Abstract
Islet-1 (Isl1) is a LIM-homeodomain (LIM-HD) transcription factor that functions in a combinatorial manner with other LIM-HD proteins to direct the differentiation of distinct cell types within the central nervous system and many other tissues. A study of pancreatic cell lines showed that Isl1 is alternatively spliced generating a second isoform, Isl1β, which is missing 23 amino acids within the C-terminal region. This study examines the expression of the canonical and alternative Isl1 transcripts across other tissues, in particular, within the retina, where Isl1 is required for the differentiation of multiple neuronal cell types. The alternative splicing of Isl1 is shown to occur in multiple tissues, but the relative abundance of Isl1α and Isl1 β expression varies greatly across them. In most tissues, Isl1α is the more abundant transcript, but in others the transcripts are expressed equally, or the alternative splice variant is dominant. Within the retina, differential expression of the two Isl1 transcripts increases as a function of development, with dynamic changes in expression peaking at E16.5 and again at P10. At the cellular level, individual retinal ganglion cells vary in their expression, with a subset of small-to-medium sized cells expressing only the alternative isoform. The functional significance of the difference in protein sequence between the two Isl1 isoforms was also assessed using a luciferase assay, demonstrating that the alternative isoform forms a less effective transcriptional complex for activating gene expression. These results demonstrate the differential presence of the canonical and alternative isoforms of Isl1 amongst retinal ganglion cell classes. As Isl1 participates in the differentiation of multiple cell types within the CNS, the present results support a role for alternative splicing in the establishment of cellular diversity in the developing nervous system.
Keywords: splice variant, isoform, gene expression, retinal ganglion cell, differentiation
Introduction
The diversity of neuronal cell types in the brain is achieved through the developmental expression of transcription factor families that control fate specification and differentiation. LIM-homeodomain (LIM-HD) proteins are a subfamily of homeodomain-containing transcription factors involved in the differentiation of distinct cell types within many tissues, including the central nervous system (Gill, 2003; Hunter and Rhodes, 2005). There are twelve mammalian LIM-HD proteins that can be grouped into six pairs of paralogues: Isl1/Isl2, Lhx1/Lhx5, Lhx2/Lhx9, Lhx3/Lhx4, Lhx6/Lhx8, and Lmx1a/Lmx1b (Failli et al., 2000). Each LIM-HD protein has a centrally located homeodomain that binds to DNA, and while most homeodomain proteins do not contain protein-binding domains, LIM-HDs have two specialized zinc finger LIM domains located towards the N-terminus. These LIM domains interact with co-factors that facilitate the formation of transcriptional complexes. The primary interacting co-factor with LIM-HDs is LIM-domain binding 1 (Ldb1) protein, which dimerizes through a self-assembly (SA) domain, and binds to LIM-containing proteins through a LIM-interacting domain (LID), forming homo- or heteromeric protein complexes (Bach, 2000; Hobert and Westphal, 2000; Jurata et al., 1998). Cell type-specific expression of various LIM-HD complexes, each with a unique set of DNA targets, drives cells to differentiate into distinct types. This LIM-combinatorial code is a mechanism that contributes to the development of cell type diversity, particularly within the central nervous system (Flandin et al., 2011; Fragkouli et al., 2009; Inoue et al., 2013; Thaler et al., 2002).
Alternative splicing, by which multiple protein products are generated from a single gene, is a potential means for increasing the number of functionally distinct proteins from a genome. For instance, while the genomes of humans and nematodes do not differ conspicuously in terms of their number of genes, the proportion of those genes that undergo alternative splicing increases from 20 to 80 percent, respectively. Additionally, the level of alternative splicing is an effective predictor of organism complexity, as measured by the number of cell types (Chen et al., 2014). Consistent with this, the human brain has the greatest number of alternatively spliced genes compared to other tissues (Lee and Irizarry, 2003; Yeo et al., 2004).
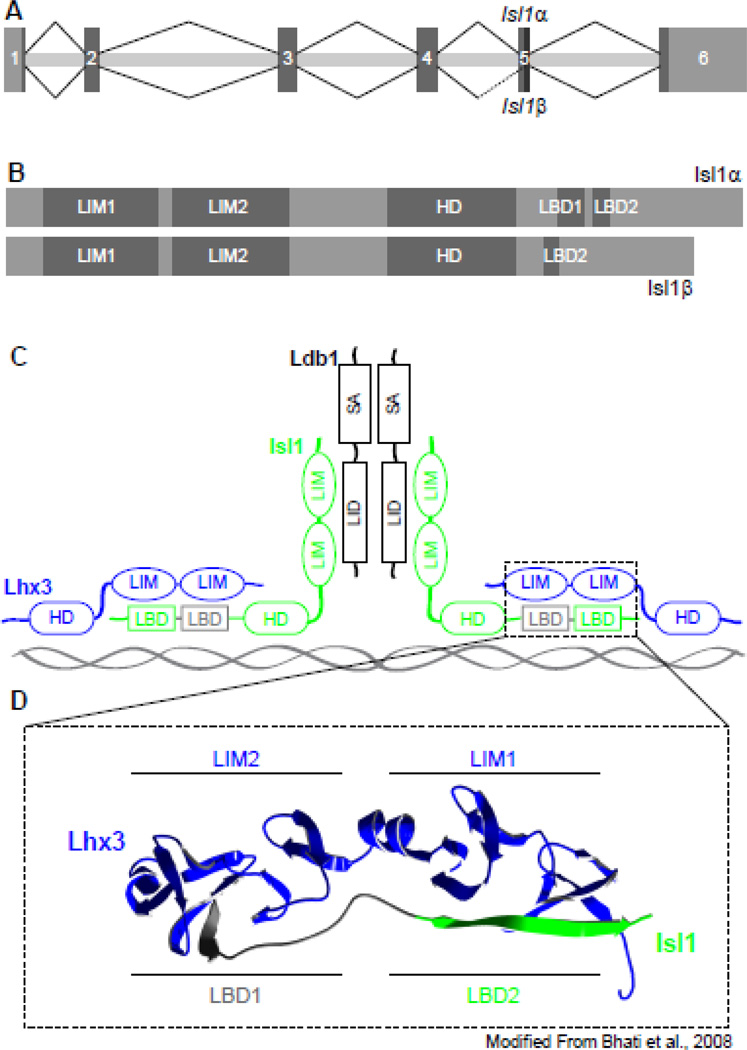
Alternative splicing events have been described for several LIM-HD genes, including Isl1, Lhx3, Lhx6, Lhx7, and Lhx9 (Ando et al., 2003; Failli et al., 2000; Grigoriou et al., 1998; Kimura et al., 1999; Sloop et al., 2001). The alternative splicing of Isl1 was identified in different pancreatic endocrine cell lines. Through the use of an alternative splice acceptor site halfway through the fifth exon, an alternative isoform, Isl1β, is generated lacking 23 amino acids towards the C-terminus relative to the canonical isoform Isl1α (Figure 1A, 1B). Interestingly, both the canonical and alternative transcripts are present in a pancreatic β cell line while only the canonical isoform is expressed in a pancreatic α cell line. This differential pattern of Isl1 splicing across these pancreatic cell lines suggests distinct roles for each isoform in the development or function of these cell types (Ando et al., 2003). The LIM and HD domains are highly conserved across all LIM-HD family members, however, the C-terminal region of these proteins is diverse and, in general, not well characterized; as such, the domains and function of the C-terminal region of Isl1 corresponding to the portion absent in Isl1β had not been described when the alternative isoform was originally identified. This region of Isl1 has since been more fully characterized, independent of an examination of the alternative isoform, and contains two protein-binding domains that are separated by a short linker sequence. These two domains bind to the two LIM domains (LIM1 and LIM2) of LIM homeobox protein 3 (Lhx3), and are accordingly named Lhx3-binding domains 1 and 2 (LBD1 and LBD2) (Bhati et al., 2008). The amino acids absent in Isl1β (aa256–278) correspond to LBD1 (aa262–273) and the short linker sequence (aa274–278) that separates it from LBD2 (Figure 1B, 1C). Scanning alanine mutagenesis of this region revealed that LBD2 is dispensable for the binding of Isl1 to Lhx3, but LBD1 is critical for this interaction (Bhati et al., 2008). The absence of LBD1 in Isl1β strongly suggests there may be functional differences between the two isoforms of Isl1, such as the transcriptional complexes in which they participate and in their subsequent gene targets.
Figure 1. Isl1β isoform lacks a critical LIM-binding domain.
(A) Isl1 undergoes alternative splicing through the use of an alternative 3’ splice acceptor site mid-way through the 5th exon. (B) The alternative isoform, Isl1β, is lacking 23 amino acids relative to the canonical isoform, Isl1α, which correspond to an Lhx3-binding domain (LBD1). (C) The interaction between the LBDs of Isl1 (green) and the LIM domains of Lhx3 (blue) are modeled highlighting the amino acids absent in Isl1β (grey) (Bhati et al., 2008).
Isl1 is expressed in multiple cell types within the retina, including bipolar cells, cholinergic amacrine cells, and most retinal ganglion cells (RGCs) (Elshatory et al., 2007a). When Isl1 is conditionally knocked-out of the embryonic retina, these cell types are all born, but subsequently die or possibly switch fates, indicating the importance of Isl1 in maintaining their differentiated state (Elshatory et al, 2007b; Mu et al., 2008; Pan et al., 2008). The presence of both a canonical Isl1α isoform and its alternatively spliced Isl1β isoform, forming distinct transcriptional complexes with other LIM-HDs across these retinal cell types, may therefore contribute to the unique differentiation patterns yielding the diversity of neuronal types.
