SUMMARY
Outbreaks of fatal leukemia-like cancers of marine bivalves throughout the world have led to massive population loss. The cause of the disease is unknown. We recently identified a retrotransposon, Steamer, that is highly expressed and amplified to high copy number in neoplastic cells of soft-shell clams (Mya arenaria). Through analysis of Steamer integration sites, mitochondrial DNA single nucleotide polymorphisms (SNPs), and polymorphic microsatellite alleles, we show that the genotypes of neoplastic cells do not match those of the host animal. Instead, neoplastic cells from dispersed locations in New York, Maine, and Prince Edward Island (PEI), Canada, all have nearly identical genotypes that differ from those of the host. These results indicate that the cancer is spreading between animals in the marine environment as a clonal transmissible cell derived from a single original clam. Our findings suggest that horizontal transmission of cancer cells is more widespread in nature than previously supposed.
INTRODUCTION
Cancers most often arise as a result of mutations accumulated in somatic cells during the lifetime of an organism, and constitute a clonal expansion of a transformed cell with a genotype derived from the host. Tumors are generally not contagious or transmitted to other individuals, being subject to immune recognition and rejection based on polymorphic surface proteins, notably the major histocompatibility complex (MHC) in vertebrates. Some tumors are induced by infectious agents such as viruses, and though these agents can be contagious, each tumor still arises in the infected individual by transformation of somatic cells. Two cases are known in which a tumor cell itself naturally spreads among individuals as a contagious cell line. These are the canine transmissible venereal tumor (CTVT) (Murgia et al., 2006), transmitted by sexual contact, and the Tasmanian devil facial tumor disease (DFTD) (Pearse and Swift, 2006), transmitted between individuals by bites. In these two cases, the tumors exhibit a genotype that does not match that of their host. Instead, the tumor cells found in all the affected animals are a single clone with a unique genotype that reflects that of its original, primordial host.
Disseminated, or hemic, neoplasia is a leukemia-like cancer occurring in many marine bivalves, including clams, mussels, cockles, and oysters, and is characterized by amplification of cells in the hemolymph, the circulatory fluid of molluscs (Barber, 2004). The disease affects soft-shell clams (Mya arenaria) along the east coast of North America and was first documented in the species in the 1970s (Brown et al., 1978; Muttray et al., 2012; Yevich and Barszcz, 1978). The neoplastic hemocytes are characterized by distinctive morphology, new surface antigens, cytoplasmic mislocalization of the p53 tumor suppressor protein, loss of phagocytic abilities, greatly increased number (Figure 1), and dissemination of the cells into tissues (Barber, 2004; Miosky et al., 1989; Smolowitz et al., 1989; Walker et al., 2006; Walker et al., 2009). Most bivalve disseminated neoplasias are aneuploid, with higher than normal DNA content, and the neoplastic cells of M. arenaria are roughly tetraploid based on flow cytometric analysis of DNA content, although there is variation between individuals (Reno et al., 1994). The disease can be transmitted to naïve animals by experimental transplantation of hemocytes (McLaughlin et al., 1992; Weinberg et al., 1997). A viral cause has been suspected but no infectious agent has been confirmed (Taraska and Bottger, 2013; Walker et al., 2009). The disease is ultimately fatal in the majority of affected clams and is contributing to depletion of the species in many areas along the east coast of North America (Barber, 2004; Cooper et al., 1982).
Figure 1. Collection of normal and leukemic soft-shell clams (M. arenaria).
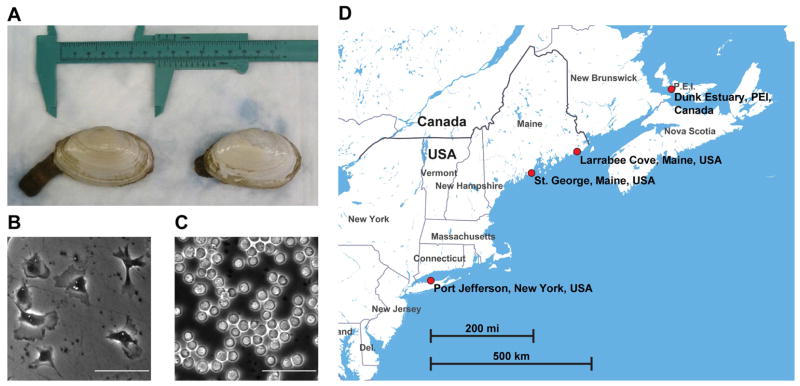
(A) Photograph of representative soft-shell clams (M. arenaria) with siphon partially extended.
(B) Hemolymph from a normal clam (NYTC-C6), showing attachment of hemocytes to the dish and extension of pseudopodia. Scale bar: 50 μm.
(C) Hemolymph from a heavily leukemic clam (NYTC-C9) showing lack of attachment and rounded, refractile morphology. Hemolymph of the leukemic clam was diluted 1:100 to allow visualization of single cells. Scale bar: 50 μm.
(D) Map of the eastern coast of North America with the locations of the clam collection sites (Made with Mapbox Studio using data from OpenStreetMaps).
The detection of reverse transcriptase activity in neoplastic cells (House et al., 1998) suggested the possibility of retroelement involvement, and we used high-throughput sequencing of clam cDNA to identify a previously unknown LTR-retrotransposon in soft-shell clams, Steamer, whose expression was found to be strongly associated with disease (Arriagada et al., 2014). In normal clams the genome contains 2–10 endogenous copies of the retrotransposon Steamer (normalized to a single copy gene), but in neoplastic hemocytes the Steamer DNA copy number is massively amplified to 150–300 copies. The finding of several common Steamer integration sites in the neoplastic cells of multiple leukemic animals prompted further investigation of the genetics of this cancer.
