (100) | 19 (100) |
The MICs for oxacillin and vancomycin are listed in Table 3. The MICs for oxacillin were between 64 to ≥ 512 μg/mL and ≤ 0.25 to 1 μg/mL for MRSA and MSSA strains, respectively. The MICs for vancomycin against both MRSA and MSSA strains were 1 μg/mL. Only, 1 MRSA strain showed MIC equal to 2 μg/mL. These strains did not fall into vancomycin resistant category according to CLSI (13).
Table 3.
Frequency and range of oxacillin and vancomycin MICs of S. aureus (MRSA and MSSA) isolated from clinical specimens by agar dilution method.
| MIC μg/ml |
Oxacillin |
MIC μg/ml |
Vancomycin |
||
|---|---|---|---|---|---|
| MRSA, n (%) |
MSSA, n (%) |
MRSA, n (%) |
MSSA, n (%) |
||
| ≤ 0.25 | - | 14 (63.6) | 1 | 18 (94.7) | 22 (100) |
| 0.5 | - | 2 (9) | 2 | 1 (5.3) | - |
| 1 | - | 6 (27.2) | |||
| 64 | 2 (25) | ||||
| 128 | 1(12.5) | ||||
| ≥ 512 | 16 (62.5) | ||||
Table 4 represents the resistance pattern of S. aureus isolates (MRSA and MSSA) to the tested antibiotics. In this study the entire S. aureus isolates were susceptible to vancomycin, mupirocin and linezolid. Among other antibiotics imipenem and co-trimoxazole showed to be the most effective antibiotics against MRSA isolates. PCR testing revealed the presence of mecA gene in all isolates (Fig 1) which were determined as methicillin resistant by the phenotypic methods. The sensitivities of oxacillin MIC test and cefoxitin disk were 100%, whereas the sensitivities of methicillin and oxacillin disks were 77.7% and 89.5% respectively.
Table 4.
Antibiotic susceptibility profiles of S. aureus strains isolated from clinical specimens by disk diffusion method.
| Antibiotic | MSSA (N = 22), n (%) |
MRSA (N = 19), n (%) |
||||
|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | Susceptible | Intermediate | Resistant | |
| Vancomycina | 22 (100) | - | - | 19 (100) | - | - |
| Penicillin | 6 (27.2) | - | 16 (72.7) | - | - | 19 (100) |
| Co-amoxiclav | 10 (45.4) | - | 12 (54.5) | 2 (10.5) | - | 17 (89.4) |
| Chloramphenicol | 22 (100) | - | - | - | - | 19 (100) |
| Tetracycline | 20 (90.9) | - | 2 (9) | 3 (15.7) | 16 (84.2) | |
| Ciprofloxacin | 22 (100) | - | - | 1 (5.2) | 5 (26.3) | 13 (68.4) |
| Ceftriaxone | 16 (72.7) | 6 (27.2) | - | 2 (10.5) | - | 17 (89.4) |
| Cefazolin | 21 (95.4) | - | 1 (4.5) | 3 (15.7) | 1 (5.2) | 15 (78.9) |
| Clindamycin | 20 (90.9) | - | 2 (9) | 1 (5.2) | - | 18 (94.7) |
| Imipenem | 22 (100) | - | - | 14 (73.6) | 3 (15.7) | 2 (10.5) |
| Co-trimoxazole | 18 (81.8) | 1 (4.5) | 3 (13.6) | 17 (89.4) | - | 2 (10.5) |
| Erythromycin | 20 (90.9) | - | 2 (9) | 3 (15.7) | - | 16 (84.2) |
| Gentamicin | 22 (100) | - | - | 4 (21) | - | 15 (78.9) |
| Rifampicin | 22 (100) | - | - | 3 (15.7) | - | 16 (84.2) |
| Pristinamycin | 18 (81.8) | 1 (4.5) | 3 (13.6) | 5 (26.3) | - | 14 (73.6) |
| Linezolid | 22 (100) | - | - | 19 (100) | - | - |
| Mupirocin | 22 (100) | - | - | 19 (100) | - | - |
Vancomycin susceptibility profile was determined using agar dilution method.
Fig1.