Results
Expression of Isl1α and Isl1β varies across tissue types
The pattern of Isl1 alternative splicing was assessed across multiple adult tissue types by RT-PCR using one primer pair capable of amplifying fragments from both transcripts (Figure 2A). Isl1α is expressed in greater abundance relative to Isl1β for all central nervous system samples: spinal cord, brain, and retina. A similar pattern of expression was also seen for samples from the thymus, adrenal gland, and testis, with Isl1α being the predominant transcript. In the kidney, however, the expression levels are closer to parity. Uniquely, Isl1α was barely detectable in the lung, but Isl1β was robustly expressed. Samples from the liver and spleen were also examined, but they did not show any Isl1α or Isl1β expression. Clearly, tissue types differentially regulate the splicing of Isl1.
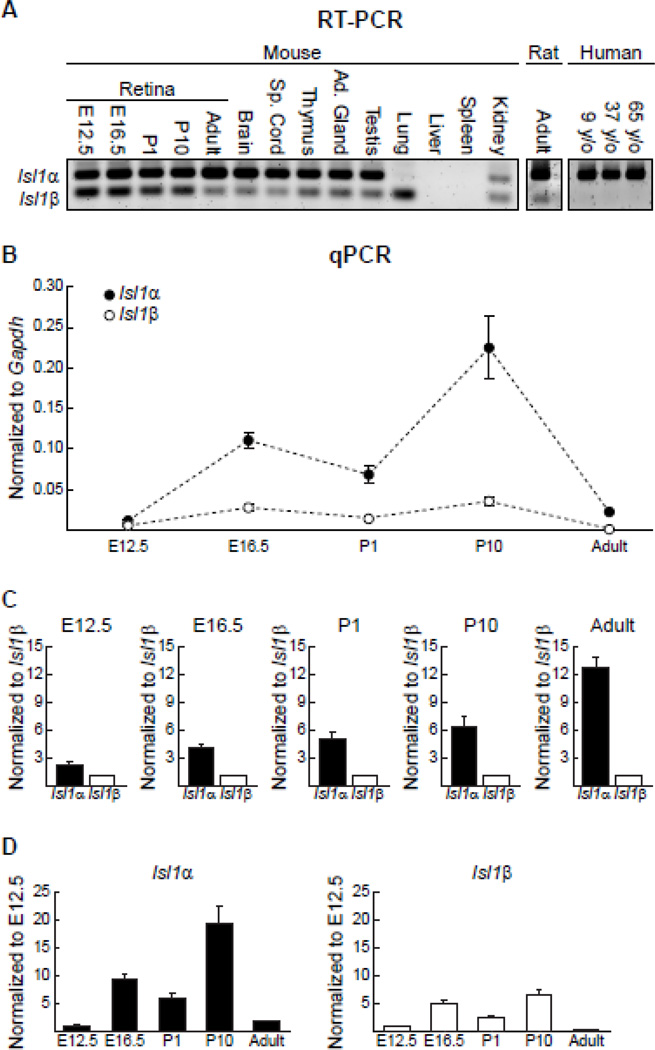
Figure 2. Isl1 alternative splicing is tissue-specific and differentially regulated across development.
(A) RT-PCR with one primer pair capable of amplifying both Isl1 transcripts shows varied abundance of each across multiple adult tissue types and across developmental stages within the retina. The alternative splicing of Isl1 was also detectable in the adult rat retina, but not in human retinal tissue (y/o = years old). (B) Unique primers were used for quantitative analysis of Isl1α and Isl1β expression in the developing and mature mouse retina. Expression levels are shown normalized to the housekeeping gene Gapdh. n = 4 biological replicates for each age, with means and standard errors indicated. (C) Expression levels are shown here normalized to Isl1β at each time point to illustrate the relative abundance of each transcript. Note the progressive difference in relative expression as a function of development. (D) To illustrate temporal changes for each transcript alone, expression levels for each time-point are shown normalized to E12.5. Both Isl1α and Isl1β have peaks in expression levels at E16.5 and P10.
The temporal pattern of Isl1 alternative splicing in the developing retina was determined using samples from multiple developmental time-points, including embryonic day (E) 12.5, E16.5, postnatal day (P) 1, and P10. Much like the adult retina, both transcripts were expressed at each developmental stage examined, with Isl1a being expressed at greater levels relative to Isl1β (Figure 2A). Isl1 mRNA is 97% homologous between mouse and rat, and RT-PCR with a similarly designed primer pair reveals the expression of both variants in the adult rat retina. While there is 100% homology of the Isl1 protein sequence between mice and humans, the homology of the mRNA sequence is slightly lower at 87%. Curiously, Isl1β is undetectable in human retinal tissue from multiple postnatal ages (Figure 2A).
Expression is dynamically and differentially regulated during retinal development
Real time RT-PCR (qPCR) using primers specific for each splice variant allows for a quantitative analysis of Isl1α and Isl1β expression in the developing and mature mouse retina. Expression was quantified at multiple ages, including E12.5, E16.5, P1, P10, and adult, revealing dynamic changes in expression levels across development and into maturity. Expression levels were normalized to the housekeeping gene Gapdh, showing conspicuous differences between the two transcripts across development (Figure 2B). To appreciate how their relative expression varies across time, they have also been normalized by the mean of Isl1β expression at each age (Figure 2C), to demonstrate the progressive enhancement of the difference in expression between Isl1α and Isl1β as a function of development. Those plots of course conceal the time-dependent changes of each mRNA relative to Gapdh, shown in Figure 2B. Expression levels for the two have also been plotted separately, now normalized by the mean expression at E12.5 for each transcript (Figure 2D), mimicking the trends shown in Figure 2B, in which both Isl1α and Isl1β exhibit temporal changes in expression, with coinciding peaks at E16.5 and at P10.
A Student’s t-test was used to identify significant differences between the canonical transcript and the alternative splice variant at each time-point, using expression levels normalized to Gapdh. A Bonferroni correction was used for multiple t-testing (p < 0.01), yielding significant differences at all ages except E12.5. To assess the change in abundance of each mRNA across time, expression levels (normalized to Gapdh) were analyzed using one-way ANOVAs for each, employing post-hoc Tukey tests (p < 0.05) to identify significant differences between the ages. Expression of Isl1α was significantly higher at P10 than at any other age, while expression at E16.5 was significantly greater than at E12.5 and in maturity. Isl1β expression was significantly greater at both E16.5 and P10 than at all three other ages, with P1 being greater than adult as the only other significant difference present. Isl1α and Isl1β, therefore, exhibit dynamic variation in their expression during the period of retinal development, with progressively greater differential expression between the canonical and alternative splice variant as a function of development.
Isl1-Pan and Isl1-Alpha antibodies can be used to discriminate the two isoforms of Isl1
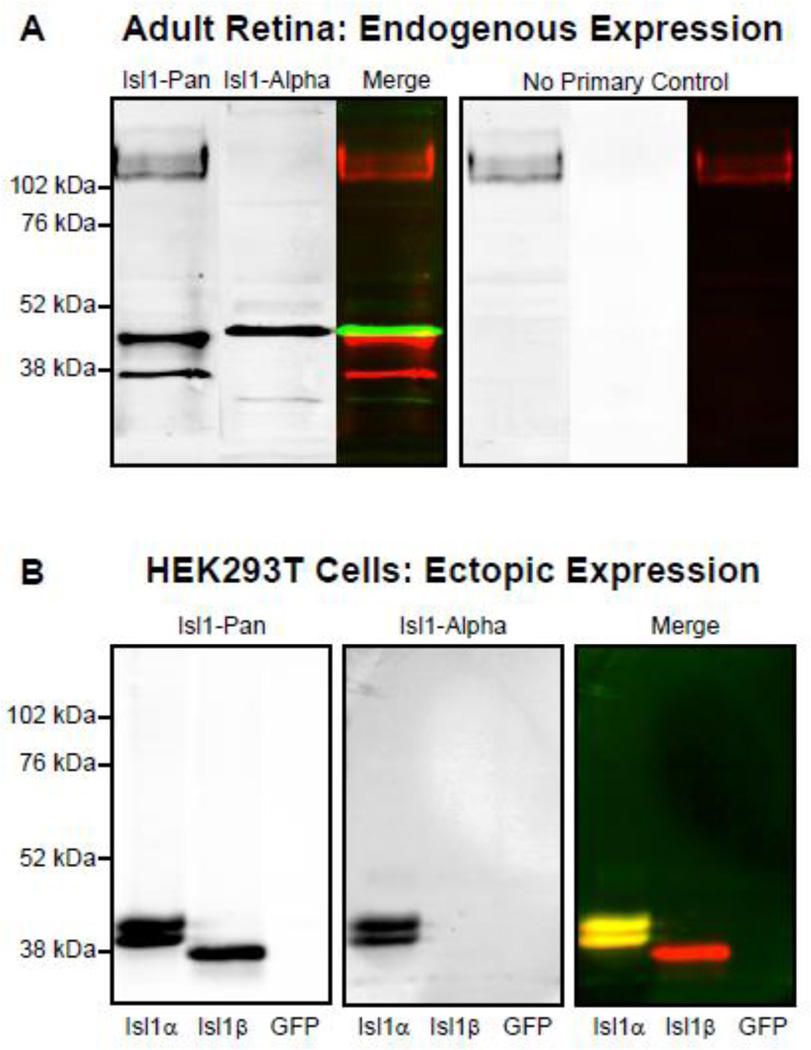
A commercially available antibody to Isl1 (39.4D5, Developmental Studies Hybridoma Bank) (Ericson et al., 1992) is capable of binding to both the Isl1α and Isl1β isoforms (Ando et al., 2003), hereafter referred to as Isl1-Pan. Western blotting with Isl1-Pan using protein lysates from adult whole retina confirms the presence of Isl1β at the protein level in the mouse retina with bands 43.75 kDA and 39.06 kDA in size corresponding to the alpha and beta isoforms, respectively (Figure 3A). To confirm the ability of this antibody (Isl1-Pan) to bind to both Isl1α and Isl1β, HEK293T cells were transiently transfected with plasmids expressing either Isl1α or Isl1β, and protein was collected from cell lysates. This particular cell line was chosen because of the absence of endogenous Isl1 expression, which has been previously shown in experiments that tested the functionality of various DNA recognition elements when Isl1 was expressed in combination with other LIM-HD transcription factors (Lee et al., 2008). Western blotting of the Isl1α-expressing sample with Isl1-Pan reveals two closely positioned bands at 42.06 and 40.59 kDa, while a single, smaller band 39.36 kDa in size is detected for the Isl1β-expressing sample (Figure 3B). The doublet seen for the Isl1α-expressing cells is consistent with results from the original paper that described the alternative splicing of Isl1 (Ando et al., 2003). That study demonstrated that the higher molecular weight band of this pair reflects phosphorylated Isl1α. Specifically, they conducted a similar experiment, expressing FLAG-tagged Isl1α or Isl1β in HIH3T3 cells and immunoblotting with the same Isl1-Pan antibody, revealing this doublet for Isl1α and a single smaller band for Isl1β. Isl1 is predicted to be phosphorylated at several sites within the 23 amino acids absent in the beta isoform, and the higher molecular weight band of the doublet was indeed abolished by treatment with calf intestinal alkaline phosphatase (Ando et al., 2003; Jurata and Gill, 1997). The lack of a doublet and the slightly larger molecular weight of Isl1α for retinal samples would suggest differences in the phosphorylation and other post-transcriptional modifications of Isl1 between the HEK293T cells and the retina.
