Abstract
Aggregate or spheroid culture has emerged as a more biologically relevant method for screening pharmaceutical compounds and understanding exact mechanism of action. Here in, the aggregate approach applied to the freshwater toxins, microcystins, further unearths exact mechanism(s) of toxicity and provides a markedly improved in vitro predictor of toxicity. Microcystins result in acute intoxication by binding covalently to protein phosphatase 1/2A, resulting in hepatocellular necrosis, hemorrhaging and death. Hepatocellular uptake by organic anion transporting polypeptides (OATPs), in addition to other intracellular sequelae, is considered essential for toxicity. In aggregate HepG2, expression of OAT1B1 and OATP1B3 significantly increased relative to monolayer culture. Uptake of two fluorescently labeled substrates significantly increased in aggregates compared with monolayer, confirmed by inhibition of uptake with known competitive substrates. Increased reaction oxygen species (ROS) production occurred following a three-hour exposure of microcystin LR at concentrations from 100 nM to 100 μM, with reversal by ROS scavengers, in contrast with no response in monolayers. These results suggest monolayer culture inadequately predict intracellular effects of microcystins and support evidence that aggregate culture more closely approximates in vivo form and function. The approach results in more reliable prediction of microcystin toxicity in vitro.
Keywords: microcystins, OATP1B1, OATP1B3, cell culture model, aggregate, cyanotoxins
1. Introduction
The acutely hepatotoxic monocyclic heptapeptide cyanotoxins, microcystins (MCs), produced worldwide by cyanobacteria, represent rigorously studied, but poorly understood toxins for which a more biologically relevant in vitro approach is needed. The identified MC congeners number well over 100 (Meriluoto and Spoof, 2008).The World Health Organization (WHO) provides a provisional drinking water guideline of 1.0 μg/l based on limited available data and risk assessment with respect to one congener, microcystin-LR. Major gaps relative to risk assessment for total MCs and exact mechanism of action remain (Dietrich and Hoeger, 2005) and must be addressed with a reproducible bioassay for toxicity to better protect public health (Monks and Moscow, 2010). Through inhibition of ubiquitously expressed protein phosphatase 1 and 2A (Runnegar et al., 1995), MCs impact numerous animal species (Campos and Vasconcelos, 2010; Dawson, 1998). Following ingestion, the toxin primarily targets the liver, resulting in cytoskeletal disruption (Hooser et al., 1991) and acute hepatic hemorrhage and necrosis (Hooser et al., 1989). In addition to the downstream effects of cytoskeletal disintegration through disruption of protein de-phosphorylation, MCs bind with cellular thiols, deplete glutathione, result in mitochondrial dysfunction, alter patterns of DNA repair and cellular proliferation and generate reactive oxygen species (ROS) in cells other than in the liver (Campos and Vasconcelos, 2010; Gehringer, 2004).
The amphiphatic 900 to 1100 Dalton, generally hydrophilic MCs do not cross cell membranes easily and rely on the transporters OATP1B1 and OATP1B3, located along the basolateral sinusoidal membranes, for hepatocellular uptake (Fischer et al., 2010; Fischer et al., 2005; Komatsu et al., 2007; Lu et al., 2008; Monks et al., 2007). Despite nanomolar sensitivities in transfected cell lines (Fischer et al., 2010; Fischer et al., 2005; Komatsu et al., 2007; Monks et al., 2007) and potent cytotoxicty in primary cells and in vivo, monolayer immortalized cultures, including HepG2, remain viable at high μM concentrations 48, 72 and 96 hours after exposure (Chong et al., 2000). Facilitated endocytosis techniques have also been employed (Jasionek et al., 2010), yet these models may exaggerate toxic effect relative to in vivo effects (Monks and Moscow, 2010).
Micro scale approaches to aggregate or spheroid culture of primary cells and immortalized cell lines provide a relatively inexpensive technique for evaluating cytotoxicity that more closely approximates in vivo function (Khademhosseini, 2007; Khademhosseini et al., 2006). To date, little literature has focused on liver OATPs despite their importance in xenobiotic absorption, distribution and elimination. Primary rat liver cell aggregates maintained expression of basolateral and apical transporters, including the murine analog OATP1B2, (Sidler Pfandler et al., 2004), and multicellular spheroid formations of HepG2 cells using peptide nanofiber hydrogels demonstrated increased structural and functional polarity (Malinen et al., 2012).
While expression of OATP1B1 and OATP1B3 has been shown to be 1.5 to 2 fold higher in fresh human liver as compared with monolayers of HepG2 (Hilgendorf et al., 2007), aggregate HepG2 cells demonstrate improved expression and functionality, including distinct cortical actin organization in spheroids concurrent with up regulation of metabolic and synthetic genes and increased cytochrome P450 activity and albumin production as compared with monolayers of the same cell type (Chang and Hughes-Fulford, 2009). Cytochrome P450 expression and inducible expression in HepG2 aggregates substantially increases (Nakamura et al., 2011), along with immune and cytokine response (Liu et al., 2011). HepG2 aggregates may prove less susceptible to pharmacological agents as compared with monolayer culture (Li et al., 2008) because of increased expression of export pumps, MDR-1 and MRP-2 (Mueller et al., 2011; Oshikata et al., 2011).
To date, toxicity of MCs has only been evaluated in monolayer culture, likely underestimating uptake and extent of toxic effects. In addition, the mechanism of effect may be altered at various delivered concentrations, thus reducing understanding of mechanism of action, particularly across congeners. Given the increasing evidence in support of the in vivo like function of aggregates, we evaluated comparative expression of relevant transporters and comparative uptake and toxicity of MCs in monolayers and aggregate HepG2. The increased OATP expression and function in addition to increased ROS generation following MC exposure in aggregate culture relative to monolayer suggests a more realistic model for MC toxicity. The approach discussed here-in can not only be utilized to further probe into mechanisms of MCs’ hepatocellular toxicity and markedly improve risk assessment, but to screen for OATP involvement in drug or toxin uptake and disposition in the liver, particularly important when evaluating elimination or drug interactions.
2. Methods & Materials
2.1 Chemicals and Reagents
HepG2, a human hepatocellular carcinoma derived cell line, was obtained from the ATCC (Manassas, VA). For cell culture and uptake studies, Dulbecco’s Modified Eagle’s Medium (DMEM) with pyruvate, Dulbecco’s Phosphate Buffered Saline (DPBS) w/Calcium and Magnesium, Penicillin-Streptomycin (100x), heat inactivated Fetal Bovine Serum (FBS), and Hank’s Buffered Saline Solution (HBSS) w/Calcium and Magnesium were purchased from Invitrogen Life Technologies (Grand Island, NY). Microcystin LR was obtained from VWR International through Enzo Life Sciences (Farmingdale, NY) and GreenWater Laboratories (Palatka, Florida) (fluorescent uptake and inhibition studies only).
For mask, wafer and PDMS mold generation, silicon prime wafers (P/Boron) were obtained from University Wafers (Boston, Massachusetts) and SU-8 Photoresist was purchased from MicroChem Corp. (Newton, MA) The Sylard 184 kit for polydimethylsiloxane (PDMS) was purchased from Dow Corning (Midland, MI). Master mask and chemicals for mask and wafer development, along with L-edit Tanner EDA software (Monrovia, CA) were obtained in kind from the Northern California Nanotechnology Center (NCNC, University of California, Davis).