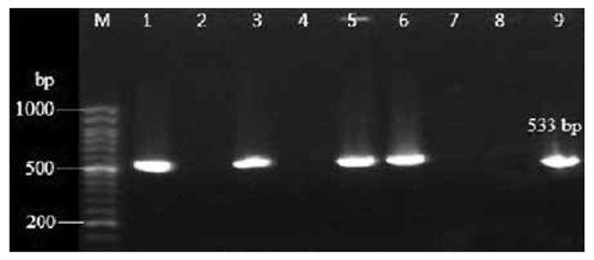
PCR detection of mecA gene among S. aureus isolates. Lane M: 50 pb DNA size marker, Lane 1: positive control strain ATCC 33591. Lane 2: negative control strain ATCC 29213. Lanes 3, 5, 6 and 9 mecA positive isolates. Lanes: 4, 7 and 8 mecA negative isolates.
DISCUSSION
Since the emergence of MRSA in 1961, there has been a steady increase in the prevalence of this type of S. aureus strains worldwide (7). Currently, more than 50% of S. aureus infections are caused by MRSA strains in the US (3). The reports from Iran also indicate the increasing incidence of MRSA in clinical specimens over the time (8-12). In this study out of 41, 19 (46.3%) of isolates were MRSA strains. Worldwide, HA-MRSA prevalence varies considerably, from <1 percent in Scandinavia to >50 percent in other countries (7). The estimated prevalence in our study locates in upper limits of the reported ranges. However there is not a uniform prevalence reported for MRSA infection in the different studies. The heterogeneity is probably due to applying different infection control measures, antibiotic administration, human population, study design and laboratory testing for determining methicillin resistance (12). In this study methicillin and oxacillin disks could not detect all MRSA isolates but cefoxitin disk and oxacillin MIC test showed the sensitivity equal to PCR. These results are similar to the previous reports (16). However, the emergence of mecA positive oxacillin susceptible and mecA negative oxacillin resistant–MRSA strains reduces the sensitivities of both the phenotypic and genotypic methods (17-19). Thus, combination of genotypic and phenotypic tests is necessary to detect the methicillin resistance in S. aureus accurately.
MRSA infections pose a significant concern for ICU patients (19). In this study, the incidence rate of MRSA infection in ICU patients was significantly higher than other wards, with an estimated prevalence as high as 68.4% and within ICU MRSA strains were responsible for about 81% of S. aureus infections. Previously it has been documented that MRSA accounted for 57% of all ICU acquired S. aureus infections (19). However, recent reports indicate declining ICU acquired MRSA infections with applying appropriate infection control measures, rapid and reliable detection of methicillin resistance and effective antibiotic therapy (2, 20).
In this study, all isolates were susceptible to vancomycin, mupirocin, linezolid. The absence of resistance to mupirocin may be related to the low usage of this antibiotic in the study setting. However, others have recently reported the incidence of high-level mupirocin resistant S. aureus strains isolated from patients in Iran (11). Mupirocin is topical agent often used to eradicate nasal carriage and control outbreaks of methicillin-resistant S. aureus (11).
The vancomycin is the drug of choice for the treatment of infections due to MRSA (21). Several studies reported emergence of vancomycin resistant clinical MRSA isolates around the world (22-24). In our study all of the isolates displayed MICs of ≤ 2 μg/ml to vancomycin and were susceptible to vancomycin.
Multiple-drug resistant characteristics of MRSA and emergence of glycopeptide resistant strains have been frequently caused treatment failure of MRSA infections (25). These findings have promoted researchers to seek new antibiotics for the treatment of MRSA infections (26). Linezolid and pristinamycin showed good activity in vitro and in vivo and are promising therapeutic options against staphylococcal infections (27-28). In this study all isolates were susceptible to linezolid. Similar to our previous study on S. aureus strains isolated from health care workers in the same setting (15), 3 (13.6%) MSSA and 14 (73.6%) MRSA strains were found to be resistant to pristinamycin. This antibiotic is not generally used in Iran for treatment of bacterial infectious. Therefore, emergence of pristinamycin resistant S. aureus strains in our hospitals could be surprising. There is also a similar finding that has been reported from India (29). Some previous studies demonstrated that antimicrobial selective pressure and microbial cross-transmission are involved in pristinamycin resistance acquisition in S. aureus (30).
The high resistance rate for most commonly used antibiotics was observed among MRSA isolates in comparison to MSSA. Most of the MRSA strains were resistant against multiple classes of commonly used antibiotics. Except for above mentioned antibiotics, co-trimoxazol and imipenem showed the lowest resistance rate for MRSA isolates. However, MRSA strains should be considered as resistant to all β-lactam agents other than the cephalosporins with anti-MRSA activity as stated by CLSI (13). Resistance of MRSA to co-trimoxazol in general is low. Several studies reported a decrease in resistance of MRSA to co-trimoxazol over the time (31-32).
