Summary
Burkholderia pseudomallei and B. mallei are bacterial pathogens that cause melioidosis and glanders, while their close relative B. thailandensis is nonpathogenic. All use the trimeric autotransporter BimA to facilitate actin-based motility, host cell fusion and dissemination. Here, we show that BimA orthologs mimic different host actin-polymerizing proteins. B. thailandensis BimA activates the host Arp2/3 complex. In contrast, B. pseudomallei and B. mallei BimA mimic host Ena/VASP actin polymerases in their ability to nucleate, elongate and bundle filaments by associating with barbed ends, as well as in their use of WH2 motifs and oligomerization for activity. Mechanistic differences among BimA orthologs resulted in distinct actin filament organization and motility parameters, which affected the efficiency of cell fusion during infection. Our results identify bacterial Ena/VASP mimics and reveal that pathogens imitate the full spectrum of host actin-polymerizing pathways, suggesting that mimicry of different polymerization mechanisms influences key parameters of infection.
Introduction
The pseudomallei group of Burkholderia species are Gram-negative bacteria that include the opportunistic human pathogens B. pseudomallei (Bp), which cause the severe disease melioidosis, and B. mallei (Bm), a clonal descendant of Bp that primarily infects equine but can cause acute human disease (Cheng and Currie, 2005; Wilkinson, 1981). Bp and Bm are of heightened concern because they are resistant to numerous antibiotics, spread via an aerosol route and exhibit low infectious doses. A third species, B. thailandensis (Bt), is closely related yet is not pathogenic to humans. All three share key virulence factors despite their differences in infectivity. Because Bt infection in animals and cells recapitulates many features of Bp and Bm virulence, it has been used as a model system to study Burkholderia pathogenesis (Galyov et al., 2010; Haraga et al., 2008; West et al., 2008). However, the basis for the dramatic differences in Burkholderia virulence remains a mystery. Understanding these differences will provide insight into the evolution of virulence among closely related bacteria.
Key features of Burkholderia virulence include their ability to invade host cells and escape from phagosomes into the cytosol where they replicate and undergo actin-based motility (Kespichayawattana et al., 2000; Stevens et al., 2005a). As with other pathogens, Burkholderia use actin-based motility to facilitate cell-to-cell spread, which enables dissemination within hosts while evading the immune system (Goldberg, 2001). However, Burkholderia spread is distinct in that it can occur through bacterial-mediated fusion of host cells (French et al., 2011; Kespichayawattana et al., 2000) induced by type VI secretion (Schwarz et al., 2014; Toesca et al., 2014), rather than the engulfment of membrane protrusions. Regardless of the spread mechanism, motility drives bacteria to the host plasma membrane to facilitate spread.
To enable actin-based motility, pathogens express proteins that mimic or activate host actin-nucleating factors to promote the polymerization of actin monomers (G-actin) into filaments (F-actin) that elongate at fast-growing barbed ends. Listeria monocytogenes and Shigella flexneri produce mimics or activators of host nucleation promoting factors (NPFs) to stimulate the host Arp2/3 complex (Welch and Way, 2013), which constructs branched actin filament networks (Campellone and Welch, 2010). Rickettsia species use an NPF that activates the Arp2/3 complex early in infection (Reed et al., 2014), and later use a mimic of host formins (Haglund et al., 2010; Kleba et al., 2010; Madasu et al., 2013), which nucleate filaments and processively bind to barbed ends to enhance elongation rates (Paul and Pollard, 2009). Vibrio and Chlamydia species, which do not undergo motility, express proteins that use tandem actin monomer-binding sequences to nucleate F-actin (Jewett et al., 2006; Liverman et al., 2007; Namgoong et al., 2011; Pernier et al., 2013; Tam et al., 2007; Yu et al., 2011). Despite this diversity, no pathogens have been shown to mimic host Ena/VASP proteins, which are weak nucleators, bind barbed ends, enhance filament elongation and bundle filaments (Barzik et al., 2005; Breitsprecher et al., 2011; Hansen and Mullins, 2010; Krause et al., 2003; Samarin et al., 2003; Winkelman et al., 2014). Within this context, it is unclear whether Burkholderia mimics or activates host nucleators or nucleates actin by an unknown mechanism. Additionally, the consequences of different nucleation mechanisms on actin-based motility and infection have not been examined.
Burkholderia actin-based motility requires the Burkholderia intracellular motility A (BimA) protein (Schell et al., 2007; Sitthidet et al., 2010; Stevens et al., 2005b), a member of the trimeric autotransporter (AT) family. Trimeric ATs contain highly conserved C-terminal sequences that mediate secretion, localization to the outer membrane, and trimerization (Cotter et al., 2005; Dautin and Bernstein, 2007). These include a β-barrel, as well as an adjacent ~35 residue α-helix (Stevens et al., 2005b), which in other trimeric ATs forms a trimeric coiled coil that is positioned inside the barrel (Meng et al., 2006). Trimeric ATs also contain N-terminal passenger domains that are exposed on the bacterial surface. Sequence comparisons suggest that the passenger domains of BimA from Bt, Bp and Bm polymerize host actin by different mechanisms (Stevens et al., 2005b). BtBimA contains putative actin-binding WASP homology 2 (WH2) and Arp2/3-binding central and acidic (CA) motifs that are conserved among NPFs that activate the Arp2/3 complex (Campellone and Welch, 2010). Previous work showed that the BtBimA CA motifs are required to activate Arp2/3 nucleation in vitro and promote actin association in host cells (Sitthidet et al., 2010). In contrast, BpBimA and BmBimA contain three (Bp) or one (Bm) putative WH2 motifs and lack CA sequences. Although two of the three putative BpBimA WH2 sequences were shown to be required for actin binding, nucleation and plaque formation (Sitthidet et al., 2011), the activity of BmBimA has not been examined. It also remains unclear what molecular mechanisms BpBimA and BmBimA use to nucleate actin and the role trimerization plays in this process.
Here, we investigated the mechanisms of actin assembly and motility driven by BimA orthologs from different Burkholderia species. Despite the conservation of key virulence factors (French et al., 2011; Galyov et al., 2010; Haraga et al., 2008), we found that Bt, Bp and Bm have evolved divergent mechanisms for actin polymerization. These differences result in Burkholderia that generate actin tails with distinct filament architectures and exhibit different efficiencies of motility and host cell fusion, providing one potential explanation for the increased virulence of Bp and Bm relative to Bt. These observations demonstrate that intracellular pathogens employ the full spectrum of actin assembly pathways and suggest that distinct mechanisms allow microbes to fine-tune motility to control spread during infection.
