SUMMARY
Exercise induces physiological cardiac growth and protects the heart against pathological remodeling. Recent work suggests exercise also enhances the heart’s capacity for repair, which could be important for regenerative therapies. While microRNAs are important in certain cardiac pathologies, less is known about their functional roles in exercise-induced cardiac phenotypes. We profiled cardiac microRNA expression in two distinct models of exercise and found microRNA-222 (miR-222) was upregulated in both. Downstream miR-222 targets modulating cardiomyocyte phenotype were identified, including HIPK1 and Homeobox-1. Inhibition of miR-222 in vivo completely blocked cardiac and cardiomyocyte growth in response to exercise, while reducing markers of cardiomyocyte proliferation. Importantly, mice with inducible cardiomyocyte miR-222 expression were resistant to adverse cardiac remodeling and dysfunction after ischemic injury. These studies implicate miR-222 as necessary for exercise-induced cardiomyocyte growth and proliferation in the adult mammalian heart and show that it is sufficient to protect the heart against adverse remodeling.
INTRODUCTION
Heart failure is a growing cause of morbidity and mortality throughout the world, and often develops after a period of abnormal growth termed pathological hypertrophy. Loss of cardiomyocytes contributes to decreased cardiac function and heart failure. While the heart has some regenerative capacity, little is known about what regulates this ability or whether it can be effectively harnessed to mitigate these processes. Thus understanding the pathways that promote cardiomyocyte survival and/or regeneration could have important fundamental and clinical implications.
Many pathways have been implicated in heart disease, but less is understood about what keeps the heart healthy. Clinical and experimental studies document the impact of exercise in both primary and secondary prevention of cardiovascular disease (Lim et al., 2013; Young et al., 2014). Prior work from our groups utilized a genome-wide analysis to compare transcriptional components involved in the exercise response versus those involved in pathological hypertrophy after pressure overload (Bostrom et al., 2010). These studies demonstrated that exercise induces a transcriptional network distinct from that seen with pathological stimuli, even at an early stage when the hearts were structurally and functionally indistinguishable. In addition, physiological growth was associated with transcriptional components linked to cell cycle progression, and exercised hearts showed an increase in proliferation markers, specifically in cells expressing cardiomyocyte sarcomere proteins (Bostrom et al., 2010). These data suggested that exercise may provide physiological cues that enhance the heart’s limited endogenous capacity for regeneration, similar to effects that have been documented in other organ systems, including the brain (van Praag et al., 1999; van Praag et al., 2005; Zhang et al., 2008). These studies also implicated a transcriptional network regulated by C/EBPβ and CITED4, which appeared central to the cardiac exercise response and protected the heart against adverse remodeling (Bostrom et al., 2010).
MicroRNAs have been shown to regulate entire gene expression networks and play important roles in cardiovascular disease (Small and Olson, 2011). Although several studies have examined microRNAs regulated by exercise (Care et al., 2007; D. A. Silva ND et al., 2012; Fernandes et al., 2011; Martinelli et al., 2014; Soci et al., 2011), less is known about their functional roles in this context. To identify microRNAs that are differentially regulated and functionally important in exercised hearts, we examined cardiac expression of all known mouse microRNAs in two distinct models of endurance exercise. Concordant changes were identified and validated before screening for functional effects in cardiomyocytes in vitro. Here we report that cardiac miR-222 is upregulated robustly in both exercise models, and induces cardiomyocyte hypertrophy and proliferation in vitro through effects on the cycle inhibitor, p27, as well as HIPK1 and Hmbox1, acting upstream of CITED4. miR-222 was necessary for exercise-induced cardiac growth in vivo and genetically mediated miR-222 expression was sufficient to protect the heart against dysfunction and adverse remodeling after ischemic injury.
RESULTS
Expression of miR-222 is induced during physiological hypertrophy
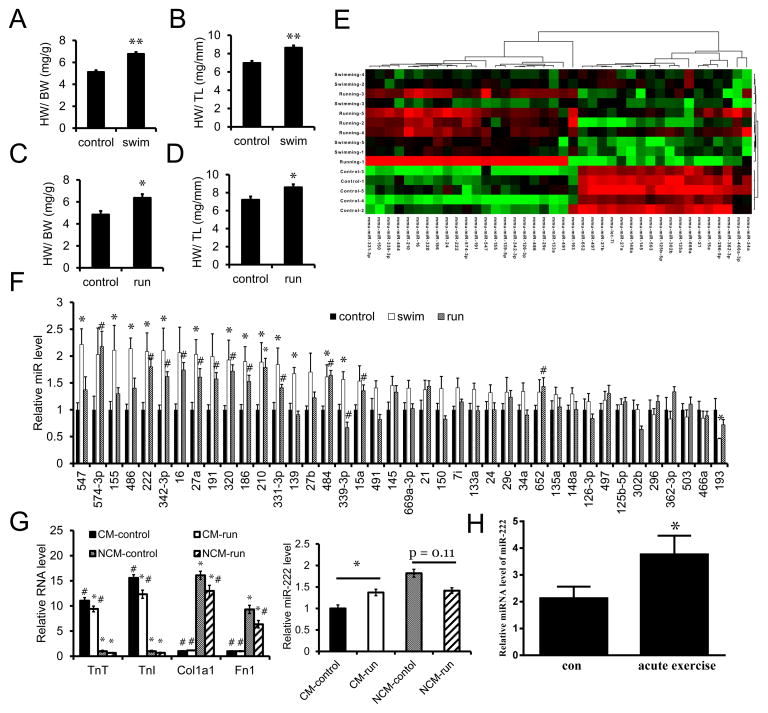
To identify microRNAs that regulate physiological cardiac growth, we subjected mice to either voluntary wheel running or a ramp swimming exercise model (Taniike et al., 2008) for three weeks. As shown in Figure 1A–D, both models induced mild cardiac hypertrophy, and microRNAs were profiled in heart samples from each exercise model in comparison to sedentary controls using the TaqMan rodent miRNAarray (A+B set v3.0), which includes 641 unique assays specific to mouse. Five cardiac samples were used from mice exercised in each of the models for three weeks in comparison to matched sedentary controls. Fifty-five microRNAs were differentially expressed in hearts from swum mice while 124 were differentially expressed in hearts from wheel-run mice, including 16 microRNAs that were concordantly regulated and robustly validated in both models (Figure 1E–F and Table S1).
Figure 1. MicroRNAs are differentially regulated by exercise.
A. and B. Heart weight/body weight (HW/BW) and heart weight/tibia length (HW/TL) ratios of sedentary control (control) and swum (swim) mice. n=9 mice per group. C. and D. HW/BW and HW/TL ratios of sedentary control (control) and voluntary-wheel running (run) mice. n=4 mice per group. E. Heat map of 40 differentially regulated miRNAs concordantly altered in hearts from swimming and running mice compared to sedentary controls. n=3 with 3 mouse hearts per pool. F. qRT-PCR analysis of the identified 40 differentially regulated miRNAs in hearts from separate cohorts. n=5 hearts per group. G. qRT-PCR analysis of miR-222 expression in isolated adult cardiomycytes and non-cardiomycytes from voluntary wheel-running mice. n=4 mouse hearts per group. H. qRT-PCR analysis of miR-222 levels in serum from heart failure patients before (con) and after acute exercise. Data are shown as fold change of miR-222 expression normalized to spiked cel-miR-39. n=28. *p<0.05, #p<0.05, **p<0.01 versus respective control using Student’s test. Data shown as mean±SEM.
