Abstract
To further contribute to the understanding of multiple myeloma, we have focused our research interests on the mechanisms by which tumour plasma cells have a higher survival rate than normal plasma cells. In this article, we study the expression profile of genes involved in the regulation and protection of telomere length, telomerase activity and apoptosis in samples from patients with monoclonal gammopathy of undetermined significance, smouldering multiple myeloma, multiple myeloma (MM) and plasma cell leukaemia (PCL), as well as several human myeloma cell lines (HMCLs). Using conventional cytogenetic and fluorescence in situ hybridization studies, we identified a high number of telomeric associations (TAs). Moreover, telomere length measurements by terminal restriction fragment (TRF) assay showed a shorter mean TRF peak value, with a consistent correlation with the number of TAs. Using gene expression arrays and quantitative PCR we identified the hTERT gene together with 16 other genes directly involved in telomere length maintenance: HSPA9, KRAS, RB1, members of the Small nucleolar ribonucleoproteins family, A/B subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins, and 14-3-3 family. The expression levels of these genes were even higher than those in human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), which have unlimited proliferation capacity. In conclusion, the gene signature suggests that MM tumour cells are able to maintain stable short telomere lengths without exceeding the short critical length, allowing cell divisions to continue. We propose that this could be a mechanism contributing to MM tumour cells expansion in the bone marrow (BM).
Keywords: cell survival, chromosomal instability, apoptosis, multiple myeloma, telomere length maintenance, telomeric associations
Introduction
Multiple myeloma (MM) is a clonal malignancy of terminally differentiated plasma cells that typically accumulate in the bone marrow (BM), producing lytic bone lesions and excessive amounts of M-protein. Progression of MM is considered a multistage and dynamic process of cell differentiation, survival, proliferation and dissemination 1–3. Expansion of the neoplastic myeloma cell clone is the result of imbalances in proliferation and the induction or inhibition of apoptosis. Previous assumptions limited the problem of clinical progression of MM to the issue of cell proliferation, not taking into account the possibility that myeloma cells may have a low rate of apoptosis 4. Some studies evaluated the contribution of the apoptotic process based on in vitro observations or animal models, and under normal conditions plasma cells survived for a variable period of time, but did not proliferate. It is thus not clear which of these mechanisms have an immediate and applicable implication for the control of the disease 5.
Telomeres are DNA and protein structures located at the ends of chromosomes, constituted by thousands of DNA tandem repeats of (TTAGGG)n, and associated proteins in vertebrates 6. Human telomerase is a ribonucleoprotein that consists of a RNA component (telomerase RNA, hTR), a reverse transcriptase subunit (telomerase reverse transcriptase, hTERT) and associated proteins 7,8. hTR anneals to the telomeric sequence TTAGGG and acts as a template for telomeric DNA synthesis, whereas hTERT catalyzes the addition of telomeric repeats onto the chromosome ends. Telomeres are needed to protect chromosome ends from degradation by capping them, to distinguish chromosome ends from double-stranded breaks 9. It has also been shown that telomeres’ length maintenance also contributes to cell immortalization 10,11. In normal human somatic cells, shortened telomeres are often associated with senescence. At some point, telomeres become so short that the genes at the end of the chromosome are in danger of being deleted. When this critical length reduction is reached, a signal is activated inside the cell to prevent further cell divisions and cells may die. Based on these observations, telomeres have been thought to be a mitotic clock that counts the number of times a cell can divide before eliciting the senescence programme 12–15.
The role of the telomeres in the mechanisms of ageing and carcinogenesis has generated a considerable interest. Most immortal cells, including germ line cells 16, some stem cells 17,18, most cancer cells 19,20 and the majority of in vitro immortalized cells, possess enzymatic activity of telomerase 19,21, which probably catalyzes de novo synthesis of telomeric repeats at chromosome ends, suggesting that telomerase and telomere maintenance may be required for unlimited cell proliferation and tumorigenesis 22–25. Thus, in MM cancer stem cells, an increase in telomerase levels/activity has been proposed to be the main mechanism of telomere maintenance that enables evasion of apoptosis and conversely, a telomerase activity inhibition may impact both by decreasing telomere length and by modulating self-renewal programmes 26,27.
Telomerase activity is mainly regulated by hTERT transcription, but it can also be regulated positively or negatively by post-transcriptional and/or post-translational modification of the enzyme 28–30, via interaction of hTERT with accessory proteins that modulate telomerase activity 31,32. Thus, telomere length is important for tumour cell growth and survival. Increases in telomere length and the appearance of telomeric associations (TAs), defined as a cytogenetic phenomenon in which the telomeres of a single chromosome or two distinct chromosomes are associated to form dicentric, multicentric and ring chromosomes without visible loss of material from either chromosome end, are considered to be biomarkers in haematological malignancies and solid tumours 33. When Cottliar et al. 34 studied telomere length measured by terminal restriction fragments (TRF) in patients with MM and monoclonal gammopathy of undetermined significance (MGUS) they observed a reduction in telomere length in MM patients, in correlation with other studies in MM 35 and other haematological malignancies 36,37, as well as a significant increase in the occurrence of chromosome instability, a critical factor in the initiation and progression of human cancers 15,38. Telomere length reduction in mantle cell lymphoma is independent of the clinical characteristics, morphology and karyotype of the disease 39, but in MM high telomerase activity and short telomere length defined a subgroup of patients with poor prognosis and shorter mean survival 35.
To further contribute to the understanding of MM and the different molecular pathogenesis, we wanted to determine the expression levels of genes involved in telomerase activity in myeloma samples. We performed gene expression profiling analysis of samples from patients with MGUS, smouldering multiple myeloma (SMM), MM and plasma cell leukaemia (PCL), and human myeloma cell lines (HMCLs). We found a gene signature in that suggests that MM cells are able to maintain stable short telomere lengths without exceeding the short critical length, allowing cell divisions to continue, and thus contributing to MM cells expansion in the BM.
Materials and methods
Culture of human myeloma cell lines
We analysed human myeloma cell lines (HMCLs: JIM-1, KMS-11, KMS-12, KMS-26, KMS-28, MM1R, RPMI8226 and U266). All cell lines were acquired from either ATCC, DSMZ or JHSF, with exception of JIM-1, which was kindly provided by Birmingham University (Birmingham, UK). All HMCLs were cultured in RPMI 1640 (Invitrogen Life Technologies, Carlsband, CA, USA) supplemented with 10% FCS (Invitrogen Life Technologies). Cultures were maintained in exponential growth phase at 37°C in a humidified atmosphere with 5% carbon dioxide.
