Abstract
Over the past few decades, actin’s presence in the nucleus has been demonstrated. Actin is a key protein necessary for different nuclear processes. Although actin is well known for its functional role in dynamic behavior of the cytoskeleton, emerging studies are now highlighting new roles for actin. At the present time there is no doubt about the presence of actin in the nucleus. A number of studies have uncovered the functional involvement of actin in nuclear processes. Actin as one of the nuclear components has its own structured and functional rules, such as nuclear matrix association, chromatin remodeling, transcription by RNA polymerases I, II, III and mRNA processing. In this historical review, we attempt to provide an overview of our current understanding of the functions of actin in the nucleus.
Keywords: Actin, Nuclear Matrix, Chromatin Remodeling, Transcription
Introduction
Actin, a globular protein with an approximately 42 kDa molecular weight, is found in all eukaryotic cells as one of the most highly-conserved proteins of the cytoskeleton. Although actin is often thought of as a single protein, in mammals it actually consists of six different isoforms encoded by distinct genes (1). All of the isoforms possess remarkably similar amino acid sequences, with no isoform sharing less than 93% identity with any other isoform. Recent studies indicate that actin isoforms carry out unique cellular functions. Four isoforms of actin including α-skeletal actin, α-cardiac actin, α-smooth actin and γ-smooth actin are expressed primarily in skeletal, cardiac, and smooth muscle, respectively. The remaining two isoforms, β-cyto actin and γ-cyto actin are basically expressed universally (2).
The fundamental roles of actin are well-known in critical biological processes such as cell migration, determination of cell shape, and vesicle trafficking (3). While actin’s roles in the cytoskeleton and cytoplasm are well established, the presence of nuclear actin has been a controversial issue for many years, owing to the lack of definitive evidence (4). Recent evidence shows the association of actin with multiple nuclear complexes, thus the existence of actin in the nucleus is slowly being accepted.
To date, actin has been implicated in various structured and functional roles in nuclears, including nuclear matrix association, chromatin remodeling, transcription and mRNA processing (5). Mass spectrometry (6) as well as immunoreactivity experiments (7,8) show that β-actin is the nuclear isoform of actin associated with heterogeneous nuclear ribonucleoproteins ( hnRNPs ) and RNA polymerase complexes. Schoenenberger et al. (9) have discovered that different polymerization states of actin co-exist in the nucleus. Studies of fluorescence recovery after photobleaching revealed that ~20% of the nuclear actin pool has characteristics of a very dynamic polymeric population (10). These results supported the existence of nuclear actin in polymeric form. It would be of interest to determine whether any of the functions of actin in the nucleus rely on polymerization/depolymerization events (3).
History of nuclear actin identification
The presence of actin in eukaryotic cell nuclei was originally reported by Ohnishi et al. (11) in 1963 when they purified actin from the nuclei of calf thymocytes. Historically, the studies that introduced actin as a nuclear protein were initially met with massive skepticism. For many years its presence in the nucleus was considered to be an artifact. The main problem was the inability to reliably detect actin protein in the nucleus by immunofluorescence microscopy (12).
Most known functions of actin in the cytoplasm encompass polymerization of actin monomers ( globular actin or G-actin ) into actin polymers ( filamentous actin or F-actin ), which is key for the diverse functions of cytoskeletal actin.The most common method to detect actin filaments in the cytoplasm is phalloidin staining which detects actin filaments with at least seven actin monomers (13,14). However, under normal conditions, nuclei and consequently nuclear actin can not be stained by phalloidin. Therefore, the actin filaments could not be visualized in the cell nucleus in the manner that they were commonly observed in the cytoplasm (15).
In 1977, Clark and Merriam (16) conclusively identified the existence of actin in the nuclei of a Xenopus oocyte by removing the nuclear envelope of manually isolated oocyte nuclei. By using a more refined method, Pederson extracted chromatin from Dictyostelium discoideum that was expected to have less than 0.3% cytoplasmic contamination in the purified chromatin fraction. He demonstrated that actin’s concentration in this chromatin fraction exceeded the cytoplasmic concentration by several orders of magnitude (17). Subsequently, actin was identified in mammalian cell nuclei by immunoelectron microscopic studies (12).