The functional effects of all sixteen candidate microRNAs were examined in cardiomyocytes in vitro using commercially available microRNA precursors. Since cardiomyocyte hypertrophy is central to the acute growth of the heart in response to exercise, we used a change in cell size as an end-point for functional effects (Figure S1A). In addition, our prior work had suggested exercise could induce a proliferative response that might contribute to cardiomyogenesis, and thus we also examined incorporation of the thymidine analogue, EdU, in response to microRNA mimic treatment (Figure S1B). As shown in Table S1, miR-222 emerged as a particularly interesting candidate that was increased 2.1- and 2.8-fold in the swimming and running exercise models, respectively (p<0.003 and 0.02). Studies in cardiomyocytes and non-cardiomyocytes isolated from sedentary and exercised hearts demonstrated that after exercise, miR-222 increased in adult cardiomyocytes (p<0.05) but showed a non-significant trend toward decreasing in non-cardiomyocytes (Figure 1G). miR-222 was one of two microRNAs (along with miR-191) that increased cardiomyocyte size in vitro, and one of four (along with miR-139, -27a and -484) that increased EdU incorporation (Figure S1A and S1B). Subsequent studies showed that only miR-222, miR-339, and miR-486 induced a physiological pattern of myosin heavy chain isoform expression (Figure S1C). Based on the unique convergence of these functional characteristics in cardiomyocytes in vitro and its exercise-induced increase specifically in cardiomyocytes in vivo, miR-222 was selected for more detailed study.
miR-222 is a highly conserved member of a microRNA cluster encoded on the X chromosome, which also includes miR-221 (Felli et al., 2005; Galardi et al., 2007). Although its function in the heart is unknown, miR-222 has documented roles in regulating cell proliferation and differentiation in vascular smooth muscle cells and some cancers (le Sage et al., 2007; Liu et al., 2009), at least in part through targeting the cell cycle inhibitor, p27 (Kim et al., 2009; Liu et al., 2009; Wurz et al., 2010). Intriguingly, miR-222 has been reported to increase in the plasma of athletes after both acute and chronic exercise (Baggish et al., 2011), suggesting potential human relevance of our observations. Of note, expression of neither miR-221 in hearts nor miR-222 in skeletal muscle was increased in our exercised mice (data not shown).
miR-222 increases after exercise in heart failure patients
Since miR-222 has been reported to increase in the peripheral blood of healthy young athletes after exercise (Baggish et al., 2011) and exercise has beneficial effects in heart failure patients (Flynn et al., 2009; O’Connor et al., 2009), we wondered whether these observations could be related. As an initial step, we examined changes in circulating miR-222 in twenty-eight heart failure patients after cardiopulmonary exercise testing using a bicycle ergometer. Subjects included chronic stable heart failure patients (NYHA Class II–IV) with both preserved and reduced systolic function (Table 1). Exercise duration ranged from 2.5 to 11 minutes on a standardized protocol (Myers, 2005). Interestingly, circulating miR-222 increased 1.8-fold (p=0.01) after exercise (Figure 1H); a similar pattern to that seen in young athletes (Baggish et al., 2011).
Table 1.
Patient information.
| Heart failure with reduced EF (HFrEF) (EF < 55%) |
Heart failure with preserved EF (HFpEF) (EF ≥ 55%) |
|
|---|---|---|
| Number | 19 | 9 |
| Sex | Male | Male |
| Age (year) | 56.71±10.35 | 63.70±16.28 |
| BMI | 26.32±2.52 | 24.07±2.34 |
| EF (%) | 39.37±6.27 | 65.22±5.54 |
| NYHA Class | 2.74±0.45 | 2.00±0 |
Values are mean ± SD
miR-222 promotes cardiomyocytes growth, proliferation, and survival in vitro
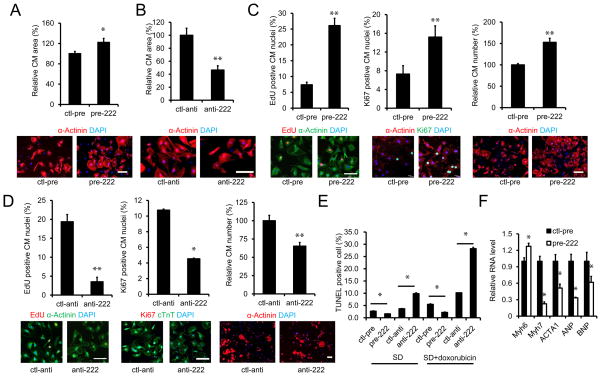
Gain- and loss-of-function were used to examine miR-222’s roles in neonatal cardiomycytes. Expression of miR-222 induced an increase in cardiomyocyte size (Figure 2A). miR-222 expression also increased markers of cardiomyocyte proliferation and cell number (Figure 2C). Conversely, a specific miR-222 antagomir inhibitor reduced cardiomyocyte size and proliferation (Figure 2B and D). In addition, miR-222 expression suppressed while miR-222 inhibition increased cardiomyocyte apoptosis induced by serum deprivation alone or in combination with doxorubicin (Figure 2E). Moreover, miR-222 induced a pattern of gene expression in cardiomyocytes consistent with physiological growth, including an increase in myosin heavy chain (MHC)-α/β ratio, as well as decreases in expression of ANP, BNP, and α-skeletal actin mRNAs (Figure 2F). These data indicate that miR-222 expression is sufficient to induce a complex pattern of physiological growth in neonatal cardiomyocytes including cellular hypertrophy and proliferation while inhibiting apoptosis.
Figure 2. miR-222 regulates cardiomyocyte hypertrophy, proliferation and apoptosis in vitro.
By using miR-222 gain- and loss-of-function in NRVMs, miR-222’s effects on cardiac cell size, proliferation, and hypertrophic markers were examined. A and B: Immunohistochemical staining for sarcomeric α-actinin followed by quantification of cardiomyocyte area as described in methods. Cells were transfected with control or miR-222 precursor in A and with control antimiR (ctl-anti) or antimiR-222 (anti-222) in B. At least 200 cells were quantified in each group. These data demonstrate miR-222 is necessary and sufficient to induce cardiomyocyte hypertrophy. C. Quantification of EdU and Ki67 staining, as well as cell number from primary NRVMs transfected with control precursor (ctl-pre) or miR-222 precursor (pre-222). D. Quantification of EdU and Ki67 staining, and cell number from NRVMs transfected with control antimiR (ctl-anti) or antimiR-222 (anti-222). These data demonstrate miR-222 is necessary and sufficient to induce proliferation of NRVMs. E. Flow cytometry analysis of TUNEL staining in cardiomyocytes treated with control precursor (ctl-pre) or miR-222 precursor (pre-222) or control antimiR (ctl-anti) or antimiR-222 (anti-222). Cardiomyocye apoptosis was induced by serum deprivation (SD) or serum deprivation plus doxorubicin (SD+doxorubicin). These data demonstrate miR-222 inhibits cardiomyocyte apoptosis. F. QRT-PCR for markers of cardiomyocyte hypertrophy and/or pathology in NRVMs treated with control (ctl-pre) or miR-222 precursor (pre-222). These data demonstrate that miR-222 induces a physiological pattern of gene expression. Data are shown as mean±SEM fold-change of gene expression normalized to U6 and reflect at least three independent experiments. Scale bar: 100 μm. *p<0.05, **p<0.01 versus respective control using Student’s test.
