Summary
SIRT3 is a member of the Sirtuin family of NAD+-dependent deacylases and plays a critical role in metabolic regulation. Organism-wide SIRT3 loss manifests in metabolic alterations, however the coordinating role of SIRT3 among metabolically distinct tissues is unknown. Using multi-tissue quantitative proteomics comparing fasted wild type mice to mice lacking SIRT3, innovative bioinformatic analysis, and biochemical validation, we provide a comprehensive view of mitochondrial acetylation and SIRT3 function. We find SIRT3 regulates the acetyl-proteome in core mitochondrial processes common to brain, heart, kidney, liver, and skeletal muscle, but differentially regulates metabolic pathways in fuel-producing and fuel-utilizing tissues. We propose an additional maintenance function for SIRT3 in liver and kidney where SIRT3 expression is elevated to reduce the acetate load on mitochondrial proteins. We provide evidence that SIRT3 impacts ketone body utilization in the brain and reveal a pivotal role for SIRT3 in the coordination between tissues required for metabolic homeostasis.
Introduction
Mammals must maintain whole-body homeostasis with inconsistent fuel intake and have developed mechanisms to manage times of fuel deprivation. The production, utilization and storage of carbohydrates, fatty acids and protein are regulated in a tissue-dependent manner, as each organ has unique functions, metabolic pathways and accessibility to fuel sources. Most tissues exhibit metabolic flexibility and are capable of using multiple types of fuels in response to shifts in nutrient availability. The regulation of fuel switching occurs at both the cellular and organismal level, and requires tissues to respond in concert to metabolic signals to maintain proper function (Stipanuk and Caudill, 2013). As the mitochondria are the energy production centers of the cell, mitochondrial metabolism is vital in the adaptation to alterations in nutrient supply.
Protein acetylation is a post-translational modification (PTM) enriched in the mitochondria of multiple tissue types and is involved in numerous cellular processes through regulation of protein interactions, activity and localization (Choudhary et al., 2009; Lundby et al., 2012). Mass spectrometry (MS) based proteomics has identified over 2500 unique acetyl sites in the mitochondria, a majority of which reside on metabolic enzymes (Choudhary et al., 2009; Hebert et al., 2013; Still et al., 2013). Much of our knowledge on mitochondrial protein acetylation comes from studies on the role of the NAD+-dependent deacetylase Sirtuin 3 (SIRT3) in liver mitochondria (Hebert et al., 2013; Rardin et al., 2013). In the liver, SIRT3 regulates mitochondrial function and specifically modulates enzymatic activity of proteins involved in fatty acid oxidation, oxidative phosphorylation, ketone body synthesis, and the urea cycle (Hebert et al., 2013; Newman et al., 2012). Additionally, SIRT3 is thought to play a key role in regulating the production of reactive oxygen species (Newman et al., 2012). SIRT3 is proposed to facilitate catabolism of fatty acids in the liver and the peripheral use of acetate during fasting (He et al., 2012; Jing et al., 2013). However, molecular details of the role of SIRT3 in extra-hepatic tissues and in managing coordinated whole-body responses to fuel availability remain unclear.
To assess the role of SIRT3 across tissues, identify tissue specific SIRT3 substrates, and regulated biological pathways, we employed a quantitative acetyl-proteomic method (Hebert et al., 2013) to study SIRT3 in five tissues (brain, heart, kidney, liver, and skeletal muscle) from fasted mice that were wild type (WT) or lacking SIRT3 (Sirt3−/−). We provide a comprehensive multi-tissue quantitative acetyl-proteome analysis comparing multiple biological conditions. We identified 6,286 acetyl sites on 1,172 proteins of which nearly 4,000 sites were localized to mitochondrial proteins, providing a compendium of acetyl sites that are dynamically altered by SIRT3 in tissues that until now, remained unexplored with respect to the function of SIRT3. A biological pathway analysis tailored to quantitative PTM data was developed and employed, allowing for an assessment of pathways regulated by acetylation and SIRT3. Bioinformatics analyses of our proteome data reveal fuel-producing (liver and kidney) and fuel-utilizing tissues (brain, heart, muscle) display unique, SIRT3-dependent alterations in their acetyl proteome, but also indicate SIRT3 regulates acetylation of proteins involved in common core mitochondrial processes among diverse tissues. We provide biochemical evidence that SIRT3 is required for utilization of ketone bodies to form acetyl-CoA in extra-hepatic tissues. We establish critical tissue-dependent roles for SIRT3 and reveal SIRT3 dictates multi-tissue coupling required for metabolic adaptation to nutrient availability.
Results
Acetyl proteomics quantifies site-specific changes in acetylation from multiple tissues in mice lacking SIRT3
We applied our quantitative acetyl-proteomic method to study the tissue specific and SIRT3 dependent alterations in the proteome and acetylome (Figure 1A) (Hebert et al., 2013). Three five-month-old WT and three germline Sirt3−/− mice were subjected to a 24 hour fast beginning the morning prior to tissue harvest. To minimize compensatory or secondary mechanisms that occur as mice age, we selected young mice to reveal primary tissue specific biochemical alterations in the acetyl-proteome that have not yet manifested as an overt phenotype. We compared, in biological triplicate, the whole brain, heart, kidney, liver, and skeletal muscle proteome and acetyl-proteome in two conditions: WT and Sirt3−/− (Figure 1A). We detected 6,286 acetyl sites and quantified 5,199 acetylation sites in five tissues (Table 1, Table S1). Nearly half of the quantified acetyl sites (2,247) were localized on mitochondrial proteins (Pagliarini et al., 2008), achieving deep quantification of the mitochondrial proteome (668 proteins) (Table 1). To confirm differences in acetylation are due to increased acetylation, rather than a result of increased protein abundance, we analyzed the un-modified proteome of each tissue (Table S2). Acetyl reporter ion intensities were corrected for protein amount in each condition, and used to calculate a protein-normalized fold change between Sirt3−/− and WT (Table S1). Of the acetyl sites quantified, 38% (1,990 acetyl sites) exhibited a statistically significant change (p ≤ 0.1, Student’s t-test) in acetyl occupancy in mice lacking SIRT3. Our experimental approach analyzed the acetyl proteome and proteome of the entire tissue, allowing for an assessment of the potential effects of SIRT3 on acetylation outside of the mitochondria. An analysis of the distribution in fold change of acetyl sites between the Sirt3−/− and WT animals reveals a normal distribution for both mitochondrial and non-mitochondrial populations and a marked shift towards hyperacetylation of mitochondrial acetyl sites when compared to non-mitochondrial acetyl sites (Figure S1A). There has been some debate on the role of SIRT3 outside of the mitochondria (Iwahara et al., 2012) however, in the tissues tested, our study indicates the primary deacetylase activity of SIRT3 resides in the mitochondria. As previously reported for the liver (Hebert et al., 2013), SIRT3 does not considerably alter the mitochondrial or non-mitochondrial proteome, thus functional changes in mitochondrial function are due primarily to alterations in acetylation and not protein abundance (Figure S1B).