Results
BtBimA activates the host Arp2/3 complex to nucleate branched filaments while BpBimA and BmBimA independently nucleate unbranched filaments
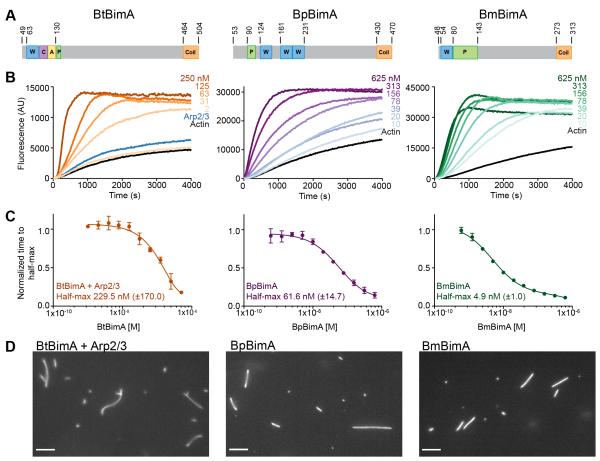
Previous studies indicated that BtBimA requires its CA motifs and the host Arp2/3 complex for actin nucleation, whereas BpBimA nucleates actin independent of Arp2/3 (Sitthidet et al., 2010; Stevens et al., 2005b). The activity of BmBimA was not investigated. In these studies, incomplete BtBimA and BpBimA passenger domains that lacked trimeric coiled coils were fused to GST, which would induce non-native dimerization (Sitthidet et al., 2010; 2011; Stevens et al., 2005b). The activity of these constructs was modest (~1.5-2.5 fold increased polymerization rates). We hypothesized that the full passenger domains in their native oligomeric state would exhibit higher nucleation activities that would be dependent (BtBimA) or independent (BpBimA, BmBimA) of Arp2/3 complex. To test this, we purified versions of each ortholog that spanned the passenger domain and extended through the trimeric coiled coil (Figure 1A) (Meng et al., 2006; Szczesny and Lupas, 2008). We tested each purified BimA for activity using pyrene actin polymerization assays. BtBimA alone had little nucleation activity (Figure S1A, B) yet it activated the Arp2/3 complex in a concentration-dependent manner (> 5 fold increase relative to actin alone), indicating that it functions as an NPF (Figure 1B, C). In contrast, BpBimA and BmBimA displayed robust nucleation activity (> 7 or 8 fold increase relative to actin alone), and Arp2/3 addition had no effect (Figure S1B). To compare their potencies, we determined the time it took to reach half maximum fluorescence over a range of BimA concentrations and fit the resulting curves (Figure 1C). All three orthologs are potent nucleators with 230 nM BtBimA (with 50 nM Arp2/3), 60 nM BpBimA and 5 nM BmBimA required for half-maximum activity. Consistent with its ability to activate the Arp2/3 complex, BtBimA and Arp2/3 generated branched actin filaments (Figure 1D). In contrast, BpBimA and BmBimA generated unbranched filaments. Thus, BtBimA is an NPF that activates Arp2/3, while BpBimA and BmBimA independently nucleate actin similar to host proteins in the formin, tandem monomer nucleator and Ena/VASP families.
Figure 1. BtBimA mimics host NPFs while BpBimA and BmBimA independently nucleate actin.
(A) Domain schematics of BimA from different Burkholderia species. Actin-related and oligomerization sequences are shown. Numbers refer to amino acid position in full-length proteins. W, WASP homology 2; C, central; A, acidic; P, proline rich; Coil, trimeric coiled coil. (B) Pyrene actin polymerization reactions with increasing BimA concentrations with Arp2/3 complex (Bt) or in its absence (Bp, Bm). (C) The time to half-maximum fluorescence of the polymerization curves in (B) normalized to actin alone. The mean ± SD are shown and fits of each dataset are reported to estimate the concentration at which half-maximum activity is observed. (D) Epifluorescence images of polymerization reactions containing BtBimA with Arp2/3 complex, BpBimA or BmBimA after 15 min that were stabilized and stained with rhodamine phalloidin. Scale bars, 5 μm. See also Figure S1.
BpBimA and BmBimA bind filament barbed ends, processively elongate filaments and remove capping protein from filaments
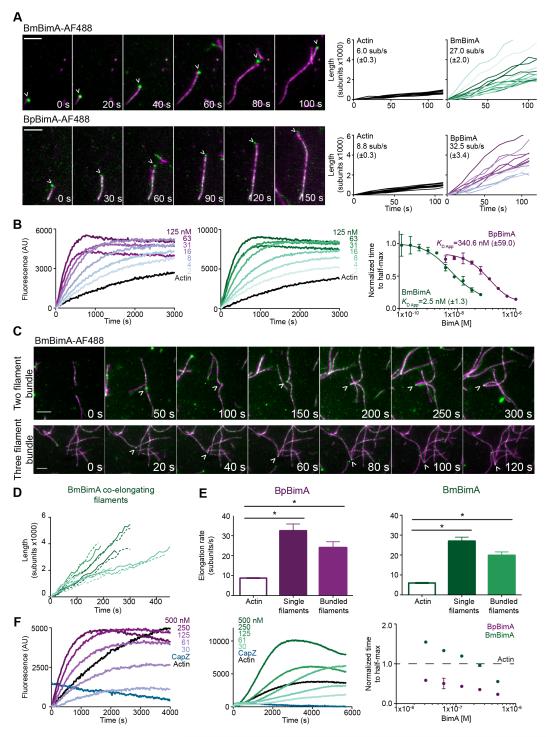
Formins and Ena/VASP proteins processively track barbed ends and alter their elongation rates (Paul and Pollard, 2009), while tandem monomer nucleators have not been shown to track growing filament ends (Carlier et al., 2011; Namgoong et al., 2011; Pernier et al., 2013). To compare BimA behavior to members of these families, we investigated how BimA orthologs interact with filament ends and tested whether they affect elongation. We labeled BpBimA and BmBimA with Alexa Fluor 488 at Cys residues that were introduced at their C termini, and monitored the effect of BimA on rhodamine-labeled actin polymerization using total internal reflection fluorescence (TIRF) microscopy. BmBimA and BpBimA bound and remained associated with barbed ends as they grew (Figure 2A; Movies S1, S2). Growth rate measurements of barbed ends in the presence or absence of BimA indicated that BmBimA and BpBimA increased elongation rates by 3.5- to 4.5-fold relative to actin alone. To measure their apparent affinities for barbed ends, we monitored elongation of pre-polymerized actin seeds with increasing BimA concentrations and fit the concentration dependence of the decrease in time to half-maximum fluorescence (Figure 2B). These measurements produced apparent affinities for barbed ends of 350 nM for BpBimA and 3 nM for BmBimA, similar to the barbed-end affinities of formin and Ena/VASP proteins (Hansen and Mullins, 2010; Harris et al., 2004; Otomo et al., 2005; Pruyne et al., 2002; Winkelman et al., 2014).
Figure 2. BmBimA and BpBimA processively bind growing filament barbed ends, increase elongation rates, bundle filaments and outcompete CapZ for barbed end binding.