miR-222 regulates growth and proliferation of cardiomyocytes in vitro through targeting p27, HIPK1, and Hmbox-1
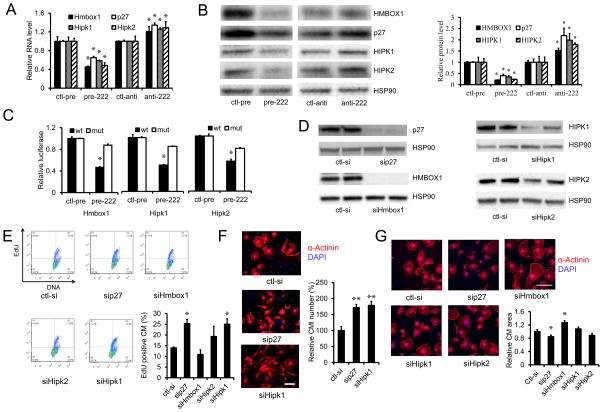
To identify the responsible downstream mechanisms for miR-222’s effects in cardiomyocytes, we performed transcript profiling on cardiomyocytes expressing miR-222 in comparison to scrambled control. These data have been deposited in the Gene Expression Ommibus (GEO accession number: GSE59641). Expression profiling revealed that miR-222 induced an increase in CITED4 mRNA levels, indicating cross-talk with the transcriptional pathway linked to exercise that we reported earlier (Bostrom et al., 2010). In GO pathway analyses, the most upregulated functional pathway was “Cell Cycle” (p<10−8). Comparison of expression profiling results with miR-222 targets predicted by two bioinformatic programs (Targetscan, Pictar) identified four relevant potential targets whose expression decreased in cardiomyocytes with miR-222 expression. These include the known target and cell cycle inhibitor, p27(kip1) (Kim et al., 2009; Liu et al., 2009; Wurz et al., 2010) as well as three additional targets: the protein kinases HIPK-1 (Isono et al., 2006; Li et al., 2005) and -2 (D’Orazi et al., 2002), and a transcriptional repressor, Hmbox1 (Chen et al., 2006). Interestingly, genetic deletion of p27 induces cardiomyocyte hyperplasia in vivo (Fero et al., 1996; Kiyokawa et al., 1996; Nakayama et al., 1996) and physiological cardiac hypertrophy (Hauck et al., 2008) but accelerates the development of heart failure after biomechanical stress (Hauck et al., 2008). The other three candidates (HIPK1, HIPK2, and Hmbox1) have not previously linked to miR-222 and have no known roles in the heart.
miR-222 expression in cardiomyocytes in vitro was sufficient to reduce expression of all four putative targets (p27, HIPK1, HIPK2, Hmbox-1) from 35 to 55 percent, while a specific miR-222 inhibitor increased their expression (Figure 3A). These changes in RNA levels were paralleled by changes in protein expression for each of the targets (Figure 3B). To determine whether HIPK1, HIPK2, and Homeobox-1, were direct targets of miR-222, we cloned either their wild-type 3′-UTRs or 3′UTRs in which the putative miR-222 binding sites had been mutated, downstream of a luciferase reporter. miR-222 had no effect on luciferase activity of the control reporter without a miR-222 binding site (data not shown). miR-222 induced a reduction in luciferase activity for each of the wild-type 3′UTR constructs but importantly, had no effect when the miR-222 binding sites were mutated (Figure 3C). These data strongly suggest that HIPK1, HIPK2 and Hmbox-1 are each direct targets of miR-222, as has been shown previously for p27 (Galardi et al., 2007).
Figure 3. miR-222 targets in cardiomyocytes.
A and B. qRT-PCR and immunoblotting were used to analyze RNA and protein levels of the four putative miR-222 targets in neonate cardiomyocytes treated with control precursor (ctl-pre), miR-222 precursor (pre-222), control antimiR (ctl-anti), or antimiR-222 (anti-222), respectively. Data are shown as fold-change in gene expression normalized to U6 in (A). These data demonstrate that miR-222 decreases RNA and protein levels for all four targets in primary cardiomyocytes. C. Luciferase assays of COS7 cells co-transfected with control precursor (ctl-pre) or miR-222 precursor (pre-222) and reporter plasmids containing 3′UTR wild-type or mutated miR-222 binding sites for each of the putative target genes. These data demonstrate all three candidates are direct targets of miR-222. D. Immunoblotting of the four genes in neonate cardiomyocytes transfected with control siRNA (ctl-si) or siRNAs of p27 (sip27), Hmbox1 (siHmbox1), Hipk2 (siHipk2) and Hipk1 (siHipk1) demonstrate effective knock-down for all. HSP90 was used as a loading control. E. Flow cytometry for EdU in neonate cardiomyocytes transfected with indicated siRNAs demonstrates that knockdown of p27 or HIPK1 increases EdU incorporation in cultured cardiomyocytes, consistent with proliferation. F and G. Neonate cardiomyocytes cultures were stained for sarcomeric α-actinin to identify cardiomyocytes, and cardiomyocyte number and area were quantified. Knockdown of p27 and HIPK1 increases cardiomyocyte proliferation (F) while Hmbox1 knockdown increases cardiomyocytes size (G). At least 200 cells or 30 images were quantified in each group. Data represent the mean±SEM from at least three independent experiments. Scale bar: 100 μm. *p<0.05, **p<0.01 versus respective control using Student’s test.
To determine how these targets contribute to miR-222’s effects in cardiomyocytes, we used siRNA knockdown for each target (Figure 3D) and examined effects on relevant cardiomyocyte phenotypes. siRNA knockdown of either p27 or HIPK1 was induced an increase in EdU incorporation (Figure 3E) and the number of cardiomyocytes (Figure 3F). Hmbox-1 knockdown did not affect cell number but increased cell size (Figure 3G). Interestingly, p27 knockdown actually decreased cell size, perhaps reflecting active cell division; HIPK2 had no significant effect in either assay. These data imply that reductions in p27 and HIPK1 are sufficient to induce the proliferative effects of miR-222 in cultured cardiomyocytes, while reduced Hmbox-1 likely contributes to the observed cellular hypertrophy. We then examined whether miR-222 and its targets intersect with the previously described pathway involving C/EBPβ and CITED4. Interestingly, siRNA knockdown of either p27 or Hmbox1 was sufficient to increase CITED4 expression without altering C/EBPβ expression (Figure S2A and S2B). In contrast, simultaneous knockdown of p27 and HIPK1 did not signficantly alter CITED4 expression and actually increased C/EBPβ expression (Figure S2C). Taken together these data suggest that the miR-222 pathway acts upstream of CITED4 but the combinatorial effects of its targets in this regard are complex.
