Abstract
We have developed a microfluidic cell culture method that allows for the formation of linear isolated myotubes organized in a parallel microarray. Attachment and spreading of cells are confined within microtracks of cell-adherent proteins separated by a protein-repellent coating. Signaling molecules or other molecules of interest can be focally delivered to the myotubes using heterogeneous microfluidic streams. We have used the method to focally deliver agrin (a molecule implicated as a postsynaptic organizer), which leads to localized acetylcholine receptor clustering. These techniques can be modified to accommodate other cell types and can be adapted to virtually any bioactive molecule such as signaling factors or drugs. This protocol features two major techniques that can be utilized simultaneously or independently to (i) micropattern cells using surface chemical modification and (ii) use a microfluidic platform for culturing and focal stimulation of cells with molecules of interest. Device design, fabrication and assembly can be completed in 3 days.
INTRODUCTION
In the development of muscle tissues, embryonic muscle cells adhere to extracellular matrices, align with each other to form linear chains, and fuse to form multinucleated muscle fibers. The formation and orientation of skeletal muscle fibers in vivo is highly regulated, resulting in uniform muscle fibers aligned in parallel. In vitro, muscle cells can be induced to fuse and form myotube-like structures, but in traditional monolayer cultures, the myotubes are randomly oriented and branched and can be interconnected into syncytia-like structures1 (Fig. 1a).
Figure 1.
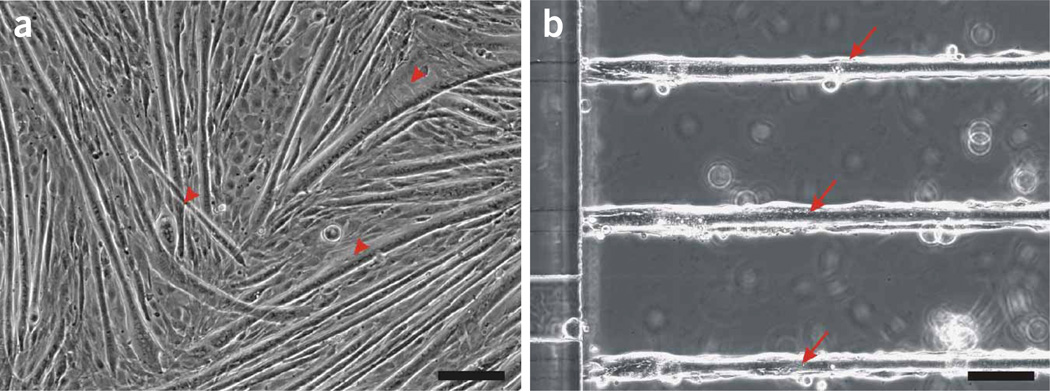
C2C12 cells in random and micropatterned microfluidic cultures. Phase-contrast images showing differences in myotube growth on polystyrene plates (a) and patterned microtracks of Matrigel on glass (b). (a) Myoblasts proliferate and differentiate into myotubes with varied sizes, orientations and structures (arrowheads). (b) Myoblasts seeded on microtracks proliferate along the cell-adhering sections and differentiate into myotubes aligned along the microtracks (arrows). Scale bar = 100 µm.
We have developed a soft-lithographic method to produce microarrays of parallel skeletal myotubes2, which better mimic muscle architecture and increase the relevance of in vitro studies involving muscle cells3 (Fig. 1b). In addition, the precise alignment of large numbers of cells with the stimulus and with the imaging field makes the results readily amenable to quantitative, statistically rich analysis. This method is compatible with conventional cell seeding protocols (e.g., medium containing high serum concentrations) and is applicable to a variety of adherent cell types2. Adhesive micropatterns of various shapes and sizes can be created using this technique. Previously, we have reported isolation of single cells, small clusters of cells and micro-co-cultures on adhesive microislands for fibroblasts, endothelial cells, smooth muscle cells and myoblasts2. In addition, in our study of axon guidance, we have micropatterned murine embryonic cortical neurons using this technique4. Here we give step-by-step instructions for fabrication of adhesive microtracks for directed myotube formation; however, this protocol can be easily modified to create desired geometries for other cell types. Micropatterned cultures have a general applicability in cell biology and biotechnology applications in which addressing and/or tailoring the microenvironment of large numbers of single cells or small clusters of cells is critical.
This protocol also includes a technique that allows focal delivery of signaling molecules or other molecules of interest to cells using heterogeneous microfluidic streams5. Others have used this approach to selectively label different subpopulations of mitochondria within a single cell, selectively disrupt cytoskeleton6, locally stimulate signaling pathways7, sort out nonmotile spermatozoids from sperm samples8 and alter embryonic patterning of Drosophila embryos9,10. We have used the method to focally deliver agrin (a molecule implicated as a postsynaptic organizer11) to subcellular domains of skeletal myotubes. This represents an attempt to mimic the presence of the neuron in the first stages of the formation of a neuromuscular synapse, leading to localized acetylcholine receptor (AChR) clustering. Even though flow is absent in the synaptic contact, our approach allows for mimicking local presentation of synaptogenic molecules and at the same time provides control over other parameters of stimulation (location, duration, and starting and ending time points) that previously could not be controlled simultaneously.
In this protocol we describe step-by-step instructions on how to produce arrays of isolated linear myotubes and focally stimulate them by means of a microfluidic device. The protocol consists of several sub-protocols, including (i) fabrication of microfluidic devices3,12 (Steps 1–30), (ii) glass substrate modification for directed cell growth (protocol in Box 1 and Steps 31–42, (iii) microfluidic micropatterned myotube cultures (Steps 43–61), and (iv) focal stimulation of myotubes with agrin (Step 62–70).
BOX 1 | GRAFTING THE PROTEIN-REPELLENT LAYER ONTO GLASS.
-
Clean glass substrates. We soak substrates for 2 h in Contrad 70 (5% solution) that contains potassium hydroxide. Rinse glass substrates with deionized water and dry. Treat with oxygen plasma (10 min, 0.75 Torr oxygen, 200 W); dip in Nano-Strip for 10 min, rinse in deionized water and dry under a stream of compressed nitrogen or air.
! CAUTION The Nano-Strip contains sulfuric acid and hydrogen peroxide; use laboratory goggles, plastic apron, and thick, chemical-grade gloves (elbow-high).
■ PAUSE POINT Keep cleaned substrates in a closed container.
! CAUTION Carry out Steps 2–8 (including weighing of chemicals and sonication) inside the fume hood.
- Quickly mix the ATC-silane with toluene (1.25% (vol/vol), denoted ’Solution A’) under ambient conditions in the fume hood.
Solution Volume ATC-silane 10 ml Toluene 790 ml -
Soak glass substrates in Solution A for 5 min. We use immersion jars and wafer holders to process the substrates in batches.
▲ CRITICAL STEP The ATC-silane and toluene must be mixed quickly. The silanes readily react with each other in the presence of water (including the water vapor in ambient air). You can continue to use the ATC-silane/toluene mix as long as it is clear.
-
Rinse substrates by soaking in pure toluene for 1 min. Use a wash bottle to rinse the glass substrates with toluene and then immediately with acetone. Finally, dip substrates in deionized water and dry under a stream of filtered air. Cure in an oven at 90 °C for 1 h. If the ATC-silane deposition was successful, glass substrates will completely de-wet after dipping in and pulling from a beaker of water.
■ PAUSE POINT Substrates can be stored for several months at room temperature in a closed container.