(A) Left, TIRF images showing BmBimA-AF488 and BpBimA-AF488 (green) and rhodamine-labeled actin (magenta). Time (s) is indicated. Scale bars, 3 μm. Right, filament length (number of subunits X 1000) over time for a minimum of 10 filaments with the mean elongation rate (sub/s ± SE) listed. (B) Left and middle, pyrene elongation assays with a range of BimA concentrations. Right, the time to half-maximum fluorescence normalized to actin alone for BmBimA (green) or BpBimA (purple) with the mean apparent KD ± SD listed. (C) TIRF images of BmBimA-AF488 (arrowhead) elongating two (top) or three (bottom) filaments (colors as in A). (D) Graph of filament length (number of subs X 1000) over time for pairs of co-elongating filaments (each pair is in matched colors of solid and dashed lines). (E) Mean elongation rates (sub/s ±SE) for actin alone or for single filaments or two- and three-filament bundles elongated with BmBimA-AF488 (left, green) or BpBimA-AF488 (right, purple). Asterisks denote paired samples that are significantly different (One-way ANOVA; P < 0.0001). (F) Left and middle, elongation reactions first incubated with 10 nM CapZ, followed by BimA and G-actin addition. Right, the time to half-maximum fluorescence normalized to actin alone (dashed black line) for BmBimA (green) or BpBimA (purple). See also Figure S2 and Movies S1-6.
Ena/VASP proteins are further distinguished from formins by the ability of at least one family member (Ena) to bundle and elongate two filaments at once (Winkelman et al., 2014). BmBimA and BpBimA also frequently gathered two or even three filaments and mediated their simultaneous elongation (Figures 2C and S2, Movies S3-S6). Bundled filaments bound by a single BimA spot grew at similar rates relative to each other (Figure 2D) and to individual BimA-elongated filaments (Figure 2E). Thus, in this regard, BpBimA and BmBimA activity closely resembles that of Ena/VASP proteins.
In cells, barbed ends are often capped by CapZ, which prevents filament elongation (Cooper and Sept, 2008). Purified CapZ displays a high affinity of ~2 nM for barbed ends (Caldwell et al., 1989) and a slow dissociation rate from barbed ends (~30 min half-life) (Schafer et al., 1996). Formins and Ena/VASP proteins compete with CapZ to prevent capping (Barzik et al., 2005; Breitsprecher et al., 2008; Paul and Pollard, 2009) but have not been reported to remove CapZ from pre-capped filaments. To test whether BpBimA and BmBimA have anti-capping activity, we monitored their ability to promote filament elongation when added to pre-formed actin seeds simultaneously with CapZ (data not shown), or when added 5 min after filaments had been pre-capped with CapZ (Figure 2F). Under both conditions, 10 nM CapZ alone completely inhibited elongation, whereas the addition of 30 nM BpBimA or BmBimA to pre-capped filaments allowed filament elongation. These results suggest that BpBimA and BmBimA have the unusual ability to remove CapZ from barbed ends to enable elongation. Together, our results demonstrate that BpBimA and BmBimA are barbed end-binding proteins that increase elongation rates, bundle filaments, and have anti-capping activity, similar to host Ena/VASP actin polymerases.
Oligomerization is required for BpBimA and BmBimA activity
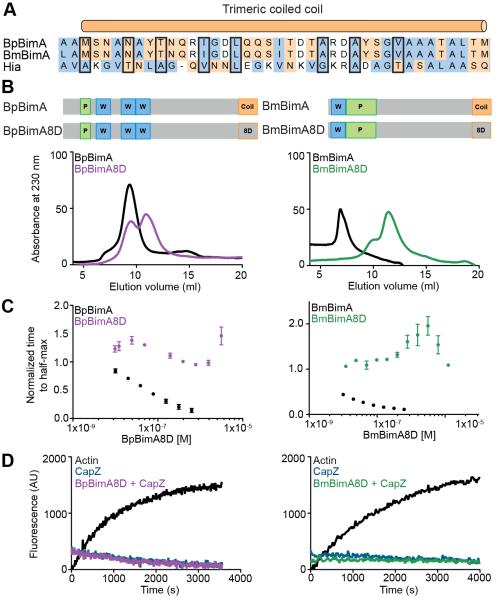
One feature of Ena/VASP proteins required for their actin polymerase activity is a tetrameric coiled coil located at their C-terminus (Breitsprecher et al., 2008; Hansen and Mullins, 2010; Kühnel et al., 2004). Based on the functional similarities of BpBimA and BmBimA with Ena/VASP proteins, we predicted that trimerization is important for BimA barbed-end-binding and elongation activities. To estimate the oligomeric state of wild-type and mutant BimA proteins we determined the behavior of wild-type BimA on a gel filtration column. BpBimA and BmBimA eluted at volumes corresponding to globular proteins of much larger molecular weight (> 500 kDa) than expected based on their sequences (127 kDa for BpBimA; 88 kDa for BmBimA; Figure 3B). However, other trimeric AT’s display aberrant behavior that is attributed to their elongated structures (Cotter et al., 2005; Hartmann et al., 2012; Mack et al., 1994). Next, we replaced eight residues of the predicted α-helix with aspartic acid (D) to disrupt the formation of the trimeric coiled coil (Figure 3A). The resulting BpBimA8D and BmBimA8D mutants eluted at a smaller size relative to their wild-type counterparts by gel filtration (Figure 3B), suggesting that the higher oligomeric state of the wild-type proteins was disrupted and that the trimeric coiled coil mediates oligomerization.
Figure 3. BpBimA and BmBimA oligomerization is required for actin nucleation and barbed end binding.
(A) Alignment of the trimeric coiled coils from BpBimA and BmBimA with the Hia trimeric coiled coil (Meng et al., 2006). Positions replaced with Asp residues in the BimA8D mutants are outlined in black. Hydrophobic residues are blue and charged residues orange. (B) Domain schematics of wild-type and BimA8D mutants in which eight positions were changed to Asp residues are shown above gel filtration elution profiles of wild-type and mutant proteins. (C) The time to half-maximum fluorescence normalized to actin alone in polymerization reactions with increasing BimA8D proteins. The means ± SD are shown with the wild-type data from Figure 1C for reference. (D) Elongation reactions with 10 nM CapZ and with our without BimA8D proteins.
To determine the importance of oligomerization in actin nucleation and elongation, we compared the activity of each BimA8D mutant with wild-type BimA in pyrene actin assembly assays. Both BpBimA8D and BmBimA8D lacked detectable nucleation activity, and at high concentrations both mutants inhibited polymerization, suggesting that they sequester G-actin (Figure 3C). Neither mutant could relieve CapZ inhibition of elongation at concentrations two orders of magnitude higher than that at which wild-type BimA relieved inhibition (Figure 3D). Thus, oligomerization of BpBimA and BmBimA is required for actin nucleation, barbed end elongation and anti-capping activity, similar to the requirement of tetramerization for Ena/VASP actin polymerase activity.
