Abstract
Objective
Coronary artery disease (CAD) is a multi-factorial and heterogenic disease with atherosclerosis plaques formation in internal wall of coronary artery. Plaque formation results to limitation of the blood reaching to myocardium leading to appearance of some problems, such as ischemia, sudden thrombosis veins and myocardial infarction (MI). Several environmental and genetic factors are involved in prevalence and incident of CAD as follows: hypertension, high low density lipoprotein-cholesterol (LDL-C), age, diabetes mellitus, family history of early-onset heart disease and smoking. According to genome wide association studies (GWAS), five polymorphisms in the 9p21 locus seem to be associated with the CAD. We aimed to evaluate the remarkable association of two polymorphisms at 9p21 locus, rs1333049 and rs10757274, with CAD.
Materials and Methods
This experimental study was conducted in Golestan, Aria Hospitals and Genetics Lab of Shahid Chamran University in the city of Ahvaz, Iran, in 2010- 2011. The collected blood samples belonging to 170 CAD patients (case group) and 100 healthy individuals (control group) were analyzed by tetra-primer amplification refractory mutation system (ARMS)-polymerase chain reaction (PCR) technique. The results were analyzed using software package used for statistical analysis (SPSS; SPSS Inc., USA) version 16. A value of p<0.05 and an odd ratio (OR) with 95% confidence intervals (CI) were considered significant.
Results
The frequencies of CC, CG and GG genotypes for rs1333049 polymorphism in patients were 18.2, 65.3 and 16.5%, while in controls, the related values were 25, 67 and 8%, respectively. GG genotypes of rs1333049 polymorphism in CAD patients were more than control cases (OR: 0.354, 95%CI: 0.138-0.912, p=0.032). The frequencies of AA, AG and GG genotypes for rs10757274 in CAD patients were 8.2, 58.3 and 33.5%, while in controls, the related values were 35, 63 and 2%, respectively. GG Genotype in rs10757274 polymorphism in CAD patients was found more than control cases (OR: 0.014, 95% CI: 0.003 -0.065, p=0.0001).
Conclusion
The rs1333049 polymorphism at 9p21 locus shows a weak association with CAD, whereas rs10757274 polymorphism reveals a significant association with CAD. These variants may help the identification of patients with increased risk for coronary artery disease.
Keywords: CAD, 9p21, Polymorphism
Introduction
Coronary artery disease (CAD) or coronary heart disease (CHD) is caused by atherosclerotic plaque formation in coronary arteries. The atherosclerotic plaque prevents blood from flowing through arteries. Approximately, 1.3 million Americans have shown CHD. The condition leading to heart attack and sudden death may be recognized in middle age or later with symptoms such as chest pain or angina. Progression of CAD in women before menopause is less than men of same age, whereas 10 years after menopause, the risk will be the same for both sexes. In addition, CAD is considered as a multi-factorial and heterogenic disease that are often associated with other heart diseases including high blood pressure (1,3).
CAD progression often occurs during a ten-year period starting with lipid accumulation, scratching the lining, and finally, hardening of coronary arteries which leads to inflammation. By the blocking of 50 to 60% of coronary arteries, CAD symptoms can be appeared (3,4).
The first description of the symptoms of CAD was introduced in 1768 (5). The factors involved in CAD disease are categorized into the following items: I. non-modifiable risk factors (age, gender and heredity), II. modifiable risk factors [hypertension, hyperlipidemia, smoking, diabetes (6,10), behavioral factors and left ventricular hypertrophy (LVH)] and III. protective factors ( high density lipoprotein-cholesterol (HDL-C), exercise and estrogen) (11,14).
Epidemiology of CAD and myocardial infarction (MI)
The cardiovascular diseases are the most common diseases worldwide. As in America, about 14 million adults show signs of CAD. One-third of the 1.5 million of the patients experiencing MI have died. The financial costs associated with CAD are approximately 50 to 100 billion dollars a year (15). As in Great Britain and the United States, CAD is the main cause of death in most countries. In addition, CAD mortality rate in Finland has showed that 500 of every 100,000 people with this disease lose their lives (16,17).
9P21 locus
The genome-wide association studies (GWAS) have showed that 9p21 locus is associated with CAD (18). In addition, Wellcome Trust Case Control Consortium (WTCCC) and other studies conducted in German, Japan, Korea and Italy have considered 9p21 locus as a risk factor for CAD (18,22).