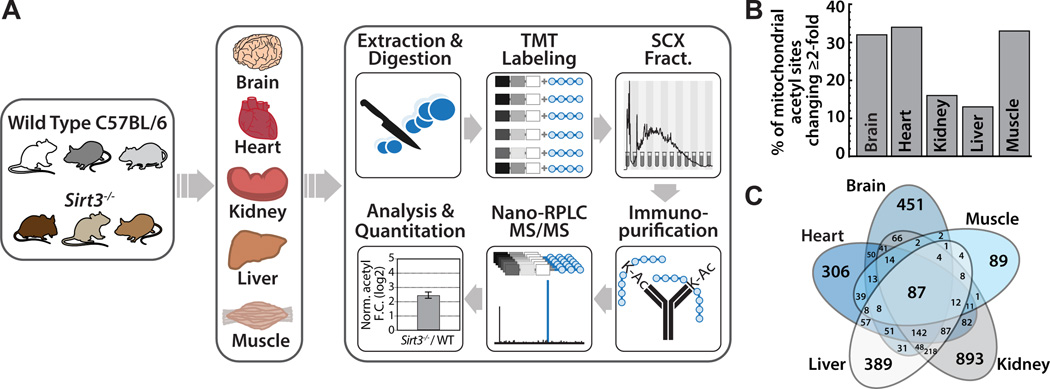
Figure 1. Multi-tissue quantitative acetylome experimental workflow and metrics, see also Figure S1, Table S1, Table S2.
(A) Wild type and Sirt3−/− mouse brain, heart, kidney, liver, and skeletal muscle were compared in biological triplicate. Tissues were disrupted by bead milling and extracts were digested. Peptides were labeled with TMT reagents, combined and fractionated by strong cation exchange chromatography. Acetyl peptides were enriched by immunoprecipitation and analyzed via nano-RPLC MS/MS on an Orbitrap Elite.
(B) Percentage of mitochondrial acetyl sites changing ≥ 2-fold in the Sirt3−/− condition for each tissue. Calculated as number of mitochondrial acetyl sites changing ≥ 2-fold per total number of quantified mitochondrial acetyl sites found in that tissue.
(C) Venn diagram displaying overlapping acetyl sites between tissues.
Table 1. Experimental metrics for each tissue.
Acetyl sites with statistically significant fold changes (p ≤ 0.1, Student’s t-test) between Sirt3−/− and WT are listed in parentheses below total number of acetyl sites. Mitochondrial proteins categorized by inclusion in the MitoCarta inventory (Pagliarini et al., 2008).
| Tissue | Brain | Heart | Kidney | Liver | Muscle |
|---|---|---|---|---|---|
| Acetyl sites | 1247 (465) | 1159 (554) | 1370 (317) | 2117 (578) | 393 (74) |
| Mitochondrial acetyl sites | 715 (339) | 897 (499) | 908 (255) | 1400 (494) | 207 (72) |
| Acetyl proteins | 523 | 338 | 500 | 647 | 152 |
| Mitochondrial acetyl proteins | 220 | 230 | 268 | 331 | 90 |
| Total proteins | 6783 | 3439 | 3977 | 4542 | 2290 |
| Mitochondrial proteins | 685 | 646 | 640 | 671 | 457 |
| Acetyl sites per protein | 2.39 | 3.43 | 2.74 | 3.28 | 2.59 |
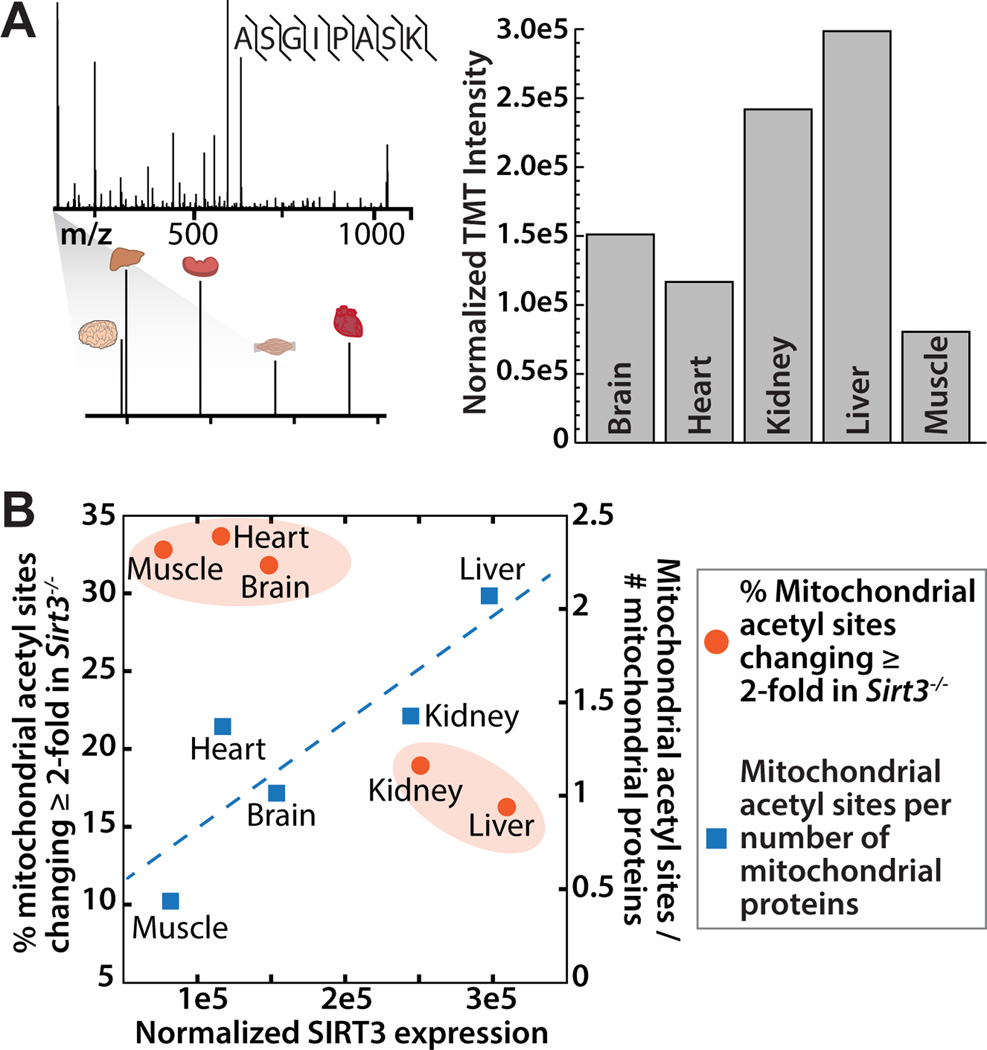
The percent of mitochondrial acetyl sites changing greater than 2-fold in mice lacking SIRT3 was calculated (Figure 1B). As SIRT3 is the only established mitochondrial deacetylase, hyperacetylated lysine residues in Sirt3−/− mice are probable SIRT3 targets. The effect of SIRT3 on mitochondrial acetylation varies in a tissue specific manner, with brain, heart and muscle displaying the greatest percentage (~33%) of acetyl isoforms increasing greater than 2-fold in the Sirt3−/− condition (Table S1, Figure 1B). Interestingly, liver and kidney contain the highest total number of mitochondrial acetyl sites (Table 1), but a lower percentage of acetyl sites that exhibit greater than 2-fold increases in acetylation in Sirt3−/−. Liver and kidney contain a number of acetyl sites that change less dramatically (between 1 and 2-fold) in the absence of SIRT3. To determine how differences in SIRT3 protein expression across tissues dictate acetylation changes during fasting, we measured SIRT3 protein abundance by targeted MS using WT samples. Peptides from the brain, heart, kidney, liver, and skeletal muscle of one WT mouse were labeled with tandem mass tags (TMT, Pierce) and combined in equal ratios. Parallel reaction monitoring (PRM) (Peterson et al., 2012) was performed to detect a single, unique peptide for SIRT3 (ASGIPASK, amino acids 171–178). The relative TMT intensities for each tissue were normalized to total intensity across the TMT channel and plotted (Figure 2A) and confirmed by western blotting of samples from three WT study animals (Figure S2). Liver and kidney have the highest protein expression levels of SIRT3, followed by brain, heart and muscle, comparable to previous literature reports (Lombard et al., 2007; Shi et al., 2005).