Culture of human embryonic stem cells lines and induced pluripotent stem cell lines
Human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) were maintained undifferentiated on allogeneic human feeders as described previously 40. Briefly, two hESCs lines (AND1, H9) and one iPSCs line (AND4) were cultured in plates on a confluent layer of inactivated human foreskin fibroblasts (HFFs) or human mesenchymal stem cells (hMSCs) with mesenchymal stem cell-conditioned medium supplemented with 8 ng/ml basic fibroblast growth factor (Invitrogen, Burlington, Ontario, Canada). Mesenchymal stem cell-conditioned medium was changed daily, and the cells were split weekly by dissociation with 200 U/ml of collagenase IV (Invitrogen).
Patient samples
Bone marrow aspirates were obtained from 42 newly diagnosed patients (20 male patients and 22 female patients) with MGUS, and 46 newly diagnosed patients (14 male patients and 32 female patients) with MM, after informed consent from all patients (Table1). The research was carried out according to the World Medical Association Declaration of Helsinki.
Table 1.
Clinical data of patients with MM and MGUS
| Gender | Median age (year) (range) | |||||||
|---|---|---|---|---|---|---|---|---|
| Groups | Female | Types of myeloma | Male | Types of myeloma | Female | Male | ||
| MGUS | 22 | 21: 46,XX | 16: MGUS 3: IgA/Kappa MGUS 3: IgG/Lambda MGUS | 20 | 19: 46,XY | 14: MGUS 1: IgM MGUS 2: IgA/Kappa MGUS 3: IgA/Lambda MGUS | 75.3 (44–87) | 64.3 (39–72) |
| 1: 46,XX/Hyperdiploid | 1: 46,XY/45,XY,+12,−13,−20,cp[16] | |||||||
| MM | 32 | 27: 46,XX | 30: MM 1: IgG MM 1: IgG/kappa MM | 14 | 11: 46,XY | 12: MM 1: IgG/Kappa MM 1: IgA MM | 73.5 (42–85) | 62 (34–70) |
| 1: 46,XX/47,XX,+3,+18,−13,t(10;13) | 1: 46,XY(15)/46,XY,del1q(5) | |||||||
| 1: 46,XX,del1q 46,XX.ish del1q,Tel10p,Tel10q,Tel15q,LSI PML(15q22) | 1: 46,XY[16]/45,XY,−13[2]/45,XY,−21[6] [cp22] | |||||||
| 1: 46, XX[17]/46, XX, −1, +i(1q)[3] | 1: 52,XY,−1,+der(1),+der(1),+3,+5,6,+r(6),+11,+15,16,+19,+22,del7q[12]/46,XY[9] | |||||||
| 1: 45∽54,XX,−1[4],+2[3],+3[4],−4[4],+5[3],+6[5],+9[4],−9[3],+10[4],−11[5],−13[7],+13[6],+15[12],+16[4],+17[7],+18[8],+19[5],+21[7],+22[8],[cp 29]. | ||||||||
| 1: 46,XX(6)/39–56,X(28) | ||||||||
MM: multiple myeloma; MGUS: monoclonal gammopathy of undetermined significance.
Plasma cells were selected as previously described 41, to a purity of >90% using CD138 microbeads and magnet-assisted cell sorting (Miltenyi Biotech, Bergisch Gladbach, Germany).
Conventional cytogenetics, fluorescence in situ hybridization and SKY analysis
Conventional cytogenetic, fluorescence in situ hybridization (FISH) and spectral karyotyping (SKY) analysis were performed in both HMCLs and patient samples. G-banding technique was done using standard approaches aimed at identifying TAs as previously described 42. The human telomere sequences were detected using the telomeric probe (Telomere PNA FISH Kits) according to the manufacturer's instructions (DAKO, Glostrup, Denmark). SKY was prepared according to the manufacturer's protocol (Applied Spectral Imaging, Migdal Ha'Emek, Israel). Images were acquired with a HiKSY system SD300-28B Spectra Cube (Applied Spectral Imaging) mounted on Leica DM 5500B microscope using a custom designed optical filter (SKY-1; Chroma technology, Brattleboro, VT, USA). Twenty metaphases were analysed for each sample, and normal BM cells were used as negative control.
Telomeric length evaluation
Telomere length was evaluated by TRF assay in HMCLs and MM patients as previously described 43. Hybridization signals were evaluated in the autoradiographs by densitometric scanning in each lane with respect to a k/HindIII molecular weight standard, and analysed using the Image Master 1D Prime (Pharmacia-Biotech, Uppsala, Sweden) software. The median telomeric length of a sample was evaluated over the range of 2–23 kb, and calculated by integrating the signal intensity over the entire TRF distribution as a function of TRF length using the formula: ∑(ODi)/∑(ODi/Li), where ODi and Li are the signal intensity, respectively, at position i on the autoradiogram. In addition, peripheral blood mononuclear cell (PBMC) from normal control (NC), K-562 cell line as internal positive control of telomere shortening, and cord blood (CB) cells as negative control of telomere shortening were used.
Microarrays hybridization
RNA from HMCLs was prepared for hybridization to the U133 Plus 2.0 expression GeneChip (Affymetrix, Santa Clara, CA, USA) according to the manufacturer′s instructions, as previously described 44–47.
Expression data analysis
The raw signal values were imported into GeneSpring GX 11.0. Data from individual microarrays were pre-processed using the MAS5 summarization algorithm and the baseline was done to the median of all samples. To analyse the especial regulation of telomere length in the haematological cancer MM, we determined the expression level of a list of 333 genes involved in regulation, inhibition, protection and maintenance of telomere length and apoptosis. We applied an anova with Tukey′s HSD and Benjamin & Hochberg FDR, P ≤ 0.05 and Fold Change ≤ 2. The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus 48 and are accessible through Gene Expression Omnibus Series accession number GSE15695.
Comparison with published expression data
We compared our results in an independent publicly available data set GSE6205 49 from HMCLs. We extend the analysis to examine primary BM samples from patients with MGUS, SMM, MM and PCL from two additional data sets GSE16122 50 and GSE6477 51,52.
