Summary
Despite recent discoveries of genetic variants associated with autoimmunity and infection, genetic control of the human immune system during homeostasis is poorly understood. We undertook a comprehensive immunophenotyping approach, analysing 78,000 immune traits in 669 female twins. From the top 151 heritable traits (up to 96% heritable), we used replicated GWAS to obtain 297 SNP associations at 11 genetic loci explaining up to 36% of the variation of 19 traits. We found multiple associations with canonical traits of all major immune cell subsets, and uncovered insights into genetic control for regulatory T cells. This dataset also revealed traits associated with loci known to confer autoimmune susceptibility, providing mechanistic hypotheses linking immune traits with the etiology of disease. Our data establish a bioresource that links genetic control elements associated with normal immune traits to common autoimmune and infectious diseases, providing a shortcut to identifying potential mechanisms of immune-related diseases.
Introduction
The immune system has evolved over millions of years into a remarkable defence mechanism with rapid and specific protection of the host from major environmental threats and pathogens. Such pathogen encounters have contributed to a selection of immune genes at the population level which determine not only host-specific pathogen responses, but also susceptibility to autoimmune disease and immunopathogenesis. Understanding how such genes interplay with the environment to determine immune protection and pathology are critical for unravelling the mechanisms of common autoimmune and infectious diseases and future development of vaccines and immunomodulatory therapies.
Studies of rare disease established major genes, and their associated pathways, that regulate pathogen specific immune responses (Casanova and Abel, 2004) and GWAS of autoimmune disease have also been productive for finding common variants (Cotsapas and Hafler, 2013; Parkes et al., 2013; Raj et al., 2014). Despite this progress, there are still major limitations in our understanding of the genetics of complex autoimmune or infectious diseases. A key missing piece is the elucidation of the genes controlling critical components of a normal human immune system under homeostatic conditions. These include the relative frequencies of circulating immune cell subsets and the regulation of cell surface expression of key proteins which we expect have strong homeostatic regulatory mechanisms.
Previous studies in humans and rodents have shown that variation in the levels of circulating blood T cells is in part heritable (Amadori et al., 1995; Kraal et al., 1983). Identifying the underlying genetic elements would help us understand the mechanisms of homeostasis – and its dysregulation. Twin studies are ideal to quantify the heritability of immune traits in healthy humans that allow adjustment for genes, early environment and important and age and cohort influences plus a number of known and unknown confounders (van Dongen et al., 2012). Early studies from our group demonstrated genetic control of CD8 and CD4 T cell levels in twins (Ahmadi et al., 2001) and others have shown similar heritable effects in non-twins and rodents and with broad white cell phenotypes (Amadori et al., 1995; Clementi et al., 1999; Damoiseaux et al., 1999; Evans et al., 1999; Ferreira et al., 2010; Hall et al., 2000; Kraal et al., 1983; Nalls et al., 2011; Okada et al., 2011). A recent study, with a family design, was the first to perform genome-wide association studies (GWAS) on a larger range of immune subtypes. The authors analysed 272 correlated immune traits derived from 95 cell types and described 23 independent genetic variants within 13 independent loci (Orru et al., 2013).
Here we report a comprehensive and high resolution deep immunophenotyping flow cytometry analysis in 669 female twins using 7 distinct 14-color immunophenotyping panels that captured nearly 80,000 cell types (comprising ~1,500 independent phenotypes), to analyse both immune cell subset frequency (CSF) as well as immune cell surface protein expression levels (SPELs). This gave us a roughly 30-fold richer view of the healthy immune system than was previously achievable. Taking advantage of the twin model we used a pre-specified analysis plan which prioritised 151 independent immune traits for genome wide association analysis and replication.
We find 241 genome-wide significant SNPs within 11 genetic loci, of which 9 are previously unreported. Importantly they explain up to 36% of the variation of 19 immune traits (18 previously unexplored). We identify pleiotropic “master” genetic loci controlling multiple immune traits, and key immune traits under tight genetic control by multiple genetic loci. In addition we show the importance of quantifying cell surface antigen expression rather than just cell type frequency.
Critically, we show overlap between these genetic associations of normal immune homeostasis with previously-established autoimmune and infectious disease associations. This rich database provides a vital, publicly accessible bioresource as a bridge between genetic and immune discoveries that will expedite the identification of disease mechanisms in autoimmunity and infection.
Results
Subjects
The discovery stage comprised 497 female participants from the UK Adult Twin Register (TwinsUK). There were 75 complete monozygotic (MZ) twin pairs, 170 dizygotic (DZ) pairs, and 7 singletons (arising from QC failures in one co-twin). The mean age was 61.4 years (range: 40–77). The replication stage comprised a further 172 participants, mean age 58.2 years (range: 32–83), with 46 MZ, 118 DZ, and 8 singletons. We stained cryopreserved PBMC from each, using a set of seven 14-color immunophenotyping panels that delineate a large range of immune subsets (Figures 1A, S1–3). Immune traits analysed included the cell subset frequency (CSF, i.e., the proportionate representation of a given phenotype) and the surface protein expression level (SPEL, i.e., a quantitative measure of gene expression on a per-cell basis). The variability of all traits was assessed using longitudinal sampling on a small cohort of individuals as described in the Methods; of the 50,000 traits meeting the first filter criterion (Figure S4), the mean covariance across samples drawn 6 months apart is 0.86. All trait values and summary analyses including variability are available for download. Data and statistical analysis of the discovery stage was completed per a pre-defined statistical analysis plan before samples from the replication stage were thawed.
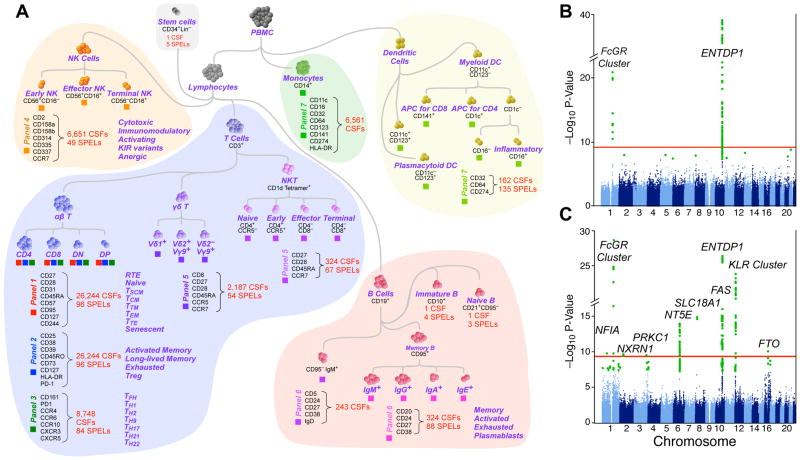
Figure 1. Schematic representation of leukocyte populations analyzed and summary Manhattan plot.
(A) This diagram illustrates the approach to analyzing the immunophenotyping data obtained by flow cytometry. It is not meant to convey differentiation stages of leukocyte populations though that property is largely reflected in this diagram. Each “lineage” of a subset of leukocytes was identified through hierarchical gating. Within each of these lineages, all possible combinations of markers with heterogeneous expression within the lineage were analyzed. The number of subsets identified by this combinatorial approach is shown in various lineages; the trait analyzed was the cell subset frequency (CSF) within its parent lineage. In addition, the cell surface protein expression level (SPEL) was quantified by the median fluorescence intensity of the antibody staining on a given cell subset; the number of SPEL traits is indicated as well. (B, C) Summary Manhattan plots: Green dots: genome-wide significant associations (P<5×10−8). The red line indicates the significance threshold of P<3.3×10−10, which corresponds to the standard genome wide threshold after further adjustment for 151 independent tests. The variants shown are MAF≥0.1, Call rate ≥0.9, HWE P-value ≥1×10−8. Shown are separate plots for SPEL associations (B) and CSF associations (C).
GWAS analysis of all 78,000 immune traits is computationally prohibitive and would require a multiple comparisons correction that dramatically reduces sensitivity. The ability to infer heritability (proportion of variance explained solely by genetic factors) by the use of twins dramatically enhanced our ability to focus on those that are most likely to be informative. Co-variation of all traits was computed; about 1,800 were independent at r < 0.7 (Figure S4).
We found no significant association of the analyzed traits with self-reported smoking or drinking, and so did not include those behaviors as covariates. We identified many traits associated with age, and included age as a covariate in all analyses. Notably, an advantage of using a twin-based cohort is to render age and other cohort effects minimally impactful. The age range of our cohort was optimal for our goal of identifying immune traits associated with genetic elements that show a risk for autoimmune diseases. Since incidence for such diseases often increases with age, the greatest power for such correlations will be obtained using samples measurements most proximal to the common onset of disease.
Heritability
Falconer’s traditional formula (twice the difference in intraclass correlations) was used to roughly estimate the heritabilities of all 78,000 immune traits; after ranking, traits were selected for further pre-specified analyses (Figure S4). Variance components analysis (ACE model) was used to more precisely estimate heritabilities of chosen traits. The heritabilities ranged widely from 0%, suggesting purely environmental or stochastic influences, to 96% (e.g. CD32 expression on dendritic cells), indicating a strong genetic effect. Figure S5 shows the range of heritabilities for selected traits, and the components of the model (additive genetic, common and specific environmental influences) are tabulated in Table S2 with full trait descriptions.
GWAS of immune traits
Single variant associations were performed on 151 immune traits selected for high heritability or biological interest, comprising cell frequency (129 CSF) and cell surface protein expression (22 SPELs). Many significant associations were found despite the stringent Bonferroni multiple testing threshold of 3.3×10−10. We also performed a conditional analysis, including the top SNP of each locus as a covariate, to identify potential independent secondary signals. This analysis did not reveal any significant evidence for additional independent signals.