- Prepare Solutions A and B. Use conical flasks. Sonicate at room temperature until dissolved (~ 20–30 min). Cover the opening with Parafilm to limit evaporation. 20 ml of grafting solution is enough to completely submerge four or five 5-cm glass wafers in a 150-mm-diameter glass Petri dish.
SOLUTION A SOLUTION B Material Amount Material Amount AAm 3.00 g PEG 0.6 g BIS 0.03 g BIS 0.9 g Benzophenone 1.09 g Benzophenone 1.09 g Acetone 20 ml Methanol 20 ml Transfer glass substrates into a 150 mm-diameter glass Petri dish. Add Solution A to the glass dish and ensure that no bubbles form on the underside of the glass substrates.
After 5 min of adsorption time, expose the substrates to UV light using the transilluminator at a high-intensity setting until a cloudy white gel of PAAm fills the thin gaps between the glass dish bottom and the glass substrates (~ 5 min). Remove the samples from the glass dish and rinse off the gel-like materials with acetone (in a wash bottle), dip the samples and rinse them with deionized water and dry them under a stream of filtered air or compressed nitrogen. The side that is in contact with the glass surface of the Petri dish is where the reaction will occur. The grafted (bottom) side will be very hydrophilic, in contrast to the hydrophobic ungrafted side.
-
Clearly mark the ungrafted (hydrophobic) side by scratching an asymmetrical reference mark (we write “4”) with a diamond pen. Transfer glass substrates into clean 150-mm-diameter glass dishes.
▲ CRITICAL STEP Make sure that the scratched side is facing up.
Add Solution B to the glass dish and ensure that no bubbles are trapped on the underside of the glass substrates. After 5 min of adsorption, expose the samples to UV light at a high setting for 5 min. The new layer of gel will not be as visible as in Step 5.
Rinse the substrates with methanol and acetone. Dip the substrates in water and dry. The grafted side will be hydrophilic in contrast to the hydrophobic ungrafted side. At this point, the substrates are now grafted with interpenetrating networks of PAAm and PEG (P(AAm-co-EG) IPN or IPN).
● TIMING Overall, this procedure takes 9–10 h.
■ PAUSE POINT These substrates can be stored for several months at room temperature in closed containers.
Briefly, we culture myogenic cells (C2C12 cell line) on microtracks of Matrigel surrounded by protein-repellent coating. The C2C12 cells adhere and spread only on the adhesive microtracks and respond to the surface chemistry constraints by fusing along the track direction into long (>1 mm) myotubes with widths approximating the width of the adhesive lines (Fig. 1b). Our experiments use multiple laminar streams for stimulating subdomains of single cells. In particular, we produce a narrow stream of agrin over micropatterned myotubes, which induces clustering of AChRs on the areas stimulated by agrin. The microfluidic cell culture device consists of a main microchannel formed by three converging inlet channels for focal delivery of signaling molecules to the myotubes that are micropatterned on the bottom of the main channel (Fig. 2). This microfluidic system provides accurate control of the perfusion rates and of the chemical microenvironment surrounding the cells and allows for characterization of synaptogenic molecules presented to the cells in a physiologically relevant manner.
Figure 2.
Schematic illustration of cellular micropatterning in the microfluidic device. (a) The bonded device is filled with medium, and cells are introduced through the outlet and allowed to adhere to the protein pattern (green). (b) Cells are perfused via the perfusion network. (c) Cells are exposed to agrin via laminar flow. The side channels deliver DM, while the central channel delivers agrin and Allura food coloring dye in DM. Only small portions of the myotubes are exposed to agrin.
General procedure notes
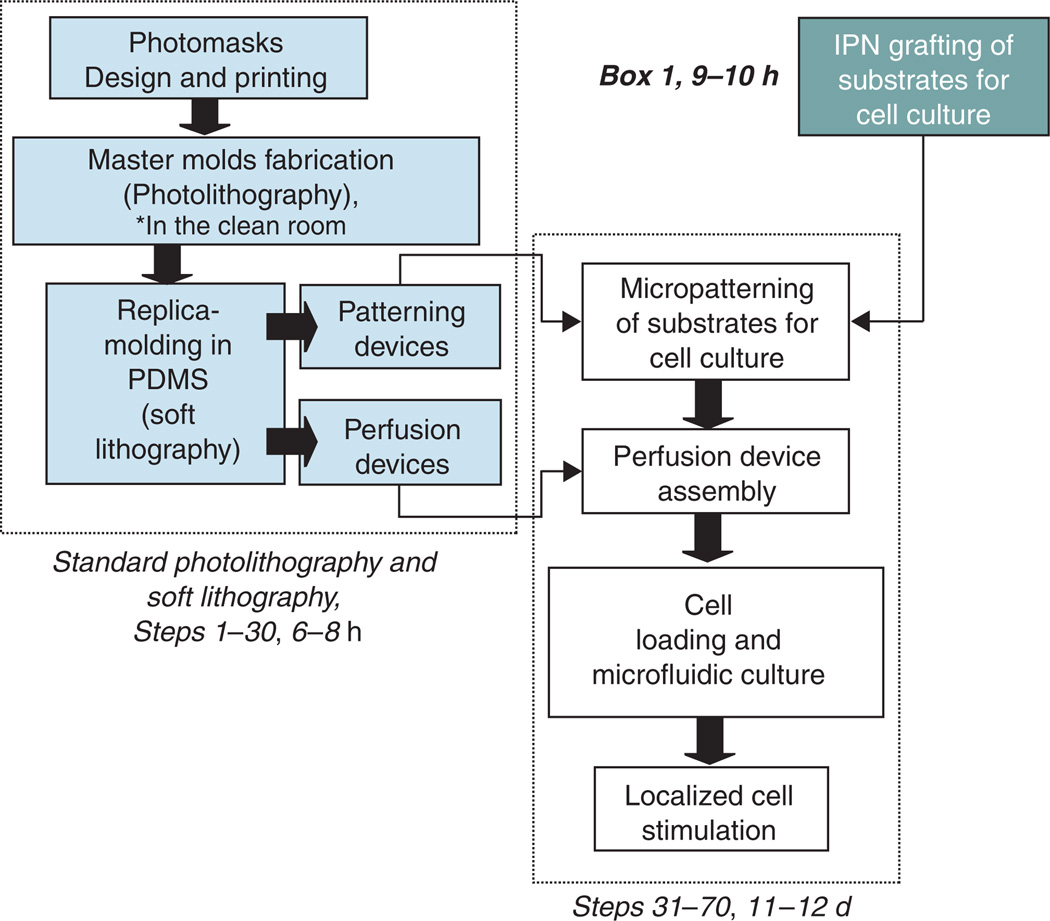
See the diagram of the protocol flow (Fig. 3) for the general outline of the procedure and the timeline.
Figure 3.
Diagram of the protocol flow.
Fabrication of the devices utilizes standard photolithography and soft lithography techniques13,14 and involves the microfabrication of two devices: a device with which the surface chemistry is patterned (hereafter referred to as “patterning device”), and a microfluidic device within which the long-term perfusion and focal exposure is performed (hereafter referred to as “perfusion device”).