Figure 3. Immunoblotting confirms the presence of each isoform in retinal tissue and in transfected HEK cells.
(A) Immunoblotting of adult retinal protein extracts with an Isl1-Pan antibody reveals two bands roughly corresponding to the predicted molecular weights of Isl1α and Isl1β. Immunoblotting with an Isl1-Alpha antibody overlaps, although not completely, with the Isl1-Pan alpha band. (B) The specificity of Isl1-Pan and Isl1-Alpha antibodies was confirmed with immunoblotting of protein extracts from HEK293T cells expressing either Isl1α, Isl1β, or GFP controls. The Isl1-Pan antibody labels a doublet for the Isl1α-expressing cells corresponding to a phosphorylated and un-phosphorylated version, and a single smaller band for the Isl1β-expressing cells. The Isl1-Alpha antibody does not label anything from the Isl1β-expressing cells, only the alpha doublet that Isl1-Pan labels.
Isoform-specific antibodies can be used to visualize the in vivo expression pattern of the Isl1 isoforms in the retina. Two attempts to generate Isl1β-specific antibodies proved unsuccessful, with the antibodies recognizing both Isl1 isoforms, likely due to partial binding to the small portion of the antigen present in both isoforms. A custom peptide antibody to Isl1α, however, was successfully made using a subset of the 23 amino acids unique to the alpha isoform. This Isl1α antibody, Isl1-Alpha, labels a single band that overlaps, though not completely, with the Isl1α band labeled by Isl1-Pan for retinal protein extracts (Figure 3A). The specificity of Isl1-Alpha was confirmed using protein extracts from Isl1α- and Isl1β-expressing HEK293T cells. Immunoblotting of the protein extracts derived from Isl1α-expressing cells shows a complete co-localization with the Isl1-Pan labeled doublet, while protein extracts from Isl1β-expressing cells showed no labeling by Isl1-Alpha (Figure 3B).
Immunolabeling of adult tissue from liver and adrenal gland with Isl1-Pan and Isl1-Alpha confirmed the in vivo specificity of these antibodies with no labeling seen in liver (Supplementary Figure 1 A) and labeling by both in adrenal gland (Supplementary Figure 1B), following the pattern of isoform expression shown above by RT-PCR. As further corroboration of antibody specificity, we show a loss of labeling by both antibodies in the GCL of an Isl1 conditional knockout (CKO) retina compared to wildtype control (Supplemental Figure 1C).
Some retinal ganglion cells express only the Isl1β isoform
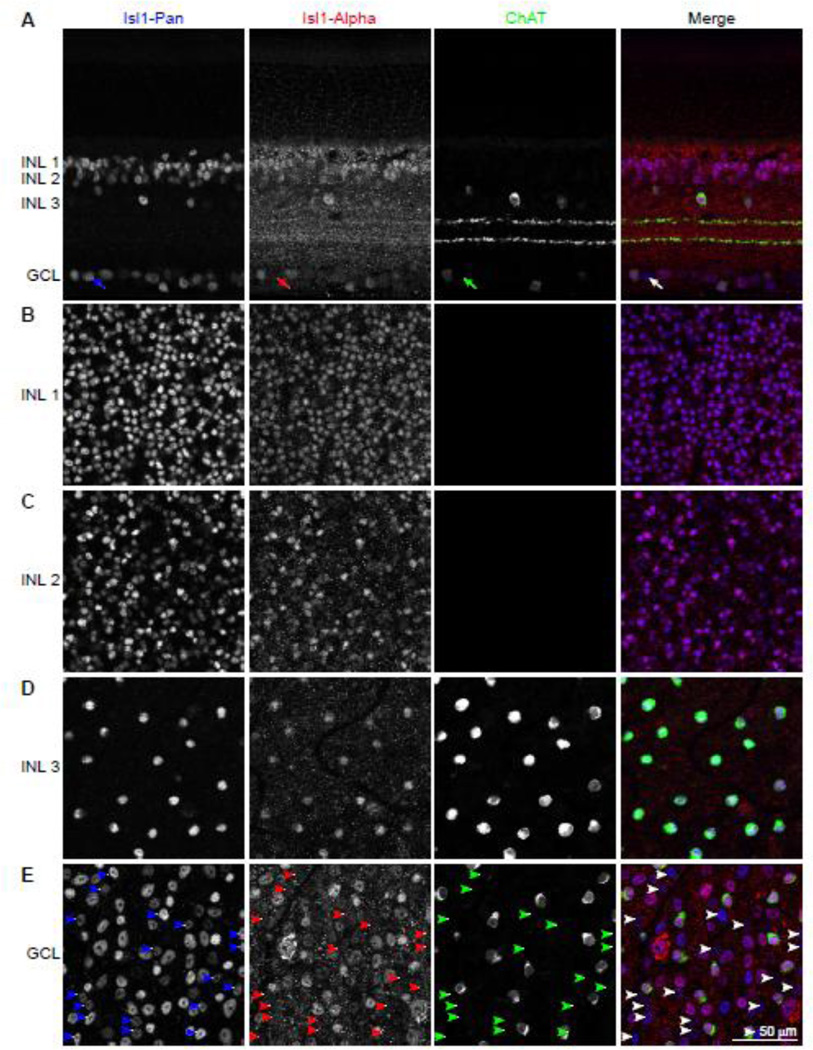
Using the Isl1-Pan and Isl1-Alpha antibodies to determine the pattern of Isl1 isoform expression within different retinal cell types has some limitations. With this combination of antibodies, cells expressing only Isl1α cannot be distinguished from cells expressing both Isl1α and Isl1β isoforms. Cells that express only Isl1β, however, can be identified, as these cells will be labeled with Isl1-Pan, but not Isl1-Alpha. In the adult retina, Isl1 is known to be expressed in ON-bipolar cells, cholinergic amacrine cells, and most retinal ganglion cells (RGCs) (Elshatory et al., 2007a). Isl1-Alpha co-localizes with most Isl1-Pan positive cells in immunolabeled retinal sections, although the speckled appearance of the Isl1-Alpha labeling makes this observation somewhat difficult (Figure 4A). A clearer visualization of Isl1-Pan and Isl1-Alpha immunolabeling can be achieved by examining retinal wholemounts, imaging each cellular layer en face where Isl1 - expressing cells are positioned. The Isl1 -positive cells in the inner nuclear layer (INL) closest to the outer plexiform layer are primarily rod bipolar cells, and all labeled cells at this depth within the retina are immunopositive for both Isl1-Pan and Isl1-Alpha (Figure 4B). Further into the INL, where cone bipolar cells are positioned, the coincidence of labeling with these two antibodies remains complete (Figure 4C). At the innermost portion of the INL, the density of labeled cells drops conspicuously, and only a single population of cells, the cholinergic amacrine cells (labeled with antibodies to choline acetyltransferase (ChAT)), are labeled with both Isl1-Pan and Isl1-Alpha antibodies (Figure 4D). The cholinergic amacrine cells that are displaced into the ganglion cell layer (GCL) are also co-labeled with Isl1-Pan and Isl1-Alpha (Figure 4E), but there are many more cells that are also Isl1-positive. While there are other amacrine cell types present in the GCL, the remaining Isl1-positive cells within the GCL are all RGCs, as cholinergic amacrine cells are the only Isl1-expressing amacrine cell subtype (Elshatory et al., 2007a). Within the population of Isl1-Pan positive RGCs, there is now present a subset of these cells that is not labeled with Isl1-Alpha, revealing a population of RGCs that only expresses Isl1β (Figure 4E, white arrowheads in merged panel; Figure 4A, white arrow). Quantification of Isl1-Pan labeled cells from eight sample fields across the GCL showed that the majority, ~74%, are also labeled with Isl1-Alpha, with the remaining ~26% being comprised of the Isl1β-only RGCs.