Here, we first extend the observation that neoplastic cells contain common Steamer integration sites that are not present in normal animals or in normal tissues of diseased animals. These results have two possible explanations: either Steamer retrotransposons exhibit unprecedented selectivity for specific integration sites in these multiple neoplasms, or these neoplasms did not arise independently but are descendants from a primordial leukemic cell carrying these common Steamer integrations, similar to the contagious cancers observed in dogs and Tasmanian devils. We therefore analyzed mitochondrial DNA (mtDNA) sequences and polymorphic microsatellite repeat loci and found that the genotypes of the neoplastic cells do not match those of their hosts. Furthermore, all neoplastic genotypes are nearly identical to each other, strongly arguing that soft-shell clam leukemia across the Atlantic coast is horizontally transmitted as a contagious cancer cell derived from a single clonal origin.
RESULTS
Common Steamer Integration Sites in Neoplastic Cells
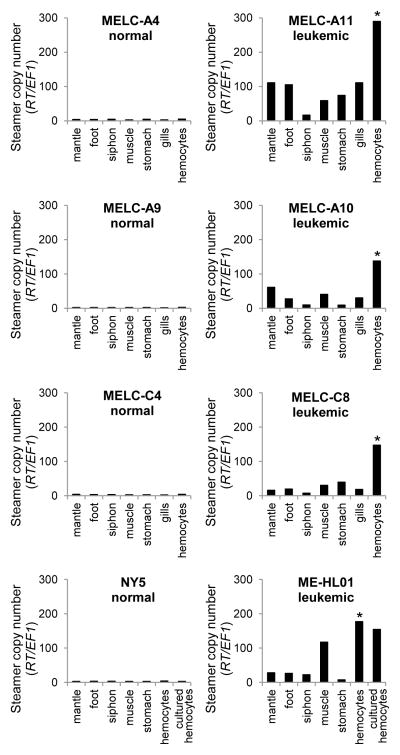
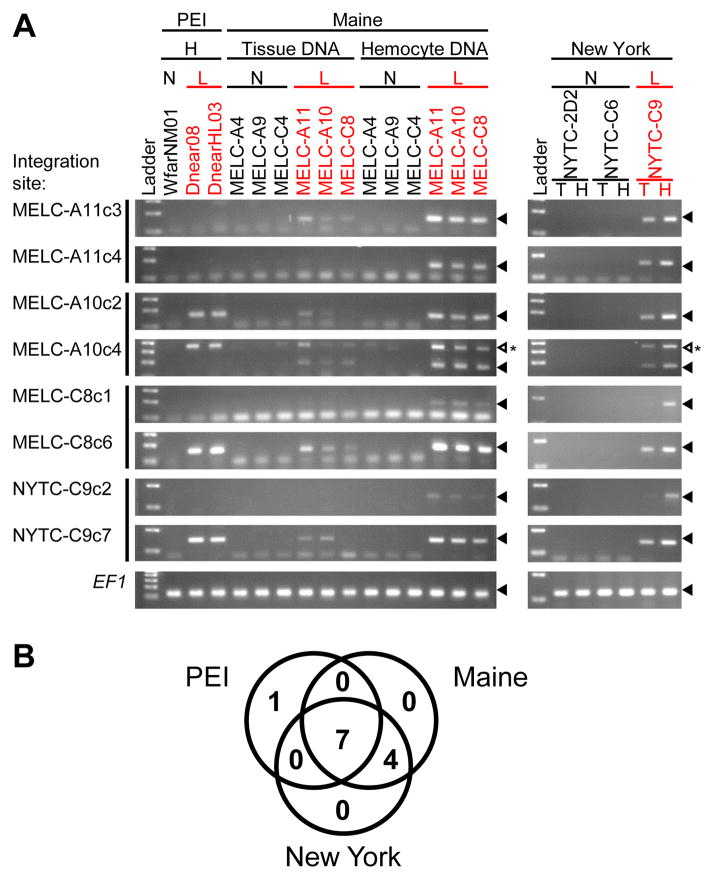
Neoplastic cells of leukemic animals have a high copy number of the Steamer retrotransposon, but this high copy number is not present in normal cells. Analysis of DNA from different tissues of diseased animals showed that Steamer copy number was low in solid tissues and highest in hemocytes, though with increases in Steamer copy number in some solid tissues, probably due to dissemination of neoplastic hemocytes (Figure 2). Preliminary sequence analysis of a small number of Steamer integration sites showed that neoplastic DNA contains new integration sites not present in normal clam DNA, and surprisingly, these new copies were often found at identical locations in different leukemic animals (Arriagada et al., 2014). We now extended this analysis to leukemic animals from independent populations in New York, Maine, and Prince Edward Island (PEI), Canada. Steamer integration sites were cloned from multiple leukemic animals by inverse PCR, and diagnostic primers were designed to amplify each specific integration site. PCR tests using these primers then allowed us to score for the presence of each insertion in neoplastic and tissue DNA (Figure 3A). Of 12 integration sites tested, seven were present in neoplastic DNA samples collected from all geographic locations, four more sites were common to the Maine and New York neoplasms but not those from Canada, and one site was unique to neoplasms from Canada (Figure 3B). These cancer-specific integration sites were not found in any healthy animals and were only weakly detected in tissue DNA from leukemic animals, likely due to infiltration of the neoplastic cells into normal tissue.
Figure 2. Steamer retrotransposon DNA copy numbers in different clam tissues.
Quantitative PCR was used to determine the copy number of the Steamer retrotransposon in genomic DNA from hemocytes and solid tissues of four representative normal (left) and four leukemic (right) clams. Steamer copy number was quantitated using primers that amplify a region in the Steamer reverse transcriptase (RT) and was normalized to the single-copy gene EF1 (Siah et al., 2011). * indicates P < 0.01 for comparisons between neoplastic hemocytes and each other tissue tested (n = 3 for gills, n = 4 for other tissues), using two-tailed paired T-test of normalized Steamer copy number.
Figure 3. Clonal Steamer integration sites in neoplastic cells.