For microwell fabrication, poly(ethylene glycol) diacrylate (PEGDA), average Mn 700, and 3-(trimethoxysilyl)propyl methacryclate (TMSPMA), sodium hydroxide solution, and 70% ethanol were purchased from Sigma Aldrich (St. Louis, MO), and glass slides were purchased from Fisher Scientific (Philadelphia, PA). The UV light source (Omnicure S1000) with filter (wavelength: 320–500 nm) was obtained from EXFO Photonic Solutions Inc (Ontario, Canada). The photo initiator Irgacure 2959 (4-(2-hydroxyethoxy)phenyl-(2-hydroxy-2-propyl)ketone) was obtained from Ciba Specialty Chemicals, Switzerland. For mRNA expression studies, TRIZOL Reagent, SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase, RNase Away, RNase free water, TAE Buffer, Ethidium Bromide, DNA ladders and relevant primers (OATP1B1, OATP1B3 and GADPH) were ordered from Invitrogen (Carlsbad, CA). Chloroform, isopropyl alcohol and ethanol were obtained from Sigma Aldrich (St. Louis, MO).
For fluorescence uptake and inhibition studies, 8-FcA, 8-2-[Fluoresceinyl]-aminoethylthio) adenosine-3′, 5′-cyclic monophosphate, was purchased from Biolog Life Science Institute (Bremen, Germany) through its North American distributor, Axxora, LLC (San Diego, CA) and fluorescein-methotrexate (FMTX) was purchased from Invitrogen (Carlsbad, CA). For the inhibition studies, Rifampicin was purchased from Sigma Aldrich (St. Louis, MO), deoxycholic acid and cyclosporine D were purchased from Calbiochemicals (Los Angeles, CA), and microcystin LR was obtained from Green Water Laboratories (Palatka, Florida). A Pierce Scientific bicinchonicnic acid (BCA) protein quantification kit was obtained from ThermoFisher (Rockford, Illinois).
For reactive oxygen species assays, 2′,7′-Dichlorofluorescein diacetate (H2DCFDA), L-absorbic acid, deferoxamine (DFO) mesylate salt, catalase from bovine liver, and hydrogen peroxide were obtained from Sigma Aldrich (St. Louis, MO). The CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay with MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) was obtained from Promega (Madison, WI). Tween-20 was obtained from Sigma, Aldrich (St. Louis, MO).
For qPCR plate preparation from Day 5 mRNA, designed primers were obtained from Eurofins MWG Operon, (http://www.eurofinsdna.com), and TaqMan probes were purchased from Roche. SuperScript® III reverse transcriptase was obtained from Invitrogen (Carlsbad, CA), and TaqMan Universal PCR Mastermix from Applied Biosystems (Carlsbad, CA).
2.2 Cell culture
HepG2 cells were cultured in Dulbecco’s Modified Eagle’s Medium with 10% (v/v) fetal bovine serum and 1% antibiotics (10,000 U/ml penicillin and 10 mg/ml streptomycin) under standard conditions at 37°C and 5% CO2 in 75 cm2 Falcon flasks. Immediately prior to all experiments, HepG2 cells were harvested with trypsin and were cultured in a) control monolayer fashion and in b) aggregates in 150 μm by 150 μm microwells formed from polyethylene glycol diacrylate (PEGDA, MW=700) UV cross-linked onto a TMSPMA pre-treated glass slide as described in section 2.4 and 2.5 below. In order to encourage aggregate formation, 200 μl of cell suspension was seeded gently onto the surface of the microwells and the solution spread with the pipette tip across the surface of the microwells. The cells aggregated into hydrophobic pockets (microwells) of the slide formed by the PEGDA polymer. The six well plates housing the microwells were then incubated for 30 minutes at 37°C and then washed once with PBS to remove unseeded cells, after which excess media was added to each well. Cells were seeded at a density of 4*10^2 cm2 (or 2* 106 cells/ml media), previously demonstrated as optimal for aggregate formation (Moeller et al., 2008).
2.3 Photomask and PDMS mold fabrication
Reusable polydimethylsiloxane (PDMS) column molds consisting of uniform posts of a diameter of 150 μm and height of 150 μm were fabricated for reproducible and easy microwell fabrication. These “reverse imprint” PDMS stamps were generated in a clean room from a light sensitive silicon wafer, containing the desired final microwell design. The “master” silicon wafer was generated using SU-8 photoresist and a photomask designed using L-Edit Tanner EDA software under the guidance of staff at the University of California, Davis, Northern California Nanotechnology Center (NCNC). The micropatterns of the master silicon wafer (150 μm in height and diameter) were initially fabricated via previously described methods with some modification (Supplemental Materials) (Moeller et al., 2008). The PDMS molds were fabricated using a 10:1 mixture of silicone elastomer base solution combined with curing agent Sylgard 184. The PDMS elastomer solution was degassed for 1 hour in a vacuum chamber, poured carefully overtop the silicon master and baked at 70 °C for 2 hours. The PDMS molds were peeled from silicon masters, resulting in the raised columns patterns, ready for use in fabrication of PEGDA microwells outside the clean room.
2.4 3-(Trimethoxysilyl) propylmethacrylate(TMS-PMA) slide pre treatment
Fisher brand plain microscope slides (25×75×1 mm) were pre- coated with 3-(Trimethoxysilyl) propylmethacrylate (TMSPMA). Briefly, slides were cleaned with 10% NaOH for at least 2 hours, rinsed three times with 70% ethanol and baked in an oven. Slides then were stacked in a clean beaker with 3 ml of TMSPMA for 10 minutes to allow capillary action to coat slides without air bubbles and then flipped to permit even coating. After 10 minutes, the TMSPMA coated slides were wrapped in aluminum foil and placed in 80°C oven over night. The slides were cleaned with 70% ethanol three times and baked for 2 hours at 80°C. Slides were cut into three and corners removed with a diamond scribe pen to fit into 6 well plates. The slides were wrapped in aluminum foil and kept cool until further use.
2.5 Microwell fabrication and sterilization
Hydrophilic non-adhesive microwells were fabricated from provided PDMS molds using UV-cross linkable polyethylene glycol diacrylate (PEGDA) 20% (w/w) prepared in DPBS with 1% photoinitiator Irgacure 2959 (w/w). 150 μl of PEGDA pre-polymer solution was pipetted on to the surface of the pre-treated glass slide and the patterned stamp carefully placed on top. The collective “sandwich” was placed under 350–500 nm wavelengths of light for 50 s at an intensity of 100 mW/cm2 using OmniCure Series 1000 curing station. After formation of solid polymer microwells, the PDMS stamp was carefully peeled back, leaving the microwells affixed to the glass slide. Any residual pieces of PDMS were cut away and the microwells rinsed with ethanol. PDMS molds were rinsed with ethanol prior to reuse and permitted to air dry. The microwells were placed in six well plates with 4 ml of 70% ethanol solution under UV light in a cell culture hood for 2 hours for sterilization purposes. Prior to cell seeding, the microwells were rinsed with 4 ml of DPBS.
2.6 OATP1B1 and OATP1B3 mRNA temporal expression
In order to determine temporal regulation of OATP expression after aggregate formation, total RNA was extracted from cell cultures using TRIZOL according to manufacturer’s instructions at day 1, 3, 5, and 7. In addition, at these same time points, cell viability and aggregate formation was assessed through microscopy and through visual observation. The concentration of mRNA was measured at A260 and quality assessed from UV absorbance A260:A280 ratio. Reverse transcriptase-polymerase chain reaction (RT-PCR) was carried out using the SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase, according to manufacturers’ instructions. cDNA synthesis was performed at 53°C for 25 min. After an initial denaturation step at 94°C, several cycles of PCR were carried out under the following conditions: 15 s denaturing at 95°C, 30 s specific primer annealing temperature, and 45 s extension at 68°C. After amplification, PCR products were loaded on 2% (w/w) agarose gels containing ethidium bromide alongside a negative control and 50 kb DNA ladder at EP 1000 V for 35 minutes in 1X TAE buffer. Digital images of the fluorescently labeled amplification products were captured using the Gel Logic 100 Imaging System with Molecular Imaging Software (Eastman Kodak Company, Rochester, NY). For quantification, images were analyzed using Image J (http://rsbweb.nih.gov/ij/) software, normalizing to loading control and GADPH housekeeping gene. Previously published primers used in fresh human liver were utilized to evaluate OATP1B1 and OATP1B3 in cell lines (Libra et al., 2006). All subsequent studies were initiated on day 5 of the growth based on temporal expression of transporters and visual observation of cells outgrowing the boundaries of the microwells on Day 7.