Generally, at the 9p21 locus, several polymorphisms exist, but only five polymorphisms, rs1333049, rs10757274, rs2383206, rs2383207 and rs10757278, have primary roles to predict CAD.
The rs1333049 have shown a good association with CAD because C allele of this single nucleotide polymorphism (SNP) was converted to G nucleotide that was first reported by Samani et al. (19) in the German population.
Other polymorphisms of the 9p21 locus are rs10757274 associated with CAD that was first reported by McPherson et al. (23) in the Canadian population. Then, many other studies have also confirmed the association of rs10757274 SNP with CAD and showed A allele is changed to G allele (21,23,24). In this study, the association of two polymorphisms at 9p21 locus (rs1333049, rs10757274) with CAD was investigated in the population living in Southwest Iran.
Materials and Methods
In this experimental study, the controls and patients were chosen among individuals referred to the Angiography and the CT-Angiography Divisions of the Golestan and Aria Hospitals, Ahwaz, Iran, in 2010. This study was conducted on 170 CAD patients and 100 healthy controls. The study included patients who were 50 years or older and who were diagnosed with coronary artery stenosis (according to cardiology tests). Approximately, 5 ml of blood from both groups were collected in falcon tubes containing 0.5 ml of 0.5 ethylene diaminetetraacetic acid (EDTA) as anti blood clotting. This study was approved by the Ethical Committee of Ahvaz Jondishapoor University of Medical Sciences. A signed informed consent was obtained from all participants.
DNA extraction
DNA extraction was done using Diatom DNA prep 100 kit (Cinna gene Co., Iran). After that, DNA quality was tested by gel electrophoresis techniques.
Tetra-primer amplification refractory mutation system (ARMS)-polymerase chain reaction(PCR)
Tetra-primer ARMS-PCR is a rapid simple technique and has more advantages in comparison to other techniques such as restriction fragment length polymorphism (RFLP) (25). Tetra-primer ARMS-PCR was used to check polymorphism. In this technique, there are four primers of which two primers external and two primers are internal that are used instead of two primers in the conventional PCR. The optimal length of tetra ARMS primers is about 26-32 bases. Two round-trip primers are complementary to each other, while their concentration must be equal, about 50 pmol. We designed primers for both polymorphisms (rs1333049 and rs10757274) using CLC Main Workbench (Qiagen, Denmark) (Table 1).
Table 1.
Primers of tetra ARMS for 9p21 locus polymorphisms including rs1333049 and rs10757274
| Type | Rs1333049 polymorphismSequence | CG% | Tm |
|---|---|---|---|
| OF primer | 5´-CGAAGTAGAGCTGCAAAGATATTTGGAA-3´ | 39.3 | 62.2 |
| OR primer | 5´-GGGCTCATAATTGCTGAATAAAACAGAA-3´ | 35.7 | 60.7 |
| IF primer | 5´-CCTCATACTAACCATATGATCAACAGATG-3´ | 37.9 | 62.4 |
| IR primer | 5´-CTTACCTCTGCGAGTGGCTGCTTATG-3´ | 53.8 | 66.4 |
| rs10757274 polymorphism | |||
| OF primer | 5´-TATGTAATGGCCTTCTTTGTCTCTTTTG-3´ | 35.7 | 60.7 |
| OR primer | 5´-AATGAATAAATGCTAACTTCTGCCTCAC -3´ | 35.7 | 60.7 |
| IF primer | 5´-GTGGGTCAAATCTAAGCTGAGTGTGGA -3´ | 48.1 | 64.9 |
| IR primer | 5´-ATCTATCTAGTGAATTTCAATTATGGCC -3´ | 32.1 | 59.2 |
ARMS; Amplification refractory mutation system, Tm; Melting temperature, OF; Outer forward, IF; Inner forward, OR: Outer reverse and IR; Inner reverse.
PCR thermal cyclers
In order to obtain the optimum temperature for PCR, a temperature gradient was carried out, and the PCR thermal cyclers were then performed as summarized in tables 2 and 3.
Table 2.
The PCR thermal cyclers of the rs1333049 polymorphism
| Stage | Number of repeat | Section | Temperature (˚C) | Time |
|---|---|---|---|---|
| First | 1 cycle | First denaturation | 95 | 5 minutes |
| Secondary denaturation | 95 | 45 seconds | ||
| Second | 30 cycles | Annealing | 62 | 90 seconds |
| First extension | 72 | 70 seconds | ||
| Third | 1 cycle | Secondary extension | 72 | 7 minutes |
PCR; Polymerase chain reaction.