(A) To determine the presence of specific integration sites in different animals, inverse PCR products were cloned and sequenced from clams from three locations (two integration sites per animal). For each integration site, a reverse primer was designed to match the flanking genomic sequence. Diagnostic PCR using a common forward primer in the Steamer LTR and an integration site specific reverse primer was used to determine the presence of each specific integration site in hemocyte (H) and tissue (T) DNA as indicated, of normal (N) and leukemic (L) clams. Sizes of the amplified DNAs were analyzed by agarose gel electrophoresis. Amplification of EF1 is shown as a control. Filled triangles mark the position of migration of the amplicon. An open triangle marks an unexpected PCR product. This band is due to amplification of a second Steamer integration site present in neoplastic cells of all populations.
(B) Venn diagram of the number of cancer-specific integration sites shared by neoplastic cells from the three locations (out of the 12 sites tested, including 4 sites cloned from PEI samples previously (Arriagada et al., 2014)). None of the 12 sites were present in any normal animal tested.
Common mtDNA SNPs in Neoplastic Cells
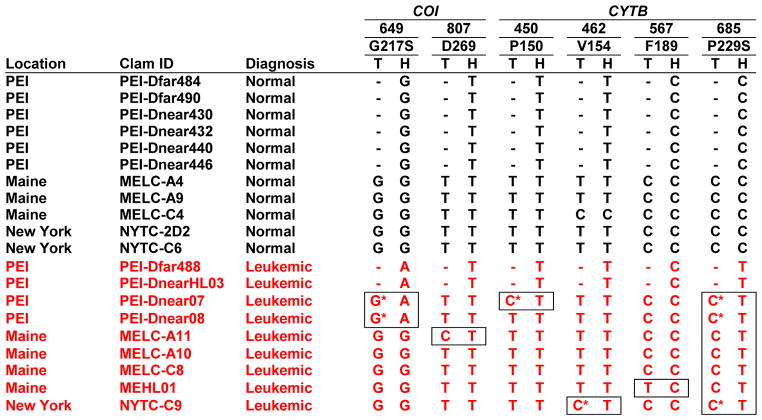
The observation of common integration sites raises the possibility of a clonal origin for all the tumors. To test the hypothesis that soft-shell clam leukemia is horizontally transmitted between animals as a clonal transmissible cancer cell, we examined DNA from leukemic and normal clams for single nucleotide polymorphisms (SNPs) in the sequences of mitochondrial genes encoding cytochrome c oxidase subunit I (COI) (Folmer et al., 1994) and cytochrome b (CYTB) (Table 1, Figure S1). DNA of all healthy animals (n=11) and the tissue DNA of all the leukemic animals contained a common allele (C685) in the CYTB gene, while the neoplastic hemocyte DNA of all leukemic animals (n=9) carried a distinctive SNP (C685T). This observation supports the hypothesis that the neoplastic cells are transferred between animals as an allograft that contains this unique SNP. Another SNP (G649A in COI) was identified in the hemocyte DNA of all four leukemic animals from PEI, but not in those from other locations and not in any of the 27 previously identified M. arenaria COI alleles (Strasser and Barber, 2009), suggesting that this mutation occurred during the evolution of the transmissible cells in this population. Additionally, seven low frequency SNPs were identified in the mitochondrial DNA of normal animals and the tissue of leukemic animals. In the four informative positions where rare SNPs were detected in tissue DNA of leukemic animals, the neoplastic hemocyte DNA did not contain that matching tissue allele but rather contained an allele common to all neoplastic cells (Table 1). These data indicate that the leukemias did not arise from their hosts, and are consistent with clonal transmission of the cancer.
Table 1. Leukemia-associated SNPs in M. arenaria COI and CYTB.
Nucleotide numbers based on the complete COI and CYTB genes in M. arenaria mtDNA genome (KJ755996). Boxes indicate discordance between hemocyte and tissue DNA. 3 additional CYTB SNPs were identified: T442C (L148) in MELC-C4 and T658C (F220L) and T696C (I232) in MELC-A4. T, Tissue; H, Hemocyte; -, Not done due to lack of availability of tissue samples.
|
Hemocyte allele can also be observed in tissue DNA, see sequencing traces in Figure S1A and Figure S1B.
Microsatellite Genotypes Confirm Clonal Origin of Clam Leukemia
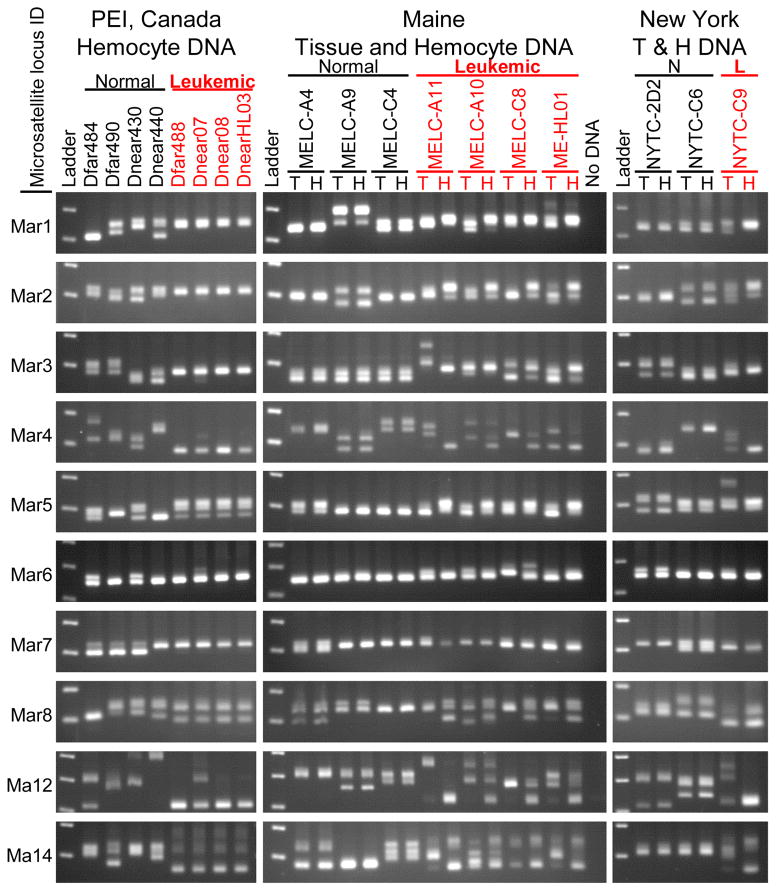
To characterize the genotype of the neoplastic cells in more detail, we analyzed ten microsatellite loci in nuclear DNA, previously identified as polymorphic in M. arenaria (Krapal et al., 2012; St-Onge et al., 2013), by PCR. The sizes of these loci were highly polymorphic among individuals, and in many cases the animals were heterozygous at the loci, with PCR products of two sizes (Figure 4). In normal animals the alleles present in hemocyte DNA always matched the alleles in the host tissue DNA. In contrast, in all leukemic animals the microsatellite genotypes in the neoplastic hemocytes were distinct from those in the tissue of the host animal. Furthermore, at nearly every locus, the microsatellite alleles were of the same lengths in the neoplastic hemocytes of all leukemic animals. That is, the genotypes of all the tumors were nearly identical to each other.