RNA isolation, cDNA synthesis and quantitative real-time PCR (qPCR)
The genes with altered expression patterns were validated using quantitative real-time PCR (qPCR) in HMCLs and CD138+ malignant plasma cells from patients with MM. For comparative analysis, PBMC from five NC, CD138− plasma cells from patients with MM, two hESCs and one iPSCs were used. For qPCR experiments, equal amounts of total RNA (3 μg) were reverse transcribed from each sample by using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's protocol. qPCR amplification was performed with Brilliant III UltraFast SYBR green qPCR Master mix (Agilent Technologies, La Jolla, CA, USA) and detection of PCR products in real time with a M × 3005 Thermocycler (Agilent Technologies) in a final volume of 20 μl with 10 pmol of each primer (Table S1). The PCR program was initiated by 3 min. at 95°C before 40 thermal cycles, each of 10 sec. at 95°C and 20 sec. at 60°C. Data were analysed according to the 2−ΔΔCt method 53 and were normalized to beta-actin expression in each sample. Melting curves for each PCR reaction were generated to ensure the purity of the amplification product, and were confirmed by a single peak and a single band in agarose gel. A no-template negative control was also included in each experiment. Analysis of all samples was carried out in duplicate, and the mean values were calculated.
Results
Conventional cytogenetics, FISH and SKY analysis
We analysed a minimum of 20 metaphases per patient in 88 patients (46 MM and 42 MGUS) and more than 50 metaphases per HMCL, in 8 HMCLs, by conventional cytogenetic analysis and telomere FISH. As shown in Table2, the percentage of TAs was 4.97 times higher in patients with MGUS (median percentage of 64.71% metaphases with TAs per MGUS patient, range, 50–81.8%); 5.68 times higher in patients with MM (median percentage of 73.86% metaphases with TAs per MM patient, range, 60–89.5%); and 2.47 times higher in HMCLs (median percentage of 32.13% metaphases with TAs per HMCL, range, 26.02–64.45%), when compared with the control group (normal BM cells), with a median percentage of 13% metaphases with TAs per healthy donor (range 12–16%) (Table2). Different types of TAs were observed in HMCLs and patient samples (Fig.1), affecting either one or two chromatids. The results obtained by FISH also identified TAs, deletions and translocations of specific telomere regions (Fig.2). SKY analysis revealed multiple nonreciprocal translocations involving several chromosomes which, in most cases, were not fully characterized by G-banding or FISH (Fig.3). Our data suggest that chromosomal translocations were common in chromosomes with loss of telomeric signals as a result of telomere shortening.
Table 2.
Median percentage of metaphases with telomeric associations (TAs) by conventional cytogenetics and fluorescence in situ hybridization (FISH), and terminal restriction fragment (TRF) values
| Cell lines, MGUS and MM patients | Median% of Metaphases with TAs | TRF (kb) |
|---|---|---|
| NC | 13.00 | 8.12 ± 0.26 |
| CB | – | 14.20 ± 2.6 |
| K-562 | – | 5.02 ± 0.30 |
| JIM-1 | 26.02 | 5.30 |
| KMS-11 | 28.89 | 5.08 |
| KMS-12 | 28.37 | 5.30 |
| KMS-26 | 64.45 | 4.99 |
| KMS-28 | 27.75 | 5.43 |
| MMIR | 26.13 | 6.07 |
| RPMI8226 | 29.03 | 4.78 |
| U266 | 26.45 | 5.97 |
| Mean HMCLs | 32.13 | 5.36 ± 0.45 |
| Mean 42 MGUS patients | 64.71 | 6.55 ± 0.89 R: 7.14–5.16 |
| Mean 46 MM patients | 73.86 | 6.39 ± 0.67 R: 7.02–4.66 |
NC (normal BM cells and PBMC) as normal control, CB (cord blood) as negative control of telomere shortening, and K-562 (a human erythroleukemia cell line) as internal positive control of telomere shortening. R as telomere length range.
Fig 1.
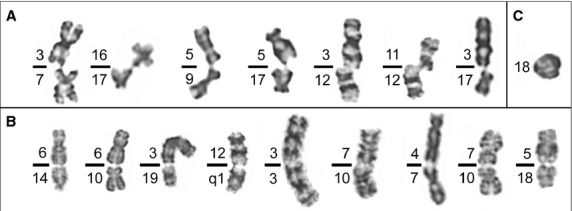
Different types of telomeric associations (TAs) were found in human myeloma cell lines, and monoclonal gammopathy of undetermined significance and multiple myeloma patients. We have divided these TAs into three types: (A) TAs between single chromatids of nonhomologous chromosomes (B) TAs between double chromatids of nonhomologous chromosomes (C) TAs between the arms of the same chromatid on a single chromosome.
Fig 2.
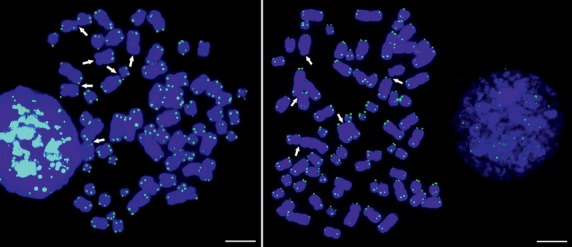
Fluorescence in situ hybridization (FISH) analysis of two metaphase spread from the human myeloma cell lines KMS26, using a PNA FISH probe specific for the telomeric repeats. The arrows indicate multiple chromosome ends showing absence or reduction of FISH signals, including those involved in telomeric associations. There is an accumulation of telomeric associations, involving one or two chromatides, probably related to a reduction in telomeric signal. Scale bars 5 μm.
Fig 3.

Spectral karyotyping (SKY) analysis from the human myeloma cell line KMS26, a hyperdiploid myeloma cell line that reveals multiple non-reciprocal translocations involving several chromosomes.
Telomeric length evaluation
Next we studied telomere length measured by TRF on eight HMCLs (Fig.4). The analysis of telomere length revealed a mean TRF peak value (5.36 ± 0.45 kb) significantly shorter than those observed in NC (8.12 ± 0.26 kb) and CB (14.20 ± 2.60 kb), and in the same range of human erythroleukemia cell line K-562 (5.02 ± 0.30 kb), and MGUS and MM patients (6.55 ± 0.89 and 6.39 ± 0.67 kb) respectively (Table2).
Fig 4.