In conclusion, the frequency of MRSA infection in our hospitals was found to be high and this finding highlights the need for applying appropriate infection control measures and effective antibiotic therapy. Moreover, results emphasize the use of cefoxitin disk diffusion or oxacillin MIC tests as accurate phenotypic methods in clinical laboratories if PCR for mecA gene detection is not feasible.
Acknowledgments
This work was performed in partial fulfilment of the requirements for M.Sc thesis in Microbiology (Solmaz Dibah, Zanjan Branch, Islamic Azad University). We gratefully acknowledge Ardabil University of Medical Sciences for providing laboratory facilities for this work.
References
- 1.Kluytmans JA, Wertheim HF. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection. 2005;33:3–8. doi: 10.1007/s15010-005-4012-9. [DOI] [PubMed] [Google Scholar]
- 2.Johnson AP. Methicillin-resistant Staphylococcus aureus: the European landscape. J Antimicrob Chemother. 2011;66(Suppl 4):43–48. doi: 10.1093/jac/dkr076. [DOI] [PubMed] [Google Scholar]
- 3.Watkins RR, David MZ, Salata RA. Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus. J Med Microbiol. 2012;61:1179–1193. doi: 10.1099/jmm.0.043513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eliopoulos GM. Microbiology of drugs for treating multiply drug-resistant Gram-positive bacteria. J Infect. 2009;59(Suppl 1):S17–24. doi: 10.1016/S0163-4453(09)60004-9. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003;36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 6.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46(Suppl 5):S350–359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, et al. Methicillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents. 2012;39:273–282. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Sadeghi J, Mansouri S. Molecular characterization and antibiotic resistance of clinical isolates of methicillin-resistant Staphylococcus aureus obtained from Southeast of Iran (Kerman) APMIS. 2013 doi: 10.1111/apm.12158. [DOI] [PubMed] [Google Scholar]
- 9.Havaei SA, Azimian A, Fazeli H, Naderi M, Ghazvini K, Mirab Samiee S, et al. Isolation of Asian endemic and livestock associated clones of methicillin resistant Staphylococcus aureus from ocular samples in Northeastern Iran. Iran J Microbiol. 2013;5:227–232. [PMC free article] [PubMed] [Google Scholar]
- 10.Hasani A, Sheikhalizadeh V, Naghili B, Valizadeh V, Nikoonijad AR. Methicillin resistant and susceptible Staphylococcus aureus: Appraising therapeutic approaches in the Northwest of Iran. Iran J Microbiol. 2013;5:56–62. [PMC free article] [PubMed] [Google Scholar]
- 11.Abbasi-Montazeri E, Khosravi AD, Feizabadi MM, Goodarzi H, Khoramrooz SS, Mirzaii M, et al. The prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolates with high-level mupirocin resistance from patients and personnel in a burn center. Burns. 2013;39:650–654. doi: 10.1016/j.burns.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Askari E, Soleymani F, Arianpoor A, Tabatabai SM, Amini A, Nasab MN. Epidemiology of mecA-methicillin resistant Staphylococcus aureus in Iran: A systematic review and meta-analysis. Iran J Basic Med Sci. 2012;15:1010–1019. [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-first informational supplement M100-MS21. Wayne[PA]: Clinical and Laboratory Standards Institute; 2011. [Google Scholar]
- 14.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulder JG. Comparison of disk diffusion, the E test, and detection of mecA for determination of methicillin resistance in coagulase-negative staphylococci. Eur J Clin Microbiol Infect Dis. 1996;15:567–573. doi: 10.1007/BF01709365. [DOI] [PubMed] [Google Scholar]
- 16.Farahani A, Mohajeri P, Gholamine B, Rezaei M, Abbasi H. Comparison of different phenotypic and genotypic methods for the detection of methicillin-resistant Staphylococcus aureus. N Am J Med Sci. 2013;5:637–640. doi: 10.4103/1947-2714.122305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hososaka Y, Hanaki H, Endo H, Suzuki Y, Nagasawa Z, Otsuka Y, et al. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: a new type of MRSA. J Infect Chemother. 2007;13:79–86. doi: 10.1007/s10156-006-0502-7. [DOI] [PubMed] [Google Scholar]