Figure 2. SIRT3 expression varies between tissues and corresponds to tissue acetylome alterations, see also Figure S2.
(A) SRM mass spectrometry and TMT quantitation were employed to assess SIRT3 protein expression in each of the five tissues studied. TMT-labeled peptides from one WT animal from each tissue were compared in a single targeted MS experiment. Spectra from a represented peptide ASGIPASK with reporter ion magnification and protein fold change is presented for each tissue.
(B) A comparison of SIRT3 expression level (x-axis) with percentage of mitochondrial sites changing ≥ 2-fold in response to Sirt3−/− (y1) and mitochondrial acetyl sites per the number of mitochondrial proteins identified in that tissue (y2).
Interestingly, the tissues with the highest levels of SIRT3 protein (liver and kidney) exhibit the lowest percentage of mitochondrial acetyl sites changing greater than 2-fold in the absence of SIRT3, but the greatest number of total mitochondrial acetyl sites identified per mitochondrial protein (Figure 2B). The percentage of mitochondrial sites changing greater than 2-fold was used in this analysis to provide a normalization factor for the differences in total number of acetyl sites found per tissue. There is a positive linear correlation between SIRT3 expression and total number of mitochondrial acetyl sites identified in each tissue (Figure 2B). However, when examining only sites that are dramatically altered, changing greater than 2-fold in Sirt3−/−, there is segregation between liver and kidney, and muscle, heart and brain (Figure 2B). Overall, in tissues with lower SIRT3 expression (brain, heart, muscle) there are fewer, but more dynamic changes in acetylation after a 24 hour fast, when compared to the corresponding tissues of the Sirt3−/− mice. These results provided a rich resource to explore the tissue specific functions of SIRT3 in whole-body metabolism.
Tissue specific patterns of acetyl site distribution
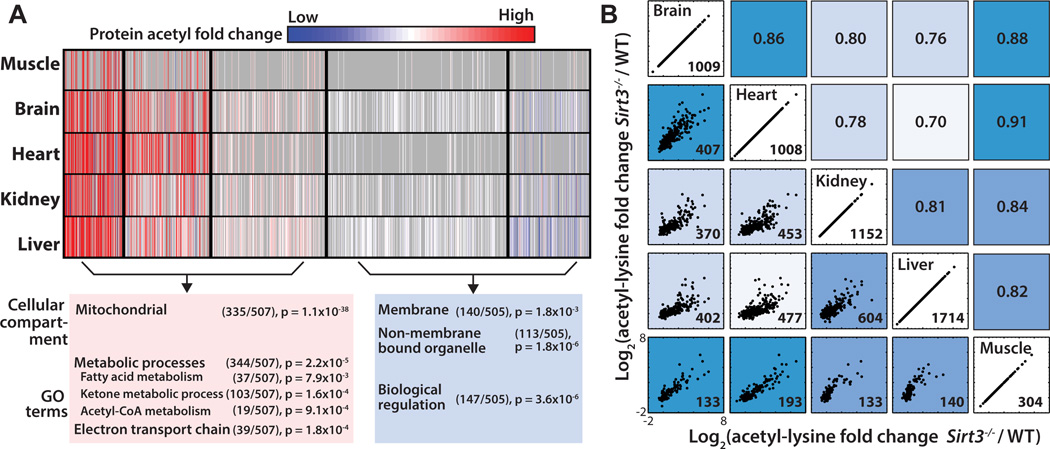
To determine whether acetylated proteins could be categorized into sub-populations based on their patterns across the five tissues, we used a probabilistic clustering algorithm based on a Gaussian mixture model (Hastie et al., 2009; Hebert et al., 2013). The algorithm used the summed acetyl fold change for each unique UniProt identifier and generated 5 clusters (Figure 3A) that segregate into two main groups: 1) acetylated proteins regulated by SIRT3 that exhibit hyperacetylation in SIRT3 null mice, and 2) proteins with minimal perturbations in acetylation due to SIRT3. We postulated that these two groups might represent functionally distinct proteins. An interrogation of the sub-cellular localization of proteins found in these groups using the functional annotation tool DAVID (Huang et al., 2009a, b) against a background of all acetyl-proteins used in the clustering analysis revealed Group 1 proteins are enriched for mitochondrial proteins (p = 1.10x10−38), while Group 2 proteins are dispersed throughout the cell (nucleus, p = 2.70x10−8, intracellular non-membrane bound organelle, p = 1.80x10−6). Acetyl-proteins in Group 1 are significantly enriched in metabolic processes (p = 2.20x10−5) in the mitochondria including carboxylic acid, fatty acid, ketone body, and acetyl-CoA metabolic processes, as well as the electron transport chain (p = 1.20x10−4). Group 2 proteins are enriched in regulation of biological processes (p = 5.20x10−7) and are not enriched in mitochondrial pathways. These results indicate Group 1 proteins comprise potential SIRT3 targets.
Figure 3. Tissue specific patterns of acetylated proteins and tissue correlation analysis, see also Figure S3.
(A) Cluster analysis of acetyl-proteins identified 5 clusters that define 2 groups based on their overall trends between Sirt3−/− and WT. Acetyl-proteins are color coded according to their normalized fold change between Sirt3−/− and WT with cluster separations are denoted by black lines. Each row represents a tissue and each column represents an acetylated protein. Red: Increased acetylation, Blue: Decreased acetylation, Gray: Missing. Pathway analysis, number of acetyl proteins and p-values for enrichment are shown for two main groups of acetyl-proteins.
(B) Normalized acetyl fold changes plotted for sites found in common between any two tissues. Scatter plots on the left side of the diagonal with the number of sites quantified and plotted in the lower right hand corner. Axes scales are equivalent for each scatter plot. Corresponding Pearson correlation coefficients are indicated on the right side of the diagonal. Tissues with acetyl sites behaving more similarly in response to Sirt3−/− are more highly correlated and are a darker shade of blue.