Table 3.
The PCR thermal cyclers of the rs10757274
| Stage | Number of repeat | Section | Temperature (˚C) | Time |
|---|---|---|---|---|
| First | 1 cycle | First denaturation | 95 | 5 minutes |
| Secondary denaturation | 95 | 45 seconds | ||
| Second | 35 cycles | Annealing | 52 | 90 seconds |
| First extension | 72 | 90 seconds | ||
| Third | 1 cycle | Secondary extension | 72 | 7 minutes |
PCR; Polymerase chain reaction.
Observation of PCR products on agarose gel
PCR products of the rs1333049 polymorphism (C/G) and of rs10757274 polymorphism (A/G) were electrophoresed on 1.5 and 2% agarose gel, respectively, using 4 ml PCR products and 2 ml loading buffer for each sample. After electrophoresis, the gel was stained with ethidium bromide for 30 minutes and were observed by a ultra violet (UV) illuminator device (UV Tec CO., Russia). Moreover, according to the type of products observed on the gel, image polymorphisms genotype were determined. The products of each genotype are shown in tables 4 and 5.
Table 4.
Comparison of size and genotype of the rs1333049 polymorphism analyzed by gel electrophoresis
| Productgenotype | 422 bp(Product along with the external primers) | 263 bp(C allele) | 214 bp(G allele) |
|---|---|---|---|
| CC | + | + | - |
| CG | + | + | + |
| GG | + | - | + |
PCR; Polymerase chain reaction.
Table 5.
Comparison of size and genotypes of the rs10757274 polymorphism analyzed by gel electrophoresis
| Product | 412 bp | 249 bp | 218 bp |
|---|---|---|---|
| genotype | (Product along with the external primers) | (G allele) | (A allele) |
| AA | + | - | + |
| AG | + | + | + |
| GG | + | + | - |
Sequencing and analyzing of the samples
The obtained PCR products were compared in terms of length and categorized into several groups (Tables 4,5), while some samples were then prepared for sequencing. The results were analyzed using software package used for statistical analysis (SPSS, SPSS Inc., USA) version16. A value of p<0.05 and an odd ratio (OR) with 95% confidence intervals (CI) were considered significant.
Results
Average age values of patients and of healthy subjects are 58.66 ± 10.4 and 53.30 ± 7.2 years, respectively. The obtained results also showed that 89.4% of patients and 83% of healthy subjects were married. In addition, 35.9% of patients had family history of heart disease among whom 28.8% of their parents (patient group) had a consanguinity marriage. The ethnicity of participants was mainly Arabs living in city of Ahvaz, Khozestan Province, Southwestern Iran.
Mean age of diabetic patients (28.2%) was 52.21 ± 4.3 years, while mean age of non-diabetics patients was 54.34 ± 5.4 years. The findings revealed that mean age of patient with high blood lipids (32.4%) is 52.94 ± 5.7 years, while none-high blood lipids patients show mean age of 53.91 ± 7.1. Mean age of patients with high blood pressure (44.7%) is 54.43 ± 4.9 years, while mean age of patients without high blood pressure is 52.06 ± 3.8. Mean age of smoker patients (24.7%) is 54.05 ± 4.1 years, while related value of non-smoker patients is 53.21 ± 5.5.
Tetra-primer ARMS-PCR products of rs1333049 and rs10757274 polymorphisms
The genotype and size of bands on gel belonging to alleles of rs1333049 and rs10757274 polymorphisms were identified as shown in figures 1 and 2. The tables 4 and 5 also show the results of gel electrophoresis analysis.
Fig.1.
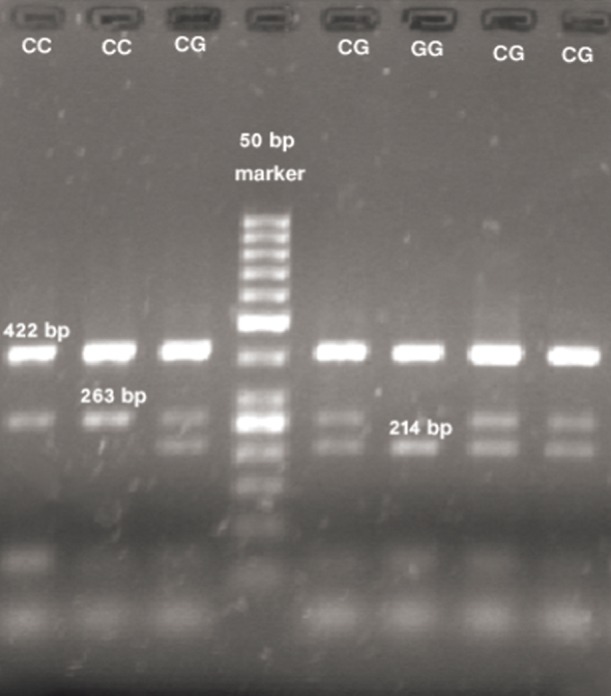
Electrophoresis results of PCR products of rs1333049 polymorphism.