- 18.Jannati E, Arzanlou M, Habibzadeh S, Mohammadi S, Ahadi P, Mohammadi-Ghalehbin B, et al. Nasal colonization of mecA-positive, oxacillin-susceptible, methicillin-resistant Staphylococcus aureus isolates among nursing staff in an Iranian teaching hospital. Am J Infect Control. 2013;41:1122–1124. doi: 10.1016/j.ajic.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Vincent JL, Bihari DJ, Suter PM, Bruining HA, White J, Nicolas-Chanoin MH, et al. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee. JAMA. 1995;274:639–644. [PubMed] [Google Scholar]
- 20.Hakan E, Selma K, Selcuk K, Tuna D, Iftihar K, Asuman I, et al. Withdrawal of Staphylococcus aureus from intensive care units in Turkey. Am J Infect Control. 2013;41:1053–1058. doi: 10.1016/j.ajic.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz FJ, Jones ME. Antibiotics for treatment of infections caused by MRSA and elimination of MRSA carriage. What are the choices? Int J Antimicrob Agents. 1997;9:1–19. doi: 10.1016/s0924-8579(97)00027-7. [DOI] [PubMed] [Google Scholar]
- 22.Aligholi M, Emaneini M, Jabalameli F, Shahsavan S, Dabiri H, Sedaght H. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med Princ Pract. 2008;17:432–434. doi: 10.1159/000141513. [DOI] [PubMed] [Google Scholar]
- 23.Askari E, Tabatabai SM, Arianpoor A, Nasab MN. VanA-positive vancomycin-resistant Staphylococcus aureus: Systematic search and review of reported cases. Infect Dis Clin Pract. 2013;21:91–93. [Google Scholar]
- 24.Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Mirab Samiee S, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol. 2012;50:3581–3585. doi: 10.1128/JCM.01727-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruef C. Epidemiology and clinical impact of glycopeptide resistance in Staphylococcus aureus. Infection. 2004;32:315–327. doi: 10.1007/s15010-004-4124-7. [DOI] [PubMed] [Google Scholar]
- 26.Lentino JR, Narita M, Yu VL. New antimicrobial agents as therapy for resistant gram-positive cocci. Eur J Clin Microbiol Infect Dis. 2008;27:3–15. doi: 10.1007/s10096-007-0389-y. [DOI] [PubMed] [Google Scholar]
- 27.Lyseng-Williamson KA, Goa KL. Linezolid: in infants and children with severe gram-positive infections. Paediatr Drugs. 2003;5:419–429. doi: 10.2165/00128072-200305060-00009. [DOI] [PubMed] [Google Scholar]
- 28.Ng J, Gosbell IB. Successful oral pristinamycin therapy for osteoarticular infections due to methicillin-resistant Staphylococcus aureus (MRSA) and other Staphylococcus spp. J Antimicrob Chemother. 2005;55:1008–1012. doi: 10.1093/jac/dki108. [DOI] [PubMed] [Google Scholar]
- 29.Keshari SS, Kapoor AK, Kastury N, Singh DK, Bhargava A. Emergence of pristinamycin resistance in India. Indian J Pharmacol. 2009;41:47–48. doi: 10.4103/0253-7613.48884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verneuil L, Marchand C, Vidal JS, Ze Bekolo R, Daurel C, Lebouvier G, Leroy D, Leclercq R. Factors associated with emergence of pristinamycin-resistant Staphylococcus aureus in a dermatology department: a case-control study. Br J Dermatol. 2010;163:329–333. doi: 10.1111/j.1365-2133.2010.09826.x. [DOI] [PubMed] [Google Scholar]
- 31.Samra Z, Gadba R, Ofir O. Antibiotic susceptibility and phage typing of methicillin-resistant and -sensitive Staphylococcus aureus clinical isolates at three periods during 1991-1997. Eur J Clin Microbiol Infect Dis. 2001;20:425–427. doi: 10.1007/pl00011284. [DOI] [PubMed] [Google Scholar]
- 32.Bishara J, Pitlik S, Samra Z, Levy I, Paul M, Leibovici L. Co-trimoxazole-sensitive, methicillin-resistant Staphylococcus aureus, Israel, 1988-1997. Emerg Infect Dis. 2003;9:1168–1169. doi: 10.3201/eid0909.020666. [DOI] [PMC free article] [PubMed] [Google Scholar]