Figure 4. Immunostaining demonstrates a discrete population of RGCs positive for only the alternative isoform.
(A) Retinal cross-sections immunolabeled with Isl1-Pan and Isl1-Alpha show prominent co-labeling, including bipolar cells and cholinergic amacrine cells in the INL, and ganglion cells and displaced cholinergic amacrine cells in the GCL. The cholinergic amacrine cells are evidenced by their being labeled with antibodies to choline acetyltransferase (ChAT). Note the presence of a single RGC that is not labeled with the Isl1-Alpha antibody (white arrow in the merged panel). (B–E) Retinal wholemounts immunolabeled with the same antibodies and imaged at progressive depths through the INL and in the GCL. All Isl1-Pan positive cells in the INL (B–D) are also Isl1-Apha positive. Only the GCL (E) shows a subset of cells that are Isl1-Pan positive but Isl1-Alpha negative (arrowheads). These are Isl1β-only RGCs. Note that the displaced cholinergic amacrine cells, like those in the INL, are not included in the Isl1β-only population in the GCL. Calibration bar = 50 µm.
Isl1β-only cells are confined to a subset of RGC types
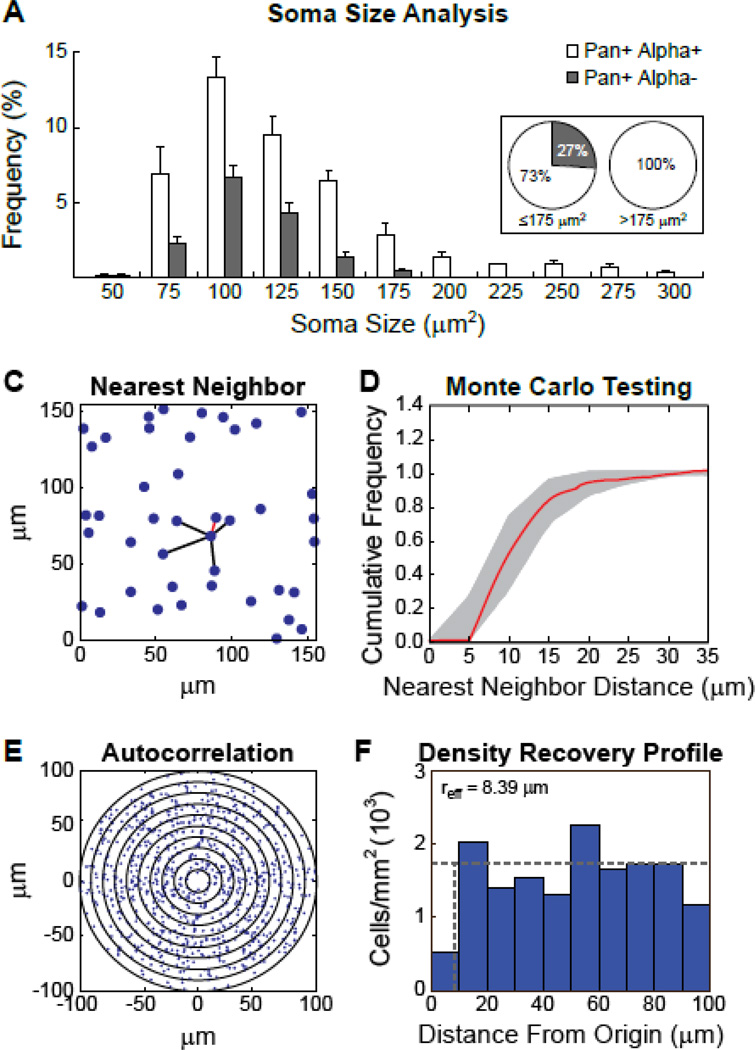
Such a large proportion of RGCs expressing only the Isl1β isoform would suggest that these cells do not comprise only a single type of RGC. Over 20 different RGC subtypes have been characterized in the mouse retina based on multiple morphological features, including dendritic area, dendritic branching, dendritic overlap, stratification depth within the IPL, and soma size (Badea and Nathans, 2004; Coombs et al., 2006; Kong et al., 2005; Sumbul et al., 2014). Indeed, soma size alone can be used to discriminate some RGC types, and particularly distinguishable are the group of large alpha RGC types (Coombs et al., 2006). Measures of soma size for the Isl1-Pan positive cells in the GCL were used to determine if the Isl1β-only cells can be found across the entire spectrum of soma sizes, or if they are restricted to a limited range, suggestive of subtype-specific expression. First, nuclei positive for Isl1-Pan in sample fields within the GCL were visually identified and outlined, these outlines were then overlaid onto the Isl1-Alpha labeling of the same fields and the two distinguishable populations of cells were identified, Pan-positive/Alpha-positive and Pan-positive/Alpha-negative, the latter being the Isl1β-only RGCs described above. To determine the range of somal areas for these two populations the fluorescent Nissl stain NeuroTrace 530/615 was used for the visualization and outlining of their cell bodies, from which somal areas were calculated. The soma sizes of 1,781 cells were sampled from eight fields within the GCL and the 1,040 Isl1-Pan positive cells identified span a range from 50 to 300 µm2 (Figure 5A). The soma sizes of the Isl1β-only RGCs, however, do not encompass this entire range, being restricted to RGCs with small and medium somas under 175µm2 (Figure 5A, grey bars), while only the Pan-positive/Alpha-positive population extends to include the larger sizes (Figure 5A, white bars). Indeed, while on average 27% of the cells less than 175 µm2 were Isl1β-only RGCs across the eight sampled fields used in this analysis, not a single cell greater than 175 µm2 was found to be Pan-positive but Alpha-negative in any of the eight sampled fields (Figure 5A insert). This soma size analysis therefore reveals a restricted expression pattern, indicating that Isl1β-only RGCs do not include the largest RGC types. Isl1β-only RGCs should therefore comprise multiple sub-types, all within small to medium soma sizes.
Figure 5. Isl1β-only cells include more than a single type of small-to-medium sized RGCs.
(A) Soma size was measured for 1,040 Isl1-Pan positive cells from four central and four peripheral fields in the GCL of one retina. The range of soma sizes for Isl1-Pan and Isl1-Alpha positive (white) versus the Isl1-Pan positive and Isl1-Alpha negative (grey) cells (i.e. the Isl1β-only RGCs) are shown as a percentage of the total population of Isl1-Pan positive cells per field, with means and standard errors indicated. These percentages are also represented in two pie charts for cells with soma sizes ≤175µm2 or >175µm2(B–E) The Isl1β-only cells exhibit a random spatial distribution consistent with multiple RGC cell types being included. (B) A sample field, in which a single cell and its five near neighbor distances are shown, the nearest neighbor distance highlighted in red. (C) The cumulative frequency histogram for the collection of nearest neighbor distances from this same field is illustrated (red trace), along with the envelope of 99 random simulations (grey region) matched in density and constrained by soma size. (D) The 2D spatial autocorrelogram derived from this same field of cells. Successive annuli plot the presence of cells at 10 µm intervals from the origin of the correlogram. (E) The density recovery profile derived from this same autocorrelogram, indicating the recovered average density as a function of distance from every cell within the field (i.e. the origin of the correlogram). The effective radius for this field (8.39 µm) approximates the effect of soma size, the average for this field being 9.21 µm.
Further confirmation that this population of Isl1β-only RGCs is not a single type of RGC comes from a consideration of their spatial statistical properties. Each type of RGC is believed to form its own independent retinal “mosaic”, within which neighboring cells of the same type space themselves apart to form regular arrays across the retina (Reese and Keeley, 2014). Spatial analysis of the Isl1β-only RGCs demonstrates them to be positioned randomly, rather than exhibiting regularity in their distribution. An analysis of their nearest neighbor distances reveals no difference in cell spacing when compared to simulations of 99 random distributions matched in density and constrained by soma size (Figure 5B). Six of the seven fields analyzed show cumulative frequency distributions that fall entirely within the 99 random simulations (e.g. Figure 5C), while the seventh showed a slight deviation from the envelope of simulations, indicating a tendency to clustering (rather than spacing). The autocorrelograms derived from such fields of cells (e.g. Figure 5D), like their density recovery profiles (Figure 5E), reveal an effective radius no larger than that for soma size alone, and Monte Carlo testing confirmed the density recovery profiles of six of the seven fields were well fit by those derived from random simulations of cells (not shown). Together, these two analyses indicate that the Isl1β-only population of RGCs is not a single type of self-spacing RGCs, even one that is undersampled (Cook, 1996). Rather, these spatial results would be expected if multiple types of RGC were included in the Isl1β-only population (Cook and Podugolnikova, 2001).