Figure 4. Amplification of microsatellite loci from tissue and hemocyte DNA from normal and leukemic clams.
PCR products using primers flanking ten microsatellite loci in hemocyte DNA and tissue DNA were displayed by electrophoresis on 2.5 % agarose gels and visualized by staining. Different alleles are determined by the sizes of the amplicons, with one band observed for animals homozygous at a particular locus, and two or more for heterozygotes and polyploid neoplastic cells. Each row of gels represents amplification from a single microsatellite locus, as labelled on the left. Hemocyte DNA is shown for normal and leukemic clams from PEI, Canada; and both Tissue (T) and Hemocyte (H) DNA is shown for Maine and New York clams. These data show that the leukemic hemocyte microsatellite alleles are identical to each other and distinct from their host tissue.
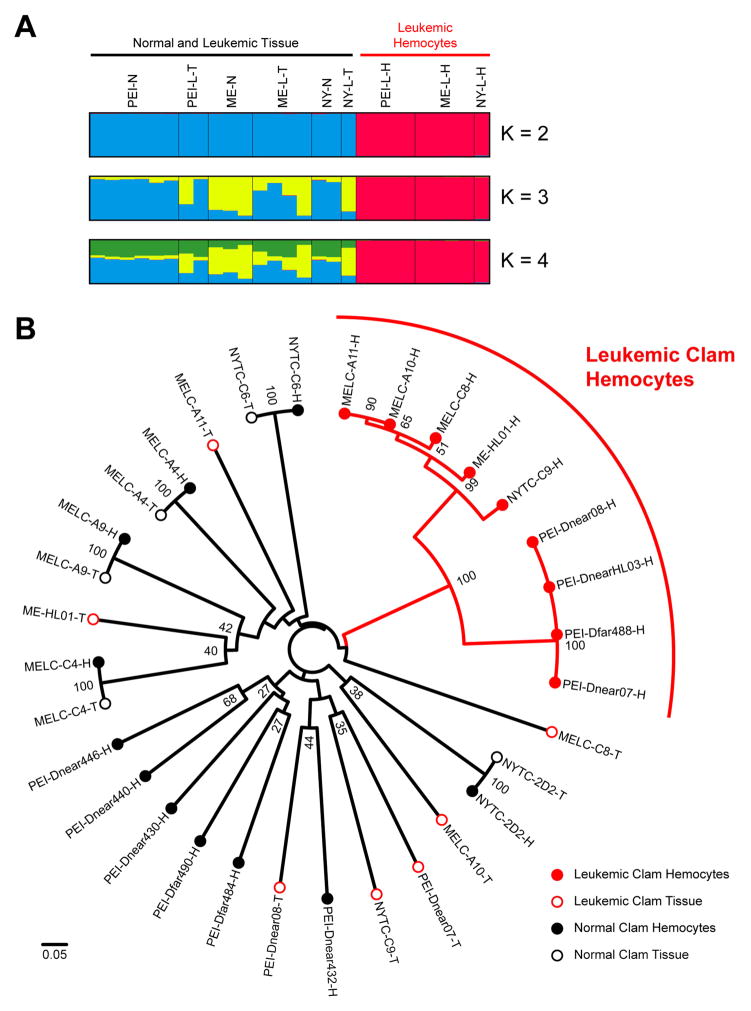
Next, fluorescently labeled primers were used to distinguish microsatellite PCR product sizes with single base resolution. While all normal animals have one allele (homozygous) or two alleles (heterozygous) for each locus, microsatellite size analysis of the leukemic DNA revealed up to four separate alleles at some of the loci. Many of the leukemic alleles were identical between independent neoplasms, but there were some small variations in repeat size, and several cases of allele loss or gain in some leukemic individuals. The model-based clustering program STRUCTURE (Pritchard et al., 2000) was used to analyze the microsatellite allele sizes, and regardless of the number of clusters (K) used, the neoplastic hemocyte genotypes clearly grouped separately from the normal animal genotypes, while the tissue of the leukemic animals clustered with normal animals (Figure 5A). A Neighbor-Joining tree based on the genetic distances between individuals (Bruvo et al., 2004; Kamvar et al., 2014) clearly demonstrated that the neoplastic hemocyte genotypes cluster as a separate group, distinct from all the normal genotypes and from the genotypes of the host tissue. Two branches were apparent within the neoplastic lineage, suggesting that the extant leukemias arose from a common ancestor that diverged into the PEI and USA cancer subgroups (Figure 5B).
Figure 5. Analysis of microsatellite alleles.