Research experiments performed in the last decade have provided convincing evidence for the presence of actin in the cell nucleus and for the roles of actin in fundamental nuclear processes. A list of some organisms and cell types where actin has been identified is shown in table 1.
Table 1.
List of some species and cell types in which nuclear actin has been reported
| Species | Cell types | References |
|---|---|---|
| Protists | Physarum polycephalum | Jockusch et al. (18) |
| Tetrahymena pyriformis | Katsumaru and Fukui (19) | |
| Dinoflagellate | Soyer-Gobillard et al. (20) | |
| Plants | Sauromatum guttatum | Skubatz et al. (21) |
| Allium cepa | Cruz et al. (22) | |
| Insects | Spodoptera frugiperda | Volkman (23) |
| Drosophila melanogaster | Sauman and Berry (24) | |
| Amphibians | Xenopus laevis | Merriam and Hill (25) |
| Rana temporaria | Parfenov et al. (26) | |
| Birds | Duck erythroblasts | Maundrell and Scherrer (27) |
| Chicken livers | Crowley and Brasch (28) | |
| Mammalian cells | Thymus cells | Ohnishi et al. (11) |
| Myoblasts | Paulin et al. (29) | |
| Ovary cells | Brunel and Lelay (30) | |
| Neural cells | Sahlas et al. (31) | |
Nucleocytoplasmic translocation of actin
The nucleocytoplasmic traffic occurs through nuclear pore complexes which form channels through the nuclear membrane. Two forms of transport occur over nuclear pores: I. passive diffusion of proteins smaller than 40 kDa and II. an active, energy-dependent traffic facilitated by specific transport receptors (32). The size of actin ( approximately 42 kDa ) is over the limit for passive diffusion through the nuclear pore. Therefore, there are some indications that actin may use this method to enter the nucleus (5).
Nuclear import of actin
Actin itself does not contain a classical nuclear localization signal ( NLS ), and until now no specific import receptor for actin has been reported. However, various actin-binding proteins ( ABPs ), such as cofilin (33), CapG (34) and megakaryocytic acute leukemia ( MAL ) (35) contain an NLS. Therefore these proteins can "piggy-back" actin into the nucleus. Cofilin is reported to be essential for nuclear accumulation of actin in response to cellular stress (36).
According to Pendleton et al. (36) latrunculin B treatment in mast cells induced formation of intranuclear filament which can be inhibited by antibodies against cofilin. Nevertheless, it is currently uncertain whether cofilin or other ABPs facilitate active transport of actin into the nucleus under physiological conditions (12). Recently, it has been shown that importin 9 works as an import factor for nuclear actins (37).
Nuclear export of actin
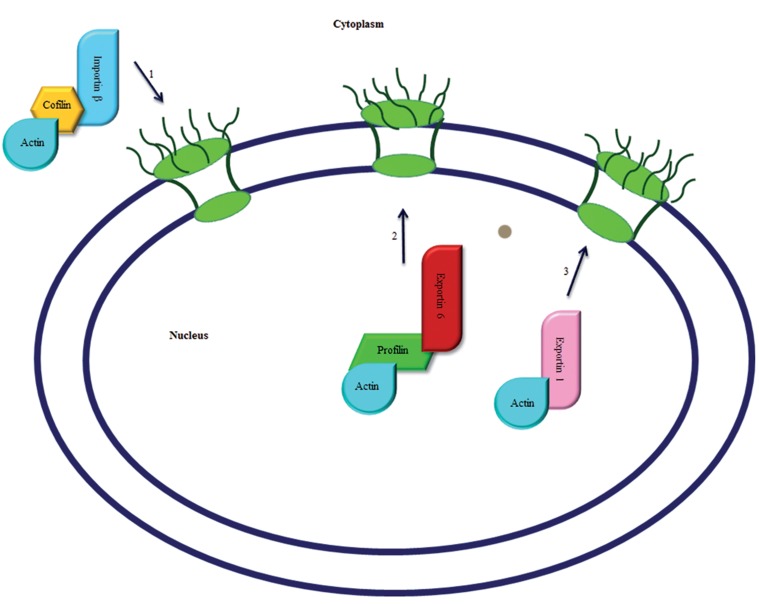
Actin appears to use an active transport mechanism to exit the nucleus. It is exported to the cytoplasm by at least two different pathways (Fig.1). Firstly, actin owns two classical leucine-rich nuclear export signals ( NES ), which exist in all of actin’s isoforms. These signals are essential for the export of actin via the export receptor exportin 1. Treatment of cells with leptomycin B, an inhibitor of exportin 1, causes actin accumulation in the nucleus (38). Secondly, the export receptor, exportin 6, appears to be responsible for the nuclear export of actin in a complex with the small ABP, profilin (39).