Transcriptional activation by the two isoforms differs
The formation of LIM-HD complexes has been well studied in two adjacent neuronal populations of the developing spinal cord, the V2 interneurons and motor neurons. Within nascent V2 interneurons, the Ldb1 co-factor forms a homodimer to which Lhx3 proteins bind, forming a symmetrical-multimeric complex. This complex is also formed in developing motor neurons, but with the additional binding of Isl1 to each Lhx3 protein (Thaler et al., 2002). The unique DNA recognition elements targeted by these two complexes (Lhx3:Ldb1 and Isl1:Lhx3:Ldb1) have been determined (Lee et al., 2008). We consequently assessed the potential effect on Isl1β function due to the absence of a critical protein-binding domain, LBD1, by comparing the transcriptional activation by Isl1:Lhx3:Ldb1 complexes containing either Isl1α or Isl1β, using a luciferase reporter with multiple copies of the hexameric Isl1:Lhx3:Ldb1 -specific DNA recognition element (Hx:RE) in the promoter region (Lee et al., 2008). Isl1α- or Isl1β-expressing plasmids were transiently transfected into HEK293T cells along with Lhx3-expressing plasmids, and the Isl1:Lhx3:Ldb1 luciferase reporter plasmid. Ldb1 is endogenously expressed in these cells so transfection of Ldb1 -expressing plasmids was not necessary (Lee et al., 2008).
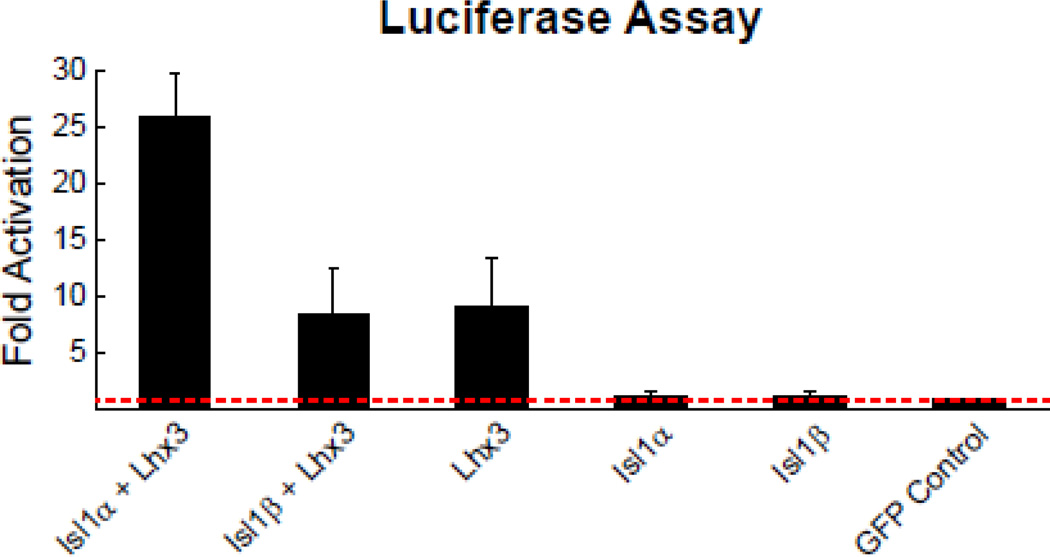
The activation of luciferase by the Isl1α:Lhx3:Ldb1 complex showed a 26-fold activation of luciferase over the GFP-control while the Isl1β:Lhx3:Ldb1 complex only achieved an 8-fold activation over the control (Figure 6). Luciferase activation by the Isl1β:Lhx3:Ldb1 complex was in fact no different from that achieved in the absence of Isl1 from the complex. For cells expressing Isl1α or Isl1β in the presence of the endogenous Ldb1 only (that is, without Lhx3), luciferase activation was no different than in the control condition expressing only GFP. A one-way ANOVA and post-hoc Tukey tests showed an effect of treatment, with the Isl1α:Lhx3:Ldb1 complex having significantly greater expression than all other conditions (p < 0.05), but with no significant differences between the other conditions being detected. These data demonstrate a functional difference between the two isoforms of Isl1, and furthermore suggest that the β isoform might not be capable of forming a complex with Lhx3 and Ldb1.
Figure 6. Luciferase assay demonstrates reduced efficiency of the alternative isoform in forming a known transcriptional complex.
Transcriptional activation by Isl1 isoforms was assessed using an Isl1α:Lhx3:Ldb1-specific luciferase reporter (Hx:RE). The cofactor Ldb1 is present in all conditions, due to endogenous expression by HEK293T cells. Isl1α:Lhx3:Ldb1 has a ~26-fold increase over the GFP-control in the ability to activate transcription of the Hx:RE luciferase reporter, while Isl1β:Lhx3:Ldb1 has only an ~8-fold increase in activation. Luciferase activation by Isl1α:Lhx3:Ldb1 is significantly different from all other conditions, while none of the other conditions were significantly different from each other. All results are normalized to control cells transfected with only GFP. Results are the mean of three independent experimental replications, with each experiment being conducted in triplicate.
Discussion
The alternative splicing of the LIM-HD transcription factor Isl1, originally identified in pancreatic cell lines, results in an in-frame deletion of 69 nucleotides due to the use of an alternative splice acceptor site within the fifth exon (Ando et al., 2003). This translates into the absence of 23 amino acids within the C-terminal region of Isl1 where a known protein-binding domain is located (Bhati et al., 2008). An analysis of multiple tissue types from adult mice revealed varying expression patterns of the canonical, Isl1α, and alternative, Isl1β, transcripts. These differences in expression patterns suggest that Isl1 alternative splicing is not simply due to the stochastic use of a less efficient splice acceptor site; rather, it is regulated by splicing factors that can enhance or suppress splicing events. Interestingly, while the alternative splicing of Isl1 is conserved between mice and rats, it does not appear to be conserved between mice and humans, despite the 100% homology of the protein sequences. It is unclear how conserved alternative splicing events are between mice and humans. A study comparing the orthologous transcripts that have been annotated between mice and humans found that 87% of the mouse splicing variants had an orthologous human splicing variant (Zambelli et al., 2010). The extent to which mouse and human splicing is conserved, however, is still unclear as not all variant transcripts have been identified. The apparent absence of Isl1β in human does not detract from the importance of studying this isoform or other non-conserved isoforms. Such events can be used to gain insight into the ways in which molecular mechanisms can be affected by alternative splicing and thereby enhance neuronal diversity.
Quantitative analysis of the changes in Isl1α and Isl1β expression across retinal development revealed two peaks in abundance. These peaks correspond to time-points when the major Isl1-expressing populations are undergoing differentiation, the first including the amacrine cells and RGCs around E16.5, followed by bipolar cells around P10. The comparable developmental change in expression suggests both isoforms are playing a role in differentiation of the major Isl1-positive populations. This is in contrast to the few examples of alternative splicing that have been shown to play a role in the development of the retina, in which the variant transcripts have more distinct temporal expression differences (Boije et al., 2013; Katyal and Godbout, 2004; Lakk et al., 2012; Liu et al., 2013).
The identification of alternatively spliced genes in the retina (Farkas et al., 2013; Gamsiz et al., 2012; Wan et al., 2011), like other regions of the CNS (Zaghlool et al., 2014), has not been difficult; showing their distribution at the cellular level, rather than the tissue level, however, has been limited within the nervous system and only recently described (Iijima et al., 2014; Norris et al., 2014). Immunolabeling of retinal tissue with Isl1-Pan and Isl1-Alpha antibodies was used to gain insight into cell type differences in the pattern of Isl1 isoform expression. Bipolar cells and cholinergic amacrine cells were immunopositive for both Isl1-Pan and Isl1-Alpha. Within the GCL, there was a conspicuous difference in the expression pattern of Isl1 isoforms across RGCs. One quarter of all Isl1-Pan positive cells in the GCL were Isl1-Alpha negative RGCs, representing an Isl1β-only expressing population. It does not appear that expression of only the beta isoform is a stochastic feature across any RGC type; rather, these Isl1β-only cells are restricted to a subset of RGC types. This inference is drawn from the fact that the Isl1β-only cells are restricted to RGC types with either small or medium sized somas. There are multiple RGC types with large cell bodies, relative to other RGCs, and these are not included in the Isl1β-only subset. The Isl1β-only RGCs are likely comprised of multiple RGC types, given their proportional occupancy amongst all Isl1-expressing RGCs, the range of their soma sizes, and their random spatial distribution. It is also likely that some retinal cells express both Isl1α and Isl1β or only Isl1α, as suggested by the pattern of Isl1 splicing across tissues types, but these distinctions could not be made with currently available antibodies.
The detection of different expression patterns of the Isl1 isoforms in combination with functional differences between the proteins further supports the idea that alternative splicing of genes encoding LIM-HD proteins could generate an expanded LIM-code. The lack of Lhx3-binding domain 2 (LBD2) within Isl1β appears to disrupt the formation of an Isl1:Lhx3:Ldb1 transcriptional complex. Lhx3 is not the only protein, however, that has been shown to bind within the C-terminal portion of Isl1 that is affected by alternative splicing. The LIM-HDs Lhx4 and Lhx8 have also been shown to form transcriptional complexes with Isl1 in a similar fashion as Lhx3, driving the differentiation of motor neurons in spinal cord and cholinergic neurons in the developing forebrain (Cho et al., 2014; Gadd et al., 2011). Additionally, the POU domain class 4 transcription factor 2 (Pou4f2), which is known to play a role in RGC differentiation (Badea et al., 2009; Mu et al., 2008; Pan et al., 2008), has recently been shown to bind to Isl1 within the C-terminal region (Li et al., 2014). While the present results suggest that the beta isoform may not be an effective partner in complexing with Lhx3 and Ldb1 to activate the He:RE, it remains a possibility that Isl1β may bind more efficiently with other proteins and have transactivational roles distinct from Isl1α not assayed in these experiments. Indeed, it is been shown be a more potent activator of the insulin gene promoter, relative to Isl1α (Ando et al., 2003).