2.7 Fluorescence Uptake and Inhibition Studies
Fluorescence uptake studies in monolayer and aggregate culture were performed as previously described for high throughput screening of inhibitors of OATP1B1 and OATP1B3 with some modification (Gui et al., 2010). On day 5 of growth, HepG2 cells were seeded onto 24 well plates at a density of 105 cells/ml, 24 hours prior to transport assays. Cells were washed with pre-warmed HBSS uptake buffer (1 ml) and allowed to equilibrate for 15 min to the uptake buffer. Uptake was started by adding 200 μl of the uptake buffer containing the indicated concentrations of fluorescent compound, and the cells were incubated at 37°C for 30 minutes. 8-FcAMP and FMTX were prepared at concentrations of 1, 10, and 50 μM. In the case of uptake inhibition assays, fluorescent substrate and inhibiting reagents were added simultaneously at final concentrations of 10 μM and 50 μM, respectively. After 30 minutes, the uptake solution was aspirated and uptake was halted with 1 ml of ice-cold PBS. Following two washes with PBS, the cell membranes were disrupted with 200 μl of 1% Triton X -100 (dissolved in PBS) and fluorescence was measured at excitation wavelength of 485 nm and emission wavelength of 528 nm. Following these readings, protein concentrations were determined using a BCA assay kit, according to manufacturer’s instructions. Fluorescent units were normalized to μg protein.
2.8 Reactive Oxygen Species (ROS) Assays
Following 5 days of aggregate or control monolayer growth, aggregate cells were harvested from the microwells through gentle pipetting of media solution over the surface of the microwells, and 2D cells were trypsinized from Falcon flasks. Cell solutions of 4*104 cells/ml were seeded into 96 well plates (100 μl added per well) and incubated at 37°C, 5% CO2 for 24 hours. Microcystin LR (MC-LR) was dissolved initially in 100% MeOH and then serially diluted 1:10 from 100 μM to 0.1μM in DMEM. 100 μl of MC-LR dilution, control (DMEM alone), positive control (0.003% H202) or solvent control (<1% MeOH) was pipetted into the appropriate well. At least 6 replicates were performed for each treatment in 3separate experiments. Cells were incubated at 37°C for 3 hours, then washed twice with DPBS prior to incubation with the fluorogenic compound 2′,7′-Dichlorofluorescein diacetate (H2DCFDA) at a final concentration of 40 μM in HBSS uptake buffer as previously described with slight modifications for HepG2 (Bouaicha and Maatouk, 2004; de Souza Votto et al., 2007). Each well received 100 μl of H2DCFDA. H2DCFDA diffuses passively through the cell membranes and is cleaved inside by esterases; the non-fluorescent compound H2DCF is oxidized by ROS to yield fluorescent DCF. The fluorescence intensity of DCF was determined on a MicroPlate Reader (Molecular Devices, Sunnyvale, CA) using excitation and emission wavelengths of 480 and 538 nm respectively. Background fluorescence was subtracted and the data were normalized to control. ROS scavenger experiments were carried out in identical fashion except cells were either incubated with 10 μM ascorbic acid, 10 μM catalase or 50 μM deferoxamine (DFO) scavengers alone or in combination with 1 μM MC-LR.
2.9 MTS cell viability studies
Following 5 days of aggregate or control 2D growth, cells were harvested as described above. Cells were seeded at 4*104 per ml into 96 well plates and incubated at 37°C, 5% CO2 for 24 hours. MC-LR concentrations were prepared as follows: 50, 30, 10, 5, 3 and 1 μM, along with a solvent control (<1% MeOH) and positive control (1% TWEEN). At least 12 replicates of control and 6 replicates of various treatments were prepared within each of 3 independent experiments. Each well received 100 μl of media with treatment. Cells were cultured for 48 hours. Media was then aspirated, the cells washed once with DPBS. The MTS and PMS (phenazine methosulfate) solution was prepared according to manufacturer’s instructions in the CellTiter 96 Aqueous Assay. 100 μl of media was added to each well along with 20 μl of the MTS/PMS solution and thoroughly mixed. The plate was then incubated for three hours at 37°C. The absorbance was then read at 490 nm using a Microplate Reader. Background was subtracted and cell viability was expressed relative to control.
2.10 Day 5 RNA extraction and Primer probe design for qPCR
Four replicates of aggregate and control 2D cells were grown as previously described for 5 days in three separate experimental studies. On day 5 of each experiment, RNA was extracted from each replicate with TRIZOL according to manufacturer’s instructions. RNA concentration was estimated from absorbance at 260 nm (NanoDrop ND 1000) and RNA purity was verified by A260/A280 ratios. RNA integrity was verified by electrophoresis on ethidium bromide-stained 1.5% agarose gels. One microgram of total RNA was used for cDNA synthesis. Primer and probes for qPCR (Table 1) were designed using Roche Universal Probe Library Assay Design Center (https://www.roche-applied-science.com).
Table 1. Primer pairs and TaqMan probes used in qPCR for Comparative Gene Expression in Aggregates and Control Monolayer HepG2.
Primer and probes for qPCR were designed using Roche Universal Probe Library Assay Design Center (https://www.roche-applied-science.com). %GC content was maintained between 45% and 55%. Designed primers were obtained from Eurofins MWG Operon (http://www.eurofinsdna.com), and TaqMan probes were purchased from Roche.
| Gene | Gene Symbol | Primer Sequence | Roche Library Probe No. | Accession No. |
|---|---|---|---|---|
| Solute carrier organic anion transporter family, member 1B1 (SLCO1B1) | OATP1B1 | gttccatcattcatatagaacggaga (f) | 35 | NM_006446.4 |
| tcaaagcttccgtcaataaaacc (r) | ||||
| Solute carrier organic anion transporter family, member 1B3 (SLCO1B3) | OATP1B3 | catgtaatttggacatgcaagaca | 74 | NM_019844.2 |
| aacatcttaatgaatcaatgcaatgttag | ||||
| Solute carrier organic anion transporter family, member 1A2 (SLCO1A2) | OATP1A2 | ctactttctggccttatgccatc | 42 | NM_134431.3 |
| agatggattccaggtttgaggat | ||||
| Solute carrier organic anion transporter family, member 2B1 (SLCO2B1) | OATP2B1 | agaggctggacttgaacaaacc | 36 | NM_001145212.2 |
| tgtttgtctccctgggtcactt | ||||
| Cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1A1) | CYP1A1 | cctgggaacatcacattcctct | 4 | NM_000499.3 |
| agaaggcagccctgtttgttc | ||||
| Glutathione peroxidase 1 | GPX | tctgttgctcgtagctgctgaa | 55 | ENST00000419349.1 |
| ctatgagtcaccgggattttgc | ||||
| GSTA1-001 glutathione S-transferase alpha 1 | GST | tgggagagaactattgagaggaaca | 85 | ENST00000334575.5 |
| tgttaaacgctgtcaccgtcc | ||||
| Human multidrug resistance protein (MDR1) gene enhancer | MDR1 | tttcctggttagtcaatgggaga | 6 | M57451.1 |
| ggtgctcttggtttgctcaca | ||||
| Human P-glycoprotein gene (MDR2), unknown exon, clone pHDR2.6 | MDR2 | tgaagggcctcaacctgaag | 81 | M28477.1 |
| tcttcccacagccactacttcc | ||||
| Multidrug resistance-associated protein (MRP)-likeprotein-2 (MLP-2) | MLP-2 | cctggctgtgctctacacct | 5 | AB010887.1 |
| tgattccagccgcttcagtt | ||||
| Glyceraldehyde-3-phosphate dehydrogenase (GADPH) | GADPH | ctctgctcctcctgttcgaca | 60 | AF261085.1 |
| aatccgttgactccgaccttc | ||||
| Topoisomerase (DNA) III beta (TOP3B) | TOP3B | tcttgctgtgggggttcag | 39 | NM_003935.3 |
| tgggagggttcgaagaagaa | ||||
| RNA polymerase II, TATA box binding protein (TBP)-associated factor | TBP | ggtaaccactgttccgaagcc | 5 | NM_005640.1 |
| actgtgacgacactggttgg | ||||
| Elongation factor 2 (EF-2) | EF-2 | aaaggaacacgcgggtagatg | 4 | M19997.1 |
| tcatggtgacaccgcacag | ||||
| Elongation factor-1 alpha (EF-1) | EF-1 | ggaccattgagaagttcgagaag | 60 | L10340.1 |
| agcacccaggcatacttgaag |
2.11 Quantitative PCR
cDNA was prepared using 1.0 μg total RNA per reaction, with random primers and SuperScript® III reverse transcriptase (200U/μl), yielding 120 μl of cDNA per original sample to generate sufficient template for qPCR analysis. The MasterCycle cDNA synthesis program was set at 50 min at 50°C, then 5 min at 95°C, ended on hold at 10°C. The cDNA was later diluted to a total of 360 μl per original sample (1:3 dilution) with nuclease free water based on previous optimization of dilution with particular primer probe combinations. The TaqMan Universal PCR Mastermix was used then to prepare reaction mixtures according to manufacturer instructions with 5 μl of cDNA sample in a final volume of 12 μl for qPCR amplification.