Fig.1.
Nucleocytoplasmic translocation of actin. 1. Nuclear import of actin. Cofilin enters the nucleus through the import receptor, importin β. 2, 3. Nuclear export of actin. Actin can exit the nucleus in coupled with the actin binding protein, profilin, through the export receptor exportin 6 and/or through the exportin 1 receptor.
Exportin 6 is a member of the importin β superfamily of transport receptors that sends actin out of the nucleus coupled with profilin (39). Although this receptor can export most types of actin isoforms (5), it is present only in higher eukaryotes (39). Chromosome region maintenance 1 ( Crm1 ), the general nuclear export receptor for cargo which has leucine-rich export signals, may be a counterpart for exportin 6 in lower eukaryotes (38). Apparently, exporting of actin coupled with actin binding proteins such as profiling is the only ligand for exportin 6 (39). This suggests that the nuclear level of actin in higher eukaryotes is specifically regulated (3).
Functional roles of nuclear actin
Actin is a well-known protein for its ability to shape the cytoplasm, target membrane growth, and ensure dynamic behavior of the cytoskeleton (40). In the nucleus, several regulators of actin polymerization are localized and/or translocated to the nucleus in a controlled manner, suggesting that at least some functions of actin in the nucleus are related to genome organization (15). Besides the structural role of actin in the nuclear matrix (41) actin is considered as an important factor in nuclear processes that range from chromatin remodeling to RNA splicing.
Actin is also a component of the chromatin remodeling complex; it associates with transcription machineries and newly synthesized ribonucleoproteins (15). Recent achievements in the field of gene transcription regulation have led to conclusions that there is a vital role for actin as a chief co-factor for all three eukaryotic RNA polymerases (42).
Actin and nuclear matrix
The interphase nucleus is thought to contain a three-dimensional filamentous protein network referred to as the nuclear matrix which is believed to be analogous to the cell cytoskeleton. The term nuclear matrix, as first introduced by Berezney and Coffey (43) in 1974, represents a highly structured residual framework obtained from rat liver nuclei by sequential salt extractions, detergent and nuclease treatments. Further ultra-structural studies have shown that the nuclear matrix is a network consisting of branched core filaments masked with a large number of hnRNPs and regulatory proteins (44). Nuclear matrix proteins ( NMPs ) make up the internal structural framework of the nucleus and are important in maintaining spatial order within the nucleus. This scaffold is an active and dynamic structure involved in different nuclear functions. The proposed functions of the nuclear matrix are mainly related to DNA replication and/or repair, gene transcription, RNA splicing and transport processes (44,49).
Field emission scanning electron microscopy experiments performed on Xenopus oocyte nuclei have clearly shown that the filamentous nuclear matrix network is basically composed of actin and protein 4.1components (50). This network may supply chromatin free channels for diffusion of ribonucleoproteins or proteins inside the nucleus. Sections of Xenopus oocyte nuclei analyzed by confocal microscopy also demonstrate bundles of actin filaments in the nuclear structure (5). It is suggested that these nuclear meshwork of actin filaments are capable of stabilizing the mechanical integrity of Xenopus oocyte nuclei (5,50).
In the context of the presence of actin in the nuclear matrix, it is possible that actin contributes to a variety of protein associations within the nuclear framework which positions them to function as organizers of the nuclear structure. Due to the fact that the body of existing data comes mainly from the Xenopus oocyte model, further studies are needed to clarify the function of actin in association with the nuclear structures of somatic cells (12).