In summary, we have determined that the two splice variants of Isl1 have tissue-specific expression patterns and the isoforms they encode are functionally distinct as transcriptional activators. The splicing of Isl1 is also differentially regulated across a major neuronal population shown by the presence of retinal ganglion cells expressing only the alternative isoform. These features support the likelihood of Isl1 alternative splicing contributing to the generation of diversity during neural development.
Experimental Methods
RT-PCR and qPCR
Retinal tissue from embryonic day (E) 12.5 and E16.5, and from postnatal day (P) 1, P10, and adult C57BL/6J mice was dissected in ice-cold RNAse-free sodium phosphate buffer (PBS) and stored in the RNA stabilization reagent, RNAlater (Ambion). The tissue was disrupted using RNase-free plastic pestles and tubes, and then completely homogenized with QIAshredder columns (Qiagen). RNA was extracted from these retinal homogenates using an RNeasy Plus Mini kit (Qiagen), which includes genomic eliminator columns to remove any DNA from the samples. Each sample was reverse transcribed into cDNA (iScript, BioRad). Adult tissue collected from other organs was processed in the same way, and cDNAs were generated for brain, spinal cord, thymus, adrenal gland, testis, lung, liver, spleen, and kidney. cDNAs were also made from RNA extracted from adult Sprague-Dawley rat retinas, as well as RNA from human retinal specimens of 9, 37, and 65 years of age. All use of animals in this study was regulated by the Institutional Animal Care and Use Committee at UCSB.
A PCR primer pair was designed to amplify both mouse Isl1α and Isl1b transcripts, generating a 139 base pair (bp) product and a 70bp product, respectively, for RT-PCR (forward 5′GCAGCAACCCAACGACAAAAC3′ and reverse 5′ CAGTACTTTCCAGGGCGGCT3′). Similar primers were also designed for rat (forward 5′ CAGCAGCAACCCAACGACAA3′ and reverse 5′ CAGGGCGGCTGGTAACTTTG 3′) and human Isl1 (forward 5′GCGGTGCAAGGACAAGAAGC3′ and reverse 5′AGGCGAAGTCGCTCAGTACTTT3′). Transcript-specific primers were designed for real time RT-PCR (qPCR). Both pairs have one primer that anneals to a portion of Isl1 that is common to both transcripts. The second primer for Isl1α anneals within the 69 nucleotides that are found only in Isl1α, and not Isl1β The second primer for Isl1β recognizes the sequence that spans the junction created by the absence of the above-mentioned 69 nucleotides (Isl1α forward 5′AAGGACAAGAAACGCAGCATC3′ and reverse 5′ CTGTAAACCACCATCATGTCTCTC3′; Isl1β forward 5′ CCCAACGACAAAACTGCTAAC3′ and reverse 5′ ATTAGAGCCTGGTCCTCCTTC3′). The specificity of these primers was determined by a restriction digest of their respective products. The restriction endonuclease, NcoI, recognizes a palindromic-hexanucleotide sequence that is present only in the Isl1α product (5′CCATGG3′). A restriction digest of these products with NcoI results in the Isl1α product being cut into two fragments, but the Isl1β product remains whole, confirming the specificity of these primers. Primers to the housekeeping gene, Gapdh, were also used (forward 5′AATGTGTCCGTCGTGGATCTGA3′ and reverse 5′AGTGTAGCCCAAGATGCCCTTC3′).
qPCR was used to determine Isl1α and Isl1β transcript abundance in the mouse retina, with an n of four for each age. Each embryonic sample contained retinas pooled from one litter, with a minimum of five embryos. The postnatal samples were made up of the retinas from one litter, with at least four pups. Each adult sample contained the retinas from one animal. These mouse retinal cDNA samples were amplified using SYBR Green I based PCR with the MyiQ Single Color Real-Time PCR Detection System to generate CT values. PCR efficiencies (EFF) were determined using linear regression software, LinReg PCR version 7.2, and used to calculate average amounts (1/EFF^Ct). The average amounts, from four experimental replicates of each sample, were adjusted for product size, and normalized to the housekeeping gene Gapdh (Whitney et al., 2009). The remaining cDNA samples, from other tissues or species, were also amplified this way. PCR products were run on an ethidium bromide-stained agarose gel to visualize the relative abundance of each product.
Isl1α and Isl1β Expressing Plasmids
Gateway cloning (Invitrogen) was used to make Isl1α- and Isl1/β-expressing plasmids. The open reading frame (ORF) of Isl1α was amplified from adult mouse retinal cDNA using primers with attB adapter sequences (forward 5′GGGG-ACAAGTTTGTACAAAAAAGCAGGCT-acc-ATGGGAGACATGGGCGATCCA3′ and reverse 5′GGGG-ACCACTTTGTACAAGAAAGCTGGGT-c-TCATGCCTCAATAGGACTGGCTACC3′). This product was gel-purified and inserted into a donor vector (pDONR221, Invitrogen), using BP Clonase to mediate the BP recombination reaction, to create an entry vector. One Shot Stbl3 chemically competent E. coli (Invitrogen) were transformed with the product of this reaction, and kanamycin resistant clones were selected for overnight culture in LB broth. Plasmid DNA was extracted from the bacteria using a QIAprep Spin Miniprep kit (Qiagen), and the successful recombination of the Isl1α ORF into the donor vector was confirmed by restriction digest and sequencing. The Isl1α-containing entry vector was then recombined with a destination vector using LR Clonase and transformed into E. coli for plasmid amplification and purification, as described above. The destination vector, pLenti CMV/TO GFP-Zeo Dest (Addgene plasmid #17431; (Campeau et al., 2009)), will express the gene of interest and GFP using two different constitutive CMV promoters. Once the sequence of the ORF in the destination vector was validated, large amounts of the plasmid were grown and purified from E. coli using an EndoFree Plasmid Maxi kit (Qiagen). The Isl1β ORF was generated using deletion PCR, to remove the 69 nucleotides from Isl1α that are not present in Isl1β (Lee et al., 2010). Once isolated, the Isl1β ORF was also cloned into the same destination vector described above, and large amounts were purified.
These two expression plasmids, in addition to a control plasmid expressing only GFP with no gene of interest inserted, were transiently transfected into HEK293T cells. The cells were grown in high glucose (4.5g/L) DMEM media plus 10% FBS, with penicillin (100 U/mL) and streptomycin (100 µg/mL), in a T75 tissue culture flask. When the cells reached 80% confluence they were passaged, and plated in 12-well tissue culture plates at 1.5×105 cells per well. Media was changed 24 hours after plating, and cells were transfected 48 hours after plating, with the proprietary transfection reagent TurboFect (Thermo Scientific). 1µg of Isl1α, Isl1β or an empty (control) plasmid was transfected into each well, and the transfectant was removed within 16 hours. Cell lysates were collected 24 hours from the start of transfection for using with western blotting.
Western Blotting
Protein samples were isolated from the transfected HEK293T cells described above, as well as from adult mouse retinas. The cell lysate or tissue was homogenized in ice-cold RIPA buffer (Thermo Scientific) with complete proteinase-inhibitors (Roche) and the supernatant was collected following centrifugation. The BCA protein assay (Thermo Scientific) was used to determine protein concentration, of all samples, relative to a set of standards at known concentrations of BSA. 2mg of protein was used for each sample from retinal tissue, and 0.1mg was used for the Isl1α-, Isl1β-, and GFP-expressing samples. Samples were run on an 8% SDS-PAGE gel, under non-reducing conditions, and transferred to PVDF membranes for immunoblotting. Membranes were blocked overnight in 5% milk at 4°C with agitation, and then rinsed with PBS before being incubated in primary antibody overnight, again at 4°C with agitation. Following another round of rinsing with PBS, membranes were incubated for two hours at room temperature with fluorescent secondary antibodies (1:5,000, LiCor Biosciences), plus 0.02% SDS and 0.1 % Tween 20. Both the primary and secondary antibody solutions were prepared in LiCor blocking buffer. After a final set of rinses in PBS, membranes were imaged using an Odyssey image scanner (LiCor Biosciences), and images were processed for brightness and contrast with Adobe Photoshop CS5. Band sizes were determined based on their relative mobility compared to a set of standards of known molecular weights.