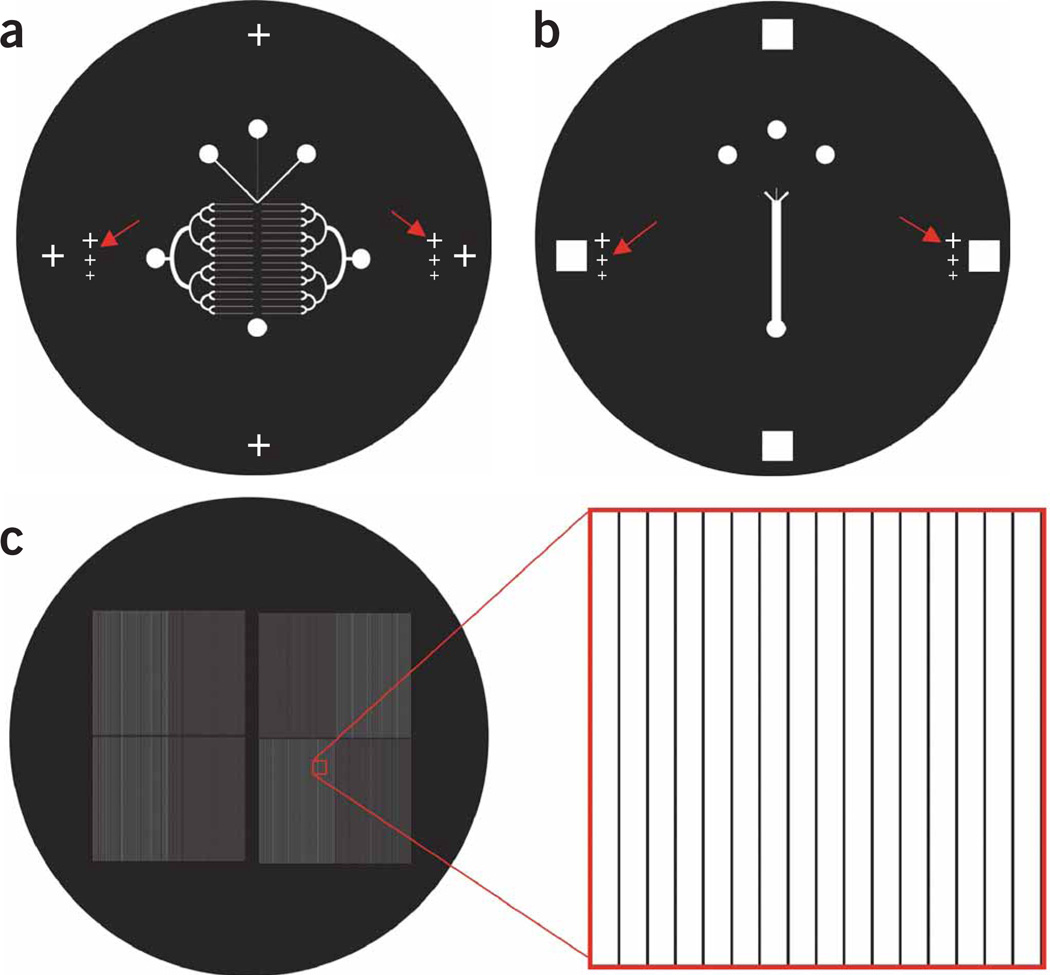
We first design two-dimensional (2D) transparency masks for our devices (Fig. 4). Once the photomasks (Fig. 4) are printed, the next steps require access to a clean room with photolithography equipment.
Figure 4.
Photomasks of the devices. (a) Layer 1 of the microfluidic device (height = 25 µm). (b) Layer 2 (250 µm) is aligned on top of the Layer 1 (red arrows point to the alignment marks). (c) The microchannels (65 µm height, 25 µm width and 300 µm separation) are used to selectively etch IPNs and deposit protein on the exposed glass.
The method for grafting interpenetrating polymer networks (IPNs) of polyacrylamide (PAAm) and polyethylene glycol (PEG) was developed in the laboratory of K. Healy15 and adapted by us with some modifications (Box 1). Parts of this procedure (oxygen plasma ashing and Nano-Strip cleaning) are performed inside a clean room. Glass substrates with homogenous protein-repellent surface chemistry (unpatterned) are prepared in advance and can be stored for several months.
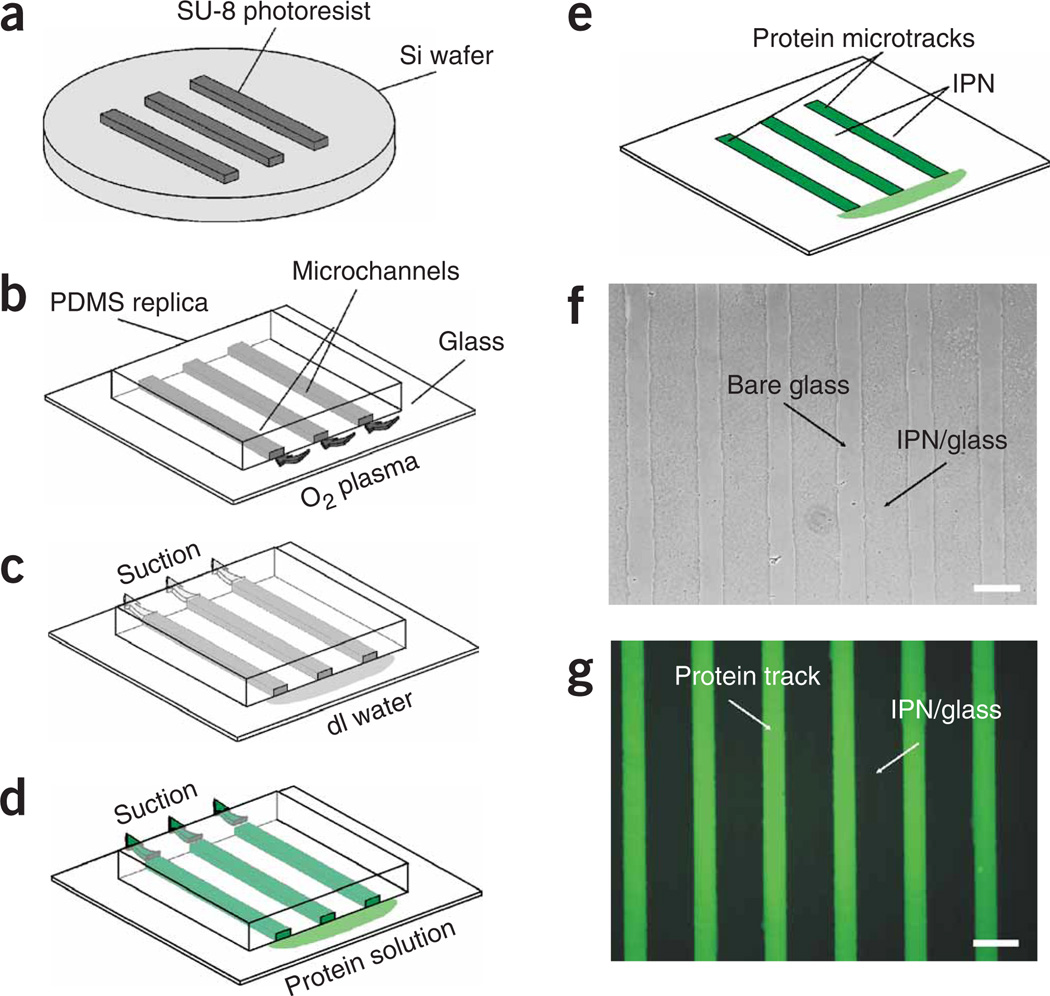
Once the patterning devices (photomask is shown in Fig. 4c; mastermold is shown in Fig. 5a and polydimethylsiloxane (PDMS) replica is shown in Fig. 5b) are created, they are used to selectively remove the IPNs from inside the microchannels and form microtracks of Matrigel (Fig. 5b–g).
Figure 5.