Six SPELs were significant (Table 1) with the strongest between MFI:516 (CD39 SPEL on CD4 T cells) and the ENTPD1 (CD39 gene) SNP rs7096317 (P=9.4×10−40). Many other variants of ENTPD1 were also associated with this trait. (Table S3). Expression of five others (MFI:189, MFI:212, MFI:231, MFI:504 and MFI:552, which include CD27 expression on B and T cell subsets, and CD161 expression on CD4 T cells) were associated with variants on chromosome 1q23, in a genetic region containing the important immune regulating genes FCGR2A, FCGR2B and FCRLA (Table 1). These associations were independently verified in the replication cohort, and the combined discovery and replication set P-values of the 6 SPELs ranged from 2.79×10−11 to 1.59×10−54 (Table 1, Figure 1B). Table S5 illustrates other examples of genetic control of cell surface expression, including the expression of CD11c, CD123, and CD274 on myeloid subsets.
Table 1. Discovery and replication results for the top significant SNPs at each locus for each immune trait.
For the Discovery stage, we used a significance threshold of p < 3.3×10−10. This threshold corresponds to the standard genome wide threshold of 5×10−8 after further adjustment for 151 independent tests. Orru et al (Orru et al., 2013) also identified Locus 1 (associated with a single trait, CD62L− dendritic cells, not measured in our panel), and Locus 9 (associated with a single trait: CD39+ CD4 T cell frequency, P2:4186 in our list). The trait ID is fully described in Table S2.
| Locus:Genes | Trait ID | Trait Phenotype | Marker | Chr | EA/NEA | EAF | Beta (SE) | P Value | Beta (SE) | P Value | Beta (SE) | P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1: FCGR2A, FCGR2B, FCRLA | MFI:189 | CD27 on IgA+ B | rs1801274 | 1 | A/G | 0.49 | 0.128 (0.02) | 6.48E-11 | 0.07 (0.03) | 3.70E-02 | 0.11 (0.02) | 2.8E-11 |
| MFI:212 | CD27 on IgG+ B | rs1801274 | 1 | A/G | 0.49 | 0.136 (0.02) | 5.38E-12 | 0.12 (0.03) | 1.11E-04 | 0.13 (0.02) | 2.9E-15 | |
| MFI:231 | CD161 on CD4 T | rs1801274 | 1 | A/G | 0.49 | 0.131 (0.02) | 2.64E-11 | 0.12 (0.03) | 2.17E-04 | 0.13 (0.02) | 2.7E-14 | |
| MFI:504 | CD27 on CD4 T | rs1801274 | 1 | A/G | 0.49 | 0.145 (0.02) | 5.42E-14 | 0.14 (0.03) | 4.20E-05 | 0.14 (0.02) | 1.1E-17 | |
| MFI:552 | CD27 on CD8 T | rs1801274 | 1 | A/G | 0.49 | 0.186 (0.02) | 1.26E-21 | 0.12 (0.03) | 1.72E-04 | 0.17 (0.02) | 4.2E-24 | |
| P7:110 | iMDC: %CD32+ | rs10494359 | 1 | C/G | 0.12 | 0.343 (0.03) | 2.52E-29 | 0.43 (0.05) | 1.05E-15 | 0.36 (0.03) | 5.9E-43 | |
| P7:224 | CD1c+ mDC: %CD32 | rs4657090 | 1 | A/G | 0.27 | −0.174 (0.02) | 1.30E-14 | −0.19 (0.04) | 3.86E-06 | −0.18 (0.02) | 2.7E-19 | |
|
| ||||||||||||
| 2: NFIA | P4:3551 | NK: %CD314-CD158a+ | rs12072379 | 1 | G/C | 0.16 | −0.131 (0.02) | 1.73E-10 | −0.10 (0.05) | 4.87E-02 | −0.13 (0.02) | 2.7E-11 |
|
| ||||||||||||
| 3: NRXN1 | P4:3551 | NK: %CD314-CD158a+ | rs17040907 | 2 | T/C | 0.07 | −0.208 (0.03) | 2.68E-10 | −0.16 (0.08) | 4.22E-02 | −0.02 (0.03) | 3.9E-11 |
|
| ||||||||||||
| 4: PRKCI | P4:3551 | NK: %CD314-CD158a+ | rs2650220 | 3 | G/A | 0.16 | −0.15 (0.02) | 3.18E-10 | −0.10 (0.05) | 4.55E-02 | −0.14 (0.02) | 6.0E-11 |
|
| ||||||||||||
| 5: NT5E, RP11-30P6 | P2:4195 | CD4 T: %CD39−CD73+ | rs9444346 | 6 | G/A | 0.19 | −0.2 (0.03) | 1.18E-14 | −0.12 (0.04) | 4.98E-03 | −0.18 (0.02) | 8.8E-16 |
| P2:4204 | CD4 T: %CD73+ | rs9444346 | 6 | G/A | 0.19 | −0.195 (0.03) | 5.85E-14 | −0.12 (0.04) | 2.82E-03 | −0.18 (0.02) | 1.8E-15 | |
|
| ||||||||||||
| 6: SLC18A1 | P4:3551 | NK: %CD314-CD158a+ | rs1390942 | 8 | T/C | 0.15 | −0.163 (0.02) | 1.39E-15 | −0.20 (0.05) | 1.70E-04 | −0.17 (0.02) | 1.4E-18 |
|
| ||||||||||||
| 7: SLC25A16 | P4:3551 | NK: %CD314-CD158a+ | rs3017072 | 10 | T/C | 0.15 | −0.153 (0.02) | 2.75E-13 | −0.15 (0.05) | 2.08E-03 | −0.15 (0.02) | 2.2E-15 |
|
| ||||||||||||
| 8: FAS, ACTA2 | P1:6601 | CD8 T: %TSCM | rs7097572 | 10 | C/T | 0.48 | −0.168 (0.02) | 8.51E-16 | −0.18 (0.03) | 2.72E-06 | −0.17 (0.02) | 1.3E-20 |
|
| ||||||||||||
| 9: ALDH18A1, ENTPD1, ENTPD1-AS1, RP11-7D5, SORBS1, TCTN3 | MFI:516 | CD39 on CD4 T | rs7096317 | 10 | G/A | 0.42 | −0.255 (0.02) | 9.40E-40 | −0.30 (0.04) | 9.92E-17 | −0.27 (0.02) | 1.6E-54 |
| P2:10491 | CD8 T: %CD39+ | rs4074424 | 10 | G/C | 0.42 | −0.219 (0.02) | 4.11E-27 | −0.19 (0.04) | 2.40E-07 | −0.21 (0.02) | 8.2E-33 | |
| P2:3460 | CD4 T:%CD39+CD38+PD1− | rs4582902 | 10 | C/T | 0.47 | −0.164 (0.02) | 4.55E-16 | −0.19 (0.03) | 2.58E-08 | −0.17 (0.02) | 9.2E-23 | |
| P2:4159 | CD4 T:%CD39+CD73− | rs6584027 | 10 | G/A | 0.47 | −0.212 (0.02) | 1.54E-25 | −0.20 (0.04) | 1.09E-08 | −0.21 (0.02) | 1.1E-32 | |
| P2:4186 | CD4 T:%CD39+ | rs6584027 | 10 | G/A | 0.47 | −0.215 (0.02) | 2.20E-26 | −0.21 (0.04) | 5.27E-09 | −0.21 (0.02) | 7.8E-34 | |
| P2:4213 | CD4 T:%CD39+CD73+ | rs10882676 | 10 | A/C | 0.47 | −0.195 (0.02) | 6.76E-22 | −0.19 (0.04) | 2.19E-07 | −0.19 (0.02) | 9.0E-28 | |
|
| ||||||||||||
| 10: KLRC1, KLRC2, KLRC4, KLRK1, RP11- 277P12 | P4:3551 | NK: %CD314-CD158a+ | rs2734565 | 12 | C/T | 0.3 | −0.144 (0.02) | 1.34E-10 | −0.18 (0.04) | 1.67E-05 | −0.15 (0.02) | 1.4E-14 |
| P4:4832 | NK: %CD314-CCR7− | rs2734565 | 12 | C/T | 0.3 | −0.233 (0.02) | 1.27E-24 | −0.33 (0.04) | 3.95E-15 | −0.26 (0.02) | 2.7E-37 | |
| P4:5538 | NK: %CD314-CD335+ | rs2734565 | 12 | C/T | 0.3 | −0.275 (0.02) | 3.40E-34 | −0.38 (0.04) | 2.27E-19 | −0.30 (0.02) | 6.4E-51 | |
|
| ||||||||||||
| 11: FTO | P4:3551 | NK: %CD314-CD158a+ | rs1420318 | 16 | A/G | 0.1 | −0.146 (0.02) | 9.34E-11 | −0.19 (0.06) | 4.05E-02 | −0.14 (0.02) | 1.2E-11 |
Overall, 241 SNP variants with a frequency above 5% were significantly associated with various SPELs (Table S3); of these, 35 SNPs were pleiotropically associated with multiple SPELs.
Genetic control of SPEL may simply be due to promoter/enhancer element variants, or more complex regulation of transcription, translation, or protein localization. In contrast, genetic control of CSF may reveal homeostatic mechanisms regulating cell subset representation in the blood. Genome-wide significant associations were identified with 13 different CSFs (Figure 1C, Table 1). Nearly all were verified in the replication cohort (Table 1) and some reached a P-value of 10−43.