Samples were prepared in 384 well plates with appropriate primer-probe combinations, and targeted gene fragments were amplified in an automated fluorometer (ABI PRISM 7900 Sequence Detection System, Applied Biosystems, Carlsbad, CA, USA). Amplification conditions were 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C and 1 min at 60°C. Fluorescence of samples was measured every 7 s and signals were considered positive if fluorescence intensity exceeded 10 times the standard deviation of the baseline fluorescence (threshold cycle, CT). SDS 2.2.1 software (Applied Biosystems, Carlsbad, CA, USA) was used to quantify transcription.
2.12 Data Analysis
A two-tailed, paired Student’s t-test was used to determine statistical significance in mRNA expression and uptake experiments (aggregate as compared with control 2D) and to determine statistical significance of inhibitors of uptake (relative to uptake in absence of inhibitor) and ROS scavengers (relative to control MC concentration). ANOVA followed by post hoc Holm-Sidak analysis was used to determine significance of the dose dependence ROS curves and MTS analysis. All statistical tests were done using GraphPad Software, Inc (La Jolla, CA).
For qPCR data, the geNorm algorithm was used to estimate the variability of reference genes transcription and determine optimal three genes for normalization of the data (Vandesompele et al., 2002). The data was analyzed using a relative quantification 2(−ΔΔCT) method (Livak and Schmittgen, 2001) and transcription was normalized relative to TATA box binding protein (TBP)-associated factor, elongation factor 2 (EF-2), and glyceraldehyde-3-phosphate dehydrogenase (GADPH) expression. Differences in transcription levels of single genes were evaluated using the Student’s t test comparing expression aggregate and 2d monolayer controls, and each gene was treated as a separate experiment. Data was analyzed in the log2-scale, representing the log2-fold change in gene transcription relative to the reference gene.
3. Results
3.1 Soft lithography fabrication of micro wells and HepG2 aggregate formation
Reusable and reproducible PDMS molds were successfully generated in the NCNC clean room from the microfabrication techniques outlined in the methods and SI materials. Figure 1 contains SEM images of the PDMS molds and PEGDA microwells generated through soft lithography processes and a schematic of the microwell fabrication process outside the clean room. Previous work had demonstrated higher MW PEGDA minimized cell adhesion to microwell surfaces but lower molecular weights avoided swelling and detachment. We also observed that <20% dilutions of prepolymer dissolved in DPBS minimized swelling and detachment from the TMSPMA pre-treated glass slide (Moeller et al., 2008), enabling a longer potential period of culture than performed in these experiments with HepG2 cells and suggests the possible use of this fabrication process with primary cells. The higher molecular PEGDA (MW=700) was selected to minimize cell adhesion and ensure aggregate formation inside the microwells. PDMS molds were reusable for over a period of several months.
Figure 1. PDMS and Microwell Images and Schematic of Soft Lithography Fabrication Process.
The SEM images were taken with a Nova™ NanoSEM Scanning Electron Microscope (SEM) (FEI, Hillsboro, OR) with an acceleration of 5 kV. The PDMS molds (1a and 1b) and micro wells (1c) were rinsed with 70% ethanol, dried under N2 gas, kept under vacuum, and then sputter coated with palladium-platinum alloy. The PDMS posts regularly measured close to 150 μm by 150 μm in height and width. Usually maintained in solution, the micro wells demonstrate shrinkage due to loss of water, but relative size was visually assessed as consistent. Figure 1d shows soft lithography process. TMSPMA pretreated glass slide is placed on platform, followed by PEGDA solution (150 μl), and finally the PDMS mold (shown top left of schematic) for the completion of the “sandwich,” the placed under UV light at 350–500 nm wavelengths of light for 50 s at an intensity of 100 mW/cm2 using OmniCure Series 1000 curing station.
Cell viability was visually assessed on day 1, 3, 5 and 7 of aggregate and 2D growth using calcein-AM (green) and ethidium homodimer (red) LIVE/DEAD staining (Invitrogen). Representative phase contrast and LIVE/DEAD images are found in Figure 2. Good cell viability was observed from day 1 through day 7 of the experiment, even at the center of 150 μm aggregates. However, increased numbers of floating dead cells in both monolayer and aggregate cultures were observed by day 7, most likely due to the high cell density. It should be noted that by day 5 and day 7 in monolayer culture, some aggregate formation was observed due to the high density of cells and tendency for HepG2 cells to form aggregates. Interestingly, aggregates harvested from microwells were observed to maintain aggregate form independent of microwells for up to a week or more of growth (data not shown).
Figure 2. Aggregate and Monolayer HEPG2 Growth at Day 1 and Day 5.
Live/Dead staining was performed with calcein-AM and ethidium homodimer. Cells were grown in six well plates as aggregates or planar culture and visually assessed on day 1, 3, 5 and 7 using a Nikon Eclipse epifluorescence microscope. Cells were incubated in 2 M calcein-AM and 4 M ethidium homodimer in DPBS for 10 minutes at 37°C prior to imaging. All images from day 1 were taken at 20x magnification. The single aggregate at day 5 was taken at 40x and the array of aggregates is taken at 2x magnification. The remaining images were taken at 20x. Note that on day 5, aggregates begin to outgrow confines of micro wells.
3.2 mRNA Expression of OATP transporters increases in HepG2 aggregates relative to 2D
Figure 3 delineates the mean mRNA expression of OATP1B1 and OATP1B3 normalized to GADPH housekeeping gene in both 2D and aggregates cultures at each time point (day 1–7). Significant differences (p<0.05) between mRNA expression levels for aggregates and 2D monolayers were observed for OATP1B1 only at day 3, 5 and 7, while significant differences (p<0.05) for OATP1B3 were found at all time points. We selected day 5 as the time point for all future experiments based on this temporal sequence and visual assessment of cells in culture.
Figure 3.
Figures 3A and 3B. OATP1B1 mRNA and OATP1B3 Expression is Increased in Aggregates as Compared with Traditional Monolayer Culture.