(A) CLUMPAK display of STRUCTURE (Pritchard et al., 2000) analysis of ten microsatellite loci showing the major population clustering with varying number of clusters (K) from K = 2 to 4. Each vertical bar represents a normal clam genotype or either tissue or hemocyte genotypes from leukemic clams with the colors representing cluster identity.
(B) Neighbor-Joining tree constructed with genetic distances based on ten microsatellite loci using Bruvo’s method for analysis of loci from individuals with variable ploidy (Bruvo et al., 2004) calculated using the poppr package with R (Kamvar et al., 2014). Bootstrap values above 25 are shown at nodes. The scale bar represents a genetic distance of 0.05 (where 0 represents completely identical genotypes and 1 represents no common alleles). Each sample is marked as Tissue (T, closed circle) or Hemocytes (H, open circle) of Normal (Black) or Leukemic (Red) clams. The leukemic genotypes all cluster together in two branches clearly apart from the normal genotypes. Allele sizes are listed in Table S2 and further information is available in Supplementary Experimental Procedures.
DISCUSSION
Three data sets (Steamer integration sites, mtDNA SNPs, and microsatellite alleles) show that the genotypes of the neoplastic cells all differ from the genotype of their host animals, and are identical or very closely related to each other. We conclude that the individual leukemias are not derived by independent oncogenic transformation of cells within each host but instead come from a single genetically unrelated parent. The data strongly argue that disseminated neoplasia is naturally spreading between animals as a transmissible cancer cell. Horizontal clonal transmission of cancer has only been observed in two other contagious cancers of mammals in the wild, the DFTD in Tasmanian devils (Pearse and Swift, 2006) and CTVT in dogs (Murgia et al., 2006).
It is remarkable that neoplastic cells from clam beds separated by hundreds of miles were found to be genetically nearly identical or very closely related. The mechanism by which the soft-shell leukemic cell line could be transferred from animal to animal in the wild is not clear. In the two previously known cases of natural cancer cell transfer (DFTD and CTVT) (Murchison, 2008), physical contact is required for transmission of cancer cells to naïve individuals (through biting or sexual contact, respectively), but adult clams are sessile and do not normally come in contact with one another. Clams do filter feed, however, raising the possibility that leukemia engraftment occurs through filtration of seawater contaminated with neoplastic cells. Each animal can filter several liters of seawater per hour, so very low concentrations of cells in seawater could be sufficient for transmission. One early study showed that hemocytes from a leukemic clam could survive in natural seawater conditions for >6 hours with minimal cell death, suggesting a plausible route for cancer cell transmission from one clam to another (Sunila and Farley, 1989). Direct surveys of seawater at the affected clam beds for tumor cells could provide support for this idea. It is currently unknown how neoplastic cells would be released by leukemic animals. Cells may be shed naturally during disease, expelled during spawning, or released after physical trauma, during predation, or at death of the leukemic individual. The stage of development when transmission might occur in nature is unknown, though experimental transmission of disease was found to be most efficient with mature animals of intermediate size (Taraska and Bottger, 2013). It is possible that its transmission has been facilitated or accelerated by human intervention, as seed stocks have been transplanted between sites along the coast at several times in recent decades (Beal and Kraus, 2002).
The length of time since the original formation of the primordial soft-shell neoplasia is not yet clear. The disease has been documented in soft-shelled clams since the 1970s (Brown et al., 1978; Yevich and Barszcz, 1978), and it is likely that all cases derive from the same clone, suggesting that it may be at least forty years old and possibly much older. Its appearance must have been sufficiently far in the past to allow for its spread widely along the North Atlantic coast. Significant divergence was observed in cancer cell subgroups specific to the USA and Canada clam populations. Several small microsatellite expansions and deletions and at least one mtDNA SNP appear to have developed separately in the PEI subgroup of cancer cells. Further analysis of the origin and continued evolution of these cancer cell lineages may allow for estimation of the time of appearance of the original leukemia. The two other examples of transmissible tumors are of very different ages: the dog tumor is thought to have arisen 10,000–13,000 years ago (Murchison et al., 2014), while the Tasmanian devil tumor is of much more recent origin, perhaps arising only 20–30 years ago (Murchison, 2008).
It is plausible that the Steamer retrotransposon had a role in the development of the transmissible disseminated neoplasia. Steamer genomic copy number is greatly expanded in neoplastic cells (from 2–10 copies to 150–300 per haploid genome), and we show that the majority of the new integrations are common to all tested neoplastic cells and therefore likely occurred early in the evolution of the cancer. One or more of these common integration events may have directly caused initial oncogenic mutations, similar to the LINE1 integration near the c-myc gene in all known cases of CTVT (Katzir et al., 1985; Murgia et al., 2006), but alternatively they may represent passenger mutations that were fixed in the genome of the leukemia. Some of the integration sites are unique to the PEI or USA subgroups of neoplastic cells, and these may represent active gain of new copies by retrotransposition, or loss of copies due to recombination or aneuploidy; these sites are unlikely to be a cause of the tumor. Further analysis of the genome or the expression profile of the neoplastic cells in comparison with those of normal hemocytes could provide clues to the cause of the transformed phenotype.
While all the M. arenaria neoplastic cells we have tested were derived from the same cell lineage, other leukemias may be derived from different clones or by more conventional mechanisms. Leukemia can be induced in clams by 5-bromodeoxyuridine (BrdU) injection (Oprandy and Chang, 1983; Taraska and Bottger, 2013) and these cases must arise by de novo transformation of host cells, and perhaps by induction of Steamer transposition. It has also been reported that clam leukemia could be transmitted between animals by a filterable agent (Oprandy et al., 1981; Taraska and Bottger, 2013) but this is controversial (AboElkhair et al., 2012; McLaughlin et al., 1992).
Transmissible disseminated neoplasias have been reported in other bivalve species, including other clams, oysters, mussels, and cockles (Barber, 2004), and these diseases may also be caused by a clonal transmissible cancer cell. A recent report (Vassilenko et al., 2010) that neoplastic hemocytes in mussels (Mytilus trossulus) on the west coast of North America share a common set of synonymous SNPs in the p53 gene is consistent with an independent development of a transmissible clonal cancer in that species.