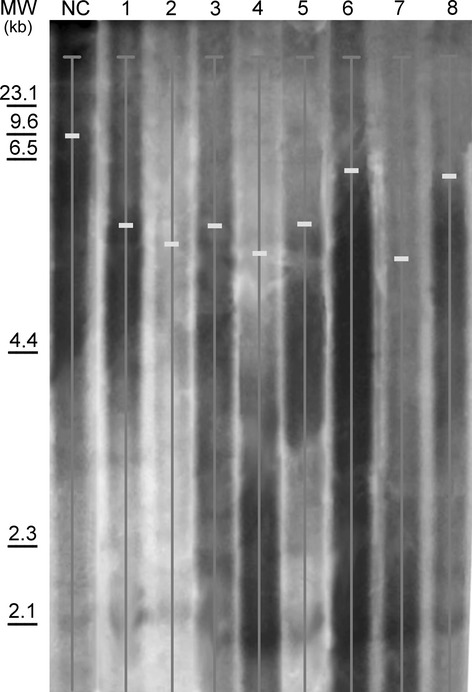
Example of Southern blot analysis used for terminal restriction fragment quantification, where hybridization signals were evaluated in the autoradiographs by densitometric scanning. The median telomeric length ratio of a sample was calculated considering the peaks observed in 1-JIM1, 2-KMS11, 3-KMS12, 4-KMS26, 5-KMS28, 6-MM1R, 7- RPMI8226, 8-U266 and NC (normal controls of peripheral blood mononuclear cell from health donors). MW: molecular weight.
On the basis of these data, we conclude that there is a strong correlation between shorter telomeres in HMCLs correlates with the percentage of telomeric association and chromosome instability (R2 = 0.9265) (Fig.5A). Human myeloma cell lines with higher number of TAs had lower telomere length (Fig.5C), with the exception of KMS26. This cell line is the only HMCL that shows an up-regulation of KRAS similar to CD138+ malignant plasma cells from patients with MM (Figure S9). Interestingly, all the patients with MGUS, SMM, MM and PCL, also show up-regulation of KRAS (Fig.6). These data also are in accord when comparing the telomere lengths with the percentage of telomeric association in mean of MGUS patients, mean of MM patients and NC, showing a R2 = 0.9969 (Fig.5B and D).
Fig 5.
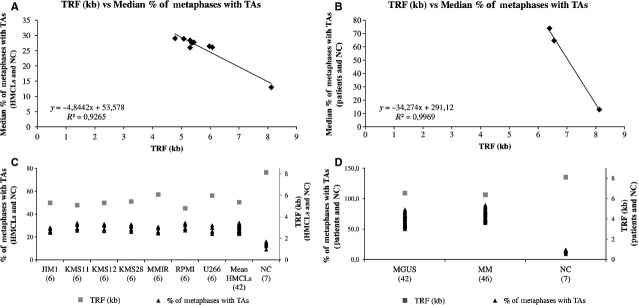
Correlation between the median percentage of metaphases with telomeric associations (TAs) versus the terminal restriction fragment (TRF) values. (A and C) in seven human myeloma cell lines (HMCLs) (JIM1, KMS11, KMS12, KMS28, MM1R, RPMI8226, U266) (six cultures of each cell type) and seven normal control (NC) (healthy donors). (B and D) in 42 monoclonal gammopathy of undetermined significance (MGUS) patients, 46 multiple myeloma (MM) patients and NC (healthy donors). There is a strong correlation between shorter telomeres correlates with the percentage of telomeric association, showing a R2 of 0.9265 (A) and 0.9969 (B) respectively. More number of TAs had lower telomere length.
Fig 6.
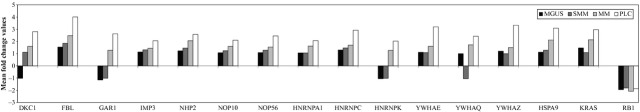
Microarray expression levels of 16 selected genes, in monoclonal gammopathy of undetermined significance (MGUS), smouldering multiple myeloma (SMM), multiple myeloma (MM) and plasma cell leukaemia (PCL). Test applied: anova with Tukey′s HSD and Benjamin & Hochberg FDR, P ≤ 0.05.
Gene expression signature of MM cells
To get an insight on the molecular mechanisms underlying these changes in telomere length and the increase in telomeric associations, we performed microarray analysis using HMCLs. To analyse the especial regulation of telomere length in MM, we determined the expression level of a list of 333 genes involved in regulation, inhibition, protection and maintenance of telomere length and apoptosis, and found 203 genes that were differentially expressed in the HMCLs. Ninety of these genes were overexpressed and 113 were underexpressed. We confirmed our results on a large independent data set (GSE6205) consisting of 23 HMCLs 49. The analysis of this list of 333 genes in patients with MGUS, SMM, MM and PCL showed 63 deregulated genes with 49 genes in common with HMCLs, including 16 genes directly involved in telomere maintenance (Table3). These sixteen genes are HSPA9, KRAS, RB1, members of the small nucleolar ribonucleoproteins family (FBL, NHP2, DKC1, NOP10, NOP56, IMP3 and GAR1), A/B subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (HNRNPA1, and HNRNPK) and the 14-3-3 family (YWHAQ, YWHAE, and YWHAZ). Most importantly, we can see an obvious progressive increase in the expression levels of these genes from MGUS to PCL, with overexpression most notably predominating (Fig.6).
Table 3.
The 16 selected genes involved in telomere maintenance and telomerase activity
| Gene Symbol | Gene Title |
|---|---|
| DKC1 | Dyskeratosis congenita 1, dyskerin |
| FBL | Fibrillarin |
| GAR1 | GAR1 ribonucleoprotein homolog (yeast) |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 |
| HNRNPC | Heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K |
| HSPA9 | Heat shock 70kDa protein 9 (mortalin) |
| IMP3 | IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| KRAS | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| NHP2 | NHP2 ribonucleoprotein homolog (yeast) |
| NOP10 | NOP10 ribonucleoprotein homolog (yeast) |
| NOP56 | NOP56 ribonucleoprotein homolog (yeast) |
| RB1 | Retinoblastoma 1 |
| YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
| YWHAQ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
Gene expression data analysis
To confirm the results from the microarray analysis we analysed gene expression by quantitative PCR. We used reverse transcribed RNA extracted from five controls of PBMC from healthy donors, eight HMCLs, six samples of CD138+ malignant plasma cells and CD138− plasma cells from patients with MM, two hESCs and one iPSCs, and measured expression levels for all sixteen deregulated genes plus four additional genes involved in telomere maintenance (TRF1, TRF2, TNKS and hTERT) 43. Log2 of the normalized microarrays ratio values were compared with log2 ratio values from qPCR experiments. The qPCR results confirmed that the relative RNA expression levels of the 16 deregulated genes were consistent with the microarray data and with public data sets, confirming the increased expression of these genes in the HMCLs and patients with respect to the hESCs, iPSCs and controls (Fig.7).
Fig 7.