WH2 motifs are required for BpBimA and BmBimA activity
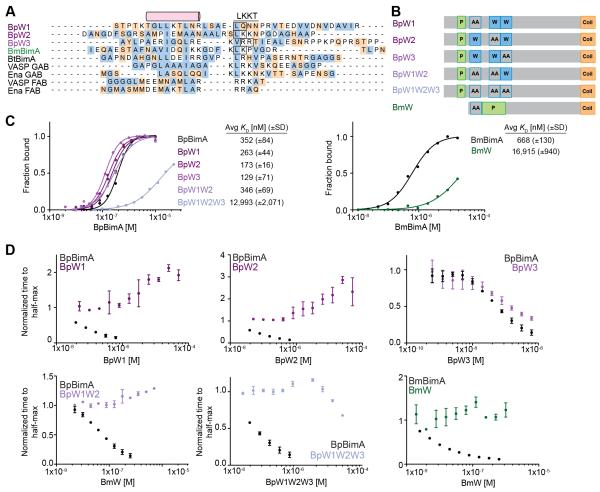
Ena/VASP barbed end binding and filament elongation requires two WH2 sequences (also called the globular actin binding or GAB and filamentous actin binding or FAB motifs) (Bachmann et al., 1999; Hansen and Mullins, 2010). BpBimA and BmBimA contain three (Bp) or one (Bm) putative WH2 sequences (Figure 1A) (Stevens et al., 2005a; 2005b) that are implicated in actin binding, and we hypothesized that each BimA would use these sequences for nucleation and elongation, similar to Ena/VASP proteins. We determined if the BimA WH2 sequences are competent for actin binding by replacing two positions of the conserved LKKT signature sequence with alanines (Figure 4A, B). To measure G-actin binding, we titrated increasing wild-type or WH2 mutant BimA into a fixed concentration of Alexa Fluor 488-labeled G-actin (actin488) in low ionic strength buffer to prevent actin polymerization (Figure 4C). Anisotropy measurements with increasing BimA produced saturable binding curves that were fit using the Hill equation. Wild-type BpBimA and BmBimA bound actin488 with apparent affinities of 350 nM and 670 nM (Figure 4C). Mutating any single predicted WH2 sequence in BpBimA (BpW1, BpW2 and BpW3 mutants) or mutating the first two WH2 motifs together (BpW1W2) had little effect on G-actin binding (Figure 4C). However, simultaneously mutating all three WH2 sequences in the BpW1W2W3 mutant severely reduced binding (KD ~13 μM). The single BmW mutant also exhibited a significant reduction in affinity relative to wild-type BmBimA (KD ~17 μM). These data demonstrate that BmBimA contains one WH2 motif capable of binding G-actin and suggests that BpBimA contains up to three WH2 motifs that bind G-actin.
Figure 4. WH2 requirements for BpBimA and BmBimA G-actin binding and nucleation.
(A) Alignment of predicted WH2 sequences from BimA orthologs with known WH2 (GAB and FAB) sequences from human VASP and Drosophila Ena. The conserved α-helix and LKKT motif are indicated. Hydrophobic residues are blue and charged residues orange. Boxed residues were mutated to AA. (B) Domain schematics of WH2 mutant BimA proteins. Predicted WH2 motifs that were mutated are outlined in blue and denoted by AA. (C) Anisotropy measurements of monomeric actin488 binding to BimA. Data are represented by circles, and Hill equation fits are shown as solid lines. The means KD ± SD from at least two experiments are listed. (D) The time to half-maximum fluorescence normalized to actin alone in polymerization reactions with BimA. The means ± SD are shown with wild-type data from Figure 1C for reference.
To determine if BimA WH2 motifs are required for filament nucleation and elongation, we tested each WH2 mutant for its ability to nucleate actin using the pyrene actin assembly assay. Although the BpW1 and BpW2 mutants bound G-actin with high affinity, they lacked nucleation activity, as evidenced by the absence of a decrease in time to half maximum fluorescence intensity (Figure 4D). Notably, these mutants inhibited polymerization at concentrations above ~1 μM, which is likely due to remaining intact WH2 motifs sequestering G-actin. In contrast, the activity of BpW3 was similar to wild-type BpBimA, indicating that the third WH2 motif is not required for activity. BpW1W2 and BpW1W2W3 were unable to nucleate actin, and BpW1W2W3 also lacked inhibitory activity, consistent with the low affinity of this mutant for G-actin (Figure 4C). Thus, while all three BpBimA WH2 motifs may bind actin monomers, only the N-terminal two WH2 motifs are required for nucleation activity. For BmBimA, the sole WH2 mutant BmW lacked detectable nucleation activity and did not inhibit actin polymerization, indicating that this WH2 motif is required for nucleation (Figure 4D). Together with the requirement for oligomerization, these results suggest that BpBimA uses up to six and BmBimA uses up to three WH2 motifs within a BimA trimer to mediate actin nucleation and elongation. Thus, like Ena/VASP proteins, BpBimA and BmBimA require oligomerization and WH2 motifs for their actin polymerase activities.
Mechanistic differences among BimA orthologs expressed in Bt result in distinct filament organization and parameters of actin-based motility
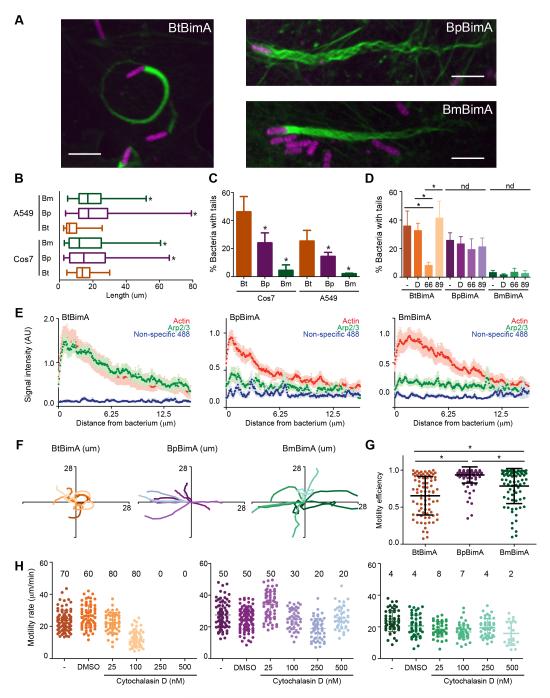
To compare how differences in the biochemical properties of BimA orthologs affect actin filament organization and actin-based motility by Burkholderia, we replaced the endogenous copy of Bt bimA with an identical copy of Bt bimA or with bimA from Bp or Bm. To assess BimA synthesis and localization, we engineered strains producing internally FLAG-tagged versions of BimA. Bacteria producing FLAG-BimA were compared with those expressing the corresponding untagged BimA orthologs in plaque assays and formed identical numbers and sizes of plaques as the untagged versions (data not shown). We found that FLAG-BtBimA and FLAG-BmBimA were produced at similar levels, whereas FLAG-BpBimA was produced at 2 to 2.5-fold higher levels than the others (Figure S3A). All displayed polar localization similar to endogenous BimA in Bp (Figure S3B) (Sitthidet et al., 2011; Stevens et al., 2005b) and also enabled actin tail formation by Bt following infection of tissue culture cells (Figure 5A) similar to previous results for BimA orthologs in Bp (Stevens et al., 2005a). Unless otherwise noted, data were obtained using strains expressing untagged BimA.