Figure 3A illustrates comparative expression of OATP1B1 in monolayer and aggregates over time, while Figure 3B contains data for OATP1B3 expression. At day 1, 3, 5 and 7, mRNA was extracted with TRIZOL and previously used primers were used with SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen). PCR products were loaded on 2% (w/w) agarose gels containing ethidium bromide. Digital images of the fluorescently labeled amplification products were captured using the Gel Logic 100 Imaging System with Molecular Imaging Software (Eastman Kodak Company, Rochester, NY). Images were analyzed using Image J software (NIH), normalized to loading control and GADPH housekeeping gene. Statistical significance (p<0.05)* between aggregates and control was determined using the student’s two-tailed t-test. Error bars refer to standard error between triplicates.
3.3 Function of OATP transporters increases in HepG2 aggregates relative to 2D monolayers
To evaluate relative functionality of the transporters and determine if an increase in expression in aggregates also correlated with increased uptake of known OATP substrates, we selected two commercially available fluorescent substrates that have been previously validated for screening for drug interactions with OATP1B1 and OATP1B3 (Bednarczyk, 2010; Gui et al., 2010). Figure 4 shows the uptake of 8FcAMP and FMTX substrates at various concentrations in aggregate cells and 2D monolayer cultures. HepG2 aggregates demonstrated significantly increased uptake of fluorescent substrates per μg protein following a 30 min incubation period relative to 2D monolayers at all concentrations with 8FcAMP and at 10 and 50μM for FMTX. Consistent with previous studies cautioning the low extinction coefficient of FMTX (Bednarczyk, 2010; Gui et al., 2010), the relative fluorescence signal of 8FcAMP was stronger and thus this substrate was selected for further uptake inhibition studies. Uptake inhibition experiments were carried out at 10 μM 8FcAMP and are presented in Figure 5. Inhibition of uptake was significant (p<0.05) in 2D monolayers for MC-LR, Cyclosporine D, and Rifampicin treatments and significant (p<0.01) for all treatments (MC-LR, deoxycholic acid, Cyclosporine D and Rifampicin) in HepG2 aggregates.
Figure 4.
Figures 4A and 4B. Transport (Uptake) of FMTX (A) and 8 FcAMP (B) in Aggregate HEPG2 and Monolayer Control HEPG2.
Cells were harvested at day 5 of growth and seeded in 24 well plates. The following day, uptake was determined by incubation with indicated concentration for 30 minutes on 24 well plates. Transport was stopped with ice cold PBS; membranes were disrupted with 1% Triton X and fluorescence was measured at an excitation wavelength of 485 nm and emissions wavelength of 528 nm. Fluorescent units were normalized to protein content determined by BCA assay. Values represent means with standard error (SE) of three independent experiments each performed with quadruplicate determinations. Significance was determined with a paired student’s t test with a p value <0.05 (*) considered significant and p<0.01 (**) very statistically significant.
Figure 5. Inhibition of Uptake of 8FcAMP by Various Competitive OATP Substrates in Monolayer and Aggregate Culture.
Uptake of 10 μM 8FcAMP co-incubated for 30 minutes with 50 μM of various known OATP substrates in both monolayer and aggregate culture. Fluorescent units were normalized to protein content determined by BCA assay. Values represent means with standard error (SE) of three independent experiments each performed with quadruplicate determinations. Significance was determined with a paired student’s t test with a p value <0.05 (*) considered significant and p<0.01 (**) very statistically significant.
3.4 Exposure to MC-LR results in increased ROS
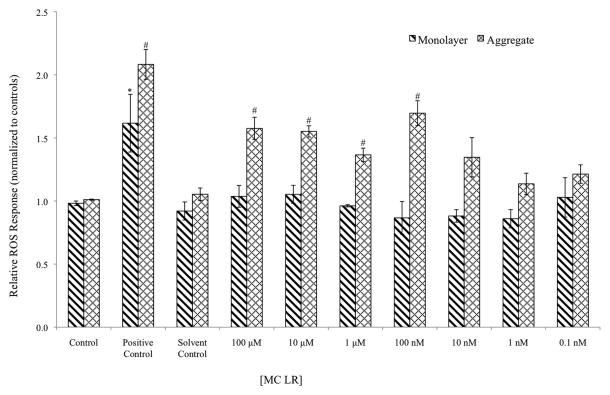
A significant increase in ROS production in aggregate cells was observed three hours following MC-LR exposure as compared with 2D monolayer cultures. The lowest concentration with significant ROS production in aggregate cells was 100 nM (p<0.001) but it did not cause significant amounts of ROS production at any concentration (0.1 nM to 100 μM) in 2D monolayer cultures. The dose dependent response is shown in Figure 6. Here we report a much higher sensitivity to MC-LR in aggregate cultures of HepG2 cells as compared with existing work using 2D HepG2 monolayers. One previous study demonstrated a ROS response following 1 mM MC-LR exposure in 2D HepG2 monolayers after 5 hours (Zegura et al., 2004). The H2DCFDA probe we used to measure ROS production is non specific for any particular oxidative stress pathway. It diffuses freely across cell membranes, is de-acetylated by intracellular esterases to DCFH, which is oxidized by a variety of free radicals to form the fluorescent compound DCF(Halliwell and Whiteman, 2004). We next tested the effect of co-incubation of MC-LR in combination with various scavengers: the antioxidant ascorbic acid, the iron-chelating agent deferoxamine (DFO), and catalase. Free radical scavenging by all three scavengers appears to be protective against MC-LR induced ROS generation inside HepG2 aggregates (Figure 7).
Figure 6. Dose Dependent ROS response in HEPG2 Response Following Microcystin LR Exposure.
At day 5 of growth, cells were seeded in 96 well plates, incubated for 24 hours, and then treated for three hours with the appropriate concentration of microcystin LR or control. Following the incubations, the cells were washed with DPBS and the incubated with 40 μM H2DCFDA in HBSS for 30 minutes. The fluorescence intensity of the final oxidized product DCF was determined at an excitation wavelength of 480 nM and emission wavelength of 538 nM. Background fluorescence was subtracted and data were normalized to control. Data represent means with standard error of three separate experiments with at least six replicates of each treatment. A two way ANOVA was performed with a post-hoc Holm Sidak analysis within groups. Significant difference of monolayer treatments as compared with monolayer control is designated by * (p<0.01), while significant difference between aggregate treatments and aggregate control is designated by # (p<0.01).
Figure 7. Co-incubation with Scavengers Reduces ROS Response in HEPG2 Aggregates Following Microcystin LR Exposure.
Cells were seeded in 96 well plates, incubated for 24 hours, and then treated with either scavengers alone or 1 μM MC-LR co-incubated with scavenger. 10 μM vitamin C and catalase and 50 μM DFO were used. Appropriate controls (solvent and positive controls) were utilized as well. Following the incubations, the cells were washed with DPBS and then incubated with 40 μM H2DCFDA in HBSS and emissions were measured as described previously. Significance of differences between MC-LR μM alone and co-incubation with scavenger were evaluated with the student’s t-test (*p<0.05).
3.5 Cell Viability as Determined by MTS in HepG2Aggregates and 2D Monolayer Cultures
Cells were treated for 48 hours with appropriate controls and with various concentrations of MC-LR ranging between 1 and 50 μM. Experiments were repeated in triplicate with at least 6 replicates. Cell viability was evaluated using the MTS assay, which assesses mitochondrial function through the reduction of the tetrazolium salt parent compound into its formazan product through cellular dehydrogenases and thus indirectly assesses cell viability. No significant difference in either cell group was observed between treatments and control, suggesting no direct change in mitochondrial function following MC-LR exposure in HEPG2 aggregates or 2D monolayer cultures.