PCR; Polymerase chain reaction.
Fig.2.
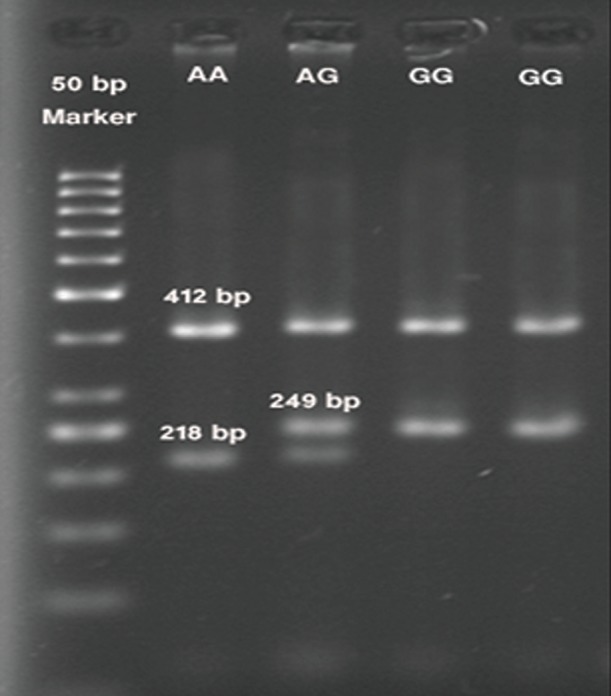
Electrophoresis results of PCR products of rs10757274 polymorphism.
PCR; Polymerase chain reaction.
Sequencing results
To confirm the tetra Arms PCR results, three samples of each polymorphism were sequenced.
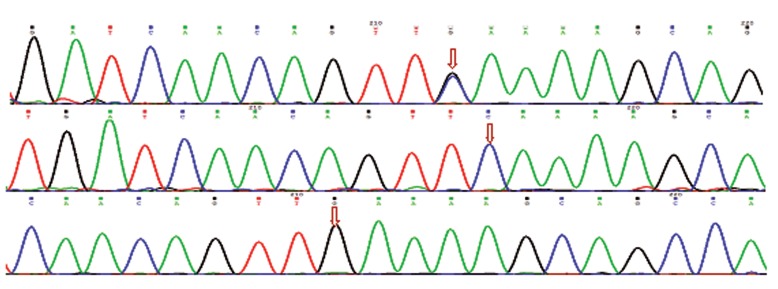
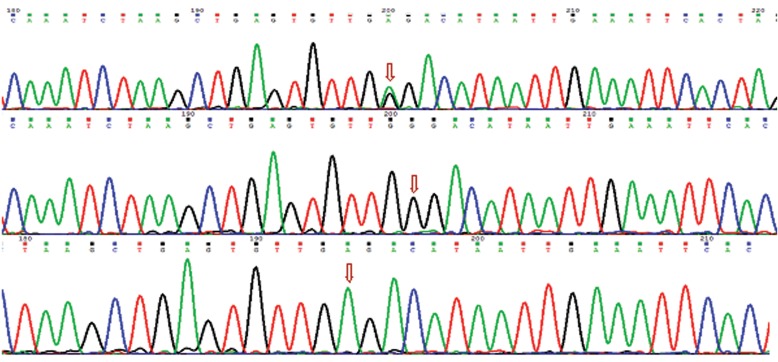
The sequencing results confirmed tetra ARMS PCR results (Figs.3,4).
Fig.3.
Sequencing analysis of rs1333049 polymorphism (arrows indicate CG, CC and GG genotypes, respectively).
Fig.4.
Sequencing analysis of rs10757274 polymorphism (arrows indicate AG, GG and AA genotypes, respectively).
Statistical results of polymorphisms
After genotyping all samples of each polymorphism, the results were statistically analyzed (Tables 6,7).
Table 6.