Procedure for protein micropatterning based on the use of PDMS microchannels.(a) SU-8 master mold. (b) IPN coating is selectively removed using microchannel mask. (c) The microchannels are flushed with deionized water. (d) The microchannels are filled with ECM protein solution. (e) The microchannels are removed, leaving microtracks of protein surrounded by IPN. (f) Phase-contrast image of lines of IPN-free glass surrounded by IPN. (g) Fluorescence microscopy image of microtracks of the adsorbed fluorescently tagged fibronectin surrounded by IPNs. Scale bars = 100 µm.
The perfusion devices must be covalently bonded to patterned substrates to protect fluids from leaking because the physical (reversible) seal can be easily broken when small pressures (or forces to the tubing) are applied to introduce liquids into the device. As explained below, exposing the PDMS device and the glass substrate to oxygen plasma and bringing the device into contact with the glass substrate bonds them irreversibly. The diagram of the device assembly and cell loading is provided in Figure 6.
Figure 6.
Schematic illustration of the device assembly and cellular micropatterning. (a) The microfluidic device is bonded to protein/IPN patterned substrate using O2 plasma activation. (b) The bonded device is filled with medium, and cells are introduced through the outlet and allowed to adhere to the protein pattern. (c) Constant perfusion of cell culture with fresh medium is established after cells adhere to the substrate.
The choice of microtrack width for single-myotube growth was based on our studies of myotube growth on microtracks of various widths, ranging from 25–250 µm. We have consistently obtained single-myotube arrays on 25- to 30-µm microtracks, and others have also reported single myotube growth on 20 µm-wide patterns16. To prevent overgrowth (“bridging”) between the myotubes growing on adjacent microtracks (Fig. 2c, right panel), the tracks should be separated by 300 µm.
We have varied all dimensions of the device (i.e., main channel width, length and height) and have found that the rate of myoblast proliferation and fusion does not depend on the main channel dimensions but is influenced by underlying substrate, seeding density and perfusion rate. In this protocol, we have provided optimal cell density and perfusion rates. The longer the main channel, the more cell tracks it can accommodate.
The absolute dimensions of converging inlet channels can also be varied; however, to provide tighter focusing of the stimulating stream, the central converging channel should be narrower than the side converging channels17. The device dimensions are also discussed elsewhere3,12.
MATERIALS
REAGENTS
SU-8 2000 resists (MicroChem). A number of different photoresists exist, but SU-8 (a negative photoresist; i.e., one that becomes insoluble when exposed to UV light) provides very good definition and vertical sidewalls for structures as high as several hundred microns. The thickness of photoresist defines the master mold height and is given primarily by photoresist viscosity and by the speed at which the wafer is spun. We use the MicroChem website (see below) to obtain the optimized spin speeds and times that yield the required heights. ▲ CRITICAL See manufacturer’s data sheets for process guidelines (http://www.microchem.com/products/su_eight.h)
SU-8 developer (1-methoxy-2-propyl acetate) (MicroChem) ! CAUTION Toxic.
Tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (TFOCS; United Chemical Technology) ! CAUTION Corrosive, toxic.
Sylgard 184 Silicone Elastomer Base and Sylgard 184 Elastomer Curing Agent (Dow Corning)
Duco Cement (Devcon)
Toluene (EMD) ! CAUTION Toxic.
Acetone (EMD) ! CAUTION Toxic.
Methanol (EMD) ! CAUTION Toxic.
Allyltrichlorosilane (ATC-silane), assay: 95% (Sigma) ! CAUTION Corrosive, toxic.
Acrylamide (AAm; Polyscience) ! CAUTION Carcinogen, toxic.
N,N′-methylene bisacrylamide (BIS; Polyscience) ! CAUTION Irritant, toxic.
Polyethylene glycol 1000 monomethyl ether monomethylacrylate (PEG; Polyscience) ! CAUTION Irritant.
Benzophenone (Fluka) ! CAUTION Irritant.
Contrad 70 (Decon Labs)
Nano-Strip solution (Cyantek Corporation) ! CAUTION Highly caustic.
α-bungarotoxin Alexa Fluor 488 conjugate (BTX*) (Molecular Probes)
Bovine serum albumin (BSA), Texas Red conjugate (Molecular Probes)
Dextran, Oregon Green 514; 70,000 MW (Molecular Probes)
Agrin (R&D Systems) (cat. #550-AG)
Growth Factor Reduced Matrigel (BD Biosciences) (cat. #354230)
Dulbecco’s Modified Eagles’ Medium (DMEM; Invitrogen) (cat. #11965-092)
Fetal bovine serum (FBS; HyClone)
Horse serum (HS; ATCC)
Penicillin/streptomycin or antibiotic-antimycotic (Invitrogen) (cat. #15140-122)
Bovine serum albumin (BSA; Sigma-Aldrich)
Food coloring dye
REAGENT SETUP
Matrigel
Prepare 30-µl aliquots. Store at −20 °C.
Growth medium
Consists of DMEM, 20% FBS (vol/vol), 1% (vol/vol) penicillin/streptomycin. Store at +4 °C.
Differentiating medium
Consists of DMEM, 5% HS(vol/vol), 1% (vol/vol) penicillin/streptomycin. Store at +4 °C.
EQUIPMENT
Photoresist spinner
UV mask aligner
Plasma processing reactor center (for example, Branson 2000, Branson Ultrasonics Corporation)
Chemical wet bench
Two hot plates
Oven set at 65 °C (for curing PDMS)
Water bath sonicator (for example, Branson 5510, Branson Ultrasonics Corporation)
Transilluminator (TFL-40, UVP, Inc.) Provides uniform and intense ultraviolet radiation. This lamp has a predominant emission peak at 365 nm.
Phase-contrast and epifluorescence microscope with a CCD camera
Laminar flow hood
CO2 water-jacketed incubator set at 37 °C and 10% CO2.
Syringe pump (for example, Genie Plus, Kent Scientific Corporation)
Glass discs or microscope slides (Erie Scientific).
Immersion jars (Wheaton).
150-mm glass Petri dishes
Silicon wafers (3-inch diameter, P/Boron; Silicon Sense, Inc.).
Silicone tubing (1.14 mm internal diameter (I.D.); Cole-Palmer Instrument Co.).
Polyetheretherketone (PEEK) tubing for agrin fluid line (35.5 cm long, 1/16” outer diameter (O.D.), 75 µm I.D., Upchurch Scientific) ▲ CRITICAL The choice of PEEK tubing is critical to prevent nonspecific adsorption of agrin (or another protein of interest) to the walls of the tubing, which reduces the effective concentration seen by the cells.
Fluorinated ethylene propylene (FEP) tubing (1/16” O.D., 250 µm I.D.; Upchurch Scientific) for medium inlet (14 cm long) and outlet fluid lines (61 cm long)
Female luer fitting (1/16”; cat. no. 06359-27, Cole-Palmer Instrument Co.)
Elbow connector (1/16”; cat. no. 06365-15, Cole-Palmer Instrument Co.)
Female luer with thread (cat. no. 30504, Cole-Palmer Instrument Co.)
Conical adapter (1/16”; cat. no. P-794, Upchurch Scientific)
Alligator clips (35 mm, RadioShack)
Kimwipes, autoclaved
Petri dishes (p35, 35 mm × 10 mm)
Flasks (T75 tissue culture flasks, polystyrene) or Petri dishes (100 mm × 15 mm tissue culture dishes, polystyrene)
Petri dishes (p150, 150 mm × 25 mm)
EQUIPMENT SETUP
Microfabrication and photolithography
The master molds should be made in a clean-room photolithography facility. Clean rooms are utilized mostly by the semiconductor industry to manufacture silicon chips. In clean rooms, air is constantly filtered to exclude dust particles that can damage the production of the chips, and temperature and humidity are controlled in order to ensure uniform materials performance over time. Working in a dust-free environment is critical because dust particles can be larger than the microstructures that we are trying to produce and would interfere with photoresist coating.