Antibodies
Two different antibodies were used for the detection of Isl1 protein by western blotting or immunofluorescence. Both Isl1 isoforms should be detected with a commercially available mouse monoclonal antibody to Isl1 (39.4D5, 1:500, DSHB), referred to as Isl1-Pan. This antibody was generated using an antigen to the entire C-terminus of Isl1, thereby including a portion common to both isoforms (Ericson et al., 1992). A custom peptide-antibody, specific to the alpha isoform, was made by 21st Century Biochemicals. Rabbits were inoculated multiple times with a synthetic peptide, 15 amino acids long, that was conjugated to the immunogenic protein KLH. This peptide (acetyl-C-ASPERHDGGLQAN-amide) is specific to part of the sequence found in Isl1α, which is not present in Isl1β. Serum from the rabbits was purified with an affinity column, containing the original synthetic peptide, to produce an affinity purified antibody (Isl1-Alpha, 1:1,000). Additionally, an antibody to choline acetyltransferase (ChAT, 1:50, Millipore) was used to label cholinergic amacrine cells in the adult mouse retina.
Immunofluorescence
Immunofluorescence was conducted on tissue from adult C57BL/6J mice or Isl1 conditional knockout mice and wildtype littermates. The latter were generated by crossing mice expressing cre recombinase under a minimal Chx10 promoter (Tg(Chx10-EGFP/cre,-ALPP)2Clc)) (Rowan and Cepko, 2004) to mice carrying floxed-Isl1 alleles (Isl1tm2Sev) (Sun et al., 2008). Briefly, mice were given a lethal dose of sodium pentobarbital (120mg/kg, i.p.) and once deeply anesthetized, were intracardially perfused with 4% paraformaldehyde, and eyes were removed and immersed in 4% paraformaldehyde for additional fixation. Retinas were dissected from the eyes, and either kept whole or embedded in 5% agarose, and sectioned into 150 µm transverse sections. Following a three hour protein block with 5% normal donkey serum, retinal tissue was incubated with primary antibodies for three nights. Finally, to label the primary antibodies fluorescent secondary antibodies were applied overnight in different combinations, including: donkey-anti goat Cy2, donkey-anti rabbit Alexa Fluor 488, donkey-anti rabbit Alexa Fluor 594, and donkey-anti mouse Cy5, (Jackson ImmunoResearch Laboratories). In some cases, the fluorescent Nissl stain NeuroTrace 530/615 (Invitrogen) was added with the secondary antibodies to label cell bodies for an analysis of somal area. Retinal wholemounts and sections were imaged with an Olympus FV1000 scanning laser confocal microscope and images were processed for brightness and contrast with Adobe Photoshop CS5. Tissue from the liver and adrenal gland was processed similarly for immunofluorescent labeling; following anesthetization, tissue was removed and immersed in 4% paraformaldehyde for 1 hour. The tissue was rinsed in phosphate buffered saline and immersed overnight in 30% sucrose at 4°C, prior to being embedded in tissue freezing medium, frozen, and cut into 20 µm transverse sections. The same protein block, primary, and secondary antibodies were used as described above but for shorter incubation times and with the addition of the nucleic acid stain Hoechst 33342 (Life Technologies) for the visualization of cell nuclei. Liver and adrenal gland sections were imaged with a Zeiss 710 laser scanning microscope.
Soma Size Analysis
A retinal wholemount, labeled with Isl1-Pan, Isl1-Alpha, and the fluorescent Nissl stain NeuroTrace 530/615, was imaged through the entire thickness of the GCL, generating a stack of optical sections 1 µm apart, for eight different fields, each 25,000 sq µm in area, taken from four central and four peripheral locations across the retina. Metamorph image analysis software (Molecular Devices) was used to analyze each feature labeled within the collected z-stacks. First, all nuclei labeled with the Isl1-Pan antibody were traced. These regions were then overlaid onto the Isl1-Alpha labeling within the same field, and all nuclei not labeled above background with Isl1-Alpha were identified. The Isl1-Pan tracings were then overlaid onto the NeuroTrace image, and the somas of every Isl1-Pan positive nucleus was outlined, from which areal measures were determined. A total of 1,781 Isl1-Pan cells were analyzed.
Spatial Statistics
Further analysis of the Isl1β-only population of RGCs was performed on seven of those eight fields (one low-density field was excluded), using specialty software designed for conducting Delaunay tessellation analysis and spatial autocorrelation analysis. The X and Y locations were identified for each cell within a field, from which the nearest neighbor distances for every cell were computed, excluding those cells closer to the boundary than to any other cell within the field. We also computed, for each field, 99 random simulations matched for density and constrained by the soma size distribution for the Isl1β-only population, and conducted the same nearest neighbor analysis. Cumulative frequency histograms for the nearest neighbor distances were constructed for each real field and its 99 simulations. Autocorrelation analysis from each field was also performed, from which density recovery profiles were constructed, to determine the size of the effective radius (Rodieck, 1991). The density recovery profiles were also constructed from the 99 random simulations, for comparison.
Luciferase Assay
HEK293T cells were transiently transfected with combinations of experimental plasmids designed to express Isl1α, Isl1β, Lhx3, or GFP (control plasmid with no gene of interest inserted). The Lhx3-expressing plasmid was also generated using Gateway cloning, as described above, starting with the amplification of the Lhx3 ORF (forward 5′GGGG-ACAAGTTTGTACAAAAAAGCAGGCT-acc-ATGGAAGCTCGCGGGGA3′ and reverse 5′GGGG-ACCACTTTGTACAAGAAAGCTGGGT-c-CTAGCCATCCACCCAGAGGC3′). Cells were also co-transfected with a luciferase reporter that has been previously shown to be specifically activated by the Isl1:Lhx3:Ldb1 complex (Hx:RE) (Lee et al., 2008), and with a β-Galactosidase (β-Gal) expressing plasmid under the control of a CMV promoter used to control for transfection efficiencies between samples. The cells were maintained in a media of high glucose (4.5g/L), DMEM plus 10% Fetal Bovine Serum, with penicillin (100 U/mL) and streptomycin (100 µg/mL), in a T75 tissue culture flask being passaged once per week or when cells reached 80% confluence. Media was changed every two days and cells were counted and plated in 12-well tissue culture plates at 1.5×105 cells per well and media was changed 24 hours later. 48 hours after plating cells were transiently transfected using TurboFect (Thermo Scientific) with 250ng of the Hx:RE luciferase reporter plasmid (Lee et al., 2008), 20ng of β-Gal, and 200ng of each experimental plasmid (Isl1α+Lhx3, Isl1β+Lhx3, Lhx3+GFP, Isl1α+GFP, or Isl1β+GFP) with the exception of the control (GFP only), in which 400ng was added. The transfectant was removed 16 hours later, and cell lysates collected 24 hours from the start of transfection.
Cell lysates were assayed for both luciferase (Luciferase Assay System, Promega) activity and β-Gal abundance on a Perkin-Elmer plate reader, in triplicate per experimental replicate. Background luciferase activation was corrected by subtraction of activation detected in untransfected well, and variation in transfection efficiency was corrected by normalization to β-Gal expression. Each bar represents mean fold induction with respect to the GFP control and error bars represent SEM. Three independent experiments were conducted at different times, with average expression levels derived from the three experiments being reported.
Supplementary Material
(A) The adult mouse liver lacks expression of Isl1 isoforms, shown by the absence of Isl1-Pan and Isl1-Alpha labeling (open arrowheads) of cell nuclei (filled arrowheads).
(B) Both Isl1-Pan and Isl1 -Alpha label all cell nuclei in the field shown from the adult adrenal gland, which expresses both Isl1α and Isl1β (filled arrowheads), as shown by RT-PCR. (C) A comparison of Isl1-Pan and Isl1 -Alpha labeling of Isl1-WT and Isl1-CKO retina shows a complete loss of labeling by either antibody in the GCL of the CKO (open arrowheads). Calibration bar = 50 µm.
Highlights.
-
-
Alternative splicing of the transcription factor Isl1 differs across tissue types
-
-
Expression of Isl1 splice variants is dynamic in the developing retina
-
-
The pattern of Isl1 alternative splicing varies across different retinal cell types
-
-
The two isoforms of Isl1 differ functionally as transcriptional activators
Acknowledgements
We thank Nils Madsen for assistance with the spatial analysis, Roxanne Croze for advice on plasmid construction and cell culturing, Kyle Ploense for providing the rat retinal tissue, Dr. Monte Radeke of the Center for Macular Degeneration at UCSB for advice on PCR and supplying human retinal RNA samples, and Dr. Soo-Kyung Lee of Oregon Heath & Science University for providing us with the luciferase reporter plasmid. We also thank Dr. Patrick Keeley for constructive comments throughout the project. This work was supported by a grant from the NIH (EY019968).
Abbreviations
- Isl1
Islet-1
- LIM-HD
LIM-homeodomain
- RGC
retinal ganglion cell
- Lhx3
LIM homeobox 3
- Ldb1
LIM domain binding 1
- SA
self-assemble
- LID
LIM interacting domain
- LBD1/2
Lhx3-binding domain 1/2
- E
embryonic day
- P
postnatal day
- ChAT
choline acetyltransferase
- INL
inner nuclear layer
- GCL
ganglion cell layer
- Hx:RE
hexameric recognition element
- ORF
open reading frame
- GFP
green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors have no competing interests for this manuscript.
Contributor Information
Irene E. Whitney, Email: irenewhitney@fas.harvard.edu.
Amanda G. Kautzman, Email: amanda.kautzman@psych.ucsb.edu.
Benjamin E. Reese, Email: benjamin.reese@psych.ucsb.edu.