3.6 Select qPCR gene expression differences between aggregate and 2D monolayer cells at day 5
We found significant increases in mRNA expression between the two culture conditions in OATP1B1, OATP1B3, CYP1A1, GPX, GST, MDR1, MDR2 and MLP2, no significant changes were observed for the non liver-specific OATPs (OATP1A2 and OAT2B1). Data were normalized to TATA box binding protein (TBP)-associated factor, elongation factor 2 (EF-2), and glyceraldehyde-3-phosphate dehydrogenase (GADPH) expression, based upon the geNorm algorithm using a relative quantification 2(−ΔΔCT) method. (Figure 8) Significant differences are shown as fold expression levels of selected genes in HepG2 aggregates relative to control. These findings were consistent with previously reported increased CYP expression in HepG2 aggregates compared to 2D cultured cells in addition to other metabolizing genes (Chang and Hughes-Fulford, 2009; Nakamura et al., 2011) and increased expression of export pumps, MDR-1 and MRP-2 (Mueller et al., 2011; Oshikata et al., 2011).
Figure 8. Significantly Increased Expression of Transporters, Metabolizing Genes and Export Pumps in HepG2 Aggregates Compared to Control Monolayer on Day 5.
Significant differences are expressed as fold expression levels of selected genes in HepG2 aggregates relative to control (*p<0.05, **p<0.01). While significant increases in mRNA expression were found for OATP1B1, OATP1B3, CYP1A1, GPX, GST, MDR1, MDR2 and MLP2 on day 5 of culture, no significant changes were observed for the other non-liver specific OATPs (OATP1A2 and OAT2B1). Data were normalized to TATA box binding protein (TBP)-associated factor, elongation factor 2 (EF-2), and glyceraldehyde-3-phosphate dehydrogenase (GADPH) expression, based upon the geNorm algorithm, using a relative quantification 2(−ΔΔCT) method.
4. Discussion
We found increased expression of OATP1B1 and OATP1B3 mRNA and increased functionality of these two transporters as demonstrated by two commercially available substrates (8FcAMP and FMTX) in HepG2 aggregate cells compared to monolayer cultured cells. These results alone are quite significant as drug disposition and elimination is highly dependent upon these transporters and effect of aggregate culture on expression and function, to our knowledge, has not been previously evaluated. Cell based assays likely have underestimated uptake when using immortalized cell lines in monolayer culture and ensuing intracellular effects. The decreased expression in monolayer may account for some of the drug interactions not predicted by in vitro assays and for discrepancies between in vitro and in vivo work in drug development.
8FcAMP has a high aqueous solubility and calculated polar surface area (Bednarczyk, 2010), suggesting it is not lipophilic and unlikely to diffuse passively through the cell membrane. FMTX and 8FcAMP may have differing affinities for OATP1B1 and OATP1B3 respectively, and substrates are not necessarily specific for OATP1B1 and OATP1B3; other transporters not identified in this work could also play a role in MC uptake. We did not find increased mRNA expression of the two other known OATPs present in the liver, but these findings do not guarantee that their function did not alter or account for part of the transport. Much still remains to be determined about the form and function of OATPs; endogenous substrates important for bi-directional transport (such as GSH) may play a role and aggregate culture of HepG2 cells may result in an altered expression profile of those substrates, enhancing function of the OATPS. Spheroids cultured on hydrogels previously have demonstrated structural and functional polarization of influx and efflux pumps (Malinen et al., 2012). These investigators did not find significant differences in expression of either transporters or metabolizing genes between spheroids and monolayers despite increased function; the exact nature of the relationship between expression and function still remains to be determined. With further characterization, aggregate culture could profoundly enhance understanding of the expression, form and function of the OATPs, but is well beyond the scope of the work presented here.
The aggregate culture approach provides a more biologically relevant tool with which to proceed forward in toxicity testing, particularly in contrast with monolayer cell line cultures transfected with specific transporters, which may overestimate delivery into the cell. A major concern with aggregate culture has been whether adequate diffusion of toxins or drugs occurs, potentially making them more resistant to drugs or toxins; the uptake data presented here combined with increased ROS production following incubation with MC-LR indicates that aggregates are able to exceed uptake of macrocyclic anionic organic compounds compared to the uptake in monolayer culture; however, we caution that a more careful characterization of the diffusion is needed to ascertain whether cells at the center of the aggregates are equally affected as compared with the cells on the periphery.
There is a dire need to develop a more realistic in vitro model with which to elucidate the exact mechanism of toxicity of MCs because they are such pervasive toxins in drinking and recreational water and huge gaps with regards to risk assessment around the well over 100 congeners remain (Monks and Moscow, 2010). Aggregate culture holds promise to address widely disputed mechanisms of action (Campos and Vasconcelos, 2010), observed variability observed between cell lines (Chong et al., 2000; de Souza Votto et al., 2007; Pichardo et al., 2005) and discrepancy with greater sensitivity in primary cells and toxicity occurring within minutes within those lines (Boaru et al., 2006; Jasionek et al., 2010). Previous work with MC-LR in HEPG2 cells has implicated its genotoxic effect (Zegura et al., 2004) and resulted in reactive oxygen species (ROS) production (Nong et al., 2007) at μM concentrations, including increased DNA strand breaks, 8-hydroxydeoxiguanosine formation, lipid peroxidation and LDH release, following 24 or 48 hour exposures to MC-LR. In contrast, we have demonstrated an increased ROS response in aggregates at nM concentrations of MC-LR after only 3 hours of exposure. Our data indicates that existing literature may underestimate the ROS effect of MC-LR exposure in HepG2 and in other cell types. While the presence and absence of MDR was identified as critical for determining susceptibility to MC-LR oxidative stress and cytotoxicity (de Souza Votto et al., 2007), the authors did not discuss the potential role of OATP uptake. Interestingly, our findings were perhaps more consistent with work done in primary rat hepatocytes that demonstrated the kinetics of ROS formation after only 60 minutes of MC-LR exposure at previously predicted non cytotoxic levels (2 and 10 nM) (Bouaicha and Maatouk, 2004).
Aggregate culture of cell lines provides a comparative tool to primary liver cells with which to examine altered expression and function and resulting change in measure of toxic effect. With respect to MCs, it provides an assay with which to evaluate comparative in vitro effects of congeners with potential to substantially enhance risk assessment. Since protein phosphatases (PP) are direct targets of MCs but protein phosphatase inhibition by congeners has not correlated consistently with congener in vitro or in vivo toxicity (Blom and Juttner, 2005; Gehringer, 2004), evaluation of PP activity in the context of MC toxicity in aggregates may help to further delineate the role of this assay in risk assessment. We did not measure the PP activity here as a toxic endpoint because of the possible wide ranging time and concentration dependent effects of MCs on both protein phosphatase inhibition and upregulation (Li et al., 2011; Liang et al., 2011).
The observed increased ROS production may be an indication of increased transport of MC into the cells due to the increased presence and function of OATP1B1 and OATP1B3. Alternatively, the increased expression and function of other intracellular processes (such as phase I metabolism) with potential to increase generation of free radicals could also play a role in altering the response of cells to the toxin. For example, the increased expression and function of cytochrome P450 or increased use of glutathione in bi-directional transport of both uptake and export pumps (Ballatori et al., 2005; Homolya et al., 2003) could result in more free radical generation through depletion of intracellular glutathione. Previous work has indicated not only increased baseline expression of P450 enzymes but also increased inducibility in HepG2 aggregates relative to control (Nakamura et al., 2011). The increased expression found of CYP1A1 in this study supports this growing body of evidence.