Figure 5. BimA from different Burkholderia species mediates the formation of distinct actin tails and parameters of actin-based motility.
(A) Merged images showing Cos7 cells infected with different Bt strains that constitutively express RFP (magenta). F-actin was stained with Alexa Fluor 488 phalloidin (green). Scale bars, 5 μm. (B) Mean actin tail lengths for the indicated strains in Cos7 and A549 cells. Boxes outline the 25th and 75th percentiles, midlines denote the medians and whiskers show minimum and maximum lengths. Asterisks denote lengths significantly different from BtBimA in that cell type (One-way ANOVA; P < 0.05). (C) Percent bacteria with actin tails (± SD). (D) Percent bacteria with actin tails (± SD) without or with 1 h treatment with DMSO, the Arp2/3 inhibitor CK-666 or the inactive compound CK-689. nd, no difference. (E) Mean actin, Arp2/3 or non-specific 488 nm fluorescence intensities (X 1000) from at least 10 actin tails (± SEM) are plotted along the first 12.5 μm of tail. (F) Tracks (ten per strain) depicting motility over 100 s for bacteria in Cos7 cells. (G) Motility efficiency from at least 80 tracks per strain, calculated as described in the text. (H) Motility velocity over 40 s for each strain in Cos7-Lifeact-EGFP cells untreated or treated with DMSO or CD. Mean frequencies of motile bacteria are listed. For all panels except (B), asterisks denote paired samples that are statistically different from one another (One-way ANOVA; P < 0.0001). See also Figure S3 and Movie S7.
Confocal microscopy of actin tail structures generated by each strain indicated that tails produced by BtBimA were curved and consisted of a dense actin network (Figure 5A; FLAG-BimA images are shown). In contrast, BpBimA and BmBimA produced longer, straighter tails that consisted of bundled F-actin strands (Figure 5B). Interestingly, the proportion of BpBimA bacteria associated with tails was moderately reduced and that of BmBimA bacteria was severely reduced relative to BtBimA bacteria (Figure 5C), suggesting differences in motility initiation. The Arp2/3 complex localized specifically to BtBimA tails, but not BpBimA or BmBimA tails, as monitored by immunofluorescence staining and by the presence of Arp3-GFP (Figures 5E and S3D-F). Furthermore, treatment of infected cells with the Arp2/3 complex inhibitor CK-666 (but not the control compound CK-689) reduced the frequency of tail formation by BtBimA, while tails formed by BpBimA or BmBimA were unaffected (Figure 5D). Thus, BtBimA requires Arp2/3 complex activity to produce shorter and curved actin tails, while the Ena/VASP mimics BpBimA and BmBimA work independent of Arp2/3 to produce longer tails with bundled filaments.
We next compared actin based-motility parameters among strains producing different BimA orthologs by time-lapse imaging. Once movement was initiated, all three strains exhibited similar average velocities of ~30 μm/min, with rates varying widely for each strain (Figure S3C). However, the movement paths of the BtBimA strain were more curved, whereas those of BpBimA and BmBimA bacteria were straighter (Figure 5F, Movie S7). To quantify path straightness and motility efficiency, we divided the linear displacement by the total distance traveled over 40 s for each bacterium. By this criteria, the BpBimA and BmBimA strains moved more efficiently than the BtBimA strain (Figure 5G). We also monitored motility following treatment with increasing concentrations of cytochalasin D (CD), a drug that binds to and inhibits growth of barbed ends. Although treatment with 250 or 500 nM CD reduced the frequency of motility for all strains, it completely blocked movement by the BtBimA strain, whereas some BpBimA and BmBimA bacteria resisted treatment and moved at rates similar to those in untreated cells (Figure 5H). This is consistent with the ability of these proteins to relieve CapZ inhibition. Overall, these results demonstrate that Arp2/3-dependent and Ena/VASP-like mechanisms of actin nucleation and elongation can drive similar rates of actin-based motility. However, distinct motility mechanisms also result in differences in motility initiation, paths, efficiency and susceptibility to inhibition by actin-disrupting drugs.
The mechanism of BimA actin assembly impacts the efficiency of host cell fusion
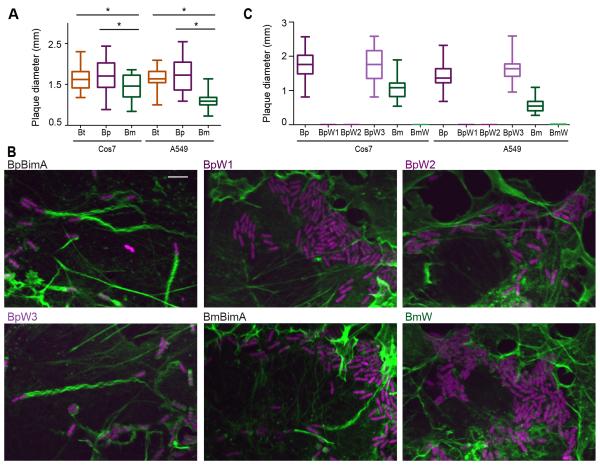
Bacterial-mediated host cell fusion is essential for Burkholderia virulence and fusion depends on actin-based motility (French et al., 2011; Schwarz et al., 2014). We therefore hypothesized that differences in actin polymerization mechanisms and motility impact Burkholderia-mediated fusion. To compare fusion efficiencies, strains producing different BimA orthologs were assessed for their ability to form plaques on host cell monolayers. BtBimA and BpBimA strains formed similar sized plaques, but the BmBimA strain produced smaller plaques in both Cos7 and A549 cells (Figure 6A). This plaque defect did not correlate with a reduction in BmBimA expression, a defect in localization (Figure S3A, B), intracellular replication (data not shown), slower rate of motility (Figure 5D), or differences in movement paths (Figure 5F). Instead, the reduced plaque size strongly correlated with a low frequency of actin tail association (< 5%; Figure 5C), suggesting that the initiation of actin-based motility is a crucial parameter for Burkholderia host cell fusion.
Figure 6. BimA mechanisms of actin nucleation impact actin tail formation and host cell fusion.