Suggestive associations, which did not meet the conservative significance threshold of 3.3×10−10, were also identified for numerous SPELs and CSFs (Tables S3, S4). The associations that were independently replicated (replication P<0.05) as well as meta-analysed variants reaching P<5×10−8 are reported in Table S5.
Genetic Control of TREG Cells
One of the most heritable traits identified from our staining panels was the frequency of CD39+ cells within the CD4 compartment (Figure 2), as previously reported (Orru et al., 2013). CD39+CD4 T, as well as CD73+ CD4 T cells, have been identified functionally as T regulatory (TREG) cells, a key subset in the modulation of immune responses (Antonioli et al., 2013).
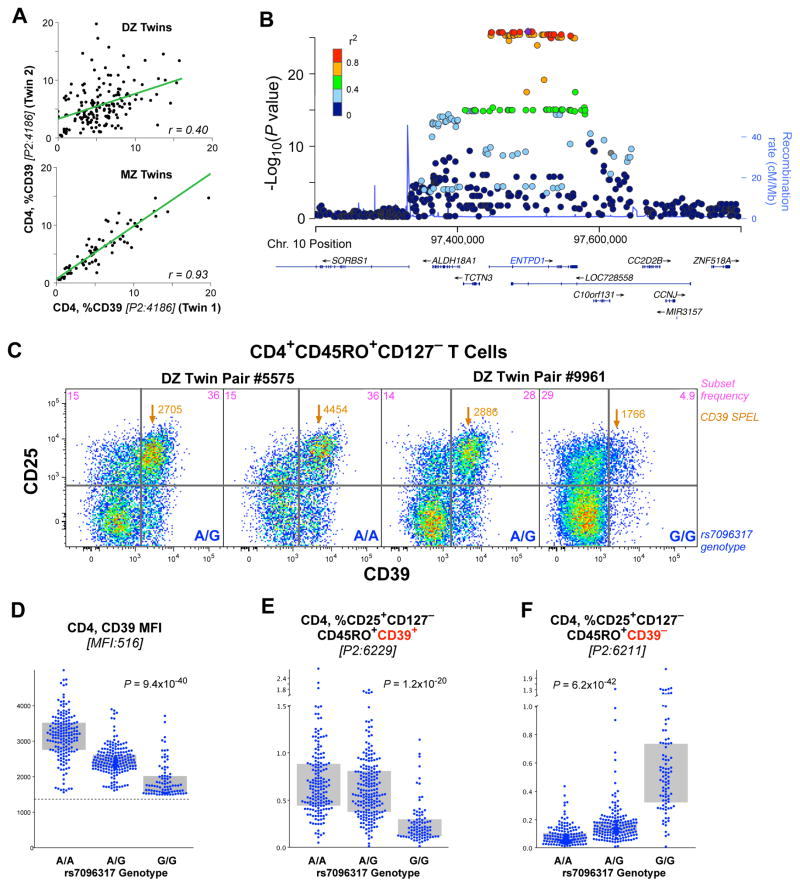
Figure 2. Genetic associations with Treg phenotype cells.
(A) The correlation of the fraction of CD4 T cells that are CD39+ in dizygotic twins (upper) and monozygotic twins (lower). The linear correlation, r, is shown for each comparison. (B) Locus-plot showing significant effect of individual SNPs on CD39 expression on CD4 T cells. (C) Shown are the expression profiles of CD39 and CD25 for the subset of CD4 T cells that are CD45RO+CD127−, for two pairs of dizygotic twins discordant for the rs7096317 allele (in the CD39 gene locus). Within each graphic is shown the fraction of cells in the upper two quadrants and the surface protein expression level (SPEL) of CD39 for the cells in the upper right quadrant, as well as the genotype of each individual. (D) The CD39 SPEL of CD39 positive cells is graphed by the genotype of rs7096317; the dotted line indicates the threshold of positivity above which a cell was considered CD39+. In the C/C genotype, relatively few cells are above this threshold and the median fluorescence intensity values are not robust. (E, F) The fraction of CD4 T cells of the designated phenotype is graphed by the rs7096317 genotype. Bars indicate interquartile range.
The heritability of CD39+CD4 T frequency was 89% (95% CI: 66–93%) (Figure 2A). GWAS analysis revealed a single locus on chromosome 10 that was highly associated with the trait (Figure 2B); this locus maps to the CD39 gene itself. Quantification of the expression of CD39 on a per cell basis (i.e., SPEL) revealed that the basis for this association was an “on/off” control of the expression of the CD39 molecule on the cells, rather than a homeostatic regulation of the circulating levels of these cells (i.e., CSF). Specifically, individuals who are homozygous for rs7096317A express the highest amount of this protein on the cell surface; heterozygotes expressed half as much; and rs7096317G homozygotes expressed virtually none (Figure 2C, D). While the A/G heterozygotes have a significantly decreased CD39 SPEL, the cells express enough so as to remain CD39+. Thus, in the analysis of the CD39 CSF by genotype, only the G/G homozygotes have a reduced frequency of this population (Figure 2E). This illustrates the power of the SPEL analysis to de-convolute potential mechanisms of genetic control that are missed by simple analysis of CSF.
Similarly, the frequency of CD4 T cells that are CD25+CD127−CD45RO+ but do not express CD39 was also strongly associated with this same locus, showing the opposite association (Figure 2F). In other words, genetic control is not over the frequency of TREG (CD4+CD25+CD127−CD45RO+), but over the quantitative expression of CD39 (the cell phenotype). Notably, this genetic control also extends to lymphocytes that are not TREG: a similar genotypic association was found for the relatively rare CD8+ and CD4−CD8− T cells expressing CD39 (Figure S7A, B). Finally, CD73 is an ectonuclease similar to CD39, and its expression has also been associated with TREG cells (Antonioli et al., 2013). The expression of CD73 was also found to be genetically controlled (Figure S7C) in the same way, and associated with a single locus on chromosome 6 mapping to the CD73 gene itself.
Thus, the main genetic control of CD39+ TREG appears to originate from a transcriptional or post-transcriptional regulation leading to the presence or absence of this protein on the cell surface of TREG cells; for those TREG defined on a basis independent of CD39, we found no evidence of genetic control over their representation in blood.
Genetic Influences on Leukocyte Differentiation
In virtually every leukocyte population, we found examples in which the frequency of certain differentiation stages was heritable (Figure 3). In some cases, despite a very high heritability, we were unable to identify genetic variants that correlated with the trait. For example, the frequency of a CD4 transitional memory (TTM) phenotype (CD28+CD127−), that comprise 15 to 20% of CD4 T cells, was very strongly heritable (Figure 3A) but did not correlate with any SNP genotypes. We found similar examples of strong heritability without genetic associations for other T cell stages, including recent thymic emigrants (RTE) and central memory (TCM). This was not unexpected: our study was only powered to find large effect sizes of gene variants, unusual for most traditional disease GWASs that need thousands of subjects per association. This suggests that possibly multiple genes of modest influence act on these phenotypes. Despite the lack of defined genetic association, the observation of strong heritability indicates that these cell types play an important and unique role in immunity, such that their numbers are strongly regulated.
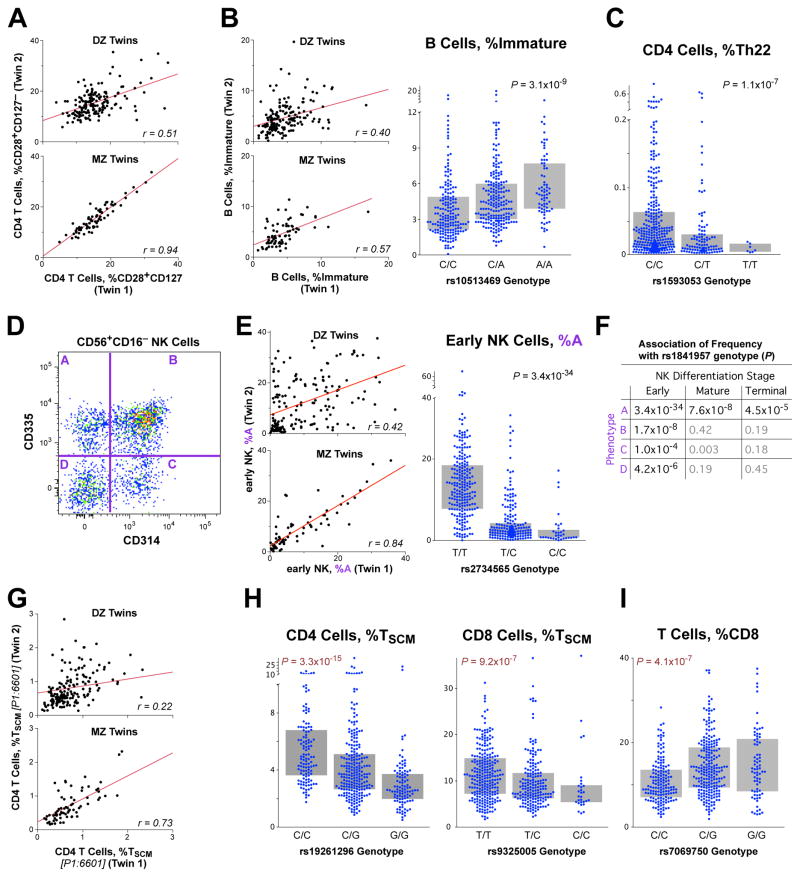
Figure 3. Genetic associations with lymphocyte differentiation.