ROS production following MC-LR exposure may not correlate directly with mitochondrial function and cell death, as evidence marked ROS generation in the absence of cell death. Our work supports previous work in HepG2 and Jurkat cells that demonstrated changes in oxygen consumption patterns following facilitated endocytosis down to 0.1 nM of MC-LR with no effect in untreated Jurkat and HepG2 (Jasionek et al., 2010). Changes in “respiratory response” did not correspond with changes in viability, total cellular ATP, extracellular acidification, ROS formation, or protein phosphorylation. Our work did not demonstrate mitochondrial toxicity up to 50 μM in HepG2 either in aggregates or monolayer. Previous work in HepG2 has shown resistance to changes in cell viability up to similar μM concentrations. One study did observe cell death at 48 and 72 hours following 30 and 100 μM (Nong et al., 2007), but a contradictory study showed no change in cell viability for 48, 72 and 96 hours for a similar concentration range (Chong et al., 2000). Despite ROS production, microcystin may have not reached the mitochondria or perhaps was being metabolized or eliminated through export pumps prior to resulting in further toxic effect. We have shown increased expression of phase II genes (GPX and GST) and export pumps (MDR1, MDR2, MLP2) in the aggregates relative to monolayer, supporting previous work that also demonstrated increased function of export pumps, resulting in reduced efficacy of pharmaceutical compounds (Mueller et al., 2011; Oshikata et al., 2011). Aggregate cultured HepG2 in fact may provide an excellent model for screening for therapeutic compounds for targeting tumors because of the ability to tolerate higher ROS. A more realistic model for in vivo toxicity of compounds like MCs may rely on the use of non tumor cell lines or non-differentiated hepatocytes. HEPA RG have emerged in recent years as a cell line with more in vivo like metabolism and excretion pathways (Le Vee et al., 2006). Culturing HEPA RG as aggregates may provide a structure and function even more closely mimicking in vivo. Regardless, the data presented here introduces caution in interpretation of previous work performed in monolayer culture and a platform with which too proceed forward with further characterization and validation.
In conclusion, we have demonstrated increased expression of the two primary liver OATPs (OATP1B1 and OATP1B3) in aggregate HepG2 control and apparent increased function as evidenced by two fluorescently tagged established substrates. We have also demonstrated a significantly increased ROS response of HepG2 aggregate cultured cells following a three-hour MC-LR exposure in concentrations as low as 100 nM, while no response was observed in 2D monolayer cultures. Further characterization and validation of aggregate culture can provide a means to address major risk assessment challenges surrounding MCs in freshwater systems. The substantially altered phenotype and function of these aggregates challenges interpretation of previous data and encourages a more rigorous investigation into the comparative risk from the over 80 MC congeners.
Supplementary Material
Method 1: PhotoMask Generation from L-Edit File.
Method 2: Generation of Silicon Wafer Master
Acknowledgments
Funding information.
This work was supported by the United States Environmental Protection Agency STAR Fellowships for Graduate Student Environmental Study, the National Institutes of Health T32 Training Grant 5TL1RR024145-02, and the ARCS (Achievement Rewards for College Scientists) Foundation of Northern California.
Special thanks to the Khademhosseini laboratory, Harvard MIT Health Sciences for initial training and development of the microwell fabrication and aggregate culture. Additional thanks to Michael Irving at the UC Davis Northern California Nanotechnology Center (NCNC) for further process design of silicon wafers, masks and PDMS molds.
References
- Ballatori N, Hammond CL, Cunningham JB, Krance SM, Marchan R. Molecular mechanisms of reduced glutathione transport: role of the MRP/CFTR/ABCC and OATP/SLC21A families of membrane proteins. Toxicol Appl Pharmacol. 2005;204:238–255. doi: 10.1016/j.taap.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Bednarczyk D. Fluorescence-based assays for the assessment of drug interaction with the human transporters OATP1B1 and OATP1B3. Analytical biochemistry. 2010;405:50–58. doi: 10.1016/j.ab.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom JF, Juttner F. High crustacean toxicity of microcystin congeners does not correlate with high protein phosphatase inhibitory activity. Toxicon. 2005;46:465–470. doi: 10.1016/j.toxicon.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Boaru DA, Dragos N, Schirmer K. Microcystin-LR induced cellular effects in mammalian and fish primary hepatocyte cultures and cell lines: a comparative study. Toxicology. 2006;218:134–148. doi: 10.1016/j.tox.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Bouaicha N, Maatouk I. Microcystin-LR and nodularin induce intracellular glutathione alteration, reactive oxygen species production and lipid peroxidation in primary cultured rat hepatocytes. Toxicol Lett. 2004;148:53–63. doi: 10.1016/j.toxlet.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Campos A, Vasconcelos V. Molecular mechanisms of microcystin toxicity in animal cells. Int J Mol Sci. 2010;11:268–287. doi: 10.3390/ijms11010268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TT, Hughes-Fulford M. Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue engineering Part A. 2009;15:559–567. doi: 10.1089/ten.tea.2007.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MW, Gu KD, Lam PK, Yang M, Fong WF. Study on the cytotoxicity of microcystin-LR on cultured cells. Chemosphere. 2000;41:143–147. doi: 10.1016/s0045-6535(99)00402-6. [DOI] [PubMed] [Google Scholar]
- Dawson RM. The toxicology of microcystins. Toxicon. 1998;36:953–962. doi: 10.1016/s0041-0101(97)00102-5. [DOI] [PubMed] [Google Scholar]
- de Souza Votto AP, Renon VP, Yunes JS, Rumjanek VM, Marques Capella MA, Neto VM, Sampaio de Freitas M, Alicia Geracitano L, Monserrat JM, Trindade GS. Sensitivity to microcystins: a comparative study in human cell lines with and without multidrug resistance phenotype. Cell Biol Int. 2007;31:1359–1366. doi: 10.1016/j.cellbi.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Dietrich D, Hoeger S. Guidance values for microcystins in water and cyanobacterial supplement products (blue-green algal supplements): a reasonable or misguided approach? Toxicol Appl Pharmacol. 2005;203:273–289. doi: 10.1016/j.taap.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Fischer A, Hoeger SJ, Stemmer K, Feurstein DJ, Knobeloch D, Nussler A, Dietrich DR. The role of organic anion transporting polypeptides (OATPs/SLCOs) in the toxicity of different microcystin congeners in vitro: a comparison of primary human hepatocytes and OATP-transfected HEK293 cells. Toxicol Appl Pharmacol. 2010;245:9–20. doi: 10.1016/j.taap.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Fischer WJ, Altheimer S, Cattori V, Meier PJ, Dietrich DR, Hagenbuch B. Organic anion transporting polypeptides expressed in liver and brain mediate uptake of microcystin. Toxicol Appl Pharmacol. 2005;203:257–263. doi: 10.1016/j.taap.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Gehringer MM. Microcystin-LR and okadaic acid-induced cellular effects: a dualistic response. FEBS Lett. 2004;557:1–8. doi: 10.1016/s0014-5793(03)01447-9. [DOI] [PubMed] [Google Scholar]