We also observed patterns between tissues within the clusters (Figure 2A). To determine tissue similarities of proteins whose acetylation is regulated by SIRT3, we performed Euclidean distance hierarchical clustering on each cluster using an algorithm that was adapted to take into consideration missing data values resulting from incomplete overlap of acetyl sites and proteins (Figure 1C) across all five tissues. Tree diagrams displaying tissue similarities are presented in Figure S3. Functionally similar tissues, like liver and kidney, and muscle and heart, are more closely related to each other in every cluster of acetyl-proteins (Figure S3). The segregation of fuel utilizing (brain, heart, muscle) and fuel producing tissues (liver and kidney) in the Sirt3−/− condition suggest SIRT3 regulates protein acetylation in a protein and organ-dependent manner.
We predicted that acetyl sites would also display tissue similarities and would further reveal tissue dependent response to a loss of SIRT3 expression. To assess tissue similarities at the acetyl site level, normalized acetyl fold changes for sites found in common between any two tissues were plotted (Figure 3B). The Pearson correlation coefficient was calculated and is presented on the right side of the diagonal of the scatter plots in Figure 3B. The greater the Pearson correlation coefficient, the more similar acetyl sites are changing due to SIRT3 in any two tissues. Tissue similarities emerge that are consistent with our protein level clustering analysis, with heart, brain, and muscle more similar to each other than liver and kidney.
Pathway analysis tool identifies biological pathways regulated by acetylation and SIRT3
The adaptation to diet and nutrient availability requires a coordinated change in mitochondrial metabolism. These metabolic shifts are tissue specific but require exquisite coupling of tissues to allow for fuel switching and metabolic homeostasis. The extent to which SIRT3 regulates divergent pathways of nutrient utilization and mediates metabolic coupling to respond to fuel availability is unknown. To identify biological pathways and understand the tissue specific functions of SIRT3-controlled acetyl sites, we developed and applied a biological pathway analysis we termed quantitative site set functional score analysis (QSSA).
Enrichment analysis based on gene ontologies is a steadfast companion in determining the biological significance of a particular physiological perturbation and is used to generate hypotheses on the functional relevance of the observed data. Most established enrichment methods were developed for gene expression data and later applied to proteomics data. Therefore the information contained in typical over-representation analysis and newer approaches such as gene set enrichment analysis (GSEA) is gene-product centric (Irizarry et al., 2009). Using available methods with quantitative PTM data necessitates discarding site-specific modification information. Over-representation analysis, where an arbitrary cutoff often based on fold change is established, entails further discarding the magnitude of the effect and all information about proteins that fall below the cutoff. GSEA and its derivatives offer an improvement in that they allow for the incorporation of protein fold changes and inclusion of an entire proteomic dataset in assigning significance to each set of genes, but are still incapable of accounting for the wealth of quantitative MS data on PTMs. QSSA shifts the focus of the enrichment analysis from sets of genes to sets of PTM sites. We start from the same pre-determined categories of genes used in previous approaches (Gene Ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000), Reactome, etc.) and incorporate site-specific data by integrating protein sequence data from the UniProt database. QSSA is not a rigorous method for determining the statistical significance of enrichment, but in accordance with the exploratory nature of proteome-wide datasets, is a tailored approach incorporating the most salient factors in quantitative PTM data to facilitate generation of biological hypotheses. Our method is broadly applicable to diverse PTM data sets, including quantitative phospho-proteomics, and the scoring algorithm is not limited to the KEGG database. Any well-annotated gene ontology or molecular function database could be employed.
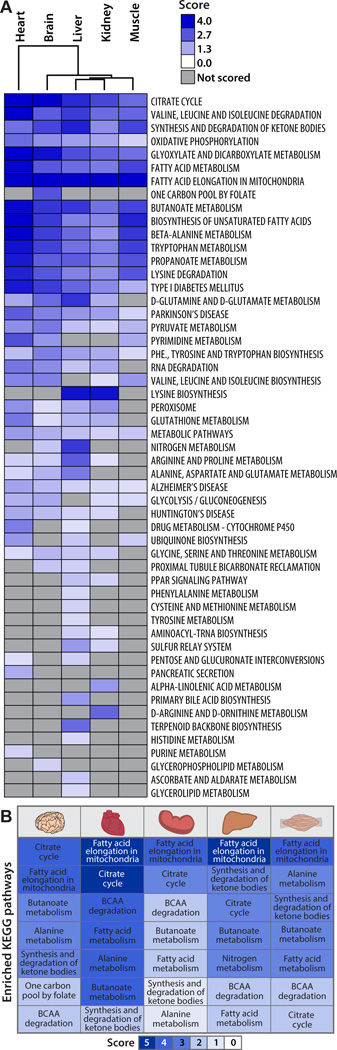
Since our focus is lysine acetylation, we consider each pathway background as the set of lysines contained in proteins identified in our proteomics analysis belonging to a gene category. Acetylation may be important to the function of a biological process through the dynamic acetylation of multiple weakly regulated sites on a protein or through the regulation of one keystone site. To account for both of these possibilities, we assessed the acetyl-coverage of a pathway and the fold change in acetylation of each acetyl site in the Sirt3−/− condition (Table S1). To identify biological pathways that may be differentially regulated by SIRT3, QSSA (described in detail in the Supplemental Information) was performed to calculate a standard score for each KEGG pathway, with higher scores representing pathways likely regulated by acetylation (Figure 4).
Figure 4. Pathway enrichment analysis tool identifies biological pathways regulated by SIRT3, see also Figure S4.
(A) QSSA performed with all quantified acetyl sites identifies high scoring biological pathways enriched with acetyl sites and controlled by SIRT3. Hierarchical clustering groups biological pathways between fuel-utilizing and fuel-producing tissues. Dark blue: Highly scoring pathway likely regulated by SIRT3; Light blue: Low scoring pathway. (B) High scoring KEGG pathways are listed with their corresponding scores color coded according to scale.
Biological pathway analysis indicated SIRT3 and acetylation regulate core mitochondrial pathways common to most tissues studied and pathways specific to individual or groups of metabolically similar tissues. Fundamental mitochondrial processes, including oxidative phosphorylation, the citrate cycle and fatty acid metabolism are enriched with acetyl sites sensitive to SIRT3 expression in brain, heart, kidney, liver, and skeletal muscle (Figure 4A). These core pathways are essential for cellular function and represent common regulatory targets of SIRT3 across tissues. In support of this observation, of the 87 acetyl sites found in all tissues (Figure S4A), nearly half are members of core mitochondrial pathways. These include multiple subunits of ATP synthase, complex I and II of the electron transport chain and numerous enzymes of the citrate cycle (citrate synthase, malate dehydrogenase, aconitase, succinate dehydrogenase, and isocitrate dehydrogenase). To illustrate the utility of the QSSA method, we directly compared KEGG pathway enrichment scores from QSSA with KEGG pathway fold enrichment scores from the commonly used functional annotation tool, DAVID (Huang et al., 2009a, b) (Figure S4B). Validating the general applicability of QSSA, a number of high scoring QSSA pathways were also identified in the DAVID analysis, including the citrate cycle and branched chain amino acid catabolism (Figure S4B). A side-by-side comparison of the scores provided for each pathway (Figure S4B) reveals that QSSA provides information on the extent of enrichment for many pathways for which DAVID provides no information, thus providing a more nuanced and fine-grained hypothesis generating tool for quantitative MS-based PTM analysis. In fact, ketone body synthesis and utilization, a pathway for which we present detailed biochemical follow up in the brain, was not identified as enriched using DAVID.