Flow cytometric analyses of DNA content of disseminated neoplasias in many bivalve species have identified characteristic ploidy levels—indeed, polyploidy has been used as diagnostic for disease (Delaporte et al., 2008). In M. arenaria specifically, the observation of roughly tetraploid DNA content in neoplastic cells (Delaporte et al., 2008; Reno et al., 1994) is consistent with a clonal contagious cancer and with the observation of up to four unique microsatellite alleles at a single locus in neoplastic cells. Given the current findings, previous observations of abnormal ploidy in bivalve disseminated neoplasia suggests that many of these diseases in other species may represent independent contagious cancer lineages. For example, mussel disseminated neoplasia has been reported to come in two types, tetraploid or pentaploid (Moore et al., 1991), consistent with either two independent cancer lineages or one which has evolved into subgroups with divergent ploidy, although a more recent study found a wider distribution of aneuploidy between individuals (Vassilenko and Baldwin, 2014). Multiple abnormal ploidy levels have also been observed in disseminated neoplasia in cockles (Cerastoderma edule) (da Silva et al., 2005). We are currently investigating the hypothesis suggested by the current findings that disseminated neoplasia in other bivalve species represent independent lineages of contagious cancer. Preliminary mtDNA sequencing of cockles with disseminated neoplasia suggests a cockle-derived contagious cancer line (data not shown).
It is possible that the disseminated neoplasia of M. arenaria could transmit to other species, though there is as of yet no evidence for this. There may be mechanisms by which a host animal can reject colonization by neoplastic cells of another species. Molluscs are not known to have a self/nonself recognition system similar to the MHC system of vertebrates, and their lack of MHC may make them highly susceptible to this form of infectious malignancy. Indeed, in the cases of CTVT and DFTD, there are unique mechanisms by which the cancers avoid MHC recognition. In CTVT, expression of MHC I and II are down-regulated during active growth of the tumor and their expression leads to immune recognition and clearance of the tumor in most dogs (Yang et al., 1987). DFTD cells also down-regulate MHC expression and the low genetic diversity of devils limits the ability of hosts to recognize DFTD as foreign (Siddle and Kaufman, 2015; Siddle et al., 2013). Contagious cancer is a serious threat to marine invertebrates, leading to severe mortalities during outbreaks in soft-shell clams and possibly leading to mass mortalities in many other species. The disease represents a significant selective pressure and supports the hypothesis that histocompatibility could have evolved in part due to selective pressure to prevent malignancy (Murgia et al., 2006), rather than simply being a secondary consequence of pressure by infectious diseases. Despite the lack of MHC, molluscs and other invertebrates may employ other self/nonself recognition mechanisms which can combat this type of disease, perhaps similar to the fusion/histocompatibility (Fu/HC) system of colonial ascidians, which protects ascidians from stem cell parasitism which can occur when unrelated individuals fuse (De Tomaso et al., 2005; Voskoboynik et al., 2013). Bivalves may utilize a histocompatibility system which may be unique to molluscs or evolutionarily related to Fu/HC or MHC.
Natural horizontal transmission of cancer between individuals has been considered a rare phenomenon, restricted to two exceptional cases in mammals. Our finding of the horizontal transmission of a clonal clam leukemia extends the phenomenon to the marine environment, and demonstrates that this mechanism is more widespread in nature than previously supposed.
EXPERIMENTAL PROCEDURES
Sample Collection
Soft-shell clams (M. arenaria) were collected from several locations (Figure 1). Clams from the Dunk estuary on Prince Edward Island (PEI), Canada, were collected and diagnosed as described previously (Arriagada et al., 2014). Clams from Larrabee Cove, Maine, USA (MELC) were collected by Dr. Brian Beal (University of Maine at Machias) as described previously, and one additional leukemic clam (ME-HL01) was collected by a commercial source near St. George, Maine, with the help of Dr. Charles Walker (University of New Hampshire). Clams from New York (NYTC) were acquired from a commercial market and collected from the north shore of Long Island, New York, USA near Port Jefferson. Individual clam IDs reflect the location and the ID number assigned at the time of collection.
Diagnosis of clams from New York and Maine was conducted by extracting hemolymph from the pericardial region using a 26 gauge needle. Six to eight drops of hemolymph were placed in a well of a 96-well plate and left undisturbed at 10° C for one hour before morphological analysis using phase-contrast microscopy. Normal clam hemocytes attached to the dish (Figure 1A), and clams with >50% rounded, refractile cells were considered leukemic (Figure 1B).
PCR and qPCR
Primers and conditions used for qPCR, diagnostic PCR for specific integration sites, COI (Folmer et al., 1994) and CYTB PCR, and PCR of microsatellite loci are provided in Supplementary Experimental Procedures and Table S1.
Microsatellite Analysis
STRUCTURE (Pritchard et al., 2000) was used to determine the population clustering of the clam tissue and hemocytes DNA. As up to 4 unique alleles were detected in leukemic DNA, loci were considered to be tetraploid, and clams with lower ploidy were considered to have missing data for the additional alleles, which are ignored in analysis. Cluster values of K = 1 to 10 were used with an admixture model with a burnin of 50,000 and 50,000 replicates after burnin. CLUMPAK was used to graphically analyze the major clusters across 10 STRUCTURE runs. Data were also recoded and reanalyzed as binary absent/present alleles (Rodzen and May, 2002), with nearly identical results (data not shown).