(A) The proportion of CD4 T cells that are “transitional memory” (CD28+CD127−) is shown for DZ and MZ twins. (B) The proportion of B cells that are immature is shown for twins (left) and is strongly associated with the genotype of rs10513469 (MME gene) (right). (C) The proportion of CD4 T cells that are Th22 (CXCR3−CCR4+CCR6+CCR10+) is associated with the genotype of rs2019604. (D) A frequency of four phenotypes within NK cells (designated as “A”…”D” based on the expression of CD314 (KLRC4) and CD335 is shown for “early” (CD56+CD16+) differentiated NK cells. (E) The proportion of early NK cells that are CD314−CD335+ (population “A”) is shown for DZ and MZ twins (left). (Right) The genotypes of rs1841957 (near the KLRC4/CD314 locus) strongly associates with the frequency of CD314−CD335+ cells amongst early NK. (F) The associations of rs1841957 with all four phenotypes within differentiation stages of NK cells is shown by P-value. (G) The proportion of CD4 T cells that are “stem cell memory” (TSCM: CD45RA+CD95+CD27+CD28+CD127+CD57−) is shown for DZ and MZ twins. (H, I) The genotypes of rs7069750 (FAS gene) are associated with the proportion of CD4 and CD8 T cells that are TSCM, as well as the proportion of all T cells that are CD8.
For a number of leukocyte lineages, we were able to identify genetic associations with differentiation stages, and illustrate four examples. (1) Within B cells, the proportion that are immature (CD10+) is associated with the genotype of the mRNA-membrane metallo-endopeptidase (MME) gene (Figure 3B). (2) The proportion of Th22 CD4 T cells (CXCR3−CCR4+CCR6+CCR10+) is strongly associated with a single locus on chromosome 16, mapping to the SPG7 gene, which codes for paraplegin, an important protein in mitochondrial function (Figure 3C). (3) Within natural killer (NK) cells, the proportion of cells that express CD335 but not CD314 (Figure 33D) maps to the KLRC4 gene (Figure 3E). This association is much more profound for NK cells that are in an early differentiation stage (CD56+CD16−), and becomes less strong as the cells mature (Figure 3F). This indicates that, a mechanism evinced in a differentiation stage-specific manner. (4) The proportion of T cells that are “stem cell memory” cells (TSCM; the earliest memory stage) is heritable (Figure 3G) and associated with a genetic locus containing FAS (CD95) (Figure 3H). This association was much stronger for CD4 than for CD8 T cells. TSCM are precursor cells that have tremendous proliferative capacity and can regenerate all other memory T cell populations (Gattinoni et al., 2011; Lugli et al., 2013). Interestingly, this same locus also has a significant association with the fraction of T cells that are CD8 (Figure 3I), demonstrating multiple (pleiotropic) effects of the FAS gene on T cell differentiation.
Pleiotropic Impact of the FcRG2 Locus
The locus with the widest range of impacts on leukocyte subset phenotype and frequency was on chromosome 1, a region including the FcRG2 gene. This locus is well known for its association with a variety of autoimmune and inflammatory diseases, including systemic lupus erythematosus (SLE), Kawasaki’s disease, inflammatory bowel disease, Crohn’s disease, Type 1 diabetes, and HIV disease progression. Despite the genes in this locus being primarily expressed on myeloid and B lymphoid cells, many of these diseases are traditionally associated with T cell dysregulation.
The strongest association (e.g. SNP rs1801274) we identified for this locus was with the expression of CD32 (FcRG2a and/or FcRG2b: these are indistinguishable by the monoclonal antibody used in our panel) on the surface of inflammatory myeloid dendritic cells (imDC; Figure 4A, B). The heritability of this trait was extremely high at 96% (CI: 81–97%).
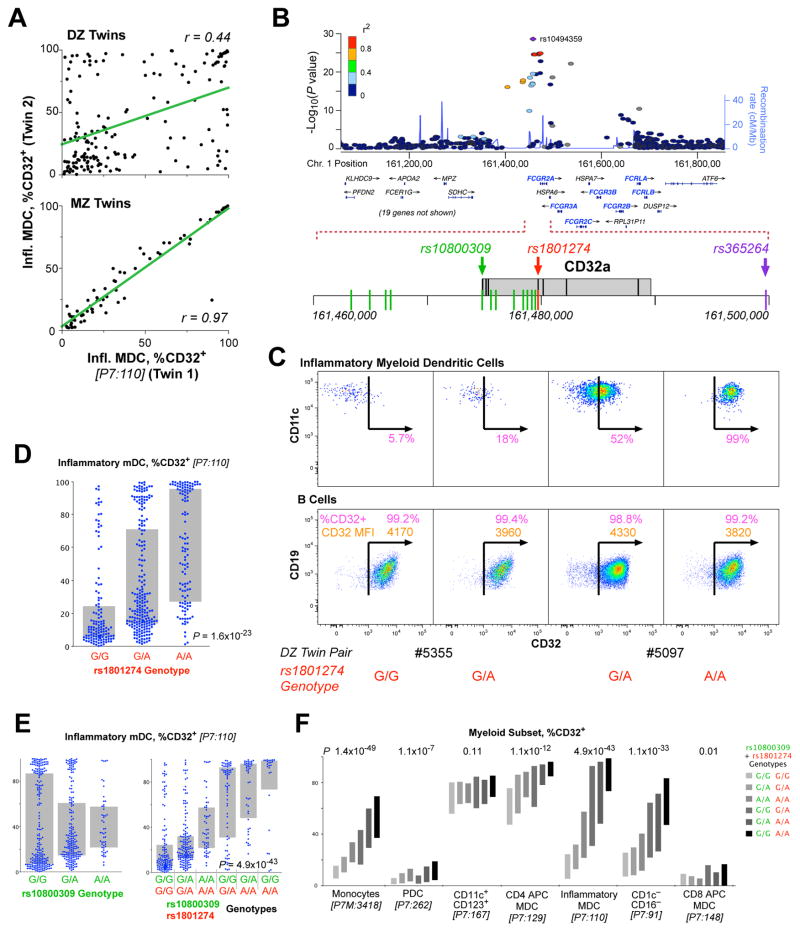
Figure 4. Genetic associations of the FcR locus with myeloid immunophenotypes.
(A) The correlation of the fraction of imDCs that are CD32+ for dizygotic twins (upper) and monozygotic twins (lower). The linear correlation, r, is shown for each comparison. (B) Organization of the FcR locus of chromosome 1 showing the position of immunologically relevant genes (shown in gray boxes). The positions of three SNPs are highlighted in color; rs1801274 and rs10800309 are the two that are most closely associated to susceptibility to SLE. SNP’s shown in green were in complete linkage disequilibrium within the samples analyzed in our cohort. (C) Sample expression profiles of CD32 on imDCs (upper) and B cells (lower). Shown are the fraction of cells that are CD32+ (in pink) and, for the B cells, the CD32 SPEL (in orange). Two pairs of dizygotic twins discordant for the genotype at rs1801274 are shown. (D) The distribution of expression of CD32+ imDCs is shown by genotype at rs1801274. (E) The expression of CD32 on the imDCs is not significantly associated with the genotype at rs10800309 by standard ANOVA (p = ns); however, the distributions are clearly different by genotype. The combination of the genotype at rs10800309 and rs1801274 provides a dramatic distinction for the expression of CD32 on imDC. (F) The expression profile of CD32 on seven different myeloid populations is shown, broken down by the combined genotype of the two SNPs. Bars indicate interquartile range.
The genetic control of the expression of CD32 on imDCs was not seen in all cell populations. For example, B cells showed no control (Figure 4C), whereas the expression of CD32 on imDCs is associated with the number of rs1801274 “T” alleles (Figure 4C, D).
The rs1801274 genotype has been strongly associated with susceptibility to SLE, as well as another SNP in the same locus, rs10800309. This latter SNP has also been associated with ulcerative colitis. The frequency of CD32+ iMDCs is strongly affected by the genotypes at both of these loci (Figure 4D, E); however, the distribution of expression for either locus is not uniform: high, intermediate, and low expressors can be found within all genotypes with differing frequencies. However, when the two genotypes are taken together as a diplotype, a powerful and replicated association becomes evident for CD32+ imDCs (Figures 4E, S8C). The impact of this diplotype on CD32 expression extends to other myeloid subsets (Figure 4F, S8A). Statistical significance of the association is greatest for monocytes, although the range in the expression levels is not as wide as it is for imDCs. Other subsets, such as the professional antigen presenting mDC, show a muted control of expression; CD11c+CD123+ DC, like B cells, show no differential regulation of CD32 expression at all.
Given the profound impact of these genotypes on particular subsets, it raises the possibility that part of the increased susceptibility to associated autoimmune diseases may be a consequence of the altered function of cells like imDCs by virtue of a differential expression of the activating (CD32a) or repressing (CD32b) proteins that we identify here. This is perhaps driven by a SNP in the promoter/enhancer areas in high linkage disequilibrium to the commonly-studied coding SNP rs1801274.