Plasma processing reactor center
Also referred to as a “barrel etcher” (the chamber has the shape of a barrel) or a “plasma asher”, a vacuum chamber where oxygen radicals are generated to remove (“ash”) organic matter. The products generated during ashing are pumped away by the vacuum system. We use plasma to clean surfaces and to bond PDMS substrates to glass or to PDMS. Unfortunately, the working settings for any particular application are highly dependent on the geometry of the chamber, the radio frequency coil size, etc., so the parameters given below (power, time, and oxygen pressure) are used only for guidance. These parameters should be optimized empirically for each type of experiment.
Transparency mask manufacture
Also called a photomask, a high-resolution, laser-printed sheet of polyethylene terephthalate (PETE) that contains the desired opaque features designed by the user and used to generate the photoresist masters. Typically, a high-resolution printer (5,060 dpi, nominal dot size of 5 µm) can resolve 30-µm line widths. Smaller features require printers with higher resolution. The edges that require the best resolution should be designed horizontally whenever possible, because for mechanical reasons the printer scan head is more accurate on any given printing line than across different lines. We design our photomasks using drawing software CorelDraw (Corel Corporation) and AutoCAD (Autodesk, Inc.). The photomasks are then electronically sent to the printing service. Examples of such services are PageWorks in Boston (http://www.pageworks.com/) or CAD Art Services in California (http://www.outputcity.com/). The latter offers 20,000 dpi printouts.
PROCEDURE
Fabrication of SU-8 master molds
▲ CRITICAL STEP Perform Steps 1 through 7 in a clean room.
-
1|
Immerse silicon wafers in Nano-Strip for 10 min, wash for 5 min in distilled or deionized water and dry. Place wafers onto a 205 °C hot plate for 10 min in order to “singe” or completely evaporate any water on the surface.
-
2|
Quickly place the wafer on a resist spinner vacuum holder (Figs. 7 and 8). (The heat capacity of silicon is so low that the wafer will cool down within a few seconds of removing it from the hot plate.) Once the wafer is centered on the holder, apply the photoresist (SU-8 50) and spin (Fig. 7a–c). Two spin phases are required. The first low-rpm phase is called the “spread” phase, which allows the resist to cover the wafer and spread from the center. We always spread photoresist at 500 rpm for 10 s. The high-speed “spin” phase removes the excess resist and evenly distributes it into a film of the desired thickness.
-
3|
Transfer the wafer to a hot plate and soft bake at 65 °C: bake Layer 1 (of the perfusion device) for 3 min and bake patterning device for 6 min. Immediately transfer to a 95 °C hot plate to “set” the photoresist: allow Layer 1 to set for 15 min and the patterning device for 20 min.
-
4|
Place the wafer onto a UV aligner and bring the photomask (emulsion side facing the wafer) into contact with the resist (Figs. 7d and 9). Once the photomask and wafer are aligned, expose to a near-UV light (350–400 nm) (Fig. 7e). We exposed Layer 1 to 200 mJ/cm2 and patterning device to 400 mJ/cm2. Refer to MicroChem supplemental information to determine exposure times based on film thickness and UV power output.
▲ CRITICAL STEP Exposure for too short a time will result in features that are attacked by the developer, whereas exposure for too long will broaden the features. If feature size is not critical, overexposure is preferred.
-
5|
Place the wafer onto a 65 °C hot plate for 1 min and then onto a 95 °C hot plate (Layer 1 for 4 min and the patterning device for 5 min). Refer to MicroChem supplemental information for bake times. The exposed areas are now cross-linked and insoluble in the SU-8 developer.
-
6|
Allow the wafer to cool to room temperature (20–25 °C) and immerse the wafer in a crystallizing glass dish filled with the SU-8 developer and covered with a watch glass. Agitate to speed up the development. The development will take about 5 min for the patterning device. For processing of the Layer 1 of the perfusion device, skip to Step 11.
-
7|
Once the unexposed photoresist has been washed away, rinse the wafer with fresh SU-8 developer with a wash bottle. Place the wafer on a piece of absorbent paper and gently blow nitrogen or clean air over the surface to dry off the SU-8 developer.
Ensure that both sides are dried. This is the SU-8 master. Once baked, a “latent” pattern should be visible on the resist. Unexposed areas of the resist should be completely removed by the developer. The silicon surface (unexposed areas) should be uniformly shiny (that is, there should be no streaks or shadows).
■ PAUSE POINT Store your SU-8 master in a dust-free environment.
-
8|
Place the master in the vacuum jar (we use a desiccator jar without the drying pellets) attached to a vacuum source. Place a small portion of absorbent paper towel inside the desiccator chamber. Add a drop of TFOCS to the paper towel and evacuate the air from the chamber.
! CAUTION The desiccator chamber must be located inside a chemical fume hood owing to the corrosive nature of TFOCS vapors.
-
9|
Apply vacuum for 1 min and turn off. This will vaporize the TFOCS and deposit a monolayer onto the masters. Close the vacuum and allow 40 min for deposition. Once the master has been coated with TFOCS, place it in a weighing boat where it will be covered with PDMS precursor. The weighing boat is preferred to a rigid dish because it can be cut to facilitate release of the PDMS from the master.
▲CRITICAL STEP Silicon surfaces exposed to air are covered by a thin layer of SiO2 (glass). Very clean SiO2 can bind to PDMS during PDMS curing. To prevent bonding, we passivate the oxide layer with a self-assembled monolayer of fluorosilane, TFOCS. TFOCS is a liquid, but the reaction is most efficiently carried on in the gas phase; i.e., by exposing the master to the TFOCS vapor. To create a gas volume saturated with TFOCS vapors, we put TFOCS and the wafer in a vacuum jar.
■ PAUSE POINT Master mold preparation is completed. Keep the masters in closed containers to protect them from dust.
Figure 7.

Process flow in photolithographic fabrication of the master mold. (a) Silicon wafer is placed on wafer holder. (b) Photoresist is dispensed. (c) Photoresist is spun on the wafer. (d) After the photoresist is soft baked, a transparency mask is placed in contact with the photoresist. (e) The photoresist is exposed to UV light. UV-exposed regions undergo cross-linking and become insoluble in developer.
Figure 8.
Wafer spin-coater. The spin-coater sits inside a fume hood. A wafer is placed in the middle of the wafer holder (insert) held in place by vacuum, and a Teflon shield captures the excess resist that is spun off the wafer. The control board (far right) sets the spin and time parameters.
Figure 9.
UV aligner. The mask needs to be in direct contact (emulsion down) with the surface of the photoresist. The wafer is held in place by a vacuum holder while the mask is taped over a glass plate. The wafer is raised to be in contact with the mask, and a shutter is opened to expose the photoresist to UV light. Once the second layer of photoresist is applied, the aligner is used to match the second-layer mask to the exposed features. The aligner has micron scale adjustment capabilities that enable precise alignment.
Fabrication of two-layer molds
▲ CRITICAL STEP Perform Steps 10 through 15 in a clean room.
-
10|
Fabricate Layer 1 (Fig. 4a) as described in Steps 2–6.
-
11|
Develop ONLY the alignment marks (Fig. 4a, arrows) by standing the wafer on its end so only the alignment marks are immersed in the developer. Repeat for both sides.
-
12|
Cover the alignment marks with a small piece of Scotch tape.
▲ CRITICAL STEP The tape prevents the developed alignment marks from being recoated and is removed after the second layer is spun and BEFORE the soft-bake.