(A) Plaque diameters in Cos7 or A549 cell monolayers infected with Bt expressing wild-type BtBimA, BpBimA or BmBimA. (B) Merged images of Cos7 cells infected with wild-type or mutant BimA-expressing Bt strains that constitutively express RFP (magenta). F-actin was stained with Alexa Fluor 488 phalloidin (green). Scale bars, 5 μm. (C) Plaque diameters in Cos7 or A549 cell monolayers infected with Bt expressing wild-type or mutant BimA. (A, C) Boxes outline the 25th and 75th percentiles, midlines denote the medians and whiskers show minimum and maximum tail lengths. Asterisks denote significantly different sample pairs (One-way ANOVA; P < 0.05). See also Figure S4.
We investigated the mechanisms used by BpBimA and BmBimA to enable intracellular motility and fusion by testing the prediction that each BimA would require one or more WH2 motifs to drive movement during infection. We generated Bt strains in which we replaced endogenous bimA with genes encoding WH2 mutants (including BpW1, BpW2, BpW3 and BmW). The synthesis and localization of BimA in each strain were similar to that of wild-type BimA as determined by immunofluorescence staining against internal FLAG tags (Figure S4). Consistent with their lack of polymerization activity, the BpW1 and BpW2 mutants had no detectable association with F-actin, while the BpW3 mutant generated actin tails similar to wild-type BpBimA (Figure 6B). The BmW mutant similarly lacked actin association. We next examined the efficiency of fusion by measuring the plaque-forming ability of the mutants. The BpW1, BpW2 and BmW strains were completely defective in plaque formation while the BpW3 strain generated a similar number and size of plaques as the wild-type BpBimA strain (Figure 6C). Thus, BpBimA requires WH2 motifs W1 and W2, and BmBimA requires its sole WH2 motif for actin-based motility and fusion during Bt infection. These results demonstrate that molecular mimicry of host Ena/VASP proteins is crucial for actin-based motility driven by BpBimA and BmBimA in host cells.
Discussion
Microbe-driven actin-based motility plays a crucial role in spread and virulence of many pathogens and is also used as a model system to understand actin dynamics in host cells. Almost all motile pathogens use an actin polymerization mechanism that depends on the host Arp2/3 complex (Truong et al., 2014; Welch and Way, 2013) with the exception of Rickettsia species that also use a formin-like mechanism late in infection (Haglund et al., 2010; Madasu et al., 2013; Reed et al., 2014). Here we show that virulent Burkholderia species employ a previously undescribed mechanism to co-opt actin for motility, mimicking Ena/VASP actin polymerases to nucleate, elongate and bundle filaments.
In contrast to BtBimA activation of the Arp2/3 complex, we found that BpBimA and BmBimA function independently of Arp2/3. Both directly nucleate actin, bind processively to filament barbed ends and increase the elongation rate of bundled filaments (Figure 7). Moreover, they have the unusual ability to dissociate CapZ from barbed ends, a property they share with the Vibrio cholerae VopF protein (Pernier et al., 2013), as well as to resist the action of the drug CD during infection. These activities most closely mirror those of the Ena/VASP family of actin polymerases (Krause et al., 2003), which similarly bind to filament barbed ends, processively elongate filaments, and under some conditions protect growing barbed ends from CapZ inhibition (Barzik et al., 2005; Bear et al., 2002; Breitsprecher et al., 2008; Hansen and Mullins, 2010; Schirenbeck et al., 2006; Winkelman et al., 2014). Perhaps the most striking parallel between the activities of these proteins is their shared ability to gather and elongate multiple filaments simultaneously, which is a recently described property of Drosophila Ena (Winkelman et al., 2014). This enables BpBimA and BmBimA to generate networks of bundled filaments similar to those formed by Ena/VASP proteins in mammalian cells (Krause et al., 2003).
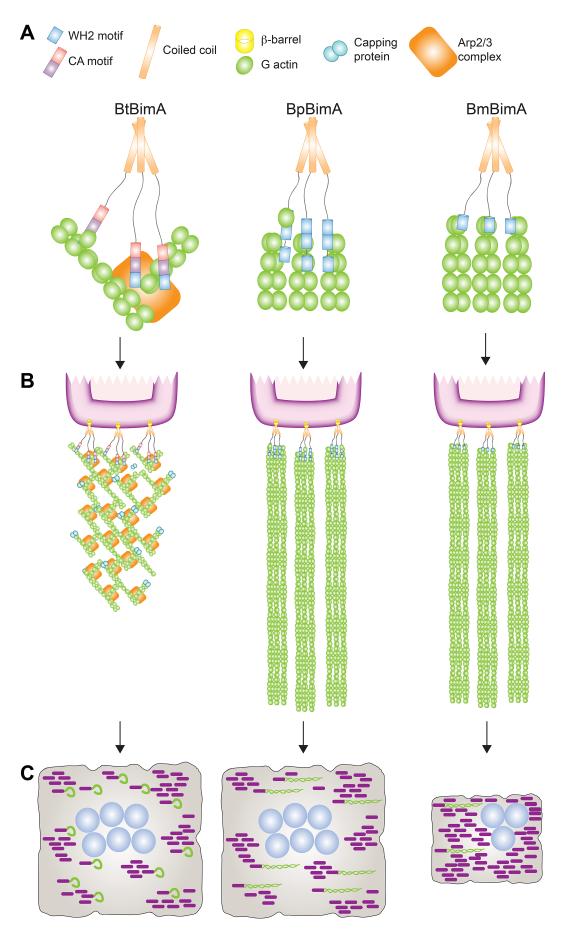
Figure 7. A model for BtBimA, BpBimA and BmBimA nucleation and actin tail and MNGC formation.
(A) BtBimA activates host Arp2/3 complex using its WCA domain to nucleate branched actin networks. BpBimA nucleates and elongates filaments from barbed ends using its two N-terminal WH2 motifs, while BmBimA exhibits the same activities using its single WH2 motif to generate bundled filaments. (B) BtBimA and Arp2/3 form shorter tails with branched actin networks. BpBimA and BmBimA produce longer tails of bundled filaments. (C) Burkholderia synthesizing BtBimA or BpBimA form more actin tails and generate larger MNGCs compared with bacteria producing BmBimA.
BpBimA and BmBimA also display similar sequence requirements as Ena/VASP for their activity, including both WH2 and coiled coils. BimA trimerization would result in the close positioning of multiple WH2 sequences (three for BmBimA and up to nine for BpBimA), which may promote elongation through the proximity of G-actin and barbed end binding sites (Figure 7). Higher-order oligomerization of BpBimA or BmBimA may also contribute to their ability to bundle and elongate multiple filaments, similar to the tetramerization requirement and role of clustering in Ena/VASP protein processivity (Bachmann et al., 1999; Breitsprecher et al., 2008; Hansen and Mullins, 2010; Samarin et al., 2003).