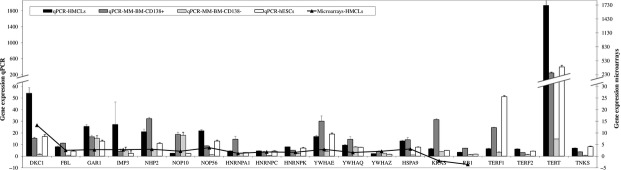
Expression levels in the eight human myeloma cell lines, six samples of CD138+ malignant plasma cells and CD138− plasma cells from patients with multiple myeloma, two human embryonic stem cells (hESCs) and one induced pluripotent stem cells (iPSCs) using qPCR, confirming the gene expression levels by microarray. Data were analysed according to the 2−ΔΔCt method and normalized to beta-actin expression. Y values are fold change, and controls of peripheral blood mononuclear cell (PBMC) from healthy donors are fold change = 1.
Discussion
Telomeres are maintained by telomerase, a specialized ribonucleoprotein complex that includes an RNA template (hTR) and a reverse transcriptase catalytic subunit (hTERT). Telomerase activity is low or absent in most human somatic tissues, although it is acting in tumoural, germ and stem cells. Telomeric DNA is progressively lost with each cell division, and when telomeres become critically short, the cells undergo senescence or apoptosis 54. This process is irreversible in most human somatic cells, and they normally die by apoptosis. However, MM tumour cells maintain stable short telomere lengths without reaching the short critical length. This suggests that telomere length maintenance is necessary for continued cell division and immortalization of normal and malignant cells 55.
In the present work, we found a distribution of chromosome ends involved in TAs that was obviously non-random. We have demonstrated an increase in the percentage of TAs and chromosomal instability in MGUS and MM patients, and HMCLs respect to NC (normal BM cells) (Figs1, 2 and 3). Both this instability and the increase in TAs numbers are obviously correlated with telomere length (Fig.5), with an inverse correlation between telomeric association and telomeric length. Therefore, it is possible to establish an association between telomere length and the probability of TAs to occur as a cause of chromosomal instability, to ensure the stability of chromosome ends, suggesting some selective advantage. Moreover, an increase in telomere stability facilitated by the increased expression of telomere maintenance proteins may constitute a possible strategy to avoid senescence or apoptosis.
Theoretically, FISH should be able to provide information on the telomere length of individual chromosomes. The telomeric FISH signals in MGUS and MM patients and HMCLs indicated a dramatic reduction of telomeric signals in some chromosomes, as compared to NC, probably because of a reduction in the number of (TTAGGG)n repeats. The lost of telomeric signal can affect either isolated chromosomes or those implicated in TAs (Fig.2). The analysis of telomere length revealed significantly shorter TRF values in the HMCLs, but in the same range of other tumoural cell lines, MGUS and MM patients (Fig.4), with values strongly correlated with the percentage of telomeric association and chromosome instability (Fig.5). This stability in telomere length maintenance may be a critical component that enables evasion of apoptosis, being necessary for telomere protection without reducing their size 56.
Telomerase and telomere maintenance is required for unlimited cell proliferation and tumorigenesis, and in tumoural cells an increase in telomerase levels/activity has been proposed to be the main mechanisms that enable evasion of apoptosis. We decided to establish the expression levels of genes involved in telomere length maintenance process in MM through microarray analysis, which were subsequently validated by qPCR (Fig.7). In this study, we have found other genes importantly involved in telomere maintenance in MM. We have identified 16 deregulated genes implicated in mechanisms for the post-transcriptional and/or post-translational modification of telomerase, which modulates telomerase activity and telomere maintenance: DKC1, FBL, NHP2, NOP10, NOP56, IMP3, GAR1, HNRNPC, HNRNPA1, HNRNPK, YWHAQ, YWHAE, YWHAZ, HSPA9, KRAS and RB (Fig.8). Our analysis showing alteration in the expression of these 16 human telomere maintenance genes in MM indicates that other genes in addition to hTERT, TRF1, TRF2 and TNKS are involved in these mechanisms. The analysis also showed a significant difference in the expression levels from MGUS to PCL (Fig.6), with overexpression most notably predominating and correlating with disease progression. These proteins seem to be mainly responsible for tumour cells survival and proliferation, both important factors in the progression of MM.
Fig 8.
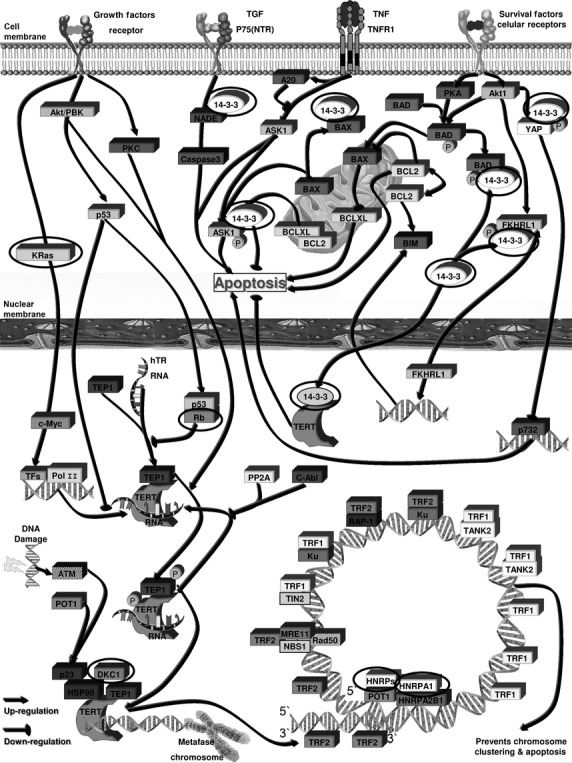
Molecular pathways involved in telomere maintenance, where we can see the 16 deregulated genes implicated directly in telomere length maintenance activity in clonal plasma cells identified in this study highlighted with an oval shape. See Table2 for the list of genes.
FBL, NHP2, DKC1, NOP10, NOP56, IMP3 and GAR1 are members of the H/ACA snoRNPs gene family. Mutations in the components of the H/ACA snoRNP result in a reduction in physiological hTR levels, which are strongly correlated with the poor telomere maintenance and the pseudouridylation of rRNAs and snRNAs 57. DKC1 gene (Dyskeratosis congenita 1, dyskerin) is required for ribosome biogenesis and telomere maintenance through the correct processing and intranuclear trafficking of hTR 58–60. It has been reported in the literature that this gene is overexpressed in prostate cancers and is also necessary for extensive tumour growth 61. In MM, their expression in patients and HMCLs is higher than in NC, hESCs and iPSCs, which have unlimited proliferation capacity, which could ensure the correct processing and intranuclear trafficking of hTR, and therefore telomere maintenance, explaining why tumour plasma cells have a higher survival rate than normal plasma cells.