-
13|
To fabricate Layer 2 (Fig. 4b), coat Layer 1 made in Steps 1–6 with another layer of SU-8: spread at 500 rpm for 10 s and spin at 1,200 rpm for 30 s. “Soft bake” by incubating for 10 min at 65 °C and then 30 min at 95 °C. Coat again to yield 250 µm: apply photoresist, spread at 500 rpm for 10 s and spin at 1,200 rpm for 30 s. “Soft bake” by incubating for 10 min at 65 °C and then 30 min at 95 °C. Thicker photoresist (SU-8 100) can be used to achieve 250 µm thickness in one coat. However, it cannot be used for thinner coats, and you will need two different photoresists, which can be expensive. For our procedure, we needed only one type of photoresist, but it required multiple coating.
-
14|
Align the transparent alignment windows on the Layer 2 photomask (Fig. 4b) to the alignment marks in the Layer 1 (for example, crosses of matching size) using the contact aligner optics.
-
15|
After UV exposure (600 mJ/cm2) and post-baking (65 °C for 1 min, 95 °C for 10 min), develop the wafer following the procedure given in Steps 6 and 7. The development will take up to 20 min. The master features should look like those in Figure 10.
-
16|
Silanize the wafer as in Steps 8 and 9.
■ PAUSE POINT Master mold preparation is completed. Keep the masters in closed containers protecting from dust.
Figure 10.
The microfluidic perfusion device. (a) Schematics of the microfluidic device. The perfusion network (inlets 1–2) is shown in dark gray; arrows denote the flow direction. Three converging inlet channels (inlets 3–5) that are used to generate heterogeneous laminar flow streams are shown in light gray; “6” is the outlet. (b) Schematics of the SU-8 two-layer master mold showing two layers: Layer 1 is shown in dark gray, and Layer 2 is shown in light gray.
Molding of the patterning devices
-
17|
Mix the PDMS with the curing agent at a 10:1 ratio (PDMS:curing agent, wt/wt). A weighing boat is a convenient receptacle for pouring the PDMS. Place the weighing boat on a scale and tare. Add the desired amount of PDMS, followed by the curing agent.
-
18|
Thoroughly mix the compounds. Do this for at least 3 min with, e.g., a wooden tongue depressor to stir PDMS. The mixing of these two compounds generates bubbles that need to be evacuated. Place the PDMS mix in a vacuum jar (e.g., a desiccator without the drying pellets). Evacuate the air from the chamber. Vent the chamber 2–3 times for the first 15 min. Degassing is complete when there are no longer bubbles visible in the mixture. Eventually, all the bubbles will surface and pop (20–30 min).
-
19|
Slowly pour PDMS over the patterning device master to a height of approximately 5 mm.
-
20|
Re-degas the PDMS (15–20 min) to remove any bubbles that may have been created.
-
21|
Once bubbles are removed, place the PDMS-covered master into a 65 °C oven and allow curing for 2 h.
-
22|
Put on a clean pair of gloves.
-
23|
Using a clean, sharp scalpel or a razor blade, cut PDMS from the master. Do not drag the scalpel over photoresist areas; cut safely around the photoresist features (leave about a 2-mm edge). Make sure that end of the blade reaches the silicon wafer.
-
24|
Carefully peel the PDMS replica off the master using tweezers and transfer it to a clean surface for further cutting. Cut the PDMS replica to make 7-mm-wide (this will define the length of the microchannels for protein patterning) and 2-cm-long individual patterning devices.
■ PAUSE POINT Replica molding of patterning devices is completed. Keep the devices in a dust-free environment.
Molding of the perfusion devices
-
25|
Repeat Steps 17–18 using the perfusion device molds. Cut silicone tubing into 1-cm-long pieces. Apply a small amount of Duco Cement to one end of the tubing and affix the tubing to the raised, circular inlet pads of the master (Fig. 7b). Filling the end of the tubing with a small amount of glue prevents PDMS from entering the tube and blocking the port. The glue plug can easily be pushed out with a blunt syringe needle after curing. Let the glue sit for 5 min at room temperature.
-
26|
Once the tubing is affixed firmly and the PDMS has been debubbled, pour the PDMS over the photoresist master. Do not pour PDMS over the tubing, as it will seal the access port you are trying to create.
-
27|
Re-degas PDMS (15–20 min).
-
28|
Place the PDMS-covered master into a 65 °C oven and allow curing for 2 h.
-
29|
Cut the device from the master. Leave about 4 mm around the photoresist-defined features.
-
30|
Autoclave the devices (e.g., 20 min at 121 °C, wet cycle).
■ PAUSE POINT Replica molding of patterning devices is completed. Keep the devices in a dust-free environment.
? TROUBLESHOOTING
Micropatterning of culture substrates
-
31|
Take the patterning devices (microchannels) from Steps 17–24 (Fig. 5b) and clean the surface with a piece of tape, gently applying and lifting the sticky tape to remove the dust particles.
-
32|
Take the IPN-coated glass substrates (see protocol in Box 1) and gently remove any dust particles with a few pulses of filtered compressed air or nitrogen.
▲ CRITICAL STEP PDMS will not seal around the dust particles, which will create defects in micropatterns.
-
33|
Apply the microchannels to the IPN-coated glass substrate (Fig. 5b); it will seal spontaneously. When PDMS makes a conformal contact with a substrate, it minimizes reflection of light at the interface, so when viewed at an angle, the areas of PDMS that contact the surface appear slightly darker than the areas that do not make contact.
-
34|
Place the assembly inside the oxygen plasma asher (Fig. 11). Set the power to 150 W and the oxygen pressure to 0.75 Torr, and etch the surface for 6 min. The expansion coefficient of PDMS is much larger than that of glass, which causes PDMS to lose contact with the surface for large temperature increases. Since the plasma causes heating, stopping the plasma at short time intervals prevents the PDMS and glass from heating. You can monitor the chamber temperature on LCD temperature display of the plasma asher. The temperature should not rise above 50 °C.
▲ CRITICAL STEP Power, flow rate and time settings will vary with the type of machine and will need to be established for your equipment.
-
35|
Turn off the power and oxygen flow, break the vacuum and allow enough time for the chamber to cool off (about 5 min).
-
36|
Repeat this full cycle (Steps 34 and 35) three more times.
-
37|
Remove the assembly (patterning device + substrate) from the chamber and place in a sterile Petri dish. Etch pattern is shown in Figure 5f.
-
38|
Gently wash the surface of the glass with sterile deionized water (Fig. 5c). Apply a few drops of deionized water to one side of the microchannels. Gently apply suction (using a Pasteur pipette and vacuum bottle trap) to the other side to draw the water through the microchannels. Suction water through the microchannels three or four times.
-
39|
Remove water from the device by applying suction as in the previous step.
▲ CRITICAL STEP Steps 38–41 of the protocol require keeping the PDMS device sealed against the substrate without any translation. If the device is dislodged from the glass substrate, discard this sample. Perform these steps in a biological safety cabinet.
-
40|
Thaw a 30-µl Matrigel aliquot on ice and add ice-cold serum-free DMEM (Matrigel:DMEM ratio = 1:2 (vol/vol)).
-
41|
Apply 90 µl of Matrigel to one side of the microchannels. Draw the solution through the microchannels (Fig. 5d) for 2–3 min. Carefully remove excess Matrigel from the areas around the patterning device. Allow the solution to dry in the biological cabinet at room temperature overnight.