Genetic distance was calculated using Bruvo’s band sharing method assuming infinite alleles (Bruvo et al., 2004), as it was created specifically for comparisons of individuals with different ploidy levels, and a Neighbor-Joining tree was constructed (with 100 bootstraps) using the poppr package for R (Kamvar et al., 2014) and displayed using Figtree. Alternate analyses using the genome addition and combined genome addition/genome loss models generated nearly identical results (data not shown). Further details of the microsatellite analysis can be found in Supplementary Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Clam leukemia genotypes are distinct from the host and nearly identical to each other
The transmissible cancer clone likely arose in a single individual
Clam leukemia is transmitted horizontally between animals as contagious cancer cell
Contagious cancer cell transmission may be widespread in the marine environment
Acknowledgments
We thank Gloria Arriagada for work on the Steamer element, Brian Beal (University of Maine at Machias) for M. arenaria samples from Maine, Annette Murray for help in collection and diagnosis of M. arenaria from PEI, and Charles Walker and Sarah Mercier (University of New Hampshire) for help in acquisition of leukemic sample ME-HL01. We also thank Anne Gifford for putting the laboratories in contact to initiate the studies. This work was supported by the Howard Hughes Medical Institute and by the National Institutes of Health (NIH) Training Grant T32 CA009503 (to M.J.M.).
Footnotes
Supplemental Data include Supplemental Experimental Procedures, two figures, and two tables.
Author Contributions
SPG and MJM planned the experiments and wrote the paper. MJM conducted the experiments. CR and JS collected and diagnosed clams from PEI.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AboElkhair M, Iwamoto T, Clark KF, McKenna P, Siah A, Greenwood SJ, Berthe FC, Casey JW, Cepica A. Lack of detection of a putative retrovirus associated with haemic neoplasia in the soft shell clam Mya arenaria. J Invertebr Pathol. 2012;109:97–104. doi: 10.1016/j.jip.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Arriagada G, Metzger MJ, Muttray AF, Sherry J, Reinisch C, Street C, Lipkin WI, Goff SP. Activation of transcription and retrotransposition of a novel retroelement, Steamer, in neoplastic hemocytes of the mollusk Mya arenaria. Proc Natl Acad Sci U S A. 2014;111:14175–14180. doi: 10.1073/pnas.1409945111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber BJ. Neoplastic diseases of commercially important marine bivalves. Aquatic Living Resources. 2004;17:449–466. [Google Scholar]
- Beal BF, Kraus MG. Interactive effects of initial size, stocking density, and type of predator deterrent netting on survival and growth of cultured juveniles of the soft-shell clam, Mya arenaria L., in eastern Maine. Aquaculture. 2002;208:81–111. [Google Scholar]
- Brown RS, Wolke RE, Saila SB, Brown CW. Prevalence of neoplasia in 10 New England populations of the soft-shell clam (Mya arenaria) Ann N Y Acad Sci. 1978;298:522–534. doi: 10.1111/j.1749-6632.1977.tb19287.x. [DOI] [PubMed] [Google Scholar]
- Bruvo R, Michiels NK, D’Souza TG, Schulenburg H. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Molecular ecology. 2004;13:2101–2106. doi: 10.1111/j.1365-294X.2004.02209.x. [DOI] [PubMed] [Google Scholar]
- Cooper KR, Brown RS, Chang PW. The Course and Mortality of a Hematopoietic Neoplasm in the Soft-Shell Clam, Mya-Arenaria. Journal of Invertebrate Pathology. 1982;39:149–157. [Google Scholar]
- da Silva PM, Soudant P, Carballal MJ, Lambert C, Villalba A. Flow cytometric DNA content analysis of neoplastic cells in haemolymph of the cockle Cerastoderma edule. Dis Aquat Organ. 2005;67:133–139. doi: 10.3354/dao067133. [DOI] [PubMed] [Google Scholar]
- De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, Weissman IL. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaporte M, Synard S, Pariseau J, McKenna P, Tremblay R, Davidson J, Berthe FC. Assessment of haemic neoplasia in different soft shell clam Mya arenaria populations from eastern Canada by flow cytometry. J Invertebr Pathol. 2008;98:190–197. doi: 10.1016/j.jip.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular marine biology and biotechnology. 1994;3:294–299. [PubMed] [Google Scholar]
- House ML, Kim CH, Reno PW. Soft shell clams Mya arenaria with disseminated neoplasia demonstrate reverse transcriptase activity. Dis Aquat Organ. 1998;34:187–192. doi: 10.3354/dao034187. [DOI] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grunwald NJ. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzir N, Rechavi G, Cohen JB, Unger T, Simoni F, Segal S, Cohen D, Givol D. “Retroposon” insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc Natl Acad Sci U S A. 1985;82:1054–1058. doi: 10.1073/pnas.82.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapal AM, Popa OP, Iorgu EI, Costache M, Popa LO. Isolation and Characterization of New Microsatellite Markers for the Invasive Softshell Clam, Mya arenaria (L.) (Bivalvia: Myidae) Int J Mol Sci. 2012;13:2515–2520. doi: 10.3390/ijms13022515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SM, Farley CA, Hetrick FM. Transmission studies of sarcoma in the soft-shell clam, Mya arenaria. In Vivo. 1992;6:367–370. [PubMed] [Google Scholar]
- Miosky DL, Smolowitz RM, Reinisch CL. Leukemia cell specific protein of the bivalve mollusc Mya arenaria. J Invertebr Pathol. 1989;53:32–40. doi: 10.1016/0022-2011(89)90071-2. [DOI] [PubMed] [Google Scholar]
- Moore JD, Elston RA, Drum AS, Wilkinson MT. Alternate pathogenesis of systemic neoplasia in the bivalve mollusc Mytilus. J Invertebr Pathol. 1991;58:231–243. doi: 10.1016/0022-2011(91)90067-z. [DOI] [PubMed] [Google Scholar]