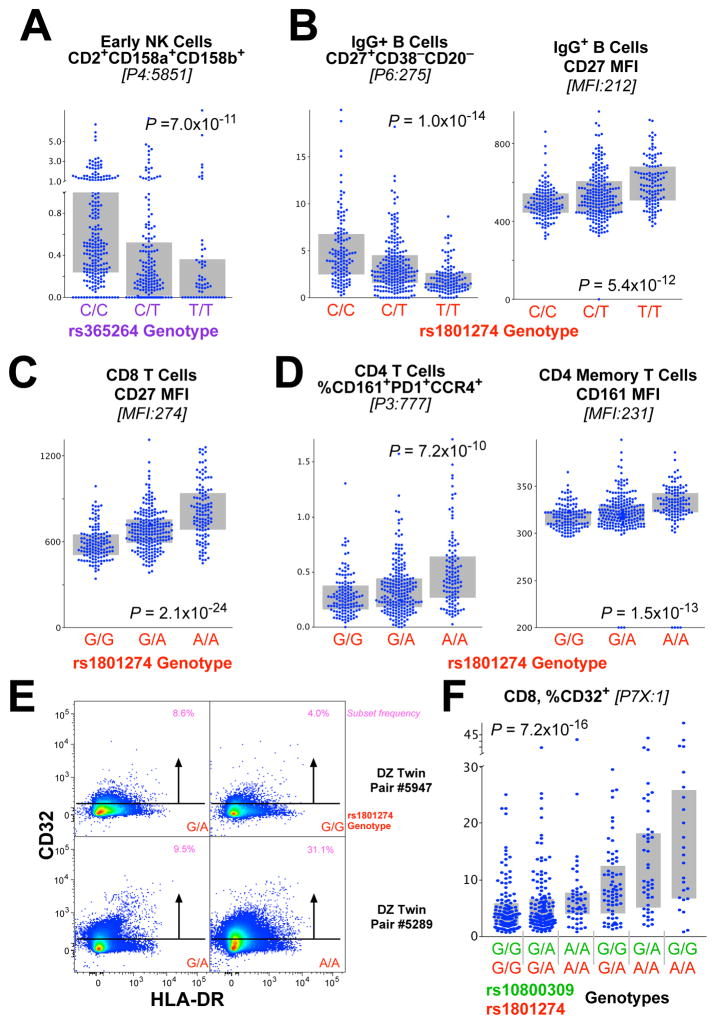
We also found a remarkable range of effects of the FcRG2 locus on a variety of lymphocyte subsets (Figure 5). For example, the proportion of early NK cells that are CD2+CD158a+CD158b+ is strongly associated with SNP rs365264 (Figure 5A), located between CD32a and CD16 (Figure 4B). The rs1801274 coding SNP in the locus was associated with phenotypes on both B cells and T cells, including the fraction of memory IgG+ (Figure 5B) or IgA+ (Figure 57) B cells that express CD27, as well as the CD27 expression level on a per cell basis. Interestingly, in this case, the higher surface expression levels of CD27 (SPEL) are associated with lower frequencies of cells that express CD27 (CSF). Thus, in contrast to the example of CD39+ TREG (Figure 2), differential regulation of CD27 protein expression does not account for differential frequency of these cell subsets.
Figure 5. Genetic associations of the FcR locus with lymphoid immunophenotypes.
(A) The genotype of rs365264 (close to CD16a on chromosome 1) is strongly associated CD56+CD16− (“early”) NK cells that are CD2+CD158a+CD158b+. (B) The genotype of rs1801274 is associated with the frequency of memory IgG+ B cells that are CD27+CD38−CD20− as well as the fraction of CD27+ cells. Note that, for this case, a lower frequency of the subset (left) is associated with higher protein expression (right). (C) Similarly, the genotype of rs1801274 is strongly associated with the cell surface expression of CD27 on CD8 T cells. (D) The genotype of rs1801274 is strongly associated with the cell surface expression of CD161 on CD4 T cells, as well as CD4 T cells that are CD161+PD1+CCR4+. (E) CD8 T cells express low levels of CD32 depending on genotype as shown by flow cytometry. (F) The fraction of CD8 T cells that express CD32 is strongly associated with the rs10800309:rs1801274 diplotype (see Figure 4).
T cells, FcRG2 and autoimmune disease
Similar to the case for IgG+ B cells, CD8 T cells also exhibited higher CD27 expression in association with rs1801274T allele (Figure 5C); this was also true for other T cell lineages (Figure S8). Furthermore, a population of CD4 T cells that express CD161 is also strongly associated with this same genotype (Figure 5D). Importantly, CD161+ T cells are either Th17 or mucosal-associated innate T (MAIT) cells, important for maintenance of mucosal integrity. Thus, we define an impact of specific gene variants on important T cell phenotypes closely related to their activation potential, which may underlie the associations with T cell-based autoimmune diseases.
Finally, Harty et al. recently demonstrated that CD8 T cells can express CD32b, and that this expression was functionally important in modulating cytolytic T cell responses (Starbeck-Miller et al., 2014). Here, we demonstrate that expression of CD32 on CD8 T cells is low and variable between individuals (Figure 5E). Notably, this expression shows a very strong association with the rs1801274:rs10800309 diplotype of the FCRG2 gene locus (Figure 5F). This suggests that the regulation of surface expression of this negative regulatory molecule on CD8 T cells (Starbeck-Miller et al., 2014) has a common expression mechanism to that in myeloid populations. Gene variants increasing susceptibility to SLE also associate with lower levels of this negative regulatory protein on CD8 T cells and imDC. Together, these data provide a possible direct link between the SNPs highly associated with autoimmune diseases and T cell phenotypes that might account for the pathogenesis.
Overlap with disease associations
The Catalog of Published Genome-wide Association Studies (http://www.genome.gov/gwastudies/) and ImmunoBase (http://immunobase.org) were used to evaluate the overlap between genetic variants associated with CSFs and SPELs in our study and those reported to be suggestively or statistically significant in candidate SNP, candidate gene or genome-wide association studies of complex and infectious diseases..
SNPs which were highly correlated variants (r2>0.8) with our significant immune traits were also interrogated for overlap with reported disease associations using the appropriate thresholds for the number of tests. A number of gene variants significantly and suggestively (P<10−3) correlated with CSFs and SPELs have been reported in associations of complex and infectious diseases; as shown in Table 2. The different gene variants of FCGR2A, associated with a range of myeloid and T cell phenotypes in our data, were reported to be associated with increased risk of a number of diseases, including inflammatory bowel disease, ulcerative colitis, SLE, Kawasaki disease, ankylosing-spondylitis and HIV progression (Table 2). An additional variant of FCGR2A, rs10494359, (associated with P7:100 (CD64−CD274− imDCs)) is closely correlated (in linkage disequilibrium) with rs10494360 (r2=0.941), and has been associated with rheumatoid arthritis. Juvenile idiopathic arthritis and chronic lymphocytic leukaemia susceptibility loci in the ACTA2/FAS region of chromosome 10q23.31 were also associated with the frequency of P1:6601 (CD4 TSCM) (P=4.1×10−12; Table 2). The Behçet’s disease susceptibility variant in the killer cell lectin-like receptors gene region corresponded with the frequencies of P4:3551, P4:4832 and P4:5538 (all three are CD314− subsets of NK cells). The correlations of variants association with tuberculosis, malaria, leprosy, HIV and hepatitis B and C our immune traits are presented in Table 2.
Table 2. Overlapping associations with complex diseases.
Association results from the discovery cohort immune trait analyses are reported in the first six columns. The trait ID is fully described in Table S2. The disease-associated variant, pathology, the disease-associated SNP’s best-reported P-value are indicated in the seventh-tenth columns, respectively. The risk alleles presented correlate with increased disease susceptibility and the r2 between disease SNP and immune trait, if different, are reported in parentheses after the reported SNP.