References
- Ando K, Shioda S, Handa H, Kataoka K. Isolation and characterization of an alternatively spliced variant of transcription factor Islet-1. Journal of molecular endocrinology. 2003;31:419–425. doi: 10.1677/jme.0.0310419. [DOI] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mechanisms of development. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009;61:852–864. doi: 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. The Journal of comparative neurology. 2004;480:331–351. doi: 10.1002/cne.20304. [DOI] [PubMed] [Google Scholar]
- Bhati M, Lee C, Nancarrow AL, Lee M, Craig VJ, Bach I, Guss JM, Mackay JP, Matthews JM. Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. The EMBO journal. 2008;27:2018–2029. doi: 10.1038/emboj.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boije H, Ring H, Shirazi Fard S, Grundberg I, Nilsson M, Hallbook F. Alternative splicing of the chromodomain protein Morf4l1 pre-mRNA has implications on cell differentiation in the developing chicken retina. Journal of molecular neuroscience : MN. 2013;51:615–628. doi: 10.1007/s12031-013-0034-4. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PloS one. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bush SJ, Tovar-Corona JM, Castillo-Morales A, Urrutia AO. Correcting for differential transcript coverage reveals a strong relationship between alternative splicing and organism complexity. Molecular biology and evolution. 2014;31:1402–1413. doi: 10.1093/molbev/msu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HH, Cargnin F, Kim Y, Lee B, Kwon RJ, Nam H, Shen R, Barnes AP, Lee JW, Lee S, et al. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS genetics. 2014;10:e1004280. doi: 10.1371/journal.pgen.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JE. Spatial properties of retinal mosaics: an empirical evaluation of some existing measures. Visual neuroscience. 1996;13:15–30. doi: 10.1017/s0952523800007094. [DOI] [PubMed] [Google Scholar]
- Cook JE, Podugolnikova TA. Evidence for spatial regularity among retinal ganglion cells that project to the accessory optic system in a frog, a reptile, a bird, and a mammal. Visual neuroscience. 2001;18:289–297. doi: 10.1017/s0952523801182131. [DOI] [PubMed] [Google Scholar]
- Coombs J, van der List D, Wang GY, Chalupa LM. Morphological properties of mouse retinal ganglion cells. Neuroscience. 2006;140:123–136. doi: 10.1016/j.neuroscience.2006.02.079. [DOI] [PubMed] [Google Scholar]
- Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. The Journal of comparative neurology. 2007a;503:182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, Gan L. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007b;27:12707–12720. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Thor S, Edlund T, Jessell TM, Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science (New York, NY) 1992;256:1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- Failli V, Rogard M, Mattei MG, Vernier P, Retaux S. Lhx9 and Lhx9alpha LIM-homeodomain factors: genomic structure, expression patterns, chromosomal localization, and phylogenetic analysis. Genomics. 2000;64:307–317. doi: 10.1006/geno.2000.6123. [DOI] [PubMed] [Google Scholar]
- Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein JL. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development (Cambridge, England) 2009;136:3841–3851. doi: 10.1242/dev.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd MS, Bhati M, Jeffries CM, Langley DB, Trewhella J, Guss JM, Matthews JM. Structural basis for partial redundancy in a class of transcription factors, the LIM homeodomain proteins, in neural cell type specification. The Journal of biological chemistry. 2011;286:42971–42980. doi: 10.1074/jbc.M111.248559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsiz ED, Ouyang Q, Schmidt M, Nagpal S, Morrow EM. Genome-wide transcriptome analysis in murine neural retina using high-throughput RNA sequencing. Genomics. 2012;99:44–51. doi: 10.1016/j.ygeno.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill GN. Decoding the LIM development code. Transactions of the American Clinical and Climatological Association. 2003;114:179–189. [PMC free article] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development (Cambridge, England) 1998;125:2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends in genetics : TIG. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Rhodes SJ. LIM-homeodomain genes in mammalian development and human disease. Molecular biology reports. 2005;32:67–77. doi: 10.1007/s11033-004-7657-z. [DOI] [PubMed] [Google Scholar]
- Iijima T, Iijima Y, Witte H, Scheiffele P. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. The Journal of cell biology. 2014;204:331–342. doi: 10.1083/jcb.201310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J, Ueda Y, Bando T, Mito T, Noji S, Ohuchi H. The expression of LIM-homeobox genes, Lhx1 and Lhx5, in the forebrain is essential for neural retina differentiation. Development, growth & differentiation. 2013;55:668–675. doi: 10.1111/dgd.12074. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Gill GN. Functional analysis of the nuclear LIM domain interactor NLI. Molecular and cellular biology. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Pfaff SL, Gill GN. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. The Journal of biological chemistry. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- Katyal S, Godbout R. Alternative splicing modulates Disabled-1 (Dab1) function in the developing chick retina. The EMBO journal. 2004;23:1878–1888. doi: 10.1038/sj.emboj.7600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Ueno M, Nakashima K, Taga T. A brain region-specific gene product Lhx6.1 interacts with Ldb1 through tandem LIM-domains. Journal of biochemistry. 1999;126:180–187. doi: 10.1093/oxfordjournals.jbchem.a022420. [DOI] [PubMed] [Google Scholar]
- Kong JH, Fish DR, Rockhill RL, Masland RH. Diversity of ganglion cells in the mouse retina: unsupervised morphological classification and its limits. The Journal of comparative neurology. 2005;489:293–310. doi: 10.1002/cne.20631. [DOI] [PubMed] [Google Scholar]
- Lakk M, Szabo B, Volgyi B, Gabriel R, Denes V. Development-related splicing regulates pituitary adenylate cyclase-activating polypeptide (PACAP) receptors in the retina. Investigative ophthalmology & visual science. 2012;53:7825–7832. doi: 10.1167/iovs.12-10417. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Irizarry K. Alternative splicing in the nervous system: an emerging source of diversity and regulation. Biological psychiatry. 2003;54:771–776. doi: 10.1016/s0006-3223(03)00375-5. [DOI] [PubMed] [Google Scholar]
- Lee J, Shin MK, Ryu DK, Kim S, Ryu WS. Insertion and deletion mutagenesis by overlap extension PCR. Methods in molecular biology (Clifton, NJ) 2010;634:137–146. doi: 10.1007/978-1-60761-652-8_10. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Developmental cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Wu F, Ruonala R, Sapkota D, Hu Z, Mu X. Isl1 and Pou4f2 form a complex to regulate target genes in developing retinal ganglion cells. PloS one. 2014;9:e92105. doi: 10.1371/journal.pone.0092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kim SY, Fu Y, Wu X, Ng L, Swaroop A, Forrest D. An isoform of retinoid-related orphan receptor beta directs differentiation of retinal amacrine and horizontal interneurons. Nature communications. 2013;4:1813. doi: 10.1038/ncomms2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Gao S, Norris ML, Ray D, Ramani AK, Fraser AG, Morris Q, Hughes TR, Zhen M, Calarco JA. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Molecular cell. 2014;54:946–959. doi: 10.1016/j.molcel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development (Cambridge, England) 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese BE, Keeley PW. Design principles and developmental mechanisms underlying retinal mosaics. Biological reviews of the Cambridge Philosophical Society. 2014 doi: 10.1111/brv.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW. The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Visual neuroscience. 1991;6:95–111. doi: 10.1017/s095252380001049x. [DOI] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Developmental biology. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Sloop KW, Dwyer CJ, Rhodes SJ. An isoform-specific inhibitory domain regulates the LHX3 LIM homeodomain factor holoprotein and the production of a functional alternate translation form. The Journal of biological chemistry. 2001;276:36311–36319. doi: 10.1074/jbc.M103888200. [DOI] [PubMed] [Google Scholar]
- Sumbul U, Song S, McCulloch K, Becker M, Lin B, Sanes JR, Masland RH, Seung HS. A genetic and computational approach to structurally classify neuronal types. Nature communications. 2014;5:3512. doi: 10.1038/ncomms4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nature neuroscience. 2008;11:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Wan J, Masuda T, Hackler L, Jr, Torres KM, Merbs SL, Zack DJ, Qian J. Dynamic usage of alternative splicing exons during mouse retina development. Nucleic acids research. 2011;39:7920–7930. doi: 10.1093/nar/gkr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney IE, Raven MA, Ciobanu DC, Williams RW, Reese BE. Multiple genes on chromosome 7 regulate dopaminergic amacrine cell number in the mouse retina. Investigative ophthalmology & visual science. 2009;50:1996–2003. doi: 10.1167/iovs.08-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome biology. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghlool A, Ameur A, Cavelier L, Feuk L. Splicing in the Human Brain. International review of neurobiology. 2014;116c:95–125. doi: 10.1016/B978-0-12-801105-8.00005-9. [DOI] [PubMed] [Google Scholar]
- Zambelli F, Pavesi G, Gissi C, Horner DS, Pesole G. Assessment of orthologous splicing isoforms in human and mouse orthologous genes. BMC genomics. 2010;11:534. doi: 10.1186/1471-2164-11-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The adult mouse liver lacks expression of Isl1 isoforms, shown by the absence of Isl1-Pan and Isl1-Alpha labeling (open arrowheads) of cell nuclei (filled arrowheads).
(B) Both Isl1-Pan and Isl1 -Alpha label all cell nuclei in the field shown from the adult adrenal gland, which expresses both Isl1α and Isl1β (filled arrowheads), as shown by RT-PCR. (C) A comparison of Isl1-Pan and Isl1 -Alpha labeling of Isl1-WT and Isl1-CKO retina shows a complete loss of labeling by either antibody in the GCL of the CKO (open arrowheads). Calibration bar = 50 µm.