-
42|
Once the Matrigel is dried, you can remove the PDMS microchannels (Fig. 5e). Carefully wipe the areas around the protein pattern with ethanol and then with sterile deionized water. At this stage, you should be able to see a film of Matrigel on the microtracks under the microscope. Matrigel will look more opaque than adjacent IPNs.
▲ CRITICAL STEP The glass surface needs to be clean for bonding to occur.
Figure 11.
Oxygen plasma device. The Faraday cage (insert) sits inside the vacuum chamber. The device requires the plasma chamber to be evacuated of air and replaced with pure oxygen. The devices sit inside the cage.
Perfusion device assembly over micropatterned protein/IPN substrates
-
43|
Take the perfusion device from Steps 25–30 and clean the surface with a piece of tape, gently applying and lifting the sticky tape to remove the dust particles.
▲ CRITICAL STEP To protect the protein micropattern created in Step 42 from being degraded by the O2 plasma, cover the pattern with a slab of PDMS. We make this PDMS slab 3 cm long, 1 cm wide and 8 mm thick. We mold it off a thin, 1-mm-tall stack of sticky tape cut to the appropriate size; this way, the slab will seal around the protein pattern but will not be in contact with it.
-
44|
Place the microfluidic device and the micropatterned (IPN/protein) glass substrate in the plasma asher. Expose to O2 plasma at 300 W and 0.75 Torr for 30 s.
-
45|
Remove the PDMS device and the glass substrate. Remove PDMS slab from the glass substrate. Surfaces will remain reactive for 10–20 min.
-
46|
To move the device from one area to another, place the PDMS device and the glass substrate in a covered Petri dish. Prevent activated surfaces from contact with each other and other surfaces. We flip the device upside down and rest it on the silicone tubing.
-
47|
The IPN/protein micropatterns are visible under phase contrast (Fig. 5f). We use an inverted tissue culture phase contrast microscope to properly align the device with the micropattern on the glass substrate. Align the pattern orthogonally to the main channel (Fig. 6a) and lower the device onto the glass substrate, pressing it down slightly. Holding the glass substrate carefully, place the assembly onto a flat surface. The device will spontaneously form a conformal contact with the substrate, but make sure the entire surface of the device is in contact with the substrate. You can gently press on the device to help it seal.
▲ CRITICAL STEP The protein pattern area is bigger (wider and longer) than the main channel, so the bonding will not affect this area because it was not exposed to O2 plasma. It will seal reversibly. Avoid applying pressure to the microchannels, it might break the seal, and leakage between the inlet channels will occur. Use vacuum instead to fill the channels and introduce the cells.
-
48|
After 10 min, check that the bonding is complete by lifting the edge of the device by applying a small amount of force, upward and toward the center of the device, with a finger. If the device is bonded, there should be no separation from the surface.
Loading cells in the devices
-
49|
After bonding the device to the glass substrate, fill the device with BSA (1.5 mg/ml in PBS) inside a biological safety cabinet. The microchannels are filled with BSA to prevent cells from attaching to the “roof” of the microchannel. Utilize a 25.5-gauge needle and a 3- to 5-ml syringe. Insert the needle into the outlet port 6 (Fig. 10a) (without piercing the inner walls of the tubing) until the syringe contacts the glass. Slowly fill the tubing of port 6, and then start applying suction (using a Pasteur pipette) to port 3 until you fill the tubing. Next, apply suction to port 5 and then 4. Last, apply suction to ports 1 and 2.
▲ CRITICAL STEP Make sure the tubing at the outlet always has liquid; otherwise, air will get sucked in the microchannels. Steps 56–69 should be done in a biological safety cabinet using proper sterile technique to prevent contamination.
? TROUBLESHOOTING
-
50|
Place the BSA-filled device in the incubator overnight at 37 °C.
-
51|
Replace BSA with cell culture medium.
? TROUBLESHOOTING
-
52|
After filling the device with cell culture medium, close ports 1 and 2 with sterile alligator clips (Fig. 6b).
-
53|
Prepare C2C12 cell suspension as follows: add 3 ml of trypsin to flask/Petri dish with subconfluent C2C12 cells and incubate for 1 min at room temperature. Remove trypsin and incubate cells under a residual layer of trypsin in the 37 °C incubator for 1 min. Add 2 ml of growth medium to suspend the cells, count cells and dilute (if needed) with growth medium to yield the desired concentration (~1 × 106 cell/ml). You will need approximately 200 µl of cell suspension.
-
54|
Using a 1-ml syringe fitted with a Luer connector or a needle, slowly inject cell suspension into port 6 (Fig. 6b). Apply gentle suction to ports 3 and 5, filling the microchannel with cell suspension.
? TROUBLESHOOTING
-
55|
Place the filled device in a p150 dish. Allow the cells to adhere for 30 min in the incubator. Cells will adhere and spread strictly on Matrigel microtracks (see Fig. 10b). There should be no need to wash excess cells out of the channel, as all the cells should have migrated over to the microtracks.
-
56|
Remove the alligator clips. Place a p35 dish inside the p150 and fill it with 2 ml of fresh growth medium (Fig. 12, “medium”). This p35 will act as a reservoir for the medium.
-
57|
The medium will be fed into the main channel through the perfusion channels (ports 1 and 2, Fig. 12) using siphoning. Attach autoclaved sterile silicone tubing (prefilled with medium) to port 1 via an elbow connector (Fig. 12).
-
58|
Place the other end of the tubing into the filled p35 (Fig. 12, “medium”).
-
59|
Insert one end of the autoclaved piece of Kimwipe (about 4 cm long) into port 2 (Fig. 12). This will serve as a conduit to move the fluid from the device into the waste reservoir. Place the other end of the Kimwipe inside a p35 lid. This p35 lid will act as a waste reservoir (Fig. 12, “waste”). Thus, the perfusion of the main channel is achieved through siphoning from medium container (Fig. 12, “medium”) into the waste container (Fig. 12, “waste”). Add another p35 dish filled with sterile deionized water to minimize evaporation of medium. Around 500 µl/d should be expelled from this device to maintain healthy culture. Replenish feeding reservoir every 2–3 d.
▲ CRITICAL STEP Make sure no bubbles form at the interface or inside the tubing; otherwise, there will be no flow.
-
60|
Cover the p150 dish with a lid and place inside the incubator. Leave overnight.
-
61|
When cells become confluent (usually the day after seeding), switch the growth media to reduced serum media (differentiation medium (DM)) to inhibit proliferation and induce fusion of myoblasts. Remove the feeding silicon tubing and the Kimwipe “wick” from the device, wash the cells and the perfusion channels with DM, refill the tubing with DM and reconnect the tubing and the wick. The cells will start fusing together within 48 h, and by 8–9 d after seeding, myotubes will form. Fusion can be detected straightforwardly because it entails a dramatic change in cell morphology: cells form multinucleated tubular assemblies (myotubes), and single-cell borders are absent (see Fig. 1).
? TROUBLESHOOTING
Figure 12.
Perfusion of the cells in the device using siphoning.
Localized delivery to C2C12 cells
-
62|
Fill syringe reservoirs with medium and agrin solution and equilibrate in the incubator for at least 15 min.
-
63|
Use alligator clips to close up ports 1 and 2. Connect the pump to the device: insert one end of PFE tubing directly into the outlet of the device (Fig. 10, port 6) as shown in Figure 13. The outlet of the device has an embedded piece of silicone tubing. Connect the other end of the PFE tubing to the pump as shown in Figure 13.
-
64|
Connect the inlets of the device with appropriate medium and agrin reservoirs (syringes, Fig. 13; ports 3–5 in Fig. 10a). Add a food coloring dye to the agrin solution for visual validation of the stream’s position. We used Allura red food coloring dye and have not observed any deleterious effects on cells (Fig. 2c).