Our analysis also revealed that Bt producing different BimA orthologs generate actin tails with distinct organization (Figure 7) and move differently within host cells. BtBimA bacteria generate shorter and curvier tails that are densely packed with actin. Although they move at velocities similar to bacteria expressing other orthologs, they exhibited lower motility efficiency due to their curvier paths. These characteristics are similar to those of L. monocytogenes, which use the NPF ActA to activate Arp2/3 and polymerize actin (Welch et al., 1998). In contrast, Bt synthesizing BpBimA or BmBimA produce longer, straighter tails consisting of bundled filaments and exhibit a higher motility efficiency due to straighter paths. The bundled filament organization in these tails is consistent with the Ena/VASP-like activities of BpBimA and BmBimA in vitro. Thus, virulent Burkholderia species have evolved a distinct molecular mechanism for efficient actin-based motility.
Actin-based motility driven by BimA orthologs also differs in promoting host cell fusion. In particular, the BmBimA strain exhibited reduced fusion efficiency relative to the BtBimA and BpBimA strains. Reduced fusion did not correlate with altered velocity, directionality of movement or even with the underlying polymerization mechanism. Fusion did, however, correlate with a ~5 to 10-fold reduction in the frequency of BmBimA-driven tail formation and motility initiation. This defect is surprising considering that BmBimA possesses more potent nucleation and elongation activity than BpBimA in vitro. Thus, despite its increased activity in isolation, BmBimA appears to be a less efficient actin nucleator in cells. Differences between the activity of BimA outside versus inside cells may be due to differential requirements for other bacterial or host factors that play important roles in motility. For example, BmBimA may require additional host actin regulators that are present in equine cells but absent from human cells. Regardless, these findings suggest that the ability of BimA to initiate actin-based motility is a particularly crucial parameter to enable host cell fusion.
Why has BimA evolved distinct mechanisms of generating actin filaments? Bp is a soil saprophyte that infects a broad range of mammals (Galyov et al., 2010), and Bm is a clonal descendent of Bp that has undergone reductive genome evolution and cannot survive outside of a host (Galyov et al., 2010; Godoy et al., 2003; Nierman et al., 2004). Bt is a soil saprophyte like Bp, but its natural host is unknown. The sequence and mechanistic similarities between BpBimA and BmBimA reflect their evolutionary lineage, and it is interesting that the length and complexity of BmBimA has decreased relative to BpBimA, mirroring changes in the size of their respective genomes. The evolution of BtBimA is less clear. We speculate that distinct BimA mechanisms evolved in response to different host cell environments, where BpBimA is optimized for infection of various mammals, BmBimA is fine-tuned to function in equines, and BtBimA has adapted to unknown hosts in the soil environment.
As a whole, our results highlight that even closely related species have evolved to mimic a diverse spectrum of host actin-polymerizing pathways, and that mimicry of different polymerization mechanisms may influence key parameters of infection such as host cell fusion and bacterial dissemination. The acquisition of distinct polymerization mechanisms may be relevant for the evolution of virulence, as both highly virulent Burkholderia species evolved mimics of host Ena/VASP proteins that nucleate, elongate and bundle actin filaments, as well as remove filament capping proteins. However, the ramifications of these actin regulatory capabilities on virulence remain to be appreciated. Studying the divergent actin-based motility mechanisms of closely related species represents a powerful approach to unravel the evolution of pathogenic strategies for exploiting actin and to reveal new principles that govern the generation, dynamics and regulation of distinct actin networks in cells.
Experimental Procedures
Protein purification and fluorescent labeling
SUMO-6XHis-BimA orthologs were expressed in E. coli and isolated by affinity, anion exchange and/or gel filtration chromatography. Alexa Fluor 488 C5 maleimide (Life Technologies) was used to label Cys-containing BpBimA and BmBimA. Details of the purification and labeling methods are in the Extended Experimental Methods.
Bulk actin assembly assays
Pyrene actin polymerization reactions contained rabbit skeletal muscle G-actin (1 or 3 μM, 10% pyrene labeled) and BimA. Elongation assays were performed by mixing F-actin with BimA, and initiated by adding G-actin (250 nM). For CP competition, 10 nM CP was mixed with F-actin seeds, BimA was added after 5 min, and reactions were initialized with G-actin (250 nM). For buffers and further details see Extended Experimental Methods.
BimA gel filtration chromatography
BimA proteins were run over a Superdex 200 10/300 GL gel filtration column (GE Healthcare) and detected by absorbance at 230 nm. See Extended Experimental Methods for further details.
Epifluorescence and TIRF microscopy to visualize actin filaments
For epifluorescence microscopy, polymerization reactions were stabilized by adding 1 μM rhodaminephalloidin (Life Technologies) and F-actin was visualized as described previously (Haglund et al., 2010). TIRF microscopy was performed as previously described (Kovar et al., 2006) except that reactions contained 1 μM ATP-actin (33% rhodamine-labeled). At least 10 filaments without BimA, or 20 filaments with 100 pM BpBimA or 10 nM BmBimA, were measured over 240 s by manually tracing filaments. See Extended Experimental Methods for further details.
G-actin binding anisotropy
BimA proteins were added to 100 nM Alexa Fluor 488-labeled G-actin under non-polymerizing conditions. Binding curves were fit using the Hill equation, and the means of at least two titrations are reported. See Extended Experimental Methods for further details.
Bacterial strain construction
Parental Bt strain E264 was a gift from P. Cotter (University of North Carolina, Chapel Hill). Bt bimA and Bt motA2 strains were gifts from C. French and J. F. Miller (University of California, Los Angeles). Details of strain construction are in the Extended Experimental Methods and in Tables S1 and S2.
B. thailandensis infection of mammalian cells
A549, COS7, HEK293T and U2OS cells were from the University of California, Berkeley tissue culture facility. Polyclonal cell lines stably expressing Lifeact-EGFP (COS7) or F-Tractin-Wasabi (A549) were generated by lentiviral transduction. For infections, bacteria grown in LB broth were added to pre-seeded mammalian cells for 1 h at 37°C at high MOIs (100-200) before adding media with gentamicin (Gm; 0.5 mg/ml). Fixed and live cell analysis was performed with samples at 8-15 hpi. For plaque assays, cells were infected (MOI of 0.1) for 1 h and overlaid with a mixture of agarose in media with Gm and imaged by neutral red staining at 36 hpi. See Extended Experimental Methods for further details.
Image analysis
Image analysis details are in the Extended Experimental Methods.
Supplementary Material
Acknowledgements
We thank Peggy Cotter, Jeffrey Iwig and Rebecca Lamason for comments on the manuscript, and Tiago Barros for expertise in model design. We are grateful to Melissa Anderson, Erin Garcia, Peggy Cotter, Todd French and Jeff F. Miller for reagents and technical advice and to Taro Ohkawa for imaging assistance. E.L.B. was supported by NIH/NIGMS NRSA F32 GM093652. M.D.W. is supported by NIH/NIGMS R01 GM059609, NIH/NIAID R01 AI109044 and NIH/NIAID R21 AI109270.