HNRNPA1, HNRNPC and HNRNPK belong to the A/B subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). This family of proteins are RNA- and DNA-binding proteins localized primarily in the nucleus where they bind telomerase RNA and telomeric DNA, and may thus be capable of recruiting telomerase to form part of the protective complex on telomere ends, which participates in telomere maintenance 62. The hnRNPs family appears to be important for telomere biology because hnRNP A1 and hnRNP C1/C2 proteins are capable of interacting with the telomere and the telomerase, and may regulate telomere length 63–66. Moreover, hnRNPs A1 can associate with the single-stranded telomeric repeat sequence in vivo 67–73, whereas short telomeres in hnRNP A1 deficient mouse CB3 cells are elongated after reconstituting hnRNP A1 expression 66. In MM we have found that the hnRNPs gene family has a similar expression in patients and HMCLs with the same ratio than in hESCs and iPSCs, forming the protective complex of telomerase RNA and telomeric DNA at the telomere ends, regulating telomere length and increasing cell survival.
YWHAQ, YWHAE and YWHAZ genes (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta, epsilon and zeta respectively) belong to the 14-3-3 family that is involved in metabolism, protein trafficking, signal transduction, evasion of apoptosis, cell cycle regulation, cell death and mitogenesis 74,75. Members of the 14-3-3 protein family antagonize the activity of proteins that promote cell death and senescence, such as Bad, Bim, Bax and SFN 76. This family of proteins are hTERT-binding partners and its interaction is required for efficient accumulation of hTERT in the nucleus. In MM tumour cells, the expression of these genes in patients is higher than in HMCLs, hESCs and iPSCs, and especially that in NC. These observations suggest that the 14-3-3 family is a post-translational modifier of telomerase that functions by controlling the intracellular localization and the antiapoptotic action of hTERT, suppressing the DNA damage-related signal, and enabling evasion of apoptosis.
Regarding the other differentially expressed genes, HSPA9 (heat shock 70 kD protein 9, mortalin) is a gene that plays a role in the control of cell proliferation and may act as a chaperone. Mortalin is functioning in intracellular trafficking 77,78, stress response 79–81, inactivation of p53 82 and growth arrest in human immortalized cells that have compromised p53 and pRB functions 83. Also, along with hTERT, in MM permitted bypass of senescence, a substantial extension of lifespan, and possibly immortalization.
KRAS is an oncogene involved in various malignancies, including MM 84. It is implicated in telomere maintenance by the stimulation of telomere-telomere fusions (T-TFs) 85, demonstrating a high degree of genomic instability 86. Its high expression in KMS12 and patients could be the cause of the high increase in frequency of TAs. Retinoblastoma 1, RB1 has an established role in the implementation of cellular senescence 87. It has been demonstrated that the disruption of members of the Rb1 pathway results in rapid telomere elongation, which, in turn, may help to sustain immortal cells. The abnormally elongated telomeres retain their end-capping function, as shown by the normal frequency of chromosomal fusions. This deregulation of telomere length is not associated with increased telomerase activity 88. These observations suggest that plasma tumour cells have other mechanisms of telomere maintenance more developed than hESCs and iPSCs, and not just the action of hTERT.
In conclusion, telomere length stabilization is required for cellular immortality in most cells, and in MM tumour plasma cells have a higher survival than normal plasma cells, which may be a possible mechanism for their accumulation in the BM. Our data suggest that in MM disease, tumour cells acquire telomere maintenance capability through the alteration of three gene families with well-defined functions in telomere maintenance, together with other genes directly involved in telomere length such as hTERT and the RNA component (hTR). These genes are involved in the process of regulation, inhibition, protection, telomere length maintenance, telomerase activity, cell signalling and regulation of apoptosis. These genes with altered expression levels could be used as targets for cancer therapy in MM. However, future studies to determine the protein expression patterns together with the use of siRNA that identify the function and the importance of these genes will be performed.
Acknowledgments
This work was supported by a grant from ISCIII-CSJA, Spain: EMER07/054 to P.E.L. We are indebted to all members of the BBSSPA for their support and technical assistance. R.D. designed and performed experiments, analysed, and interpreted the data and wrote the article. J.P., P.C., C.E., A.P., M.B.L., V.A., G.L., and I.S., performed research. P.E.L. analysed the data and interpreted the results, supervised the study and wrote the article. We thank Dr Santiago del Castillo, Dr Eduardo Ríos-Herranz, Dr Jesús Martín-Sánchez, Dra Isabel Blancas López-Barajas, and Dr José Luis García-Puche for providing BM aspirates obtained from newly diagnosed patients.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1 Real-time qPCR gene expression data in sixteen deregulated genes in multiple myeloma (MM), implicated in mechanisms for the post-transcriptional and/or post-translational modification of telomerase (DKC1, FBL, NHP2, NOP10, NOP56, IMP3, GAR1, HNRNPC, HNRNPA1, HNRNPK, YWHAQ, YWHAE, YWHAZ, HSPA9, KRAS, and RB) in addition to hTERT, TRF1, TRF2 and TNKS, in eight human myeloma cell lines (JIM-1, KMS-11, KMS-12, KMS-26, KMS-28, MM1R, RPMI8226 and U266), CD138+ malignant plasma cells and CD138− plasma cells from six patients with MM, 2 hESCs lines (AND1, H9), and 1 iPSCs line (AND4). PBMC from five healthy donors were used as normal control NC.
Table S1 Primers used for qPCR analysis.