Actin and chromatin modification/ remodeling
Chromatin is the combination of DNA and structural/regulatory proteins in eukaryotic cells. For processes that require accession to DNA to proceed, the chromatin structure needs to be changed; this is of extreme importance in gene regulation. Chromatin modification is a key process during all genome-related processes including DNA duplication and repair, regulation of gene expression and cell differentiation. A wide variety of different multi-subunit complexes are responsible for displaying these chromatin modifications (51,52). They are capable of moving and/or replacing nucleosomes on DNA strands ( chromatin remodeler ) or mark nucleosomes with covalent modifications ( chromatin modifier ). Nucleosomes are comprised of double-stranded DNA that has complexed with small proteins called histones. Nucleosomes, which constitute the smallest units in the chromatin structure, are the chief targets in the remodeling process. They include histone core proteins around which 146 bp of DNA is wrapped and the core proteins consist of dimers of the four histone molecules H2A, H2B, H3, and H4, respectively (53). Alterations in chromatin structure can result from different interactions between DNA and histones, the process mainly regulated by chromatin remodeling and chromatin modifying enzymes (54).
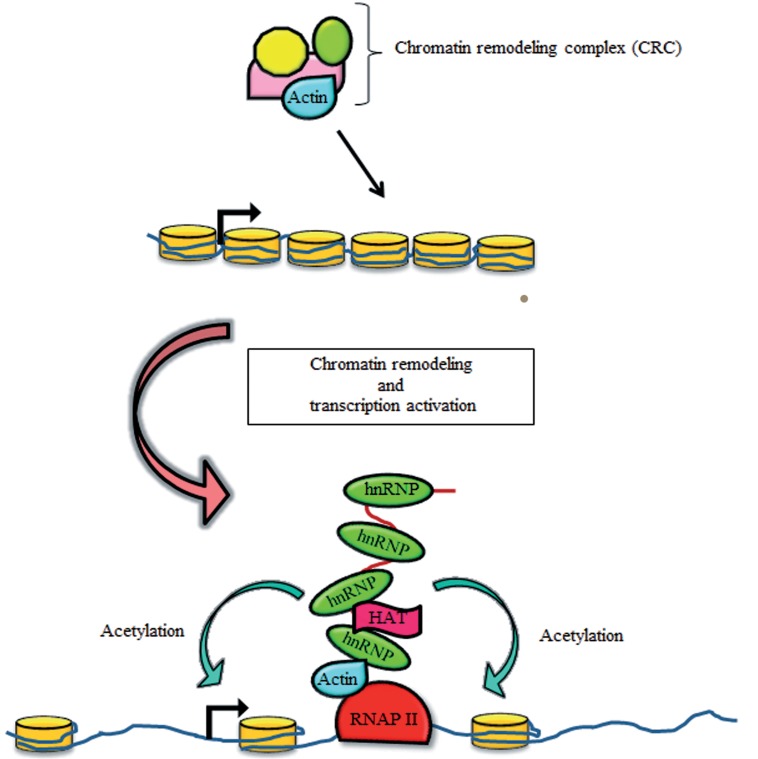
Recently investigators have shown that actin and actin-related proteins are involved in both chromatin remodeling and chromatin modifying complexes; this association is conserved from yeast to humans (52,54,55). It is well established that actin and a large number of actin-related proteins are vital subunits for the function of chromatin-remodeling complexes during transcriptional activation (56,59). Nuclear actin and its related proteins are also found in chromatin modifying enzymes such as histone acetyltransferase ( HAT ) complexes in a wide range of organisms that range from yeast to humans (Fig.2) (60,63).
Fig.2.
Models for the function of actin in chromatin regulation. A. Actin is a component of ATP-dependent chromatin remodeling complexes ( CRC ) involved in transcriptional activation and B. Co-transcriptional recruitment of histone modifier elements to RNA polymerase II ( RNAP II ), mediated by the presence of heterogeneous nuclear ribonucleoproteins ( hnRNPs ). During transcription, actin can be recruited to the elongating transcription machinery and facilitate recruitment of histone acetyltransferase ( HAT ) to the active gene, enhancing the processivity of RNAP II.