Footnotes
Author Contributions E.L.B. and M.D.W. designed the experiments, E.L.B. and C.M.N. generated the data, E.L.B. analyzed the data and E.L.B. and M.D.W. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. Embo J. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TEB, Small JV, Curth U, Dickinson RB, Faix J. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. Embo J. 2011;30:456–467. doi: 10.1038/emboj.2010.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell JE, Heiss SG, Mermall V, Cooper JA. Effects of CapZ, an actin-capping protein of muscle, on the polymerization of actin. Biochemistry. 1989;28:8506–8514. doi: 10.1021/bi00447a036. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M-F, Husson C, Renault L, Didry D. Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu. Int. Rev. Cell Mol. Biol. 2011;290:55–85. doi: 10.1016/B978-0-12-386037-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 2008;267:183–206. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter SE, Surana NK, St Geme JW., III Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 2005;13:199–205. doi: 10.1016/j.tim.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- French CT, Toesca IJ, Wu T-H, Teslaa T, Beaty SM, Wong W, Liu M, Schröder I, Chiou P-Y, Teitell MA, et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc. Natl. Acad. Sci. USA. 2011;108:12095–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galyov EE, Brett PJ, Deshazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 2001;65:595–626. doi: 10.1128/MMBR.65.4.595-626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 2010;12:1057–1063. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 2010;191:571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect. Immun. 2008;76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. The mouse formin, FRL alpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 2004;279:20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- Hartmann MD, Grin I, Dunin-Horkawicz S, Deiss S, Linke D, Lupas AN, Hernandez Alvarez B. Complete fiber structures of complex trimeric autotransporter adhesins conserved in enterobacteria. Proc. Natl. Acad. Sci. USA. 2012;109:20907–20912. doi: 10.1073/pnas.1211872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Fischer ER, Mead DJ, Hackstadt T. Chlamydial TARP is a bacterial nucleator of actin. Proc. Natl. Acad. Sci. USA. 2006;103:15599–15604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kespichayawattana W, Rattanachetkul S, Wanun T, Utaisincharoen P, Sirisinha S. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 2000;68:5377–5384. doi: 10.1128/iai.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun. 2010;78:2240–2247. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Kühnel K, Jarchau T, Wolf E, Schlichting I, Walter U, Wittinghofer A, Strelkov SV. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc. Natl. Acad. Sci. USA. 2004;101:17027–17032. doi: 10.1073/pnas.0403069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liverman AD, Cheng H-C, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, Rosen MK, Orth K. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. USA. 2007;104:17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Heesemann J, Laufs R. Characterization of different oligomeric species of the Yersinia enterocolitica outer membrane protein YadA. Med. Microbiol. Immunol. 1994;183:217–227. doi: 10.1007/BF00194174. [DOI] [PubMed] [Google Scholar]
- Madasu Y, Suarez C, Kast DJ, Kovar DR, Dominguez R. Rickettsia Sca2 has evolved formin-like activity through a different molecular mechanism. Proc. Natl. Acad. Sci. USA. 2013;110:E2677–E2686. doi: 10.1073/pnas.1307235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Surana NK, St Geme JW, Waksman G. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. Embo J. 2006;25:2297–2304. doi: 10.1038/sj.emboj.7601132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S, Boczkowska M, Glista MJ, Winkelman JD, Rebowski G, Kovar DR, Dominguez R. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat. Struct. Mol. Biol. 2011;18:1060–1067. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Deshazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, et al. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. USA. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernier J, Orban J, Avvaru BS, Jégou A, Romet-Lemonne G, Guichard B, Carlier M-F. Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat. Struct. Mol. Biol. 2013;20:1069–1076. doi: 10.1038/nsmb.2639. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Reed SC, Lamason RL, Risca VI, Abernathy E, Welch MD. Rickettsia Actin-Based Motility Occurs in Distinct Phases Mediated by Different Actin Nucleators. Curr. Biol. 2014;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarin S, Romero S, Kocks C, Didry D, Pantaloni D, Carlier M-F. How VASP enhances actin-based motility. J. Cell Biol. 2003;163:131–142. doi: 10.1083/jcb.200303191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrázek J, Nierman WC, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TEB, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc. Natl. Acad. Sci. USA. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Singh P, Robertson JD, LeRoux M, Skerrett SJ, Goodlett DR, West TE, Mougous JD. VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 2014;82:1445–1452. doi: 10.1128/IAI.01368-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthidet C, Korbsrisate S, Layton AN, Field TR, Stevens MP, Stevens JM. Identification of motifs of Burkholderia pseudomallei BimA required for intracellular motility, actin binding, and actin polymerization. J. Bacteriol. 2011;193:1901–1910. doi: 10.1128/JB.01455-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthidet C, Stevens JM, Field TR, Layton AN, Korbsrisate S, Stevens MP. Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J. Bacteriol. 2010;192:5249–5252. doi: 10.1128/JB.00608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JM, Ulrich RL, Taylor LA, Wood MW, Deshazer D, Stevens MP, Galyov EE. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 2005a;187:7857–7862. doi: 10.1128/JB.187.22.7857-7862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Stevens JM, Jeng RL, Taylor LA, Wood MW, Hawes P, Monaghan P, Welch MD, Galyov EE. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 2005b;56:40–53. doi: 10.1111/j.1365-2958.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- Szczesny P, Lupas A. Domain annotation of trimeric autotransporter adhesins--daTAA. Bioinformatics. 2008;24:1251–1256. doi: 10.1093/bioinformatics/btn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Toesca IJ, French CT, Miller JF. The Type VI Secretion System Spike Protein VgrG5 Mediates Membrane Fusion during Intercellular Spread by Pseudomallei Group Burkholderia Species. Infect. Immun. 2014;82:1436–1444. doi: 10.1128/IAI.01367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D, Copeland JW, Brumell JH. Bacterial subversion of host cytoskeletal machinery: hijacking formins and the Arp2/3 complex. Bioessays. 2014;36:687–696. doi: 10.1002/bies.201400038. [DOI] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Welch MD, Way M. Arp2/3-mediated actin-based motility: a tail of pathogen abuse. Cell Host Microbe. 2013;14:242–255. doi: 10.1016/j.chom.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West TE, Frevert CW, Liggitt HD, Skerrett SJ. Inhalation of Burkholderia thailandensis results in lethal necrotizing pneumonia in mice: a surrogate model for pneumonic melioidosis. Trans. R. Soc. Trop. Med. Hyg. 2008;102(Suppl 1):S119–S126. doi: 10.1016/S0035-9203(08)70028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson L. Glanders: medicine and veterinary medicine in common pursuit of a contagious disease. Med. Hist. 1981;25:363–384. doi: 10.1017/s0025727300034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc. Natl. Acad. Sci. USA. 2014;111:4121–4126. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Cheng H-C, Brautigam CA, Tomchick DR, Rosen MK. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat. Struct. Mol. Biol. 2011;18:1068–1074. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.