Comparison of genotype frequencies of the rs1333049 polymorphism between two groups in order to verify its association with CAD
| Genotype | Controls | Patients | OR | 95% CI for OR | P value | |
|---|---|---|---|---|---|---|
| Low | High | |||||
| CC | 25 | 31 | 0.806 | - | - | - |
| CG | 67 | 111 | 0.747 | 0.408 | 1.375 | 0.350 |
| GG | 8 | 28 | 0.354 | 0.138 | 0.912 | 0.032 |
CAD; Coronary artery disease, OR; Odd ratio and CI; Confidence intervals.
Table 7.
Comparison of genotype frequencies of the rs10757274 polymorphism in order to verify its association with CAD
| Genotype | Controls | Patients | OR | 95% CI for OR | P value | |
|---|---|---|---|---|---|---|
| Low | High | |||||
| AA | 35 | 14 | 2.500 | - | - | - |
| AG | 63 | 99 | 0.255 | 0.127 | 0.510 | 0.0001 |
| GG | 2 | 57 | 0.014 | 0.003 | 0.065 | 0.0001 |
CAD; Coronary artery disease, OR; Odd ratio and CI; Confidence intervals.
The overall result
The rs1333049 polymorphism showed a poor association (X2=4.343, df=1; p value=0.032), while the rs10757274 polymorphism showed a strong association with CAD (X2=53.476, df =1; p value=0.0001).
Discussion
Comparison of the obtained results between other studies and the present study is given in table 8 (18-21, 23, 24, 26).
Table 8.
Comparison of the other studies results with the present study
| Polymorphism | Source | P value | Country |
|---|---|---|---|
| Rs1333049 | Samani et al. (19) | 0.0001 | Germany |
| Hinohara et al. (18) | 0.00027 | Japan and South Korea | |
| Schunkert et al. (20) | 0.0079 | Germany and UK | |
| Shen et al. (21) | No association | America | |
| Present study | 0.032 | Iran | |
| Rs10757274 | McPhersson et al. (23) | <0.025 | Canada |
| Shen et al. (21) | 0.037 | Italy | |
| Talmud et al. (24) | 0.01 | UK | |
| Dehghan et al. (26) | No association | Netherlands | |
| Present study | 0.0001 | Iran | |
As our results indicate, presence of a group of risk factors, such as family history of heart disease, consanguineous marriage, age, diabetes mellitus, smoking, hypertension and high blood lipid, increases incidence of CAD.
The average age of onset of CAD in patients with family history of heart diseases (51.54 ± 3.6 years) was lower in comparison with patients without family history (55.48 ± 4.2 years). It was also suggested that the family history of heart diseases may predispose individual to CAD. As supposed, the average age of onset of CAD in patients whose parents had consanguinity marriage was lower by more than 2 years in comparison with patients whose parents were not relative. Our results also show diabetes, high triglycerides and low density lipoprotein cholesterol (LDL-C) increase the risk factors of CAD. High blood pressure and smoking are the other risk factors, whereas this study did not confirm the effects of these two factors on CAD.
The present study showed a weak association between rs1333049 polymorphism and CAD (p=0.032) as reported by Shen et al. (21), while a good association was observed between CAD and rs1333049 in Arab patients (p=0.023), which is consistent with the studies by Samani et al. (19), Hinohara et al. (18) and Schunkert et al. (20). The difference in our results may be due to racial or life styles differences of Arab ethnic group.
Conclusion
The present study showed a significant association between rs10757274 polymorphism and CAD (p=0.0001). This polymorphism may be a marker for the diagnosis of CAD, but more research is needed to reveal existence of involved genes in CAD prevalence.
Acknowledgments
We would like to thank the personnel of Angiography and CT-Angiography Sections of Golestan and Aria Hospitals of Ahvaz for their helps. We also appreciate the deputy of research and technology of Shahid Chamran University, Ahvaz, for financially supporting the project. There is no conflict of interest in this study.