Actin in association with transcription machinery
The connection of actin with transcription was first mentioned by Smith et al. (64) in 1979, when they reported that actin was co-purified with RNA polymerase II ( RNAP II ) from the slime mold P. polycephalum. Egly et al. (65) in 1984 identified actin in transcriptionally active extracts from human HeLa and calf thymus cells. A direct role of actin in transcription was reported by Scheer et al. (66) in 1984 when they showed that microinjection of antibodies directed against actin and actin-related proteins such as fragmin caused transcriptional inhibition of chromosome loops in the nuclei of amphibian oocytes in sites where active transcription occurred. Sauman and Berry (24) showed an active transcriptional role of actin in Drosophila melanogaster cell nuclei. They have suggested that actin recruits the RNA polymerase complex to target DNA. In 2001, actin was found in Balbiani ring localization of nascent pre-mRNA in Chironomus alivary gland polytene chromosomes (67). Since numerous studies have shown the functional role of nuclear actin in transcription (68,72), it is clear that actin has more than one role as an epigenetic factor involved in the process of transcription.
Actin in nascent transcripts
In eukaryotic cells the nascent transcripts synthesized by RNAP II undergo extensive processing steps before a functional mRNA is produced. These mRNA precursors are named heterogeneous nuclear ribonucleicacids ( hnRNAs ), a historical term based on their size, heterogeneity and cellular location (73). When hnRNAs mature into mRNAs, they associated with a variety of proteins, as a group, named hnRNPs (73,74). These proteins function in an array of cellular activities such as pre-mRNA processing, mRNA export, localization, translation and stability (75). Although actin by itself is not capable of binding to RNA, its association with hnRNPs has been reported (12). In this way actin is a component of hnRNPs in a variety of mammalian cells (30), avian erythroblasts (27), and Xenopus oocytes (76).
Whereas the functional role of actin in RNP complexes is not clearly defined, studies in Chironomustentans have suggested that the cotranscriptional incorporation of actin into newly assembled pre-mRNPs affects chromatin architecture at transcribed genes (77). It is proposed that actin plays a role in this case via coupling with pre-mRNA-binding proteins, as a platform for the localization of a HAT activity alongside active transcription units (Fig.2).
Conclusion
After approximately 70 years since actin’s discovery in the cytoplasm, it is now undoubtedly accepted that actin has major roles in the cytoskeleton of eukaryotic cells and is a new member of nuclear proteins. The data mentioned in this article strongly support the idea that actin and actin-related proteins are dynamic nuclear factors involved in a multitude of nuclear functions. As discussed, nuclear actin plays functional roles in genome regulation in at least one of the three following ways: i. as a component of all three RNA polymerases in the eukaryotic cell nucleus, ii. as a constituent of nuclear ribonucleoprotein particles and iii. as a regulatory component of chromatin remodeling complexes. It is mentioned that nuclear actin can also function as a signal responsive regulator of specific transcription factors. Based on these data, a controversial issue will be the relationship between the dynamics of actin in the cytoplasm and nucleus. Due to the fact that actin in the cytoskeletal structure is very dynamic and interacts with a wide range of extracellular signals, it is suggested that actin may play a role as a sensor of extracellular signals in order to transduce signals to the chromatin state and moderate genome organization of living cells. To date, investigators suggest that as nuclear actin levels respond to different cellular stresses, it may therefore play a role in the pathology of different diseases. Also, identification of nuclear actin as a component of chromatin remodeling complexes is a current, hot topic between molecular cell biologists. Since chromatin is the critical site for maintaining the balance between gene activation and repression, scientists hope to identify potential ways by which nuclear actin can be involved in cancer.
Together, it can be concluded that while the biological significance of nuclear actin seems to be as versatile and important as in the cytoplasm, we are only beginning to understand the mechanisms that lie behind the regulation of nuclear actin.