References
- Drewinko B, Alexanian R, Boyer H, et al. The growth fraction of human myeloma cells. Blood. 1981;57:333–8. [PubMed] [Google Scholar]
- Asosingh K, De Raeve H, Van Riet I, et al. Multiple myeloma tumour progression in the 5T2MM murine model is a multistage and dynamic process of differentiation, proliferation, invasion, and apoptosis. Blood. 2003;101:3136–41. doi: 10.1182/blood-2002-10-3000. [DOI] [PubMed] [Google Scholar]
- Bataille R, Robillard N, Pellat-Deceunynck C, et al. A cellular model for myeloma cell growth and maturation based on an intraclonal CD45 hierarchy. Immunol Rev. 2003;194:105–11. doi: 10.1034/j.1600-065x.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- Bataille R. A cellular model for myeloma cell growth and differentiation. Hematology Journal. 2002;2:48. [Google Scholar]
- Scudla V, Ordeltova M, Bacovsky J, et al. The relationship between proliferation and apoptosis in patients with monoclonal gammopathy of undetermined significance or multiple myeloma. Haematologica. 2005;90:1713–4. [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumour cells and during immortalization. Cell. 1997;90:785–95. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–9. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–40. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601–7. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–65. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–60. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Dempster M, Dunlop MG, et al. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–8. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- Allsopp RC, Vaziri H, Patterson C, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–8. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–9. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, et al. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–9. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Chiu CP, Dragowska W, Kim NW, et al. Differential expression of telomerase activity in hematopoietic progenitors from adult human bone marrow. Stem Cells. 1996;14:239–48. doi: 10.1002/stem.140239. [DOI] [PubMed] [Google Scholar]
- Harle-Bachor C, Boukamp P. Telomerase activity in the regenerative basal layer of the epidermis inhuman skin and in immortal and carcinoma-derived skin keratinocytes. Proc Natl Acad Sci USA. 1996;93:6476–81. doi: 10.1073/pnas.93.13.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci USA. 1995;92:9082–6. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–91. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Bacchetti S, Counter CM. Telomeres and telomerase in human cancer. Int J Oncol. 1995;7:423–32. [PubMed] [Google Scholar]
- Holt SE, Wright WE, Shay JW. Multiple pathways for the regulation of telomerase activity. Eur J Cancer. 1997;33:761–6. doi: 10.1016/S0959-8049(97)00066-X. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–9. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Brennan SK, Wang Q, Tressler R, et al. Telomerase inhibition targets clonogenic multiple myeloma cells through telomere length-dependent and independent mechanisms. PLoS ONE. 2010;5:e12487. doi: 10.1371/journal.pone.0012487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA, Koley H, Bertheau RC, et al. Telomerase inhibitor GRN163L inhibits myeloma cell growth in vitro and in vivo. Leukemia. 2008;22:1410–8. doi: 10.1038/leu.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RA, Allsopp RC, Chin L, et al. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–30. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- Liu K, Schoonmaker MM, Levine BL, et al. Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc Natl Acad Sci USA. 1999;96:5147–52. doi: 10.1073/pnas.96.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya H, Sawada H, Muramatsu Y, et al. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–61. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–52. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Counter CM, Hahn WC, Wei W, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–8. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson U, Roos G. Telomere length as a biological marker in malignancy. Biochim Biophys Acta. 2009;1792:317–23. doi: 10.1016/j.bbadis.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Cottliar A, Pedrazzini E, Corrado C, et al. Telomere shortening in patients with plasma cell disorders. Eur J Haematol. 2003;71:334–40. doi: 10.1034/j.1600-0609.2003.00157.x. [DOI] [PubMed] [Google Scholar]
- Wu KD, Orme LM, Shaughnessy J, Jr, et al. Telomerase and telomere length in multiple myeloma: correlations with disease heterogeneity, cytogenetic status, and overall survival. Blood. 2003;101:4982–9. doi: 10.1182/blood-2002-11-3451. [DOI] [PubMed] [Google Scholar]
- Engelhardt M, Ozkaynak MF, Drullinsky P, et al. Telomerase activity and telomere length in pediatric patients with malignancies undergoing chemotherapy. Leukemia. 1998;12:13–24. doi: 10.1038/sj.leu.2400889. [DOI] [PubMed] [Google Scholar]
- Ohyashiki K, Ohyashiki JH, Iwama H, et al. Telomerase activity and cytogenetic changes in chronic myeloid leukemia with disease progression. Leukemia. 1997;11:190–4. doi: 10.1038/sj.leu.2400560. [DOI] [PubMed] [Google Scholar]
- Riboni R, Casati A, Nardo T, et al. Telomeric fusions in cultured human fibroblasts as a source of genomic instability. Cancer Genet Cytogenet. 1997;95:130–6. doi: 10.1016/s0165-4608(96)00248-8. [DOI] [PubMed] [Google Scholar]
- Cottliar AS, Panero J, Pedrazzini E, et al. Analysis of telomere length in mantle cell lymphoma. Eur J Haematol. 2009;83:433–8. doi: 10.1111/j.1600-0609.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- Cortes JL, Sanchez L, Ligero G, et al. Mesenchymal stem cells facilitate the derivation of human embryonic stem cells from cryopreserved poor-quality embryos. Hum Reprod. 2009;24:1844–51. doi: 10.1093/humrep/dep107. [DOI] [PubMed] [Google Scholar]
- Davies FE, Dring AM, Li C, et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood. 2003;102:4504–11. doi: 10.1182/blood-2003-01-0016. [DOI] [PubMed] [Google Scholar]
- Paz-y-Mino C, Sanchez ME, Del Pozo M, et al. Telomeric association in women with breast and uterine cervix cancer. Cancer Genet Cytogenet. 1997;98:115–8. doi: 10.1016/s0165-4608(96)00420-7. [DOI] [PubMed] [Google Scholar]
- Panero J, Arbelbide J, Fantl DB, et al. Altered mRNA expression of telomere-associated genes in monoclonal gammopathy of undetermined significance and multiple myeloma. Mol Med. 2010;16:471–8. doi: 10.2119/molmed.2010.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Leone PE, Jenner MW, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–43. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- Jenner MW, Leone PE, Walker BA, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- Leone PE, Walker BA, Jenner MW, et al. Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res. 2008;14:6033–41. doi: 10.1158/1078-0432.CCR-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens NJ, Walker BA, Leone PE, et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin Cancer Res. 2010;16:1856–64. doi: 10.1158/1078-0432.CCR-09-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L, Poretti G, Mattioli M, et al. Molecular characterization of human multiple myeloma cell lines by integrative genomics: insights into the biology of the disease. Genes Chromosom Cancer. 2007;46:226–38. doi: 10.1002/gcc.20404. [DOI] [PubMed] [Google Scholar]