Mitochondrial metabolism must also be regulated in a tissue dependent manner to allow for metabolic flexibility. In response to a fast, fuel producing pathways, such as fatty acid oxidation and ketone body generation, must be upregulated in hepatic tissues, while fuel utilizing pathways, like acetyl-CoA production for the citrate cycle, are necessary in extra-hepatic tissues. To determine whether SIRT3 regulated biological processes in a tissue dependent manner, calculated pathway scores were visualized by Euclidean distance hierarchical clustering (Figure 4A), identifying segregation in pathway scoring patterns between fuel-utilizing and fuel-producing tissues. We find a number of SIRT3-regulated pathways in brain, heart and muscle that promote alternative fuel utilization, including degradation of short chain carbon molecules (propanoate, butanoate metabolism) and metabolism of amino acids (Figure 4B). Skeletal muscle is a predominant location for branched chain amino acid catabolism, providing succinyl-CoA for the citrate cycle and alanine that is shuttled to the liver to provide carbon for gluconeogenesis (Suryawan et al., 1998). Both branched chain amino acid degradation in the skeletal muscle and alanine metabolism in the liver and skeletal muscle were identified as probable metabolic pathways controlled by SIRT3, suggesting SIRT3 may be an important regulator of the glucose-alanine cycle during fasting metabolism. During fasting, another essential metabolic transition is the generation of ketone bodies by the liver and their utilization as an alternative fuel source by extra-hepatic tissues. Ketone body synthesis and utilization was identified as one of the highest candidate SIRT3-controlled pathways in brain, heart, skeletal muscle, and liver (Figure 4B). SIRT3 is known to regulate ketone body production in the liver through the deacetylation and activation of hydroxymethylglutaryl-CoA synthase 2 (HMGCS2) (Hirschey et al., 2010; Shimazu et al., 2010) however, whether SIRT3 mediates ketone body utilization in extra-hepatic tissues was unclear.
SIRT3 mediates cross tissue coupling for ketone body synthesis and degradation
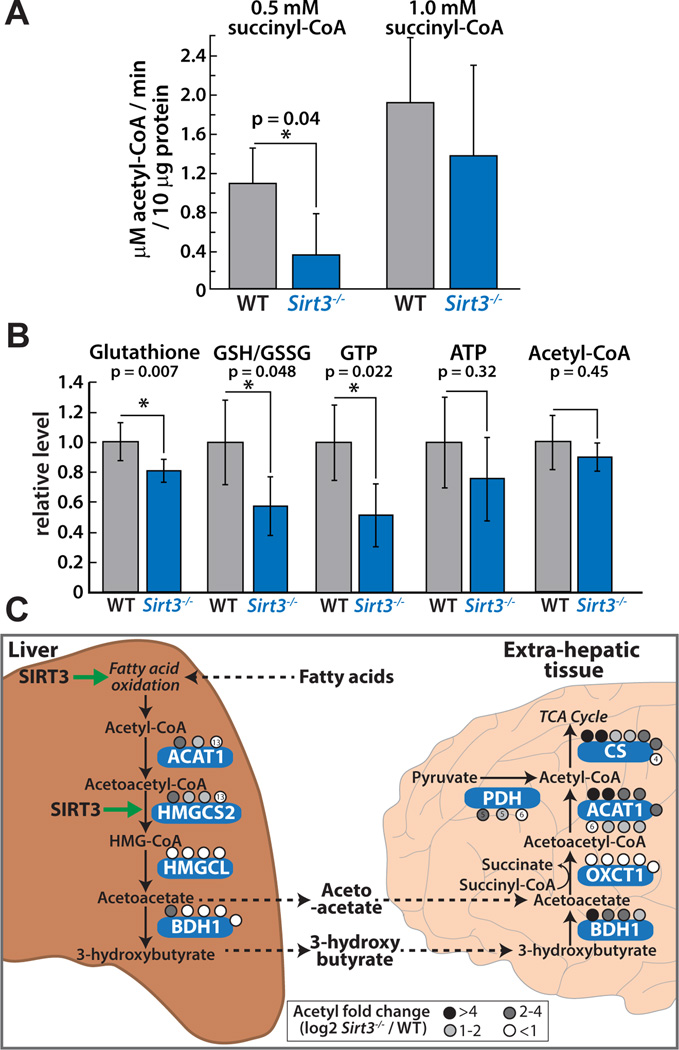
To provide evidence that SIRT3 differentially affects ketone body metabolism in hepatic and extra-hepatic tissues, we determined acetoacetate utilization and acetyl-CoA generation in the brain. This was accomplished by monitoring acetoacetate-dependent acetyl-CoA production in homogenates of brain cortices of 8-month old WT and Sirt3−/− mice that were fasted for 24 hours. The brain is an extra-hepatic tissue that relies solely on glucose and ketone bodies during a fast, therefore the brain serves as the model tissue to study the effects of SIRT3 controlled protein acetylation on ketone body degradation. Using brain cortex homogenate from fasted WT or Sirt3−/− mice and quantitative LC-MS based analysis, we monitored the production of acetyl-CoA from the addition of ketone body acetoacetate (Figure 5A and Figure S5). Succinyl-CoA and CoA were included in the reaction to ensure that flux through the pathway was not initially limited by the presence of required co-substrates. We find a 3-fold decrease in acetoacetate-dependent acetyl-CoA production in Sirt3−/− brain cortex homogenates at low succinyl-CoA levels (Figure 5A), suggestive of a defect in acetyl-CoA production through the ketone body utilization pathway in mice lacking SIRT3 (Figure 5C). We performed a targeted LC-MS based metabolite analysis on WT and Sirt3−/− brain. Consistent with an alteration in ketone body utilization, a slight decrease in whole cell acetyl-CoA was measured in the Sirt3−/− condition (Figure 5B). Although this trend did not reach statistical significance, the observation is similar to our previously published report that acetyl-CoA levels were lower in liver of Sirt3−/− mice compared to WT (Hebert et al., 2013). The observation that reduced activity in ketone body utilization presented in Figure 5A is more dramatic than the slight reduction in whole tissue acetyl-CoA levels (Figure 5B) is not surprising and suggests that other compensating activities, including amino acid or fatty acid metabolism, reduction in acetyl-CoA consuming processes, or other non-mitochondrial metabolic processes may prevent cellular acetyl-CoA from further depletion.