- Gui C, Obaidat A, Chaguturu R, Hagenbuch B. Development of a cell-based high-throughput assay to screen for inhibitors of organic anion transporting polypeptides 1B1 and 1B3. Current chemical genomics. 2010;4:1–8. doi: 10.2174/1875397301004010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? British journal of pharmacology. 2004;142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf C, Ahlin G, Seithel A, Artursson P, Ungell AL, Karlsson J. Expression of thirty-six drug transporter genes in human intestine, liver, kidney, and organotypic cell lines. Drug Metab Dispos. 2007;35:1333–1340. doi: 10.1124/dmd.107.014902. [DOI] [PubMed] [Google Scholar]
- Homolya L, Varadi A, Srakadi B. Multidrug resistance-associated proteins: Export pumps for conjugates with glutathione, glucoronate or sulfate. Biofactors. 2003;17:103–114. doi: 10.1002/biof.5520170111. [DOI] [PubMed] [Google Scholar]
- Hooser SB, Beasley VR, Lovell RA, Carmichael WW, Haschek WM. Toxicity of microcystin LR, a cyclic heptapeptide hepatotoxin from Microcystis aeruginosa, to rats and mice. Vet Pathol. 1989;26:246–252. doi: 10.1177/030098588902600309. [DOI] [PubMed] [Google Scholar]
- Hooser SB, Beasley VR, Waite LL, Kuhlenschmidt MS, Carmichael WW, Haschek WM. Actin filament alterations in rat hepatocytes induced in vivo and in vitro by microcystin-LR, a hepatotoxin from the blue-green alga, Microcystis aeruginosa. Vet Pathol. 1991;28:259–266. doi: 10.1177/030098589102800401. [DOI] [PubMed] [Google Scholar]
- Jasionek G, Zhdanov A, Davenport J, Blaha L, Papkovsky DB. Mitochondrial toxicity of microcystin-LR on cultured cells: application to the analysis of contaminated water samples. Environ Sci Technol. 2010;44:2535–2541. doi: 10.1021/es903157h. [DOI] [PubMed] [Google Scholar]
- Khademhosseini A. Experimental approaches to tissue engineering. Journal of visualized experiments : JoVE. 2007:272. doi: 10.3791/272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Furukawa T, Ikeda R, Takumi S, Nong Q, Aoyama K, Akiyama S, Keppler D, Takeuchi T. Involvement of mitogen-activated protein kinase signaling pathways in microcystin-LR-induced apoptosis after its selective uptake mediated by OATP1B1 and OATP1B3. Toxicol Sci. 2007;97:407–416. doi: 10.1093/toxsci/kfm054. [DOI] [PubMed] [Google Scholar]
- Le Vee M, Jigorel E, Glaise D, Gripon P, Guguen-Guillouzo C, Fardel O. Functional expression of sinusoidal and canalicular hepatic drug transporters in the differentiated human hepatoma HepaRG cell line. Eur J Pharm Sci. 2006;28:109–117. doi: 10.1016/j.ejps.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Li CL, Tian T, Nan KJ, Zhao N, Guo YH, Cui J, Wang J, Zhang WG. Survival advantages of multicellular spheroids vs. monolayers of HepG2 cells in vitro. Oncology reports. 2008;20:1465–1471. [PubMed] [Google Scholar]
- Li T, Huang P, Liang J, Fu W, Guo Z, Xu L. Microcystin-LR (MCLR) induces a compensation of PP2A activity mediated by alpha4 protein in HEK293 cells. International journal of biological sciences. 2011;7:740–752. doi: 10.7150/ijbs.7.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Li T, Zhang YL, Guo ZL, Xu LH. Effect of microcystin-LR on protein phosphatase 2A and its function in human amniotic epithelial cells. J Zhejiang Univ Sci B. 2011;12:951–960. doi: 10.1631/jzus.B1100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libra A, Fernetti C, Lorusso V, Visigalli M, Anelli PL, Staud F, Tiribelli C, Pascolo L. Molecular determinants in the transport of a bile acid-derived diagnostic agent in tumoral and nontumoral cell lines of human liver. The Journal of pharmacology and experimental therapeutics. 2006;319:809–817. doi: 10.1124/jpet.106.106591. [DOI] [PubMed] [Google Scholar]
- Liu J, Abate W, Xu J, Corry D, Kaul B, Jackson SK. Three-dimensional spheroid cultures of A549 and HepG2 cells exhibit different lipopolysaccharide (LPS) receptor expression and LPS-induced cytokine response compared with monolayer cultures. Innate immunity. 2011;17:245–255. doi: 10.1177/1753425910365733. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu H, Choudhuri S, Ogura K, Csanaky IL, Lei X, Cheng X, Song PZ, Klaassen CD. Characterization of organic anion transporting polypeptide 1b2-null mice: essential role in hepatic uptake/toxicity of phalloidin and microcystin-LR. Toxicol Sci. 2008;103:35–45. doi: 10.1093/toxsci/kfn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinen MM, Palokangas H, Yliperttula M, Urtti A. Peptide nanofiber hydrogel induces formation of bile canaliculi structures in three-dimensional hepatic cell culture. Tissue engineering Part A. 2012;18:2418–2425. doi: 10.1089/ten.TEA.2012.0046. [DOI] [PubMed] [Google Scholar]
- Meriluoto JA, Spoof LE. Cyanotoxins: sampling, sample processing and toxin uptake. Adv Exp Med Biol. 2008;619:483–499. doi: 10.1007/978-0-387-75865-7_21. [DOI] [PubMed] [Google Scholar]
- Moeller HC, Mian MK, Shrivastava S, Chung BG, Khademhosseini A. A microwell array system for stem cell culture. Biomaterials. 2008;29:752–763. doi: 10.1016/j.biomaterials.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks NR, Liu S, Xu Y, Yu H, Bendelow AS, Moscow JA. Potent cytotoxicity of the phosphatase inhibitor microcystin LR and microcystin analogues in OATP1B1- and OATP1B3-expressing HeLa cells. Molecular cancer therapeutics. 2007;6:587–598. doi: 10.1158/1535-7163.MCT-06-0500. [DOI] [PubMed] [Google Scholar]
- Monks NR, Moscow JA. Addressing the design of in vitro models for the investigation of microcystins: the essential role of transporter-mediated uptake. Cell Biol Int. 2010;34:907–909. doi: 10.1042/CBI20100244. [DOI] [PubMed] [Google Scholar]
- Mueller D, Koetemann A, Noor F. Organotypic Cultures of HepG2 Cells for In Vitro Toxicity Studies. J Bioengineering & Biomedical Sciences. 2011:S2. [Google Scholar]
- Nakamura K, Kato N, Aizawa K, Mizutani R, Yamauchi J, Tanoue A. Expression of albumin and cytochrome P450 enzymes in HepG2 cells cultured with a nanotechnology-based culture plate with microfabricated scaffold. The Journal of toxicological sciences. 2011;36:625–633. doi: 10.2131/jts.36.625. [DOI] [PubMed] [Google Scholar]
- Nong Q, Komatsu M, Izumo K, Indo HP, Xu B, Aoyama K, Majima HJ, Horiuchi M, Morimoto K, Takeuchi T. Involvement of reactive oxygen species in Microcystin-LR-induced cytogenotoxicity. Free Radic Res. 2007;41:1326–1337. doi: 10.1080/10715760701704599. [DOI] [PubMed] [Google Scholar]
- Oshikata A, Matsushita T, Ueoka R. Enhancement of drug efflux activity via MDR1 protein by spheroid culture of human hepatic cancer cells. J Biosci Bioeng. 2011;111:590–593. doi: 10.1016/j.jbiosc.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Pichardo S, Jos A, Zurita JL, Salguero M, Camean AM, Repetto G. The use of the fish cell lines RTG-2 and PLHC-1 to compare the toxic effects produced by microcystins LR and RR. Toxicol In Vitro. 2005;19:865–873. doi: 10.1016/j.tiv.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Runnegar M, Berndt N, Kong SM, Lee EY, Zhang L. In vivo and in vitro binding of microcystin to protein phosphatases 1 and 2A. Biochem Biophys Res Commun. 1995;216:162–169. doi: 10.1006/bbrc.1995.2605. [DOI] [PubMed] [Google Scholar]
- Sidler Pfandler MA, Hochli M, Inderbitzin D, Meier PJ, Stieger B. Small hepatocytes in culture develop polarized transporter expression and differentiation. J Cell Sci. 2004;117:4077–4087. doi: 10.1242/jcs.01279. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegura B, Lah TT, Filipic M. The role of reactive oxygen species in microcystin-LR-induced DNA damage. Toxicology. 2004;200:59–68. doi: 10.1016/j.tox.2004.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Method 1: PhotoMask Generation from L-Edit File.
Method 2: Generation of Silicon Wafer Master