- Agnelli L, Mosca L, Fabris S, et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosom Cancer. 2009;48:603–14. doi: 10.1002/gcc.20668. [DOI] [PubMed] [Google Scholar]
- Chng WJ, Kumar S, Vanwier S, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–9. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- Tiedemann RE, Zhu YX, Schmidt J, et al. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 2010;115:1594–604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ju Z, Lenhard Rudolph K. Telomere dysfunction and stem cell ageing. Biochimie. 2008;90:24–32. doi: 10.1016/j.biochi.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Gilson E, Londono-Vallejo A. Telomere length profiles in humans: all ends are not equal. Cell Cycle. 2007;6:2486–94. doi: 10.4161/cc.6.20.4798. [DOI] [PubMed] [Google Scholar]
- Dewar JM, Lydall D. Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. EMBO J. 2010;29:4020–34. doi: 10.1038/emboj.2010.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahan C, Martel C, Dragon F. Effects of dyskeratosis congenita mutations in dyskerin, NHP2 and NOP10 on assembly of H/ACA pre-RNPs. Hum Mol Genet. 2009;19:825–36. doi: 10.1093/hmg/ddp551. [DOI] [PubMed] [Google Scholar]
- Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–79. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Chilla A, Trere D, et al. Increased mortality rate and not impaired ribosomal biogenesis is responsible for proliferative defect in dyskeratosis congenita cell lines. J Invest Dermatol. 2002;118:193–8. doi: 10.1046/j.0022-202x.2001.01634.x. [DOI] [PubMed] [Google Scholar]
- Vulliamy TJ, Knight SW, Mason PJ, et al. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol Dis. 2001;27:353–7. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- Sieron P, Hader C, Hatina J, et al. DKC1 overexpression associated with prostate cancer progression. Br J Cancer. 2009;101:1410–6. doi: 10.1038/sj.bjc.6605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Jones K, Wayman L, Kennedy DD, et al. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res. 2005;33:486–96. doi: 10.1093/nar/gki203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Wright WE, Shay JW. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21:580–3. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]
- Eversole A, Maizels N. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol Cell Biol. 2000;20:5425–32. doi: 10.1128/mcb.20.15.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Suh JM, Wright WE, et al. Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol Cell Biol. 2000;20:9084–91. doi: 10.1128/mcb.20.23.9084-9091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche H, Dupuis S, Ben-David Y, et al. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- Dallaire F, Dupuis S, Fiset S, et al. Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J Biol Chem. 2000;275:14509–16. doi: 10.1074/jbc.275.19.14509. [DOI] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–15. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlitzki R, Fry M. Sequence-specific binding protein of single-stranded and unimolecular quadruplex telomeric DNA from rat hepatocytes. J Biol Chem. 1997;272:15881–90. doi: 10.1074/jbc.272.25.15881. [DOI] [PubMed] [Google Scholar]
- Ishikawa F, Matunis MJ, Dreyfuss G, et al. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–10. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SD, Lew JE, Berman J. Gbp1p, a protein with RNA recognition motifs, binds single-stranded telomeric DNA and changes its binding specificity upon dimerization. Mol Cell Biol. 1999;19:923–33. doi: 10.1128/mcb.19.1.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Cooke H. hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res. 1992;20:6461–4. doi: 10.1093/nar/20.24.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarig G, Weisman-Shomer P, Erlitzki R, et al. Purification and characterization of qTBP42, a new single-stranded and quadruplex telomeric DNA-binding protein from rat hepatocytes. J Biol Chem. 1997;272:4474–82. doi: 10.1074/jbc.272.7.4474. [DOI] [PubMed] [Google Scholar]
- Aitken A. 14-3-3 proteins on the MAP. Trends Biochem Sci. 1995;20:95–7. doi: 10.1016/s0968-0004(00)88971-9. [DOI] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, et al. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–28. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- Nomura M, Shimizu S, Sugiyama T, et al. 14-3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J Biol Chem. 2003;278:2058–65. doi: 10.1074/jbc.M207880200. [DOI] [PubMed] [Google Scholar]
- Mizukoshi E, Suzuki M, Loupatov A, et al. Fibroblast growth factor-1 interacts with the glucose-regulated protein GRP75/mortalin. Biochem J. 1999;2:461–6. [PMC free article] [PubMed] [Google Scholar]
- Mizukoshi E, Suzuki M, Misono T, et al. Cell-cycle dependent tyrosine phosphorylation on mortalin regulates its interaction with fibroblast growth factor-1. Biochem Biophys Res Commun. 2001;280:1203–9. doi: 10.1006/bbrc.2001.4225. [DOI] [PubMed] [Google Scholar]
- Merrick BA, Walker VR, He C, et al. Induction of novel Grp75 isoforms by 2-deoxyglucose in human and murine fibroblasts. Cancer Lett. 1997;119:185–90. doi: 10.1016/s0304-3835(97)00270-x. [DOI] [PubMed] [Google Scholar]
- Sadekova S, Lehnert S, Chow TY. Induction of PBP74/mortalin/Grp75, a member of the hsp70 family, by low doses of ionizing radiation: a possible role in induced radioresistance. Int J Radiat Biol. 1997;72:653–60. doi: 10.1080/095530097142807. [DOI] [PubMed] [Google Scholar]
- Carette J, Lehnert S, Chow TY. Implication of PBP74/mortalin/GRP75 in the radio-adaptive response. Int J Radiat Biol. 2002;78:183–90. doi: 10.1080/09553000110097208. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Robert M, et al. Inactivation of tumour suppressor p53 by mot-2, a hsp70 family member. J Biol Chem. 1998;273:29586–91. doi: 10.1074/jbc.273.45.29586. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Taira K, et al. Reduction in mortalin level by its antisense expression causes senescence-like growth arrest in human immortalized cells. J Gene Med. 2004;6:439–44. doi: 10.1002/jgm.530. [DOI] [PubMed] [Google Scholar]
- Liu P, Leong T, Quam L, et al. Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: analysis of the Eastern Cooperative Oncology Group Phase III Trial. Blood. 1996;88:2699–706. [PubMed] [Google Scholar]
- Siveke JT, Schmid RM. Chromosomal instability in mouse metastatic pancreatic cancer–it's Kras and Tp53 after all. Cancer Cell. 2005;7:405–7. doi: 10.1016/j.ccr.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Khleif SN, DeGregori J, Yee CL, et al. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci USA. 1996;93:4350–4. doi: 10.1073/pnas.93.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, Gonzalo S, Dean D, et al. A role for the Rb family of proteins in controlling telomere length. Nat Genet. 2002;32:415–9. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Real-time qPCR gene expression data in sixteen deregulated genes in multiple myeloma (MM), implicated in mechanisms for the post-transcriptional and/or post-translational modification of telomerase (DKC1, FBL, NHP2, NOP10, NOP56, IMP3, GAR1, HNRNPC, HNRNPA1, HNRNPK, YWHAQ, YWHAE, YWHAZ, HSPA9, KRAS, and RB) in addition to hTERT, TRF1, TRF2 and TNKS, in eight human myeloma cell lines (JIM-1, KMS-11, KMS-12, KMS-26, KMS-28, MM1R, RPMI8226 and U266), CD138+ malignant plasma cells and CD138− plasma cells from six patients with MM, 2 hESCs lines (AND1, H9), and 1 iPSCs line (AND4). PBMC from five healthy donors were used as normal control NC.
Table S1 Primers used for qPCR analysis.