Figure 5. SIRT3 mediates cross-tissue link for ketone body production and utilization, see also Figure S5, S6.
(A) Average rate of acetoacetate-dependent acetyl-CoA production in brain cortex homogenates from four WT (grey) or four Sirt3−/− (blue) mice indicates a defect in acetyl-CoA production in Sirt3−/− at low succinyl-CoA concentrations. Error bars represent standard deviation (* indicates statistical significance, p<0.05). (B) Targeted metabolite analysis of WT and Sirt3−/− brain cortex (15 month old, n=4 per condition) identifies altered energetic status in Sirt3−/− brain. (C) A model for the differential regulation of ketone body production and utilization by SIRT3 in hepatic and extra-hepatic tissues. In the liver, SIRT3 is required for the synthesis of the ketone bodies acetoacetate and 3-hydroxybutyrate, through regulation of HMGCS2 enzymatic activity and pathways that generate acetyl-CoA in response to nutrient deprivation. This counters the role of SIRT3 in the brain, where SIRT3 is necessary for the utilization of acetoacetate to form acetyl-CoA to be used for energy production.
To confirm defects in ketone body generation in hepatic tissues, we measured a ketogenesis reaction in liver homogenates of WT and Sirt3−/− mice by monitoring the rate of acetyl-CoA dependent CoA formation by LC-MS. In support of the hypothesis that SIRT3 is involved in tissue-specific fuel switching, we find a decrease in the activity of a ketogenesis pathway in Sirt3−/− liver (Figure S5). Consistent with our results, previous metabolomics analyses in plasma and liver identified alterations in the ketone body 3-hydroxybutyrate during fasting and in hydroxybutyryl carnitine levels (Hallows et al., 2011; Hirschey et al., 2010; Shimazu et al., 2010) in mice lacking SIRT3.
Acetyl-CoA generated through ketone body degradation is combined with oxaloacetate to form citrate by the enzyme citrate synthase, entering the citrate cycle to provide reducing equivalents required for adenosine triphosphate (ATP) production (Figure 5C). In a separate reaction, we monitored citrate synthase activity by combining cortex homogenate with oxaloacetate and acetyl-CoA and measuring citrate formation over time to determine whether decreased acetyl-CoA production in the Sirt3−/− condition was due to accelerated utilization of acetyl-CoA by citrate synthase (Figure S5). This was not the case; in fact we detected a decrease in oxaloacetate-dependent citrate formation in Sirt3−/−, which is consistent with prior results suggesting citrate synthase is also a target of SIRT3 (Hebert et al., 2013). However, we cannot rule out the possibility that acetyl-CoA or another co-substrate, like succinyl-CoA, is consumed by an undefined metabolic pathway in the Sirt3−/− condition and competes with the assayed activity.
Taken together, the decrease in acetoacetate-dependent generation of acetyl-CoA at low succinyl-CoA concentrations represents a deficiency in ketone body utilization in the brain of mice lacking SIRT3, supporting a critical role for SIRT3 in this pathway. Consistent with the metabolite flux analysis, two central enzymes in acetyl-CoA production are highly probable targets for SIRT3 regulation: Succinyl-CoA:3-ketoacid-CoA transferase (OXCT) and acetyl-CoA acetyltransferase 1 (ACAT1). OXCT transfers the CoA from succinate to acetoacetate, forming acetoacetyl-CoA that is then converted by ACAT1 to two molecules of acetyl-CoA. Both enzymes are hyperacetylated in the brain of Sirt3−/− animals (Figure 5C, Table S1). Importantly, OXCT is only expressed in extra-hepatic tissues to avoid non-productive cycling of acetoacetate in the liver and is exclusively acetylated in the brain (5 acetyl sites) and heart (3 acetyl sites) in this analysis. Our previous report on ACAT1 enzymatic function identified a defect in catalysis of acetyl-CoA formation due to acetylation of K260 and K265 (Still et al., 2013), providing direct enzymatic evidence that supports our current findings. Numerous acetyl sites, including K260 and K265 were identified in all tissues in this study (Table S1). The observation that the magnitude of the defects in acetoacetate-dependent acetyl-CoA production were dependent on succinyl-CoA levels (Figure 5A), suggested that hyperacetylation of OXCT decreased catalytic efficiency, likely by affecting the Vmax/Km kinetic parameter for succinyl-CoA. Consistent with this observation, lysine 451 (K451), a site identified and quantified only in the brain, displays the largest magnitude acetyl fold change (67% increase, p = 1.1x10−4, Student’s t-test) in the Sirt3−/− condition, when compared to other acetyl sites quantified on OXCT. This acetyl residue lies within the CoA binding pocket of OXCT (PDB 3OXO), approximately 4 Å from the ribosyl-phosphate group of CoA, thus acetylation of this site would be predicted to disrupt the binding or orientation of succinyl-CoA and serve as a mechanism for regulating OXCT function. To provide in vitro evidence for the effect of acetylation, we performed a biochemical analysis of recombinant OXCT, demonstrating that acetylation of K451 resulted in low catalytic activity that could be rescued in part by pre-incubation with SIRT3 (Figure S6). Using site-specific incorporation of acetyllysine, we expressed and purified recombinant OXCT that is fully acetylated at K451 (Neumann et al., 2009). In vitro OXCT activity assays across several concentrations of succinyl-CoA were performed and revealed that SIRT3 induced a 2.5 fold increase in the catalytic activity of acetylated OXCT (Figure S6). Minimal changes in activity were observed with identically treated wild type OXCT (data not shown). We note that upon expression and purification, acetylated OXCT displayed extremely low activity and likely formed an oligomeric structure that partially occluded the acetyl site. This necessitated limited proteolysis of acetylated OXCT to demonstrate that SIRT3 was capable of removing the acetyl mark, as indicated by LC-MS measurement of the reaction product O-acetyl-ADP-ribose (Figure S6).
The end product of ketone body utilization, acetyl-CoA, is the oxidation substrate for the citrate cycle, a critical pathway for energy generation in the mitochondria. To determine whether the deficiency in ketone body utilization in the brain and the alteration in mitochondrial acetylation manifests in an overt energetic state or redox state, we performed a targeted LC-MS based metabolite analysis on brain cortex of WT and Sirt3−/− mice (Figure 5B). We observed a significant decrease of GSH/GSSG and glutathione in Sirt3−/− mice, consistent with prior studies showing that SIRT3 loss leads to increased ROS production and oxidative stress (Someya et al., 2010), indicating that our current biological analysis reflects established phenotypes of SIRT3 knockout. Chemically, cellular energy is mainly produced in the form of two triphosphate compounds, GTP, which is directly formed in the citrate cycle by succinyl-CoA synthetase, and ATP, produced by the electron transport chain. The high-energy cofactors, GTP and ATP showed approximately a 50% and 20% decrease in the brains of mice lacking SIRT3 (Figure 5B), respectively, with the change in GTP being statistically significant. Collectively, these data are consistent with a defect in the ability of Sirt3−/− mice to switch fuel under a prolonged fast, including decreased ketone body generation in the liver and reduced ketone body utilization in extra hepatic tissues such as brain. The magnitude of decreased ATP and acetyl-CoA levels in the Sirt3−/− brain may be indicative of organism-wide compensatory mechanisms that act to maintain metabolic and energetic homeostasis. This study reveals multi-tissue homeostatic effects and captures early stage biochemical dysregulation within five individual tissues resulting from whole body loss of SIRT3 expression.