| Immune trait association | Disease association | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Gene (Chr) | Marker | Trait ID | Trait Phenotype | Beta | P Value | Reported SNP | Disease | Best P | Reference |
| FCGR2A (1) | rs1801274 | P7:110 | iMDC: %CD32+ | 0.2 | 1.6E-23 | rs1801274 (A) | IBD | 2.1E-38 | (Jostins et al., 2012) |
| MFI:189 | CD27 on IgA+ B | 0.13 | 6.5E-11 | Kawasaki disease | 7.4E-11 | (Khor et al., 2011) | |||
| MFI:212 | CD27 on IgG+ B | 0.14 | 5.4E-12 | Ulcerative colitis | 2.2E-20 | (Anderson et al., 2011) | |||
| MFI:231 | CD161 on CD4 T | 0.13 | 2.6E-11 | Ankylosing-spondylitis | 1.4E-09 | (Cortes et al., 2013) | |||
| MFI:504 | CD27 on CD4 T | 0.15 | 5.4E-14 | SLE | 6.8E-07 | (Harley et al., 2008) | |||
| MFI:552 | CD27 on CD8 T | 0.19 | 1.3E-21 | HIV progression | 1.0E-04* | (Forthal et al., 2007) | |||
| P7:224 | CD1c+ mDC: %CD32 | 0.12 | 4.4E-09 | Lymphoma | 0.006* | (Wang et al., 2006) | |||
| Malaria | 0.013* | (Sinha et al., 2008) | |||||||
|
| |||||||||
| FCGR2A (1) | rs10494359 | P7:110 | iMDC: %CD32+ | 0.34 | 2.5E-29 | rs10494360 (r2=0. 94) | Rheumatoid arthritis | 9.3E-05* | (Eyre et al., 2012) |
|
| |||||||||
| ZNF804A (2) | rs6755404 | P2:3367 | Effector CD4: CD127-PD1− | 0.09 | 3.8E-04 | rs6755404 (A) | Malaria | 1.2E-06 | (Band et al., 2013) |
|
| |||||||||
| MIR216B (2) | rs6751715 | P3:5372 | CD8 T: %CXCR3-R4+R6+R10− | −0.09 | 1.5E-05 | rs6751715 | HIV | 1.1E-06 | (Fellay et al., 2009) |
| P3:5661 | CD8 T: %CD161+ | −0.07 | 5.1E-04 | ||||||
| P6:197 | IgA+ B: %CD27-CD38− | 0.07 | 7.0E-04 | ||||||
| P3:5658 | CD8 T: %CD161+PD1− | −0.07 | 7.8E-04 | ||||||
|
| |||||||||
| LOC100505836 (3) | rs2593321 | Lin:19 | %NKT | 0.08 | 5.5E-04 | rs2593321 | HIV-1 control | 7.7E-06 | (Pelak et al., 2010) |
|
| |||||||||
| MICA (6) | rs4418214 | P6:112 | IgE+B: %CD20-CD27-CD38+ | 0.12 | 7.6E-04 | rs4418214 (C) | HIV-1 control | 1.4E-34 | (Pereyra et al., 2010) |
|
| |||||||||
| BTNL2 (6) | rs3817963 | P2:12609 | CD8 T: %CD25+CD38+45RO+ | −0.08 | 3.5E-04 | rs3817963 (A) | Hep C liver cirrhosis | 1.3E-08 | (Urabe et al., 2013) |
| P2:10486 | CD8 T: %CD25+CD38+ | −0.08 | 4.1E-04 | ||||||
|
| |||||||||
| HLA-DQB1 (6) | rs2856718 | P2:12609 | CD8 T: %CD25+CD38+45RO+ | −0.08 | 5.3E-04 | rs2856718 (A) | Hepatitis B | 4.0E-37 | (Mbarek et al., 2011) |
| P2:10486 | CD8 T: %CD25+CD38+ | −0.07 | 7.8E-04 | ||||||
|
| |||||||||
| EHMT2 (6) | rs652888 | MFI:578 | CD3 on CD8 T | −0.08 | 5.7E-04 | rs652888 | Chronic hepatitis B | 7.1E-13 | (Kim et al., 2013) |
|
| |||||||||
| MTCO3P1 - HLADQA2 (62) | rs4273729 | P2:10486 | CD8 T: %CD25+CD38+ | 0.08 | 1.4E-04 | rs4273729 | Chronic Hepatitis C | 1.7E-16 | (Duggal et al., 2013) |
| P2:12609 | CD8 T: %CD25+CD38+45RO+ | 0.08 | 2.3E-04 | ||||||
|
| |||||||||
| ADGB (6) | rs2275606 | P1:12906 | CD8 T: %TSCM | 0.13 | 6.3E-04 | rs2275606 (A) | Leprosy | 3.9E-14 | (Zhang et al., 2011) |
| ABO (9) | rs8176722 | MFI:1 | CD123 on mDC | −0.14 | 1.4E-04 | rs8176722 (A) | Malaria | 8.9E-10 | (Band et al., 2013) |
|
| |||||||||
| ACTA2, FAS (10) | rs7069750 | P1:6601 | CD8 T: %TSCM | −0.14 | 4.1E-12 | rs7069750 (C) | Juv. idiopathic arthritis | 2.9E-08 | (Hinks et al., 2013) |
| rs2147420 (r2=1) | CLL | 3.1E-13 | (Berndt et al., 2013) | ||||||
|
| |||||||||
| KLRC4-KLRK1 (12) | rs1049172 | P4:3551 | NK: %CD314-CD158a+ | −0.13 | 8.8E-09 | rs2617170 (r2=0.922) | Behçet’s disease | 1.3E-09 | (Kirino et al., 2013) |
| P4:4832 | NK: %CD314-CCR7− | −0.23 | 7.6E-22 | ||||||
| P4:5538 | NK: %CD314-CD335+ | −0.27 | 1.4E-30 | ||||||
|
| |||||||||
| MMP16 (8) | rs160441 | P4:3551 | NK: %CD314-CD158a+ | −0.08 | 2.3E-05 | rs160441 | Tuberculosis | 8.4E-06 | (Thye et al., 2010) |
|
| |||||||||
| ARHGAP20 (11) | rs1469170 | P4:3551 | NK: %CD314-CD158a+ | −0.08 | 5.6E-04 | rs1469170 (A) | Malaria | 8.0E-08 | (Band et al., 2013) |
Disease association is significant with candidate SNP method/gene approach.
Discussion
Understanding the fundamental principles of how the immune system protects the host from infection yet also contributes to autoimmunity and other disease pathogenesis is essential for the development of novel diagnostics and medicines. There remains a major gap in our understanding of genetic determinants of a normal human immune system and its main coordinates such as the frequency of immune cells and expression of relevant proteins. Using 669 twins and the richest immunophenotyping performed to date we investigated the genetic architecture of immune traits. We describe multiple independent genetic variants at several genetic loci explaining a substantial proportion (up to 96%) of the genetic variation. We identify both pleiotropic genetic loci that control multiple immune traits, and single immune traits under genetic influence by multiple loci. For certain canonical immune traits, genetic control is exerted at the level of immune cell surface protein expression (i.e., a consequence of promoter/enhancer or signalling mechanisms) rather than at the level of cell subset frequency (i.e., homeostasis or differentiation mechanisms). We further describe multiple genetic associations with common canonical immune traits related to leukocyte lineage and differentiation of major immune cell subsets such as B cells, T cells and natural killer cells. Finally, we identify genetic elements associated with both immune traits and autoimmune and infectious diseases. Providing the heritabilities of thousands of cell subtypes plus a basis to uncover the genetic architecture of the numerous gene-immune associations establishes this dataset as an essential bioresource for researchers. The remarkably strong associations we find for genetic traits linked to disease illustrates the power of our approach by using twins and optimized high quality immune phenotyping.
Some limitations of our study should be noted: the cohort used is all female, it is (for GWAS) a relatively limited sample size, and it is relatively homogeneous in terms of environmental exposure. The low numbers of genetic associations on chromosome 6 (the major histocompatibility region) is possibly explained by the considerable complexity and polymorphism in this gene region, which would require larger sample sizes to obtain statistically significant genetic associations. With regard to immunological traits, it should be noted that our discovery cohort ranges in age from 41–77. It is possible that analysis of a younger cohort, for which less environmental pressure on the immune system has occurred, would reveal stronger associations; on the other hand, it is likely that the greatest power to detect immune correlates related to disease will come from measurements at a time most proximal to the typical onset. Nevertheless, our success in identifying a large number of genetic variants with genome-wide significance validates our approach of focussing on well-defined and curated immune phenotypes. It should be noted that the 297 SNPs we report (Table S5) are those that attained genome wide significance (with a conservative correction for multiple comparisons) in both the discovery and replication cohorts. Many more associations are evident in the dataset (e.g., Tables S2 & S3) which can serve to formulate new testable hypotheses and genetic studies.
An example of the power of the resource was in distinguishing two important mechanisms that lead to differential representation of immune cell phenotypes (e.g., CD39+ CD4 T cells). The first is homeostatic: i.e., mechanisms which control the recirculation, proliferation, and elimination of a certain cell type in the blood – a cellular mechanism expressed at the whole body level. Such mechanisms, while unidentified, are known to exist for regulating major subset numbers (such as CD4 T cell numbers). A second, completely independent mechanism is the molecular regulation of the protein expression at the cell level itself (e.g., promoter/enhancer variants). Thus, even in the presence of intact homeostatic mechanisms regulating that cell type, a reduction in the number of cells expressing a protein (perhaps part of the defining phenotype) may be due to a specific promoter variant which simply abrogates the expression of the gene. Here we show examples of both mechanisms.
An example of a major immune trait under genetic control is the phenotype, but not frequency, of regulatory T (TREG) cells. TREG cells are essential for maintenance of immune homeostasis, and their dysregulation might lead to autoimmunity (Sakaguchi et al., 2010). A pleiotropic locus containing the ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1, CD39) gene controls several phenotypic features of CD39+ TREG. While we confirm an apparent association of this gene locus with the frequency of circulating CD39+ TREG (Orru et al., 2013), we show here that this is a consequence solely of altered phenotype. Quantitative analysis of CD39 protein expression on the cell surface demonstrates that the genetic of control of CD39+ TREG is exerted at the level of surface protein expression, rather than cell frequency (homeostasis). This establishes a paradigm for future studies of immune traits and points to the necessity of including both cell frequency and cell protein expression analyses in immunogenetic studies of the human immune system. We also describe the association of a genetic locus containing the ecto-5′-nucleotidase (NT5E, CD73) gene with a population of CD73+ TREG. From a functional perspective it is of interest that both CD73 and CD39 are ectonucleotidases involved in the generation of immunosuppressive adenosine that alters T cell and NK cell activities (Deaglio et al., 2007). Thus, it appears that the CD39 and CD73 adenosine immunosuppressive pathway has been under evolutionary selection and might therefore be a critical determinant of functional TREG activity (Bastid et al., 2013) and establishment of tissue homeostasis. Importantly, we conclude that there is no genetic control of the frequency of TREG (i.e., CD127−CD25+ memory CD4 cells), but rather, control is evinced at the level of their specific phenotype and presumably function.
We discovered several genetic associations with immune traits relevant to lymphocyte lineage and differentiation. A genetic locus containing the cell surface death receptor FAS (CD95) was associated with the frequency of circulating T stem cell memory cells (TSCM). TSCM are a recently described infrequent and functionally important lymphocyte subset (Gattinoni et al., 2011); these cells have a largely naïve T cell phenotype but are able to self-renew while displaying functional attributes of memory cells. Genetic control at the level of CD95 suggests a potential role of CD95 in the control of T stem cell homeostasis, for not only differentiation stages such as TSCM but total CD8 as well (Figure 3). It further provides a possible link between human autoimmune syndromes based on genetic CD95 deficiency (Strasser et al., 2009) and TSCM.