- Murchison EP. Clonally transmissible cancers in dogs and Tasmanian devils. Oncogene. 2008;27(Suppl 2):S19–30. doi: 10.1038/onc.2009.350. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Wedge DC, Alexandrov LB, Fu B, Martincorena I, Ning Z, Tubio JM, Werner EI, Allen J, De Nardi AB, et al. Transmissible [corrected] dog cancer genome reveals the origin and history of an ancient cell lineage. Science. 2014;343:437–440. doi: 10.1126/science.1247167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Clonal origin and evolution of a transmissible cancer. Cell. 2006;126:477–487. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muttray A, Reinisch C, Miller J, Ernst W, Gillis P, Losier M, Sherry J. Haemocytic leukemia in Prince Edward Island (PEI) soft shell clam (Mya arenaria): spatial distribution in agriculturally impacted estuaries. Sci Total Environ. 2012;424:130–142. doi: 10.1016/j.scitotenv.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Oprandy JJ, Chang PW. 5-bromodeoxyuridine induction of hematopoietic neoplasia and retrovirus activation in the soft-shell clam, Mya arenaria. J Invertebr Pathol. 1983;42:196–206. doi: 10.1016/0022-2011(83)90062-9. [DOI] [PubMed] [Google Scholar]
- Oprandy JJ, Chang PW, Pronovost AD, Cooper KR, Brown RS, Yates VJ. Isolation of a viral agent causing hematopoietic neoplasia in the soft-shell clam, Mya arenaria. Journal of Invertebrate Pathology. 1981;38:45–51. [Google Scholar]
- Pearse AM, Swift K. Allograft theory: transmission of devil facial-tumour disease. Nature. 2006;439:549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reno PW, House M, Illingworth A. Flow cytometric and chromosome analysis of softshell clams, Mya arenaria, with disseminated neoplasia. Journal of Invertebrate Pathology. 1994;64:163–172. [Google Scholar]
- Rodzen JA, May B. Inheritance of microsatellite loci in the white sturgeon (Acipenser transmontanus) Genome. 2002;45:1064–1076. doi: 10.1139/g02-083. [DOI] [PubMed] [Google Scholar]
- Siah A, McKenna P, Danger JM, Johnson GR, Berthe FC. Induction of transposase and polyprotein RNA levels in disseminated neoplastic hemocytes of soft-shell clams: Mya arenaria. Dev Comp Immunol. 2011;35:151–154. doi: 10.1016/j.dci.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Siddle HV, Kaufman J. Immunology of naturally transmissible tumours. Immunology. 2015;144:11–20. doi: 10.1111/imm.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle HV, Kreiss A, Tovar C, Yuen CK, Cheng Y, Belov K, Swift K, Pearse AM, Hamede R, Jones ME, et al. Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc Natl Acad Sci U S A. 2013;110:5103–5108. doi: 10.1073/pnas.1219920110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolowitz RM, Miosky D, Reinisch CL. Ontogeny of leukemic cells of the soft shell clam. J Invertebr Pathol. 1989;53:41–51. doi: 10.1016/0022-2011(89)90072-4. [DOI] [PubMed] [Google Scholar]
- St-Onge P, Sevigny JM, Strasser C, Tremblay R. Strong population differentiation of softshell clams (Mya arenaria) sampled across seven biogeographic marine ecoregions: possible selection and isolation by distance. Mar Biol. 2013;160:1065–1081. [Google Scholar]
- Strasser CA, Barber PH. Limited genetic variation and structure in softshell clams (Mya arenaria) across their native and introduced range. Conserv Genet. 2009;10:803–814. [Google Scholar]
- Sunila I, Farley CA. Environmental Limits for Survival of Sarcoma-Cells from the Soft-Shell Clam Mya-Arenaria. Diseases of Aquatic Organisms. 1989;7:111–115. [Google Scholar]
- Taraska NG, Bottger SA. Selective initiation and transmission of disseminated neoplasia in the soft shell clam Mya arenaria dependent on natural disease prevalence and animal size. J Invertebr Pathol. 2013;112:94–101. doi: 10.1016/j.jip.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Vassilenko E, Baldwin SA. Using flow cytometry to detect haemic neoplasia in mussels (Mytilus trossulus) from the Pacific Coast of Southern British Columbia, Canada. J Invertebr Pathol. 2014;117:68–72. doi: 10.1016/j.jip.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Vassilenko EI, Muttray AF, Schulte PM, Baldwin SA. Variations in p53-like cDNA sequence are correlated with mussel haemic neoplasia: A potential molecular-level tool for biomonitoring. Mutat Res. 2010;701:145–152. doi: 10.1016/j.mrgentox.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Voskoboynik A, Newman AM, Corey DM, Sahoo D, Pushkarev D, Neff NF, Passarelli B, Koh W, Ishizuka KJ, Palmeri KJ, et al. Identification of a colonial chordate histocompatibility gene. Science. 2013;341:384–387. doi: 10.1126/science.1238036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, Bottger S, Low B. Mortalin-based cytoplasmic sequestration of p53 in a nonmammalian cancer model. Am J Pathol. 2006;168:1526–1530. doi: 10.2353/ajpath.2006.050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, Bottger SA, Mulkern J, Jerszyk E, Litvaitis M, Lesser M. Mass culture and characterization of tumor cells from a naturally occurring invertebrate cancer model: applications for human and animal disease and environmental health. Biol Bull. 2009;216:23–39. doi: 10.1086/BBLv216n1p23. [DOI] [PubMed] [Google Scholar]
- Weinberg JR, Leavitt DF, Lancaster BA, Capuzzo JM. Experimental field studies with Mya arenaria (Bivalvia) on the induction and effect of hematopoietic neoplasia. J Invertebr Pathol. 1997;69:183–194. doi: 10.1006/jipa.1996.4641. [DOI] [PubMed] [Google Scholar]
- Yang TJ, Chandler JP, Dunne-Anway S. Growth stage dependent expression of MHC antigens on the canine transmissible venereal sarcoma. Br J Cancer. 1987;55:131–134. doi: 10.1038/bjc.1987.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yevich PP, Barszcz CA. Neoplasia in soft-shell clams (Mya arenaria) collected from oil-impacted sites. Ann N Y Acad Sci. 1978;298:409–426. doi: 10.1111/j.1749-6632.1977.tb19281.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.