References
- 1.Katz AM. Physiology of heart. 5th ed. Philadelphia: Wolters Kluwer, Lippincot Wiliams and Wilkins; 2011. pp. 3-33, 297-343. [Google Scholar]
- 2.Velodaver Z, Wilson RF, Garry DJ. Coronary heart disease: clinical, pathological, imaging and molecular profiles. 1st ed. New York: Springer Science; 2012. pp. 187-219, 471-496. [Google Scholar]
- 3.Shah PK. Risk factor in coronary artery disease. 1st ed. New York: Taylor & Francis Group; 2006. pp. 169–187. [Google Scholar]
- 4.Bhagavatula MR, Fan C, Shen GQ, Cassano J, Plow EF, Tropol EG, et al. Transcription factor MEF2A mutations in patients with coronary artery disease. Hum Mol Genet. 2004;13(24):3181–3188. doi: 10.1093/hmg/ddh329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaret BL, Moser M, Cohen LS. Yale University School of Medicine Heart book. 1st ed. New York: Yale University School of Medicine; 1992. pp. 133–147. [Google Scholar]
- 6.Randall OS, Segerson NM, Romaine DS. The encyclopedia of the heart and heart disease. 2nd ed. New York: facts and file; 2010. pp. 104–106. [Google Scholar]
- 7.Lichtlen PR. History of coronary heart disease. Z Kardiol. 2002;91(Suppl 4):56–59. doi: 10.1007/s00392-002-1409-7. [DOI] [PubMed] [Google Scholar]
- 8.Rosendorff C. Essential cardiology principles and practice. 2nd ed. Totawa: Humana Press; 2005. pp. 117-245, 409-438, 585-595. [Google Scholar]
- 9.Bronner G, Erdmann J, Mayer B, Hinney A, Hebebrand J. Genetic factors for overweight and CAD. Herz. 2006;31(3):189–199. doi: 10.1007/s00059-006-2797-7. [DOI] [PubMed] [Google Scholar]
- 10.Damani SB, Topol EJ. Future use of genomics in coronary artery disease. J Am Coll Cardiol. 2007;50(20):1933–1940. doi: 10.1016/j.jacc.2007.07.062. [DOI] [PubMed] [Google Scholar]
- 11.Yang C, Wang X, Ding H. Is coronary artery disease a multifactorial inherited disorder with a sex-influenced trait? Med Hypotheses. 2008;71(3):449–452. doi: 10.1016/j.mehy.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Robin NH, Tabereaux PB, Benza R, Korf BR. Genetic testing in cardiovascular disease. J Am Coll Cardiol. 2007;50(8):727–737. doi: 10.1016/j.jacc.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Channon KM, Watkins H. Coronary artery disease genetics: bigger is better. Eur Heart J. 2004;25(11):900–901. doi: 10.1016/j.ehj.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Hansson GK. Immune mechanisms in atherosclerosis. Arterioscler Thromb Vasc Biol. 2001;21(12):1876–1890. doi: 10.1161/hq1201.100220. [DOI] [PubMed] [Google Scholar]
- 15.Rani HS, Madhavi G, Rao VR, Sahay BK, Jyothy A. Risk factors for coronary heart disease in type II diabetes mellitus. Indian J Clin Biochem. 2005;20(2):75–80. doi: 10.1007/BF02867404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipsky MS, Mendelson M, Havas S, Miller M. Guide to preventing and treating heart disease. 1st ed. New Jersey, USA: Mc Graw Hill; 2007. pp. 117–149. [Google Scholar]
- 17.Tamburino C, Di salva ME, La manna A, Capodanno D. Left main coronary artery disease. 1st ed. Verlag: Springer; 2009. pp. 27–41. [Google Scholar]
- 18.Hinohara K, Nakajima T, Takahashi M, Hohda S, Sasaoka T, Nakahara K, et al. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008;53(4):357–359. doi: 10.1007/s10038-008-0248-4. [DOI] [PubMed] [Google Scholar]
- 19.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117(13):1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen GQ, Rao S, Martinelli N, Li L, Olivieri O, Corrocher R, et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008;53(2):144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 22.Anderson JL, Horne BD, Kolek MJ, Muhlestein JB, Mower CP, Park JJ, et al. Genetic variation at the 9p21 locus predicts angiographic coronary artery disease prevalence but not extent and has clinical utility. Am Heart J. 2008;156(6):1155–1162. doi: 10.1016/j.ahj.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 23.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talmud PJ, Cooper JA, Palmen J, Lovering R, Drenos F, Hingorani AD, et al. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008;54(3):467–474. doi: 10.1373/clinchem.2007.095489. [DOI] [PubMed] [Google Scholar]
- 25.Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29(17):E88–E88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dehghan A, van Hoek M, Sijbrands EJ, Oostra BA, Hofman A, van Duijn CM, et al. Lack of association of two common polymorphisms on 9p21 with risk of coronary heart disease and myocardial infarction; results from a prospective cohort study. BMC Med. 2008;6(30):1–7. doi: 10.1186/1741-7015-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]