miR-222 is necessary for exercise-induced cardiac growth
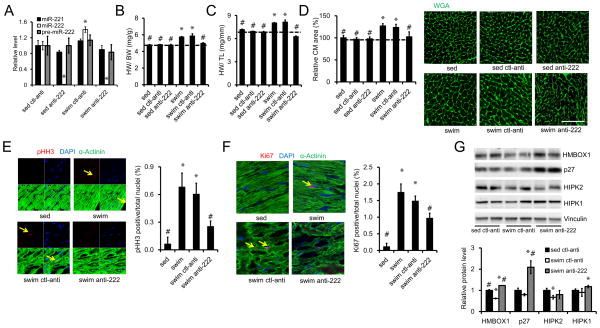
To investigate the role of miR-222 in the cardiac exercise response, we injected mice with LNA-anti-miR-222 or scrambled control LNA-anti-miR for three days prior to exercise and then weekly during the three week swim protocol. LNA-antimiR-222 decreased miR-222 expression in hearts from both sedentary and exercised animals by ≥98% without affecting the expression of pre-miR-222 or the closely related miR-221 (Figure 4A). Exercised but untreated animals showed an increase in cardiac mass reflected in ratios of heart to body weight (HW/BW) or tibial length (HW/TL) which were increased 21 and 15% respectively (p<0.001 for both) compared to sedentary controls (Figure 4B, C). Treatment of sedentary animals for three weeks with either the LNA-antimiR-222 or scrambled control antimiR had no effect on HW/BW or HW/TL (Figure 4B, C). The scrambled LNA-antimiR also did not affect exercise-induced cardiac growth. However, the specific LNA-antimiR-222 completely blocked the increase in heart size induced by three weeks of intensive exercise (Figure 4B, C). As noted earlier, short-term changes in heart size are generally related to changes in cardiomyocyte size and, indeed, examination of cardiomyocyte size on WGA-stained sections revealed a similar pattern. Treatment of sedentary animals for three weeks with LNA-antimiR-222 or scrambled control antimiR had no effect on cardiomyocyte size (Figure 4D). Exercise increased cardiomyocyte size ~20% (p<0.05) in untreated or control LNA-antimiR-treated mice (Figure 4D), while cardiomyocytes in swum mice treated with LNA-antimiR-222 were no different in size from cardiomyocytes in sedentary, untreated mice (p=NS). There was no significant difference in hypertrophic or fetal gene markers between hearts from mice treated with the control or specific LNA-antimiR-222 (Figure S3A). Our prior work had suggested that exercise increases proliferation markers in the cardiomyocyte lineage (Bostrom et al., 2010) and indeed, a substantial increase in both phospho-HistoneH3 (pHH3) and ki67 was again seen in cardiomyocytes from exercised, untreated animals (Figure 4E, F). This increase was not altered by the control antimiR but was decreased, though not reduced to baseline, by LNA-antimiR-222. Taken together, these data demonstrate that miR-222 is necessary for the short-term growth of the heart and cardiomyocytes in response to exercise. Although the change in cardiomyocyte size appears to account for the change in heart size over these short periods of time, it is interesting to note that miR-222 also appears to contribute to the increase in markers of proliferation seen in cardiomyocytes after exercise. Whether these markers of proliferation reflect productive cardiomyogenesis that has an impact over longer periods of time will require long-term cell fate-mapping experiments to address.
Figure 4. miR-222 is necessary for exercise-induced cardiac growth.
A. Sedentary or swum mice were intravenously injected with LNA-antimiR-222 or control LNA-antimiR for 3 weeks prior to quantification of cardiac miR-222 to demonstrate effective reductions of mature miR-222 not pre-miR-222. B and C. HW/BW and HW/TL ratios are shown from sedentary control (sed, n=15) and swum (swim, n=16) mice without injection, sedentary mice injected with control LNA-antimiR (sed ctl-anti, n=7) or LNA-antimiR-222 (sed anti-222, n=7), and swum mice injected with control LNA-antimiR (swim ctl-anti, n=7) or LNA-antimiR-222 (swim anti-222, n=6). These studies demonstrate that LNA-antimiR-222 completely blocks cardiac hypertrophy in response to swimming. D. Quantification of cardiomyocyte area from heart sections stained with wheat germ agglutinin (WGA) (n=5–6, ~500 cells per animal) demonstrate that LNA-antimiR-222 also blocks exercise-induced cardiomyocyte hypertrophy. Immunohistochemical staining for phospho-histoneH3 (pHH3) E and Ki67 (F) from heart sections and quantification (n=5–6 hearts in each group) demonstrate that LNA-antimiR-222 reduces markers of cardiomyocyte proliferation. G. Western blot results show the effect of LNA-antimiR-222 on miR-222 targets in hearts after exercise. Scale bar: 100 μm in D and 50 μm in E and F. Error bars stand for standard errors. *p<0.05, versus sed ctl-anti or sedentary control; #p<0.05 versus swim ctl-anti using One-way ANOVA.
We next examined whether the targets identified in vitro were regulated by exercise or LNA-antimiR-222 in vivo. Three weeks of exercise decreased cardiac Hmbox1 and HIPK2 in mice treated with the control LNA-antimiR, while p27 or HIPK1 were not significantly altered at this time point (Figure 4G). Treatment of exercising mice with LNA-antimiR-222 increased protein levels for both Hmbox1 and p27 in comparison to exercised mice treated with control LNA-anitmiR (Figure 4G). LNA-antimiR-222 did not affect HIPK1 or -2 levels in exercised mice in this assay.
miR-222 protects against cardiac dysfunction after ischemic injury
Since miR-222 specifically increases in cardiomyocytes from exercised hearts, we created inducible (doxycycline-off) cardiomyocyte-specific miR-222 transgenics (Tg-miR-222) (Figure S3A–D) to investigate the in vivo effects of cardiomyocyte miR-222 expression. Tg-miR-222 mice manifested doxycycline-regulated, cardiac-specific miR-222 expression that was ~6.5-fold increased 4 weeks after removal of doxycycline from chow (Figure S4A–C). The induced expression of miR-222 was specific to cardiomyocytes (Figure S4C). Interestingly, Tg-miR-222 mice appeared grossly normal at baseline after induction of cardiac miR-222 expression with normal heart size and cardiac function (Figure S4E and Table S4). Expression of fetal gene or hypertrophic markers was not significantly different after miR-222 induction for 4 weeks (Figure 3SB). Surprisingly, protein expression of HIPK1 and 2, as well as p27 and Hmbox1 were not significantly different in Tg-miR-222 at baseline (Figure S4F). Thus, although miR-222 is necessary for growth of the heart in response to exercise, cardiomyocyte miR-222 expression, even at levels higher than those induced by exercise, is not sufficient to recapitulate the exercised-heart phenotype.