A genetic locus within a cluster of genes referred to as the “NK complex” containing NKG2D (CD314, KLRK1) was associated with the frequency of a distinct population of CD314−CD335+ “early” (CD56+CD16−) NK cells. The NKG2D gene encodes for a C-type lectin protein preferentially expressed in NK cells. It binds to a diverse family of stress induced ligands that include MHC class I chain-related A and B proteins (MICA, MICB), essential for the activation of T cells and NK cells (Raulet et al., 2013). These data establish an unexpected link between the genetic control of the frequency of a specific subset of NK cells and a gene locus containing major genes with functional relevance to NK cell activity and their activation by stressed normal tissue cells or tumor cells.
One locus in chromosome 1q23 containing FCGR2A, FCGR2B and FCRLA was associated with multiple immune traits. FcGR genes encode immunoglobulin Fc surface receptors found on macrophages, dendritic cells and neutrophils, as well as B and NK cells, and are involved in the regulation of B-cell antibody production and phagocytosis of immune complexes. The main associated immune traits were the frequencies of CD32+ inflammatory dendritic cells and monocytes, as well as several T cell phenotypes. Genetic variation in the FcGR gene locus is associated with an increase in susceptibility to several autoimmune and infectious disease including SLE, ankylosing spondylitis, HIV progression, and several other syndromes. Given the profound impact of this gene locus on particular immune cell subsets, altered function of CD32+ dendritic cells could be key to increased susceptibility to these autoimmune and infectious diseases.
Indeed, our demonstration of an association between the FCGR2A SNP and T cell phenotypes and/or inflammatory myeloid cells (e.g., imDCs) provides a potential link between this locus and autoimmune diseases with T cell aetiology. This coding SNP results in variants of the Fc receptor that have different avidities for immunoglobulin; consequently, much current research is aimed at understanding the possible functional role of these alleles in autoimmunity. However, our data suggest a different possibility with a more proximal mechanistic link: that association is with a promoter/enhancer SNP (in strong linkage disequilibrium with the coding FCGR2A SNP) that modulates expression of the negative regulatory CD32b molecule on imDC, monocytes, and/or T cells.
In addition to the wide range of diseases associated with the FcRG locus, further examples include a SNP within a genetic locus associated with Behçet’s disease (Kirino et al., 2013) that is in tight linkage disequilibrium with a SNP controlling the frequency of CD314−CD335+ early NK cells. We also report that a genetic locus containing FAS and associated with Juvenile idiopathic arthritis (Hinks et al., 2013), is also associated with functionally important CD8+ T stem memory cells.
These findings illustrate a key value of our database and approach: the identification of candidate immune traits associated with genetic loci of relevance to autoimmunity and infection. In summary, using one of the most comprehensive immune phenotyping efforts to date, we identified numerous genetic loci controlling key parameters of a normal human immune system. This comprehensive human immune phenotyping bioresource will allow the identification of critical immune phenotypes associated with common autoimmune and infectious diseases ultimately leading to accelerated discovery of mechanisms of disease and response to therapy.
Experimental Procedures
Samples
This study was approved by the NIAID (NIH) IRB, and London-Westminster NHS Research Ethics Committee; all participants provided informed consent. The discovery stage comprised 497 female participants from the UK Adult Twin Register, TwinsUK, with full genotyping data on 460 subjects. The TwinsUK cohort is described in detail in (Moayyeri et al., 2012). Briefly, TwinsUK is a large cohort of twins historically developed to study the heritability and genetics of diseases with a higher prevalence among women. The study population is not enriched for any particular disease or trait and is representative of the British general population of Caucasian ethnicity. Selected twins were all female with an age range of 41–77 (mean 61.4), and by self-report, 100% Caucasian with most being UK ancestry. From the subsequent genotype data we excluded a few individuals showing evidence of non-European ancestry as assessed by principal component analysis comparison with HapMap3.
The replication stage included a further unrelated 172 TwinsUK participants with whole genome genotyping data on 169. The samples for the discovery samples were selected to match the characteristics (age and gender) of the discovery dataset. For this reason the replication cohort included only Caucasian women with an age range 32–83 (mean 58.2). All subjects were nominally healthy at the time of sample collection.
A total of 746 PBMC vials were analysed: PBMC from the 669 twin specimens (plus 4 replicates), and 30 healthy controls from the US (two vials of PBMC from blood drawn six months apart were analysed for 29 subjects, a replicate vial of PBMC from one of the two blood draws for 14 (of the 29), and one vial only for 1 subject). The samples in each stage were ordered such that twin or longitudinal control samples were analysed in the same experimental run (each comprising 15–30 vials), whereas replicate control samples were analysed in different experimental runs. Staining and data analyses were otherwise performed blinded to identity.
Immunophenotyping
See Supplementary Methods for cell processing details.
Flow Cytometry and data analysis
Cells were analyzed in 96 well plate format on an 18-color LSR (BD Biosciences) using an HTS unit. Each run on the flow cytometer was accompanied by a set of compensation controls of antibody stained IgG kappa beads (BD Biosciences). Data were evaluated on FlowJo software v9.7 (FlowJo, LLC). Postprocessing of data and visualizations were done with JMP v10 (SAS) and SPICE v5.3 (NIAID; (Roederer et al., 2011)).
Gating
A graphic depicting the fluorescence distribution of all samples in the discovery cohort is shown in Figure S1. Figures S2 and S3 illustrate the gating hierarchy for a single sample. The Supplementary Methods describes the generation of the ~80,000 gates analysed.
For the discovery cohort, there were 20 experimental runs for 543 samples (501 twin specimens from 497 subjects and 42 US control specimens from 14 subjects). Within each run, uniform scatter gating was used. Each sample was gated on time (to eliminate spurious events from beginning or end of sample run); this gate could vary by sample. With two major exceptions, all samples received the same fluorescence gating, After the third run, we chose to replace the CD4 reagent in Panels 1, 2, and 5 due to poor performance; this necessitated a different CD4 gating for those samples. Similarly, the reagents in the “dump” channel of Panel 7 were modified after 6 runs.
For the replication cohort, there were 8 experimental runs for 203 samples (172 twin specimens from 172 subjects and 31 US control specimens from 16 subjects). All analysis procedures were identical to the discovery cohort. Minor modifications to the panels were necessitated by unavailability of the same lots of reagents but these did not impact enumeration of subsets. Specimen processing for the replication cohort was initiated after final analysis of the discovery cohort.
Genotyping
Genotyping was conducted with a combination of Illumina arrays (HumanHap300, and HumanHap610Q) (Richards et al., 2008; Soranzo et al., 2009). The Illuminus calling algorithm (Teo et al., 2007) was used to assign genotypes. No calls were assigned if an individual’s most likely genotype was called with less than a posterior probability threshold of 0.95. Validation of pooling was achieved via a visual inspection of 100 random, shared single-nucleotide polymorphisms (SNPs) for overt batch effects. Finally, intensity cluster plots of significant SNPs were visually inspected for over-dispersion, biased no calling, and/or erroneous genotype assignment. SNPs exhibiting any of these characteristics were discarded. Stringent quality control (QC) measures were performed on the genotypes prior to data analysis. The sample exclusion criteria were: (i) sample call rate <98%, (ii) heterozygosity across all SNPs ≥2 standard deviation (SD) from the sample mean; (iii) evidence of non-European ancestry as assessed by principle component analysis comparison with HapMap3 populations; (iv) observed pairwise identity by descent (IBD) probabilities suggestive of sample identity errors; (v) misclassified monozygotic and dizygotic twins were corrected based on IBD probabilities. The exclusion criteria for SNPs were: (i) Hardy-Weinberg equilibrium (HWE) P-value < 10−6, assessed in a set of unrelated samples; (ii) minor allele frequency (MAF) <1%, assessed in a set of unrelated samples; (iii) SNP call rate <97% (SNPs with MAF ≥5%) or <99% (for 1%≤ MAF <5%). Alleles of both datasets from the genotyping arrays were aligned to HapMap2 or HapMap3 forward strand alleles. Imputation was performed using the IMPUTE v2 software package (Howie et al., 2009). After imputation of the 2,986,407 SNPs available for analysis, 1,419,558 SNPs passed further QC (call rate ≥95%, MAF ≥0.05, HWE ≥10−4) and were used for analysis.
Statistical Analyses
Selection of subsets for heritability and GWAS analysis
Full details are in the Supplementary Methods. In brief, we eliminated CSFs with frequencies below 0.1% or above 99% (Figure S4A) From these, we selected approximately 200 for in-depth analysis based on heritability or description in the literature (“canonical”).
Genome wide association analysis
Because of relatedness in the TwinsUK cohort, we utilized the GenABEL software package (Aulchenko et al., 2007) which is designed for GWAS analysis of family-based data by incorporating pair-wise kinship matrix calculated using genotyping data in the polygenic model to correct relatedness and hidden population stratification. The score test implemented in the software was used to test the association between a given SNP and the trait. Additional quality control was conducted on the association results: minor allele frequency (MAF) >0.1, Hardy-Weinberg equilibrium <10−8, SNP call rate >90%. For our results, we used a genome wide significance threshold of p<3.3×10−10. This threshold represents a standard genome-wide significance threshold (p<5×10−8) further adjusted for 151 independent tests (the number of traits analysed for GWAS in the discovery cohort). Because there is a high intercorrelation amongst some of the 151 analysed traits, this is a very conservative approach and ensures the robustness of our findings. Genome-wide validations were also performed on normalised residuals (after corrections for age) with GenABEL taking into account family structure in the model. The validation P-value threshold was set to p<0.05.
Results from the genome-wide analyses of all the analyzed traits for both the discovery and the replication cohorts were meta-analysed. Fixed effects inverse-variance weighted meta-analyses were conducted using METAL (Willer et al., 2010), with significance evaluated at P < 5×10−8.