-
65|
Set the pump to withdraw the fluid from the device. We used flow rate of 7 µl/min. Start the pump. A thin stream of Allura red–labeled cell medium should be visible in the middle of the channel.
-
66|
Place the device inside the incubator for the duration of the experiment. The pump will remain outside at all times. The outlet tubing is stiff FEP tubing, which will not collapse when pressed by the incubator door.
-
67|
After exposing cells to signaling molecule solution for an appropriate amount of time (we used 2–18 h, depending on the experimental design), disconnect the agrin fluid line from the device inlet and rinse the inlet tubing of the device using a syringe filled with cell culture medium. Aspirate rinsed solution while injecting buffer solution.
-
68|
Let the pump run for 10 min to wash agrin from the microchannel.
-
69|
Stop the pump and disconnect the device.
-
70|
The agrin flow profile can be quantitatively measured by filling the center channel with Texas red BSA or a fluorescently labeled dextran (with a molecular weight similar to that of agrin) and imaging it with fluorescence microscopy. If the camera is operated in its linear regime, after background subtraction (the PDMS areas should be calibrated to zero intensity) the height-averaged concentration at any given point is proportional to the fluorescence intensity value at that point. After stimulation, labeling with α-bungarotoxin conjugated to Alexa 488 should reveal that AChRs are clustering exclusively in stimulated areas of the myotube.
Figure 13.
Fluidic setup used for flow experiments. The setup consists of a microfluidic device placed in a holder. An external Genie Plus syringe pump is connected to the outlet of the device through FEP fluid line. The inlets of the device are connected to the reservoirs (modified syringes) via PEEK (analyte) and FEP (medium) fluid lines.
● TIMING
Fabrication of SU-8 master molds (Steps 1–7): 3–5 h
Fabrication of SU-8 master molds (Steps 8–9): 1 h
Fabrication of two-layer molds (Steps 10–16): 3 h
Molding of the patterning devices (Steps 17–24): 3 h
Molding of the perfusion devices (Steps 25–30): 3 h
Micropatterning of culture substrates (Steps 31–42): 12 h
Perfusion device assembly over micropatterned protein/IPN substrates (Steps 43–48): 20 min
Loading cells (Steps 49–54): 20 min
Loading cells (Step 55): 30 min
Loading cells (Steps 56–61): 15 min
Localized delivery to cells (Steps 62–70): 1 h
? TROUBLESHOOTING
See Table 1 for troubleshooting details.
TABLE 1.
Troubleshooting table.
| PROBLEM | POSSIBLE REASON | SOLUTION |
|---|---|---|
| When filling the device with solution, many bubbles form in the channels (Steps 50, 52). |
|
|
| When filling the device, the solution leaks from the device (Steps 50, 52, 56). |
|
|
| Cells do not respect the Matrigel/IPN pattern (Step 57). |
|
|
| Dead cells (Step 63) | ||
| No myotubes form. |
ANTICIPATED RESULTS
The described method allows production of linear isolated myotubes organized in a parallel microarray (Fig. 1b) and focal delivery of signaling molecules to the myotubes. For example, we have been studying the effects of focal agrin (a molecule implicated as a postsynaptic organizer11) on AChR aggregation12 and AChR density (unpublished data). Such focal stimulation of myotubes represents an attempt to mimic the presence of the neuron in the first stages of the formation of a neuromuscular synapse and leads to localized AChR clustering12 and changes in AChR turnover (in preparation). Individual AChR clusters can be followed with time (note: no time-lapse apparatus is needed) because the geometrical arrangement of the myotubes and their confinement to the precise locations of a cell culture surface allows for easy identification and monitoring of the myotubes. This approach can be further applied to study the effects of other synaptogenic molecules and their combinations on postsynaptic differentiation. In addition, we used the described cell-patterning technique to isolate and position single cells and cell clusters of different adherent cell types2.
ACKNOWLEDGMENTS
We thank K. Healy and T. Barber for the original IPN-grafting protocol and G. Cooksey for the photographs of microfabrication equipment.
Footnotes
AUTHOR CONTRIBUTIONS A.T.: study concept, design, development and preparation of manuscript. X.F.M.: preparation of manuscript. A.F.: study concept and preparation of manuscript.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
References
- 1.Neville C, Rosenthal N, McGrew M, Bogdanova N, Hauschka S. Skeletal muscle cultures. Methods Cell Biol. 1997;52:85–116. [PubMed] [Google Scholar]
- 2.Tourovskaia A, et al. Micropatterns of chemisorbed cell adhesion-repellent films using oxygen plasma etching and elastomeric stencils. Langmuir. 2003;19:4754–4764. [Google Scholar]
- 3.Tourovskaia A, Figueroa-Masot X, Folch A. Differentiation-on-a-chip: a microfluidic platform for long-term cell culture studies. Lab Chip. 2005;5:14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 4.Li N, Tourovskaia A, Folch A. Biology on a chip: microfabrication for studying the behavior of cultured cells. Crit. Rev. Biomed. Eng. 2003;31:423–488. doi: 10.1615/critrevbiomedeng.v31.i56.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takayama S, et al. Patterning cells and their environments using multiple laminar fluid flows in capillary networks. Proc. Natl. Acad. Sci. USA. 1999;96:5545. doi: 10.1073/pnas.96.10.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takayama S, et al. Subcellular positioning of small molecules. Nature. 2001;411:1016. doi: 10.1038/35082637. [DOI] [PubMed] [Google Scholar]
- 7.Sawano A, Takayama S, Matsuda M, Miyawaki A. Lateral propagation of EGF signaling after local stimulation is dependent on receptor density. Dev. Cell. 2002;3:245–257. doi: 10.1016/s1534-5807(02)00224-1. [DOI] [PubMed] [Google Scholar]
- 8.Cho BS, et al. Passively driven integrated microfluidic system for separation of motile sperm. Anal. Chem. 2003;75:1671–1675. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- 9.Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature. 2005;434:1134–1138. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchetta EM, Munson MS, Ismagilov RF. Characterization of the local temperature in space and time around a developing Drosophila embryo in a microfluidic device. Lab Chip. 2006;6:185–190. doi: 10.1039/b516119c. [DOI] [PubMed] [Google Scholar]
- 11.McMahan UJ. The agrin hypothesis. Cold Spring Harb. Symp. Quant. Biol. 1990;55:407–418. doi: 10.1101/sqb.1990.055.01.041. [DOI] [PubMed] [Google Scholar]
- 12.Tourovskaia A, Kosar TF, Folch A. Local induction of acetylcholine receptor clustering in myotube cultures using microfluidic application of agrin. Biophys. J. 2006;90:2192–2198. doi: 10.1529/biophysj.105.074864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia YN, Whitesides GM. Soft lithography. Angew. Chem. Int. Edn Engl. 1998;37:551. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 14.McDonald JC, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Bearinger JP, et al. P(AAm-co-EG) interpenetrating polymer networks grafted to oxide surfaces: Surface characterization, protein adsorption, and cell detachment studies. Langmuir. 1997;13:5175. [Google Scholar]
- 16.Griffin MA, et al. Patterning, prestress, and peeling dynamics of myocytes. Biophys. J. 2004;86:1209–1222. doi: 10.1016/S0006-3495(04)74195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight JB, Vishwanath A, Brody JP, Austin RH. Hydrodynamic focusing on a silicon chip: Mixing nanoliters in microseconds. Phys. Rev. Lett. 1998;80:3863–3866. [Google Scholar]