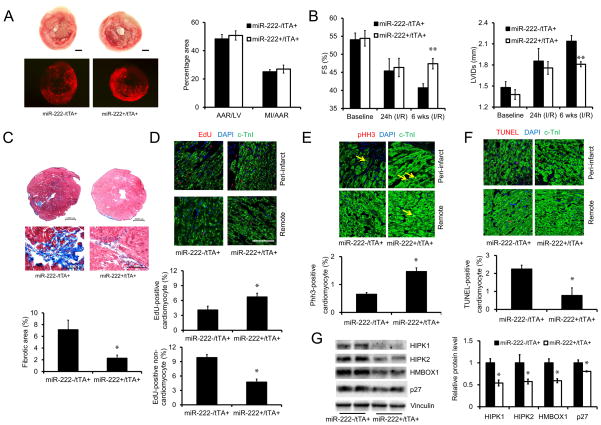
Exercise reduces adverse ventricular remodeling and cardiac dysfunction when initiated after infarction in animal models (Bansal et al., 2010; Wan et al., 2007; Xu et al., 2008; Xu et al., 2010; Yengo et al., 2012) and humans (Haykowsky et al., 2011), attenuating fibrosis, dilatation, and cardiac dysfunction. To assess the possible protective role of miR-222 expression in this context, Tg-miR-222 mice were withdrawn from doxycycline and then subjected to ischemia-reperfusion injury (IRI) induced by 30 minutes coronary ligation and reperfusion. One day after ischemic injury, Tg-miR-222 mice showed no difference in initial infarct size or degree of cardiac dysfunction compared to controls (Figure 5A, B). However, while control mice showed the progressive ventricular dilation and decline in cardiac function known as adverse remodeling, Tg-miR-222 mice did not but maintained both chamber dimension and cardiac function (Figure 5B). Six weeks after ischemic injury, miR-222-expressing mice had better function (Figure 5B) and a 70% reduction in cardiac fibrosis (Figure 5C). Interestingly, in vivo miR-222 expression after ischemic injury was associated with EdU incorporation that was increased in cardiomyocytes but decreased in non-cardiomyocytes, which are predominantly fibroblasts (Figure 5D). Another marker of proliferation, pHH3, was increased 2-fold in cardiomyocytes in hearts of miR-222-expressing mice compared one week after ischemic injury controls (Figure 5E). In contrast, cardiomyocyte apoptosis was reduced 3-fold in hearts of miR-222-expressing mice at the same time-point (Figure 5F). There was no significant difference in expression of hypertrophic markers one week after ischemic injury (Figure S3C). Taken together, these data demonstrate that miR-222 is sufficient to mitigate adverse remodeling after ischemic injury, preserving cardiac structure and function while reducing late scar formation after ischemic injury.
Figure 5. Cardiac-specific expression of miR-222 protects against cardiac remodeling and dysfunction after ischemic injury.
A. Tiphenyltetrazolium chloride (TTC) staining to delineate infarct area and fluorescent microsphere distribution to define the area-at-risk (AAR) in hearts from tTA single (miR-222−/tTA+) and double transgenic (miR-222+/tTA+) mice, 24 hours after reperfusion after ischemia, demonstrate no difference in initial infarction in miR-222 expressing hearts. Representative photographs of TTC staining and fluorescent microsphere distribution (bottom) of medial sections of cardiac tissues are shown (n = 6–7 in each group). Scale bar: 1000 μm. B. Cardiac fractional shortening and left ventricular internal dimension in systole (LVIDs) as measured by transthoracic echocardiography in tTA single (miR-222−/tTA+) and double transgenic (miR-222+/tTA+) mice at baseline, 24 hours or 6 weeks after ischemic injury (n = 8–9 mice in each group). These data demonstrate similar cardiac dysfunction at 24 hours but better function and less dilation in miR-222 expressing hearts at 6 weeks. C. Masson trichrome staining (n=6–7 hearts in each group) demonstrates less fibrosis in miR-222-expressing double transgenic hearts at 6 weeks after ischemic injury. Scale bars are shown. D and E. Immunofluorescence demonstrates increased EdU incorporation in cardiomyocytes in miR-222-expressing hearts 6 weeks after ischemic injury but reduced EdU incorporation in non-cardiomyocytes (n=5 animals in each group, ~2500 cells counted per animal) (D) and marker of cardiomyocyte proliferation phospho-histoneH3 (pHH3) a increases in miR-222-expressing hearts 1 week after ischemic injury in cardiomyocytes (n=4 hearts in each group) (E). F. TUNEL staining demonstrates reduced apoptosis in cardiomyocytes in miR-222-expressing hearts 1 week after ischemia injury (n=4 hearts in each group). G. Immunoblotting shows reduced expression of HIPK1, HIPK2, HMBOX1 and p27 in miR-222-expressing hearts 1 week after ischemic injury (n=4 hearts in each group). Scale bar: 100 μm. Data shown as mean±SEM. *p<0.05, **p<0.01 versus respective control using Student’s test.
We examined the targets identified in vitro in transgenic and control animals one week after ischemic injury. Interestingly, protein expression of all four targets was significantly reduced in Tg-miR-222 hearts after ischemic injury (Figure 5G). These data are consistent with the hypothesis that one or more of these targets could contribute to the protection observed in vivo. However, establishing such a causal link will require future studies assessing their in vivo functional contributions, individually and/or in combination.
DISCUSSION
Despite a wealth of evidence supporting the cardiovascular benefits of exercise, the mechanisms responsible, particularly in the heart, remain poorly understood. microRNAs have emerged as central regulators of cardiac gene expression and key participants in a variety of cardiac pathologies (Small and Olson, 2011). Others have also examined microRNAs that are dynamically regulated by exercise (Care et al., 2007; D. A. Silva ND et al., 2012; Fernandes et al., 2011; Martinelli et al., 2014; Soci et al., 2011), although less is known about their functional roles in exercise or their ability to mitigate cardiac pathologies. Here we provide evidence for a cardiac pathway induced by exercise and mediated by miR-222, which appears important in both. miR-222 was necessary for exercise-induced growth of cardiomyocytes and overall heart size. Interestingly, while miR-222 expression was not sufficient to recapitulate the cardiac exercise phenotype at baseline, it did protect against adverse ventricular remodeling and cardiac dysfunction after ischemic injury. Moreover, these effects were associated with inhibition of cardiomyocyte apoptosis as well as a dramatic decrease in fibrosis late after ischemic injury. Since miR-222 expression also protected isolated primary cardiomyocytes from cell death and miR-222 expression in Tg-miR-222 mice is cardiomyocyte-specific, it seems likely that improved cardiomyocyte survival is a primary contributor to the benefits observed and that reduced fibrosis (and fibroblast proliferation) is secondary to this. However, additional contributions from paracrine cross-talk cannot be excluded based on these studies.
The short-term growth of the heart in response to exercise appears to occur primarily through changes in cardiomyocyte size. Indeed the inhibition of exercise-induced cardiac growth by LNA-antimiR-222 corresponded well with inhibition of cardiomyocyte hypertrophy. Thus the increased markers of cell proliferation, specifically in the cardiomyocyte lineage, seen here with exercise, as in previous reports from our own groups (Bostrom et al., 2010) and others (Waring et al., 2012) are unlikely to have an important impact over the short time periods examined. Moreover, markers of proliferation alone cannot establish whether exercise stimulates an increase in cardiomyogenesis. Multiple lines of evidence suggest the heart has some endogenous capacity for repair (Bergmann et al., 2009; Bersell et al., 2009), and exercise has been reported to enhance endogenous regeneration in other organ systems such as the brain (van Praag et al., 1999; van Praag et al., 2005; Zhang et al., 2008). However, unambiguously demonstrating whether exercise can do similar things in the heart will require long-term cell fate-mapping experiments. Nevertheless, we believe this hypothesis is worth pursuing since identifying and learning to exploit pathways regulating this response in the heart could have important implications for the treatment of a range of cardiac diseases.