Finally, correlation against the FCRG “diplotype” shown in Figure 4 and S8 were done using JMP; ANOVA P-values without correction for family structure are reported.
Identifying traits correlated with SNPs
Once we had identified significant associations between the selected 151 traits and all SNPs based on standard analyses, we looked for pleiotropic effects of those SNPs. Approximately 1,200 SNPs from about 18 unlinked loci were correlated against all traits.
To correct for multiple comparisons, we evaluated the covariance among all traits. As shown in Figure S4B, there are less than 1,800 traits that show a covariance less than 0.7. Thus, Bonferroni correction sets a significance threshold of P < 2.8 × 10−5. In our analyses, which covered about 20 unlinked loci, we used a more conservative threshold, requiring a Wilcoxon rank association of trait values with SNP genotype to be significant if P < 10−7.
GWAS catalogue look-ups
In order to ascertain whether the variants from the associations of the immune cell subsets analyses overlapped with significant disease associations, the GWAS catalogue (http://www.genome.gov/gwastudies/), Immunobase (http://immunobase.org/page/Welcome/display), and SNPedia (www.snpedia.com) repositories were used. Gene regions and variants reaching genome-wide significance were searched on these repositories and overlapping associations or correlations with proxy SNPs or those in strong LD are reported in Table 2.
Supplementary Material
Highlights.
Resource of heritabilities and genetic associations of 80K immune traits in 669 twins
Genetic associations with immune cell frequencies and surface protein expression levels
Of the top 150 traits, 11 genetic loci explained up to 36% of variation of 19 traits
Loci include autoimmune susceptibility genes, providing etiological hypotheses
Acknowledgments
This work was supported by the Vaccine Research Center (NIAID, NIH) intramural research program, and by the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation (guysbrc-2012-1) Trust, and Dunhill Medical Trust. TwinsUK is also supported by the Wellcome Trust, and TDS is an ERC Advanced Researcher. We wish to thank Kaimei Song, Steve Perfetto, Richard Nguyen, Lynda Myles, Gabriela Surdulescu, Dylan Hodgkiss and Ayrun Nessa for technical help with the samples, as well as the TwinsUK volunteers for participation.
Footnotes
BioData Repository
Raw and summary data are available for downloading and analysis. Genotype data is available upon request to the authors. See Supplementary Methods for detailed downloading instructions.
Author Contributions. M.R., T.D.S., and F.O.N. designed and supervised the study; I.T., L.N., M.T.B., F.V. processed samples, coordinated by P.D.M.; M.H.B., Y.M., P.C., P.D.M. performed experimental work; M.R., L.Q., M.M., M.H.B. performed primary data analysis; M.R., L.Q., M.M., C.M. performed statistical analyses; M.R., L.Q., M.M., P.D.M., T.D.S., and F.O.N. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadi KR, Hall MA, Norman P, Vaughan RW, Snieder H, Spector TD, Lanchbury JS. Genetic determinism in the relationship between human CD4+ and CD8+ T lymphocyte populations? Genes Immun. 2001;2:381–387. doi: 10.1038/sj.gene.6363796. [DOI] [PubMed] [Google Scholar]
- Amadori A, Zamarchi R, De Silvestro G, Forza G, Cavatton G, Danieli GA, Clementi M, Chieco-Bianchi L. Genetic control of the CD4/CD8 T-cell ratio in humans. Nat Med. 1995;1:1279–1283. doi: 10.1038/nm1295-1279. [DOI] [PubMed] [Google Scholar]
- Antonioli L, Pacher P, Vizi ES, Hasko G. CD39 and CD73 in immunity and inflammation. Trends in molecular medicine. 2013;19:355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Bastid J, Cottalorda-Regairaz A, Alberici G, Bonnefoy N, Eliaou JF, Bensussan A. ENTPD1/CD39 is a promising therapeutic target in oncology. Oncogene. 2013;32:1743–1751. doi: 10.1038/onc.2012.269. [DOI] [PubMed] [Google Scholar]
- Casanova JL, Abel L. The human model: a genetic dissection of immunity to infection in natural conditions. Nat Rev Immunol. 2004;4:55–66. doi: 10.1038/nri1264. [DOI] [PubMed] [Google Scholar]
- Clementi M, Forabosco P, Amadori A, Zamarchi R, De Silvestro G, Di Gianantonio E, Chieco-Bianchi L, Tenconi R. CD4 and CD8 T lymphocyte inheritance. Evidence for major autosomal recessive genes. Hum Genet. 1999;105:337–342. doi: 10.1007/s004399900140. [DOI] [PubMed] [Google Scholar]
- Cotsapas C, Hafler DA. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends Immunol. 2013;34:22–26. doi: 10.1016/j.it.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JG, Cautain B, Bernard I, Mas M, van Breda Vriesman PJ, Druet P, Fournie G, Saoudi A. A dominant role for the thymus and MHC genes in determining the peripheral CD4/CD8 T cell ratio in the rat. J Immunol. 1999;163:2983–2989. [PubMed] [Google Scholar]
- Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen JF, Enjyoji K, Linden J, Oukka M, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Frazer IH, Martin NG. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- Ferreira MA, Mangino M, Brumme CJ, Zhao ZZ, Medland SE, Wright MJ, Nyholt DR, Gordon S, Campbell M, McEvoy BP, et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet. 2010;86:88–92. doi: 10.1016/j.ajhg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MA, Ahmadi KR, Norman P, Snieder H, MacGregor AJ, Vaughan RW, Spector TD, Lanchbury JS. Genetic influence on peripheral blood T lymphocyte levels. Genes Immun. 2000;1:423–427. doi: 10.1038/sj.gene.6363702. [DOI] [PubMed] [Google Scholar]
- Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, Martin P, Comeau ME, Sajuthi S, Andrews R, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet. 2013;45:664–669. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, Ozyazgan Y, Sacli FS, Erer B, Inoko H, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45:202–207. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraal G, Weissman IL, Butcher EC. Genetic control of T-cell subset representation in inbred mice. Immunogenetics. 1983;18:585–592. doi: 10.1007/BF00345966. [DOI] [PubMed] [Google Scholar]
- Kyvic K. Generalisability and assumptions of twin studies. In: Spector TD, Sneider H, MacGregor AJ, editors. Advances in twin and sib-pair analysis. London: Greenwich Medical Media; 2000. pp. 67–77. [Google Scholar]
- Lugli E, Dominguez MH, Gattinoni L, Chattopadhyay PK, Bolton DL, Song K, Klatt NR, Brenchley JM, Vaccari M, Gostick E, et al. Superior T memory stem cell persistence supports long-lived T cell memory. The Journal of clinical investigation. 2013;123:594–599. doi: 10.1172/JCI66327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke YD, Beddall MH, Roederer M. OMIP-013: Differentiation of human T-cells. Cytometry A. 2012;81:935–936. doi: 10.1002/cyto.a.22201. [DOI] [PubMed] [Google Scholar]
- Mahnke YD, Beddall MH, Roederer M. OMIP-015: Human regulatory and activated T-cells without intracellular staining. Cytometry A. 2013a;83:179–181. doi: 10.1002/cyto.a.22230. [DOI] [PubMed] [Google Scholar]
- Mahnke YD, Beddall MH, Roederer M. OMIP-017: human CD4(+) helper T-cell subsets including follicular helper cells. Cytometry A. 2013b;83:439–440. doi: 10.1002/cyto.a.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke YD, Beddall MH, Roederer M. OMIP-019: Quantification of human gammadeltaT-cells, iNKT-cells, and hematopoietic precursors. Cytometry A. 2013c;83:676–678. doi: 10.1002/cyto.a.22326. [DOI] [PubMed] [Google Scholar]
- Moayyeri A, Hammond CJ, Hart DJ, Spector TD. Effects of age on genetic influence on bone loss over 17 years in women: the Healthy Ageing Twin Study (HATS) J Bone Miner Res. 2012;27:2170–2178. doi: 10.1002/jbmr.1659. [DOI] [PubMed] [Google Scholar]
- Nalls MA, Couper DJ, Tanaka T, van Rooij FJ, Chen MH, Smith AV, Toniolo D, Zakai NA, Yang Q, Greinacher A, et al. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 2011;7:e1002113. doi: 10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M, Cardon L. Methodology for Genetic Studies of Twins and Families. Dordrecht: Kluwer Academic Publishers; 1992. [Google Scholar]
- Okada Y, Hirota T, Kamatani Y, Takahashi A, Ohmiya H, Kumasaka N, Higasa K, Yamaguchi-Kabata Y, Hosono N, Nalls MA, et al. Identification of nine novel loci associated with white blood cell subtypes in a Japanese population. PLoS Genet. 2011;7:e1002067. doi: 10.1371/journal.pgen.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orru V, Steri M, Sole G, Sidore C, Virdis F, Dei M, Lai S, Zoledziewska M, Busonero F, Mulas A, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, Feng T, Lee M, Asinovski N, Frohlich I, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, Hammond N, Stolk L, Nica A, Inouye M, Hofman A, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starbeck-Miller GR, Badovinac VP, Barber DL, Harty JT. Cutting edge: Expression of FcgammaRIIB tempers memory CD8 T cell function in vivo. J Immunol. 2014;192:35–39. doi: 10.4049/jimmunol.1302232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Inouye M, Small KS, Gwilliam R, Deloukas P, Kwiatkowski DP, Clark TG. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–2746. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J, Slagboom PE, Draisma HH, Martin NG, Boomsma DI. The continuing value of twin studies in the omics era. Nat Rev Genet. 2012;13:640–653. doi: 10.1038/nrg3243. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.