References
- 1.Pollard TD. Genomics, the cytoskeleton and motility. Nature. 2001;409(6822):842–843. doi: 10.1038/35057029. [DOI] [PubMed] [Google Scholar]
- 2.Vandekerckhove J, Weber K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J Mol Biol. 1978;126(4):783–802. doi: 10.1016/0022-2836(78)90020-7. [DOI] [PubMed] [Google Scholar]
- 3.Miralles F, Visa N. Actin in transcription and transcription regulation. Curr Opin Cell Biol. 2006;18(3):261–266. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Chen M, Shen X. Nuclear actin and actin-related proteins in chromatin dynamics. Curr Opin Cell Biol. 2007;19(3):326–330. doi: 10.1016/j.ceb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Bohnsack MT, Stuven T, Kuhn C, Cordes VC, Gorlich D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol. 2006;8(3):257–263. doi: 10.1038/ncb1357. [DOI] [PubMed] [Google Scholar]
- 6.Jonsson AP, Aissouni Y, Palmberg C, Percipalle P, Nordling E, Daneholt B, et al. Recovery of gel-separated proteins for in-solution digestion and mass spectrometry. Anal Chem. 2001;73(22):5370–5377. doi: 10.1021/ac010486h. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6(11):1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 8.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18(24):3010–3015. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenenberger CA, Buchmeier S, Boerries M, Sutterlin R, Aebi U, Jockusch BM. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol. 2005;152(30):157–168. doi: 10.1016/j.jsb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 10.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172(4):541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohnishi T, Kawamura H, Yamamoto T. Extraction of a protein resembling actin from the cell nucleus of the calf thymus. J Biochem. 1963;54:298–300. doi: 10.1093/oxfordjournals.jbchem.a127789. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann WA. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 2009;273:219–263. doi: 10.1016/S1937-6448(08)01806-6. [DOI] [PubMed] [Google Scholar]
- 13.Visegrady B, Lorinczy D, Hild G, Somogyi B, Nyitrai M. A simple model for the cooperative stabilisation of actin filaments by phalloidin and jasplakinolide. FEBS Lett. 2005;579(1):6–10. doi: 10.1016/j.febslet.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 14.Vartiainen MK. Nuclear actin dynamics--from form to function. FEBS Lett. 2008;582(14):2033–2040. doi: 10.1016/j.febslet.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2(4):a000620–a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell. 1977;12(4):883–891. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 17.Pederson T. Isolation and charaterization of chromatin from the cellular slime mold, Dictyostelium discoideum. Biochemistry. 1977;16(12):2771–2777. doi: 10.1021/bi00631a029. [DOI] [PubMed] [Google Scholar]
- 18.Jockusch BM, Brown DF, Rusch HP. Synthesis and some properties of an actin-like nuclear protein in the slime mold Physarum polycephalum. J Bacteriol. 1971;108(2):705–714. doi: 10.1128/jb.108.2.705-714.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsumaru H, Fukui Y. In vivo identification of Tetrahymena actin probed by DMSO induction nuclear bundles. Exp Cell Res. 1982;137(2):353–363. doi: 10.1016/0014-4827(82)90036-2. [DOI] [PubMed] [Google Scholar]
- 20.Soyer-Gobillard MO, Ausseil J, Geraud ML. Nuclear and cytoplasmic actin in dinoflagellates. Biol Cell. 1996;87(1-2):17–35. [PubMed] [Google Scholar]
- 21.Skubatz H, Orellana MV, Yablonka-Reuveni Z. Cytochemical evidence for the presence of actin in the nucleus of the voodoo lily appendix. Histochem J. 2000;32(8):467–474. doi: 10.1023/a:1004140215519. [DOI] [PubMed] [Google Scholar]
- 22.Cruz JR, de la Torre C, Moreno Diaz de la Espina S. Nuclear actin in plants. Cell Biol Int. 2008;32(5):584–587. doi: 10.1016/j.cellbi.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Volkman LE. Autographa californica MNPV nucleocapsid assembly: Inhibition by cytochalasin D. Virology. 1988;163(2):547–553. doi: 10.1016/0042-6822(88)90295-4. [DOI] [PubMed] [Google Scholar]
- 24.Sauman I, Berry SJ. An actin infrastructure is associated with eukaryotic chromosomes: structural and functional significance. Eur J Cell Biol. 1994;64(2):348–356. [PubMed] [Google Scholar]
- 25.Merriam RW, Hill RJ. The germinal vesicle nucleus of Xenopus laevis oocytes as a selective storage receptacle for proteins. J Cell Biol. 1976;69(3):659–668. doi: 10.1083/jcb.69.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parfenov VN, Davis DS, Pochukalina GN, Sample CE, Bugaeva EA, Murti KG. Nuclear actin filaments and their topological changes in frog oocytes. Exp Cell Res. 1995;217(2):385–394. doi: 10.1006/excr.1995.1101. [DOI] [PubMed] [Google Scholar]