Surprisingly, although miR-222 was necessary for exercise-induced cardiac growth, miR-222 expression – even at levels higher than those induced by exercise – was not sufficient to induce an increase in heart size at baseline or changes in its targets. Interestingly, after ischemic injury, miR-222 had important protective phenotypic effects, which corresponded with reductions in all four targets. Two general models could contribute to this observation. Since LNA-antimiR-222 inhibits miR-222 in all lineages, perhaps miR-222 expression in other lineages is also important in exercise-induced cardiac growth. Arguing against this explanation, we found that exercise increased miR-222 specifically in cardiomyocytes and the Tg-miR-222 mice have miR-222 expression in non-cardiomyocytes and non-cardiac tissues similar to controls. A second and intriguing possibility is that miR-222 is necessary but not sufficient because other signals – acting in cardiomyocytes – are needed in concert to recapitulate the growth response. For example, sixteen cardiac microRNAs were dynamically regulated by exercise and multiple altered cardiomyocyte size and proliferation. Future studies examining their combinatorial effects would be of great interest. In addition, microRNA effects can be regulated by a variety of binding proteins (Hammell et al., 2009; King et al., 2014) and it is possible that interactions with such regulators inhibits miR-222’s effects at baseline but this inhibition is released in response to a second stimulus, such as ischemic injury.
The current studies did not examine whether miR-222 affects pathological hypertrophy, for example in response to pressure overload. Previous work by others has suggested that some physiological growth pathways (e.g. Akt1) are activated in both settings but actually required for physiological growth and antagonistic to pathological growth (DeBosch et al., 2006). The models presented here will provide useful tools for testing whether miR-222 functions in a similar way.
A combination of experimental and bioinformatic approaches identified four candidate miR-222 targets in cardiomyocytes in vitro. These included the cell cycle inhibitor, p27, a known target in other systems (Kim et al., 2009; Liu et al., 2009; Wurz et al., 2010), and three other targets, HIPK-1 and -2 (D’Orazi et al., 2002) as well as Hmbox1 (Chen et al., 2006). Luciferase assays confirmed that all are direct targets of miR-222 in vitro. Moreover, functional studies in cardiomyocytes suggested that reductions in p27 and HIPK1 contribute to the pro-proliferative effects of miR-222 in vitro while decreased Hmbox1 is sufficient to drive cardiomyocyte hypertrophy. Thus the phenotypic effects of miR-222 appear mediated via distinct pathways and may, in principle, be dissociable. Consistent with this model, genetic deletion of p27 induces cardiomyocyte hyperplasia in vivo (Fero et al., 1996; Kiyokawa et al., 1996; Nakayama et al., 1996) and physiological cardiac hypertrophy (Hauck et al., 2008). However, in contrast to our results, p27 deletion accelerates the development of heart failure after biomechanical stress (Hauck et al., 2008). In comparison, the distinctly beneficial effects seen with miR-222 expression likely reflects the combination of less complete reduction in p27 and simultaneous modulation of other pathways. HIPK2 knockdown did not significantly affect either cardiomyocyte size or proliferation in vitro, and understanding the role of HIPK2 in this context will be of interest for future experiments. While all four putative targets were decreased in vivo by miR-222 expression after ischemic injury, we were only able to document modulation of two targets with exercise (Hmbox1, HIPK2) or LNA-antimiR-222 (Hmbox1, p27) in vivo. It’s unclear whether these differences relate to technical aspects of the experiments or the stronger signal provided in the transgenic system. More importantly, while these studies provide some support for modulation of the putative targets in vivo, they do not test their functional roles which would require additional gain- and/or loss-of-function studies for each in vivo.
Finally, the potential clinical relevance of this pathway is underscored by prior observations that circulating miR-222 increases in healthy volunteers after exercise (Baggish et al., 2011) and our finding of a similar increase in heart failure patients. Of course, these studies do not establish a mechanistic link between miR-222 and the cardiovascular benefits of exercise in humans, nor do they demonstrate that the heart is the source of circulating miR-222 although miR-222 did not increase in skeletal muscle of exercised mice (data not shown). Future studies examining the clinical implications of miR-222 as a biomarker or functional mediator would obviously be of considerable interest.
EXPERIMENTAL PROCEDURES
Please also see Supplemental Materials for additional information on experimental procedures.
Cardiomyocyte isolation, culture, and transfection
Primary neonatal rat ventricular cardiomyocytes (NRVMs) were prepared as described (Matsui et al., 2001). Isolated NRVMs were purified either by Percoll gradient centrifugation (first two graphs of Figure 2C and Figure S1) or by pre-plating (all other experiments). Before treatment, NRVMs were synchronized and cultured in serum-free (first two graphs of Figure 2C and Figure S1) or 0.2% FBS media (all other experiments). DMEM medium was used for experiments except DMEM/F12 for cell size experiments. siRNAs for p27(kip1), Hmbox1, Hipk1, and Hipk2 and negative controls as well as microRNA precursors (Pre-miR™ miRNA Precursors) and negative controls were purchased from Invitrogen. Transfection of siRNAs (20 μM), LNA-modified antimiR oligonucleotides (20 μM), and microRNA precursors (0.4 μM) were carried out using Lipofectamine RNAiMAX (Invitrogen) as recommended by the manufacturer.
Cardiomyocytes and non-cardiomyocytes were isolated from adult mice as described previously (Graham et al., 2013). Briefly, left ventricles were harvested from perfused and digested hearts, dissected into small pieces to dissociate in transfer buffer. After filtering, cell solution was settled to sedimentation for several minutes in a Falcon tube. The cell pellet and supernatant were transferred to individual Falcon tubes for further separation. The cell pellet was resuspended in transfer buffer and settled to precipitation. After the second precipitation, the cell pellet was checked for typical rod-shaped morphology before RNA and miRNA extraction to confirm expression of cardiomyocyte markers. The initial supernatant was centrifuged first at 50g (3 minutes) and then 300g (5 minutes) before confirmation of non-cardiomyocyte identity of pelleted cells by QRT-PCR for fibroblast and cardiomyocyte markers.
Mouse exercise protocols
All mice were maintained and studied using protocols approved by the Animal Care and Use Committee of Beth Israel Deaconess Medical Center. For forced exercise training, male C57BL6/J mice swam in water tanks as described (Taniike et al., 2008). Twenty-four hours after the last swimming session, exercised mice were sacrificed and tissues were collected. For voluntary exercise, male C57BL6/J mice aged 10–12 wks were housed with exercise wheels as previously described (Bourajjaj et al., 2008). Mice were sacrificed at the specified time-points, and tissues harvested for analysis.
Generation of Tg-miR-222 mice
A tetracycline-off binary α-MHC transgene system was used as previously described (Sanbe et al., 2003). For the responder mouse line, a 388bp fragment containing mmu-miR-222 was amplified from mouse genomic DNA, confirmed by sequencing, and subcloned into a vector generously given by Dr. Jeffrey Robbins. A Not I fragment was microinjected into FVB oocytes and transferred to pseudopregnant mice. After confirmation of stable Mendelian transmission, positive mice were bred to the appropriate driver line (Sanbe et al., 2003) and cardiac-specific, doxycycline-regulated miR-222 expression was confirmed in line 12, which was used for all the experiments presented. Doxycycline was administered in the food using a special diet formulated by Purina (625 mg/kg in pellets). To induce miR-222 expression, 10 to12 week old mice were fed normal chow without doxycycline for 4 weeks.
LNA-antimiR Injections
LNA-antimiR injections were performed as described (Grueter et al., 2012). Briefly, 12-week-old C57Bl6 male mice at baseline or used for swimming training were injected subcutaneously or via tail vein with 10 mg/kg of locked nucleic acid (LNA)-modified antimiR-222 (LNA-antimiR-222) or scrambled control (LNA-SC) reconstituted in saline. Both LNA-antimiR oligonucleotides were purchased from Exiqon. The sequence of LNA-antimiR-222 is +g*t*+a*+g*c*+c*a*+g*+a*t*+g*+t*a*g*+c, in which the “+” and “*” signs indicated an LNA residue and the modified phosphorothioate linkages respectively. Mice were injected for 3 consecutive days and then weekly throughout the experiments.