Discussion
Through the regulation of protein acetylation, SIRT3 controls enzymatic activity and flux of numerous metabolic pathways. Our study provides evidence that SIRT3 possesses 1.) common core functions in all tissues through the regulation of pathways like the citrate cycle and oxidative phosphorylation, 2.) tissue specific functions to allow for crosstalk between tissues that enables a response to nutrient shifts (e.g. ketone body generation in the liver and utilization in the brain) and 3.) an additional maintenance function to minimize the effects of untargeted acetylation in energy producing tissues (liver, kidney) where higher SIRT3 expression reflects the acetate-burden of each tissue.
The effect of SIRT3 on mitochondrial acetylation varies in a tissue specific manner. Brain, heart and skeletal muscle exhibit a greater percentage of sites hyperacetylated (>2x) in SIRT3 knockout mice, while liver and kidney contain the highest total number of mitochondrial acetyl sites, but a lower percentage of dynamic acetyl sites (Figure 2). In this 24 hour fasted phase, extra-hepatic tissues rely on fatty acids and ketone bodies produced by increased fatty acid oxidation in the liver (Bauer et al., 2004). Brain, heart and skeletal muscle must efficiently oxidize acetyl-CoA for energy production, while hepatic tissues produce large amounts of intra-mitochondrial acetyl-CoA. We propose that this necessary demand for acetyl-CoA production drives widespread protein acetylation in liver, creating a greater need for the maintenance deacetylase functions of SIRT3.
Our study and others reveal widespread acetylation of the mitochondrial proteome (Hebert et al., 2013; Kim et al., 2006; Still et al., 2013) however, the mechanism of acetylation is as of yet unknown, with scarce evidence for enzymatic acetyltransferase activity residing in the mitochondria (Scott et al., 2014). Protein acetylation might occur non-enzymatically (Tanner et al., 2000) with the levels and extent of acetylation reflecting acetyl-CoA concentration and conditions of the organelle (Cai et al., 2011). The slightly elevated pH (pH ~ 7.9) of the mitochondrial matrix compared to the cytosol and the millimolar intra-mitochondrial concentrations of acetyl-CoA generate a favorable condition for non-enzymatic lysine acetylation (Garland et al., 1965; Paik et al., 1970). Regardless, both enzyme-catalyzed and non-enzymatic acetylation reactions are expected to increase as acetyl-CoA increases, and therefore the tissue specific levels of protein acetylation might reflect the distinct conditions of each tissue type. Moreover, it is important to highlight that functional (targeted) acetylation and spurious (untargeted or non-regulatory) acetylation can occur via enzyme-catalyzed or uncatalyzed reactions. Here we suggest that higher SIRT3 expression in liver and kidney is required to remove both targeted and spurious protein acetylation that is a consequence of the increased demand to produce acetyl-CoA under fasting. In the brain, heart and skeletal muscle, acetyl-CoA is rapidly consumed for energy, leading to reduced levels of spurious acetylation and reduced need for high levels SIRT3, which acts in a more targeted manner by primarily deacetylating regulatory acetylation sites. Consistent with this idea, acetyl-CoA concentrations in liver are estimated to be 3 to 5-fold higher compared to those in skeletal muscle and brain (Allred and Guy, 1969; Cederblad et al., 1990; Palladino et al., 2012).
Our study reveals that SIRT3 mediates metabolic coupling between fuel producing and fuel utilizing tissues through dynamic acetylation of numerous mitochondrial pathways identified with our multi-tissue mitochondrial acetylome and innovative bioinformatics pathway analysis tool. We provide evidence that SIRT3 promotes ketone body utilization in the brain by deacetylating and enhancing flux through OXCT and ACAT1, manifesting in bioeneregetic deficiencies in aged Sirt3−/− animals. While a picture is emerging for the importance of SIRT3 in fuel switching during adaptation to altered metabolic status, our multi-tissue, quantitative acetyl proteome compendium provides a rich resource to further explore the tissue specific functions of SIRT3 and the comparative mitochondrial acetylome required to understand metabolic regulation and the etiology of human disease associated with the inability to perform efficient fuel-switching.
Experimental procedures
Sample preparation, Mass spectrometry, Data analysis
Mouse brain, heart, kidney, liver, and skeletal muscle were isolated from 5-month, age matched Sirt3−/− and WT mice that were fasted for 24 hours (9AM to 9AM) and immediately frozen in liquid nitrogen. Samples were suspended in lysis buffer and lysed by glass bead-milling (Retsch). Samples were denatured, reduced, alkylated and digested with trypsin (Promega). TMT labeling of desalted peptides was performed according to manufacturer’s instructions (Thermo Pierce). Labeled peptides were fractionated by SCX chromatography and acetylated peptides were enriched with pan-acetyl lysine antibody-agarose conjugate. Acetyl enriched and non-enriched protein fractions were analyzed by online nano reverse phase liquid chromatography (Waters) coupled to an Orbitrap Fusion (Q-OT-qIT, Thermo). All MS/MS data were analyzed using the Coon OMSSA Proteomics Software Suite (COMPASS) (Wenger et al., 2011). Results were filtered to a 1% FDR.
SIRT3 Expression
Targeted MS
Unlabeled peptide fractions from brain, heart, kidney, liver, and skeletal muscle of a wild type mouse were subject to TMT labeling. Following tagging, peptides were mixed at a 1:1:1:1:1 ratio according to BCA results. For targeted MS analysis, the Sirt3 peptide ASGIPASK (+2, m/z 594.864, aa171–178) was isolated over a 1 Th window. Results were normalized to total signal intensity for each TMT tag.
Bioinformatics
Clustering
To reduce the overall percentage of missing values to enable clustering of the entire data set, missing protein abundance values were imputed for acetyl isoforms that were identified and quantified, but lacked protein level quantitation. To impute values, the average normalized intensity for all proteins found in each tissue of each mouse was calculated. This value was then used to calculate a protein fold change, to which the acetyl fold change could be normalized. Acetyl site fold change values were collapsed onto protein by taking the sum of the acetyl fold change for all acetyl sites identified and quantified for a unique UniProt identifier. To cluster the protein data set (~1100 proteins in 5 tissues) we used a Gaussian mixture model clustering approach (Hastie et al., 2009) with consensus clustering (Nguyen, 2007). Due to the large number of missing values (85% of proteins having missing values in two or more tissues), prior to applying the clustering method we interpolate the missing value with the mean of the non-missing values of the same protein. To cluster each K cluster, we used hierarchical clustering using Euclidean metric for distance.