- 27.Maundrell K, Scherrer K. Characterization of pre-messenger-RNA-containing nuclear ribonucleoprotein particles from avian erythroblasts. Eur J Biochem. 1979;99(2):225–238. doi: 10.1111/j.1432-1033.1979.tb13249.x. [DOI] [PubMed] [Google Scholar]
- 28.Crowley KS, Brasch K. Does the interchromatin compartment contain actin? Cell Biol Int Rep. 1987;11(7):537–546. doi: 10.1016/0309-1651(87)90016-6. [DOI] [PubMed] [Google Scholar]
- 29.Paulin D, Nicolas JF, Jacquet M, Jakob H, Gros F, Jacob F. Comparative protein patterns in chromatins from mouse teratocarcinoma cells. Exp Cell Res. 1976;102(1):169–178. doi: 10.1016/0014-4827(76)90312-8. [DOI] [PubMed] [Google Scholar]
- 30.Brunel C, Lelay MN. Two-dimensional analysis of proteins associated with heterogenous nuclear RNA in various animal cell lines. Eur J Biochem. 1979;99(2):273–283. doi: 10.1111/j.1432-1033.1979.tb13254.x. [DOI] [PubMed] [Google Scholar]
- 31.Sahlas DJ, Milankov K, Park PC, De Boni U. Distribution of snRNPs, splicing factor SC-35 and actin in interphase nuclei: immunocytochemical evidence for differential distribution during changes in functional states. J Cell Sci. 1993;105(Pt 2):347–357. doi: 10.1242/jcs.105.2.347. [DOI] [PubMed] [Google Scholar]
- 32.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 33.Iida K, Matsumoto S, Yahara I. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct Funct. 1992;17(1):39–46. doi: 10.1247/csf.17.39. [DOI] [PubMed] [Google Scholar]
- 34.De Corte V, Van Impe K, Bruyneel E, Boucherie C, Mareel M, Vandekerckhove J, et al. Increased importin-beta-dependent nuclear import of the actin modulating protein CapG promotes cell invasion. J Cell Sci. 2004;117(Pt 22):5283–5292. doi: 10.1242/jcs.01410. [DOI] [PubMed] [Google Scholar]
- 35.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316(5832):1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 36.Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278(16):14394–14400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 37.Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci USA. 2012;109(9):E544–552. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17(6):1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuven T, Hartmann E, Gorlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin actin complexes. EMBO J. 2003;22(21):5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 41.Clubb BH, Locke M. Peripheral nuclear matrix actin forms perinuclear shells. J Cell Biochem. 1998;70(2):240–251. doi: 10.1002/(sici)1097-4644(19980801)70:2<240::aid-jcb10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 42.Obrdlik A, Kukalev A, Percipalle P. The function of actin in gene transcription. Histol Histopathol. 2007;22(9):1051–1055. doi: 10.14670/HH-22.1051. [DOI] [PubMed] [Google Scholar]
- 43.Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 44.Shahhoseini M, Rabbani Chadegani A. An overview to the structure and function of nuclear matrix. Yakhteh. 2009;10(4):232–241. [Google Scholar]
- 45.Berezney R, Mortillaro M, Ma H, Meng C, Samarabandu J, Wei X, et al. Connecting nuclear architecture and genomic function. J Cell Biochem. 1996;62(2):223–226. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C223::AID-JCB10%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 46.Berezney R. Regulating the mammalian genome: the role of nuclear architecture. Adv Enzyme Regul. 2002;42:39–52. doi: 10.1016/s0065-2571(01)00041-3. [DOI] [PubMed] [Google Scholar]
- 47.Hancock R. A new look at the nuclear matrix. Chromosoma. 2000;109(4):219–225. doi: 10.1007/s004120000077. [DOI] [PubMed] [Google Scholar]
- 48.Shahhoseini M, Rabbani-Chadegani A, Abdosamadi S. Identification of nonhistone protein LMG (160) as a ribonucleoprotein of the nuclear matrix with a role in transcription in vitro. Biochimie. 2007;89(11):1343–1350. doi: 10.1016/j.biochi.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Gall JG. Exporting actin. Nat Cell Biol. 2006;8(3):205–207. doi: 10.1038/ncb0306-205. [DOI] [PubMed] [Google Scholar]
- 50.Kiseleva E, Drummond SP, Goldberg M, Rutherford SA, Allen TD, Wilson KL. Actinand protein-4.1-containing filaments link nuclear pore complexes to subnuclear organelles in Xenopus oocyte nuclei. J Cell Sci. 2004;117(Pt 12):2481–2490. doi: 10.1242/jcs.01098. [DOI] [PubMed] [Google Scholar]