Ischemia reperfusion model and analyses
Transgenic FVB or C57BL/6 wild type mice were subjected to ischemia-reperfusion as previously described (Matsui et al., 2002). Briefly, the left anterior descending artery (LAD) was ligated with 7–0 silk. Five minutes into ischemia, 50μl of fluorescent microspheres (10 μm FluoSpheres, Molecular Probes) were injected into the LV cavity. Following 30 min LAD occlusion, the LAD ligature was released, and reperfusion confirmed visually. After 24 hours, 1 or 6 weeks of reperfusion, mice were sacrificed, and hearts collected for analyses. To determine the area-at-risk and myocardial infarct size, hearts were stained with 2,3,5-triphenyltetrazolium chloride (TTC) as reported previously (Matsui et al., 2002). In animals euthanized 6 weeks after IRI, EdU (50 mg/kg, SQ) was injected every two days for the first 2 weeks after reperfusion. Sham-operated mice served as controls. All surgeries and analyses were performed by investigators blinded to genotype or treatment.
Microscopy, Confocal Microscopy and Image Quantification
Images of cultured cells were taken in a Leica DM 5000 B microscope. Heart sections were imaged in a Zeiss LSM 501 Meta confocal microscope using standard procedures. All imaging was performed and analyzed by investigators blinded to treatment group. At least 30 random images were obtained from each group. Images were then were quantified using ImageJ and CellProfiler software.
Echocardiography
Echocardiography was performed on conscious mice by using a GE Vivid7 with i13L probe (14 MHZ) as described previously (Das et al., 2012). Briefly, parasternal long-axis views, short-axis views and 2-D guided M-mode images of short axis at the papillary muscle level were recorded. Echocardiography data were analyzed by investigators blinded to treatment and genotype. The average of at least three measurements was used for every data point from each mouse.
Heart Failure Patients
All human investigation conformed to the principles outlined in the Declaration of Helsinki, and was approved by the relevant institutional review committees (either Shanghai Tongji Hospital, Tongji University and/or Shanghai University). All participants gave written informed consent before enrollment in the study. Between September 2013 and March 2014, twenty-eight patients with chronic stable heart failure cared for at the Shanghai Tongji Hospital, Tongji University underwent a symptom-limited cardiopulmonary exercise test on a bicycle ergometer (GE, USA) using a standardized exercise protocol of revised Ramp10 programs (Myers, 2005). Venous blood was collected in serum tubes before and after the cardiopulmonary exercise test and was processed within 1 hour of collection. After a two-step centrifugation (820×g for 10 min and 16000×g for 10 min, both at 4°C), the supernatant was transferred to RNase/DNase-free tubes and stored at −80°C until RNA isolation and PCR were performed as described before.
Statistical analysis
Data are presented as mean±SE unless otherwise indicated. Unpaired, two-tailed Students t-test was used when indicated with p<0.05 considered significant. When assessing multiple groups, One-Way ANOVA was utilized with turkeys Post Hoc test. The statistical software used was SPSS 17.1.
Supplementary Material
Acknowledgments
We thank Dr. Ling Li for technical support and Dr. Serafima Zaltsman for managing the mouse colony. This research was supported by grants from the NIH (AR[R21HL114352, 1UH2TR000901, R01HL110733], XL[T32HL073734], FD[T32HL073734], and CP[T32GM007226]). VB was supported by grants from the NIH (T32HL007572-27A1) and a LaDue Fellowship Award from Harvard Medical School. JX was supported by the grants from the National Natural Science Foundation of China (81200169), Innovation Program of Shanghai Municipal Education Commission (13YZ014), Foundation for University Young Teachers by Shanghai Municipal Education Commission (2012), Innovation Fund from Shanghai University (sdcx2012038), and Program for the Integration of Production, Teaching and Research for University Teachers supported by the Shanghai Municipal Education Commission (2014). AR is a principal faculty member of the Harvard Stem Cell Institute.
Footnotes
Author contributions
Drs. Xiaojun Liu and Junjie Xiao contributed equally to this work. Dr. Xiao performed the initial microRNA screen in exercised mice as well as the microarray experiments in cardiomyocytes and the LNA-miR-222 exercise experiments. Dr. Liu generated the transgenic miR-222 mice and characterized their phenotype at baseline and after ischemic injury, as well as performing the detailed experiments of miR-222 targets.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, Wang TJ, Chan SY. Dynamic Regulation of Circulating MicroRNA during Acute Exhaustive Exercise and Sustained Aerobic Exercise Training. J Physiol (Lond) 2011 doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A, Dai Q, Chiao YA, Hakala KW, Zhang JQ, Weintraub ST, Lindsey ML. Proteomic analysis reveals late exercise effects on cardiac remodeling following myocardial infarction. Journal of proteomics. 2010;73:2041–2049. doi: 10.1016/j.jprot.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Mann N, Wu J, Quintero PA, Plovie ER, kova DP, Gupta RK, Xiao C, MacRae CA, Rosenzweig A, et al. C/EBPbeta Controls Exercise-Induced Cardiac Growth and Protects against Pathological Cardiac Remodeling. Cell. 2010;143:1072–1083. doi: 10.1016/j.cell.2010.11.036. (*co-senior, co-corresponding, equal contributing authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourajjaj M, Armand AS, da Costa Martins PA, Weijts B, van der Nagel R, Heeneman S, Wehrens XH, De Windt LJ. NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J Biol Chem. 2008;283:22295–22303. doi: 10.1074/jbc.M801296200. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nature medicine. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chen S, Saiyin H, Zeng X, Xi J, Liu X, Li X, Yu L. Isolation and functional analysis of human HMBOX1, a homeobox containing protein with transcriptional repressor activity. Cytogenetic and genome research. 2006;114:131–136. doi: 10.1159/000093328. [DOI] [PubMed] [Google Scholar]
- D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nature cell biology. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- DA Silva NDJ, Fernandes T, Soci UP, Monteiro AW, Phillips MI, EMDEO Swimming training in rats increases cardiac MicroRNA-126 expression and angiogenesis. Medicine and science in sports and exercise. 2012;44:1453–1462. doi: 10.1249/MSS.0b013e31824e8a36. [DOI] [PubMed] [Google Scholar]
- Das S, Aiba T, Rosenberg M, Hessler K, Xiao C, Quintero PA, Ottaviano FG, Knight AC, Graham EL, Bostrom P, et al. Pathological role of serum- and glucocorticoid-regulated kinase 1 in adverse ventricular remodeling. Circulation. 2012;126:2208–2219. doi: 10.1161/CIRCULATIONAHA.112.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBosch B, Treskov I, Lupu TS, Weinheimer C, Kovacs A, Courtois M, Muslin AJ. Akt1 is required for physiological cardiac growth. Circulation. 2006;113:2097–2104. doi: 10.1161/CIRCULATIONAHA.105.595231. [DOI] [PubMed] [Google Scholar]
- Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes T, Hashimoto NY, Magalhaes FC, Fernandes FB, Casarini DE, Carmona AK, Krieger JE, Phillips MI, Oliveira EM. Aerobic exercise training-induced left ventricular hypertrophy involves regulatory MicroRNAs, decreased angiotensin-converting enzyme-angiotensin ii, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1–7) Hypertension. 2011;58:182–189. doi: 10.1161/HYPERTENSIONAHA.110.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- Flynn KE, Pina IL, Whellan DJ, Lin L, Blumenthal JA, Ellis SJ, Fine LJ, Howlett JG, Keteyian SJ, Kitzman DW, et al. Effects of exercise training on health status in patients with chronic heart failure: HF-ACTION randomized controlled trial. JAMA. 2009;301:1451–1459. doi: 10.1001/jama.2009.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- Graham EL, Balla C, Franchino H, Melman Y, del Monte F, Das S. Isolation, culture, and functional characterization of adult mouse cardiomyoctyes. J Vis Exp. 2013:e50289. doi: 10.3791/50289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck L, Harms C, An J, Rohne J, Gertz K, Dietz R, Endres M, von Harsdorf R. Protein kinase CK2 links extracellular growth factor signaling with the control of p27(Kip1) stability in the heart. Nat Med. 2008;14:315–324. doi: 10.1038/nm1729. [DOI] [PubMed] [Google Scholar]
- Haykowsky M, Scott J, Esch B, Schopflocher D, Myers J, Paterson I, Warburton D, Jones L, Clark AM. A meta-analysis of the effects of exercise training on left ventricular remodeling following myocardial infarction: start early and go longer for greatest exercise benefits on remodeling. Trials. 2011;12:92. doi: 10.1186/1745-6215-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Nemoto K, Li Y, Takada Y, Suzuki R, Katsuki M, Nakagawara A, Koseki H. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Molecular and cellular biology. 2006;26:2758–2771. doi: 10.1128/MCB.26.7.2758-2771.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IN, Yartseva V, Salas D, Kumar A, Heidersbach A, Ando DM, Stallings NR, Elliott JL, Srivastava D, Ivey KN. The RNA-binding protein TDP-43 selectively disrupts microRNA-1/206 incorporation into the RNA-induced silencing complex. J Biol Chem. 2014;289:14263–14271. doi: 10.1074/jbc.M114.561902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. The EMBO journal. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Luo D, Park SJ, Wang Q, Kim Y, Min W. Tumor necrosis factor alpha-induced desumoylation and cytoplasmic translocation of homeodomain-interacting protein kinase 1 are critical for apoptosis signal-regulating kinase 1-JNK/p38 activation. J Biol Chem. 2005;280:15061–15070. doi: 10.1074/jbc.M414262200. [DOI] [PubMed] [Google Scholar]
- Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, Amann M, Anderson HR, Andrews KG, Aryee M, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380:2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A Necessary Role of miR-221 and miR-222 in Vascular Smooth Muscle Cell Proliferation and Neointimal Hyperplasia. Circulation Research. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli NC, Cohen CR, Santos KG, Castro MA, Biolo A, Frick L, Silvello D, Lopes A, Schneider S, Andrades ME, et al. An analysis of the global expression of microRNAs in an experimental model of physiological left ventricular hypertrophy. PloS one. 2014;9:e93271. doi: 10.1371/journal.pone.0093271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Li L, Wu JC, Cook SA, Nagoshi T, Picard MH, Liao R, Rosenzweig A. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J Biol Chem. 2002;277:22896–22901. doi: 10.1074/jbc.M200347200. [DOI] [PubMed] [Google Scholar]
- Matsui T, Tao J, del Monte F, Lee KH, Li L, Picard M, Force TL, Franke TF, Hajjar RJ, Rosenzweig A. Akt activation preserves cardiac function and prevents injury after transient cardiac ischemia in vivo. Circulation. 2001;104:330–335. doi: 10.1161/01.cir.104.3.330. [DOI] [PubMed] [Google Scholar]
- Myers J. Applications of cardiopulmonary exercise testing in the management of cardiovascular and pulmonary disease. International journal of sports medicine. 2005;26(Suppl 1):S49–55. doi: 10.1055/s-2004-830515. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- O’Connor CM, Whellan DJ, Lee KL, Keteyian SJ, Cooper LS, Ellis SJ, Leifer ES, Kraus WE, Kitzman DW, Blumenthal JA, et al. Efficacy and safety of exercise training in patients with chronic heart failure: HF-ACTION randomized controlled trial. JAMA. 2009;301:1439–1450. doi: 10.1001/jama.2009.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92:609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soci UP, Fernandes T, Hashimoto NY, Mota GF, Amadeu MA, Rosa KT, Irigoyen MC, Phillips MI, Oliveira EM. MicroRNAs 29 are involved in the improvement of ventricular compliance promoted by aerobic exercise training in rats. Physiological genomics. 2011;43:665–673. doi: 10.1152/physiolgenomics.00145.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniike M, Yamaguchi O, Tsujimoto I, Hikoso S, Takeda T, Nakai A, Omiya S, Mizote I, Nakano Y, Higuchi Y, et al. Apoptosis signal-regulating kinase 1/p38 signaling pathway negatively regulates physiological hypertrophy. Circulation. 2008;117:545–552. doi: 10.1161/CIRCULATIONAHA.107.710434. [DOI] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci. 2005;25:8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Powers AS, Li J, Ji L, Erikson JM, Zhang JQ. Effect of post-myocardial infarction exercise training on the renin-angiotensin-aldosterone system and cardiac function. The American journal of the medical sciences. 2007;334:265–273. doi: 10.1097/MAJ.0b013e318068b5ed. [DOI] [PubMed] [Google Scholar]
- Waring CD, Vicinanza C, Papalamprou A, Smith AJ, Purushothaman S, Goldspink DF, Nadal-Ginard B, Torella D, Ellison GM. The adult heart responds to increased workload with physiologic hypertrophy, cardiac stem cell activation, and new myocyte formation. European heart journal. 2012 doi: 10.1093/eurheartj/ehs338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurz K, Garcia RL, Goff BA, Mitchell PS, Lee JH, Tewari M, Swisher EM. MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: Relationship to CDKN1B, CDKNIC and overall survival. Genes Chromosomes Cancer. 2010;49:577–584. doi: 10.1002/gcc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wan W, Powers AS, Li J, Ji LL, Lao S, Wilson B, Erikson JM, Zhang JQ. Effects of exercise training on cardiac function and myocardial remodeling in post myocardial infarction rats. J Mol Cell Cardiol. 2008;44:114–122. doi: 10.1016/j.yjmcc.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhao W, Lao S, Wilson BS, Erikson JM, Zhang JQ. Effects of exercise and L-arginine on ventricular remodeling and oxidative stress. Med Sci Sports Exerc. 2010;42:346–354. doi: 10.1249/MSS.0b013e3181b2e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo CM, Zimmerman SD, McCormick RJ, Thomas DP. Exercise training post-MI favorably modifies heart extracellular matrix in the rat. Med Sci Sports Exerc. 2012;44:1005–1012. doi: 10.1249/MSS.0b013e318244bc8a. [DOI] [PubMed] [Google Scholar]
- Young DR, Reynolds K, Sidell M, Brar S, Ghai NR, Sternfeld B, Jacobsen SJ, Slezak JM, Caan B, Quinn VP. Effects of Physical Activity and Sedentary Time on the Risk of Heart Failure. Circulation Heart failure. 2014;7:21–27. doi: 10.1161/CIRCHEARTFAILURE.113.000529. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–1007. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.