Quantitative site set biological function score analysis (QSSA)
The intersection of the KEGG pathway map (Merico et al., 2010) and proteins identified with <1% FDR was used for the gene set background. Acetylation coverage for each (p) pathway was calculated as the number of acetyl sites identified (nk) over the total number of lysines in the pathway (ns), counted using protein sequences from UniProt. Dynamic response to SIRT3 was taken into account by calculating the mean magnitude fold change (FC) for each acetyl site quantified. To allow for combining acetylation coverage and fold change, the standard score of each quantity was taken. The overall pathway score was then calculated as the average of the individual z-scores:
Tissues were clustered by hierarchical clustering by calculating Euclidean distance and centroid linkage in Cluster 3.0 (de Hoon et al., 2004). Clusters were visualized with Java TreeView (Saldanha, 2004)
Ketone body utilization activity assay in brain homogenate
Brain cortices isolated from 8-month old, 24 hour fasted WT and Sirt3−/− mice (n=4) were placed in ice cold PBS supplemented with deacetylase inhibitors (1 mM sodium butyrate, 1 µM trichostatin A) and homogenized 3x1000 rpm with a glass homogenizer and Teflon pestle. The sample was centrifuged twice at 1000xg for 10 minutes at 4°C to remove insoluble material. The supernatant was used for the activity assay and protein was concentration was determined by BCA. To test acetoacetate utilization activity, homogenates were combined with 1 mM acetoacetate, 0.5 or 1 mM succinyl CoA, and 0.5 mM CoA in PBS at 37°C. A control reaction was performed without acetoacetate. Reaction time points were taken at 1, 3, 5, 8, 10, 12, 15, and 20 minutes by quenching the reaction mixture in 9 volumes of ice cold methanol. Citrate synthase activity was monitored by combining homogenates with 0.5 mM oxaloacetate and 0.5 mM acetyl-CoA in PBS at 37°C. A control reaction was performed without oxaloacetate. Reaction time points were taken at 1 and 10 minutes by immediately quenching the reaction mixture in 9 volumes of ice cold methanol. All samples were centrifuged at 21000xg for 10 minutes and supernatants were diluted and further analyzed by reverse phase separation on a Synergy Hydro-RP column (100 mm x 2 mm, 2.5 µm particle size, Phenomenex, Torrence, CA) coupled by negative mode electrospray ionization to an Orbitrap mass spectrometer, as previously described (Lu et al., 2010).
Sample preparation for metabolite analysis in brain
Brain cortices isolated from 15-month, 24 hour fasted WT and Sirt3−/− mice (n=4) were placed in 0.8 mL cold (−20°C) 80:20 methanol:H2O (v/v) and homogenized 3x1000rpm with a glass homogenizer and Teflon pestle. The sample was centrifuged at 14000xg for 10 minutes at 4°C. The supernatant was transferred to a new tube on ice. The pellet was re-extracted with 0.5 mL cold (−20°C) 80:20 methanol:H2O (v/v) twice, and the supernatants were combined. The samples were dried under nitrogen and resuspended in H2O for LC-MS analysis. The results were normalized to tissue weight.
Statistical Analysis
Proteomic data are expressed as log2 fold change between Sirt3−/− and WT. Statistical significance was calculated by Welch’s t test. Biochemical assay results are expressed as mean ± SD. Statistical significance was assessed by Student’s t test. A p-value of < 0.05 was considered statically significant for biochemical assays. Graphs were prepared and statistical analyses were performed using Origin, GraphPad Prism 6, Excel or MATLAB.
Supplementary Material
Acknowledgments
We would like to thank Daniel Amador-Noguez for assistance with metabolite studies. This work was funded by NIA grant AG038679 to J.M.D. and T.A.P, NIH grant GM065386 to J.M.D., NIH GM080148 to J.J.C. S.R. was supported by a Sloan Foundation research fellowship. K.E.D. was funded by a NSF Graduate Research Fellowship and NIH traineeship (5T32GM08349). A.L.R. was supported by a NIH-funded Genomic Sciences Training Program (5T32HG002760).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
K.E.D. designed research, isolated tissue, analyzed data, performed biochemical experiments, wrote manuscript; A.L.R. prepared MS samples, performed MS experiments; J.F. performed MS metabolite analysis, biochemical experiments, data analysis; M.J.S. designed and implemented QSSA; A.F. implemented clustering algorithms; Z.A.K. performed biochemical experiments.
References
- Allred JB, Guy DG. Determination of coenzyme A and acetyl CoA in tissue extracts. Anal Biochem. 1969;29:293–299. doi: 10.1016/0003-2697(69)90312-1. [DOI] [PubMed] [Google Scholar]
- Bauer M, Hamm AC, Bonaus M, Jacob A, Jaekel J, Schorle H, Pankratz MJ, Katzenberger JD. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol Genomics. 2004;17:230–244. doi: 10.1152/physiolgenomics.00203.2003. [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederblad G, Carlin JI, Constantin-Teodosiu D, Harper P, Hultman E. Radioisotopic assays of CoASH and carnitine and their acetylated forms in human skeletal muscle. Anal Biochem. 1990;185:274–278. doi: 10.1016/0003-2697(90)90292-h. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Garland PB, Shepherd D, Yates DW. Steady-state concentrations of coenzyme A, acetyl-coenzyme A and long-chain fatty acyl-coenzyme A in rat-liver mitochondria oxidizing palmitate. Biochem J. 1965;97:587–594. doi: 10.1042/bj0970587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning : data mining, inference, and prediction. 2nd edn. New York, NY: Springer; 2009. [Google Scholar]
- He W, Newman JC, Wang MZ, Ho L, Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab. 2012;23:467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Wang C, Zhou Y, Speed TP. Gene set enrichment analysis made simple. Stat Methods Med Res. 2009;18:565–575. doi: 10.1177/0962280209351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara T, Bonasio R, Narendra V, Reinberg D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol Cell Biol. 2012;32:5022–5034. doi: 10.1128/MCB.00822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, O'Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, et al. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62:3404–3417. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, et al. Proteomic Analysis of Lysine Acetylation Sites in Rat Tissues Reveals Organ Specificity and Subcellular Patterns. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem. 2012;287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N, Caruana R. Consensus Clusterings. Seventh IEEE International Conference on Data Mining (ICDM '07) 2007 [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik WK, Pearson D, Lee HW, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213:513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- Palladino AA, Chen J, Kallish S, Stanley CA, Bennett MJ. Measurement of tissue acyl-CoAs using flow-injection tandem mass spectrometry: acyl-CoA profiles in short-chain fatty acid oxidation defects. Mol Genet Metab. 2012;107:679–683. doi: 10.1016/j.ymgme.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, et al. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem. 2013;288:26209–26219. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanuk MH, Caudill MA. Biochemical, physiological, and molecular aspects of human nutrition. 3rd edn. St. Louis, Mo: Elsevier/Saunders; 2013. [Google Scholar]
- Suryawan A, Hawes JW, Harris RA, Shimomura Y, Jenkins AE, Hutson SM. A molecular model of human branched-chain amino acid metabolism. Am J Clin Nutr. 1998;68:72–81. doi: 10.1093/ajcn/68.1.72. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011;11:1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.