- 51.Hogan C, Varga-Weisz P. The regulation of ATP-dependent nucleosome remodelling factors. Mutat Res. 2007;618(1-2):41–51. doi: 10.1016/j.mrfmmm.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 52.Castano E, Philimonenko VV, Kahle M, Fukalova J, Kalendova A, Yildirim S, et al. Actin complexes in the cell nucleus: new stones in an old field. Histochem Cell Biol. 2010;133(6):607–626. doi: 10.1007/s00418-010-0701-2. [DOI] [PubMed] [Google Scholar]
- 53.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 54.Farrants AK. Chromatin remodelling and actin organization. FEBS Lett. 2008;582(14):2041–2050. doi: 10.1016/j.febslet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 55.Gangaraju VK, Batholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat Res. 2007;618(1-2):3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Percipalle P, Visa N. Molecular functions of nuclear actin in transcription. J Cell Biol. 2006;172(7):967–971. doi: 10.1083/jcb.200512083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grummt I. Actin and myosin as transcription factors. Curr Opin Genet Dev. 2006;16(2):191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Zheng B, Han M, Bernier M, Wen J. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009;276(10):2669–2685. doi: 10.1111/j.1742-4658.2009.06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodeling complex involved in transcription and DNA processing. Nature. 2000;406(6795):541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 60.Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, et al. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5(6):927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 61.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 62.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106(3):297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 63.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15(5):469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith SS, Kelly KH, Jockusch BM. Actin co-purifies with RNA polymerase II. Biochem Biophys Res Commun. 1979;86(1):161–166. doi: 10.1016/0006-291x(79)90395-4. [DOI] [PubMed] [Google Scholar]
- 65.Egly JM, Miyamoto NG, Moncollin V, Chambon P. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 1984;3(10):2363–2371. doi: 10.1002/j.1460-2075.1984.tb02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheer U, Hinssen H, Franke WW, Jockusch BM. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984;39(1):111–122. doi: 10.1016/0092-8674(84)90196-x. [DOI] [PubMed] [Google Scholar]
- 67.Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol. 2001;153(1):229–236. doi: 10.1083/jcb.153.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen E, Besombes D, Debey P. Immunofluorescent localization of actin in relation to transcription sites in mouse pronuclei. Mol Reprod Dev. 1998;50(3):263–272. doi: 10.1002/(SICI)1098-2795(199807)50:3<263::AID-MRD2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 69.Kysela K, Philimonenko AA, Philimonenko VV, Janacek J, Kahle M, Hozak P. Nuclear distribution of actin and myosin I depends on transcriptional activity of the cell. Histochem Cell Biol. 2005;124(5):347–358. doi: 10.1007/s00418-005-0042-8. [DOI] [PubMed] [Google Scholar]
- 70.Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6(12):1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 71.Fallah-Zadeh Kh, Shahhoseini M, Favaedi R, Afsharian P, Nabioni M. Epigenetic role of nuclear actin in Oct4 regulation of human embryonal cells. Cell J. 2012;14(Suppl 1):Ps–30. [Google Scholar]
- 72.Fomproix N, Percipalle P. An actin-myosin complex on actively transcribing genes. Exp Cell Res. 2004;294(1):140–148. doi: 10.1016/j.yexcr.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 73.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 74.Gall JG. Small granules in the amphibian oocyte nucleus and their relationship to RNA. J Biophys Biochem Cytol. 1956;2(Suppl 4):393–396. doi: 10.1083/jcb.2.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3(3):195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 76.Gounon P, Karsenti E. Involvement of contractile proteins in the changes in consistency of oocyte nucleoplasm of the newt Pleurodeles waltlii. J Cell Biol. 1981;88(2):410–421. doi: 10.1083/jcb.88.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sjolinder M, Bjork P, Soderberg E, Sabri N, Farrants AK, Visa N. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev. 2005;19(16):1871–1884. doi: 10.1101/gad.339405. [DOI] [PMC free article] [PubMed] [Google Scholar]