Abstract
Background/Aims
Although human leukocyte antigens (HLA) have been shown in association with the outcomes of HCV infection among different ethnic groups, such studies remain absent in China where the HCV prevalence is higher than the global average.
Methods
In this study, 426 HCV-infected and 709 uninfected blood donors were analyzed, among whom the HLA alleles were sequenced using a high-resolution-based genotyping method.
Results
At the 2-digit level, no alleles showed a statistical difference between the HCV-infected and uninfected groups. However, at the 4-digit level, HLA-B*15:01 and B*15:02, showed an opposite association with HCV infection, i.e. B*15:01 was significantly higher in the HCV-infected group (OR=1.561, P=0.010), while B*15:02 was significantly higher in the uninfected group (OR=0.778, P=0.016. We also identified a higher frequency of B*13:02 in the HCV-infected group (OR=1.515, P=0.009), and a higher frequency of B*07:05 in the uninfected group (OR=0.299, P=0.001).
Conclusions
The frequencies of four HLA alleles, B*07:05, B*13:02, B*15:01, and B*15:02, were found to be significantly different between the HCV-infected and uninfected blood donors in China, revealing an inverse relation of B*15:01 and B*15:02 with HCV infection. This finding suggests that the ethnic genetic variations of HLA may greatly affect the host immune responses against HCV.
Keywords: HCV, Chronic infection, HLA, alleles, Chinese population, blood donor
Introduction
Hepatitis C virus (HCV) is a blood-borne pathogen that causes a major threat to global public health [1]. Currently, an estimated 170–210 million people worldwide are infected with the virus [2]. The majority of the infection are persistent that could finally result in hepatic fibrosis, liver cirrhosis, hepatocellular carcinoma, and other end-stage liver diseases [3]. To date, interferon-α plus ribavirin has been used as a recommended regimen [3]; however, it is effective only in about half of the patients treated, and there remains a lack of reliable vaccines [4].
The host immunity can greatly influence the outcomes of HCV infection [5]. The robust and sustained immunological responses mediated by the activation of CD4+ and CD8+ cytotoxic T lymphocytes are the most important factors in determining the viral clearance or persistence [6–8]. Evidence has been shown that strong and broad T cell responses are correlated with viral clearance, while chronic HCV infection is associated with attenuated and narrowly focused CD4+ and CD8+ T cell responses [9]. Both T cells recognize the viral antigens presented on the surface of the infected hepatocytes through binding the T cell receptors to human leukocyte antigen (HLA) molecules that present the peptide fragments of the viral antigens. The HLA molecules are encoded by genes located on chromosome 6 that are among the most polymorphic region in human genome. The diversity of HLA molecules is critical in predicting the susceptibility and outcomes of natural HCV infection in human [10,11].
Previous studies have revealed associations between the HLA patterns and host immune responses among individuals infected with human immunodeficiency virus type 1 (HIV-1) and hepatitis B virus (HBV) [12–15]. Studies also showed the importance of HLA molecules to the natural outcomes of HCV infection. Although it has been reported in a few sufficiently sampled and well-designed studies that some alleles have strong associations with HCV infection [10,16–18], many other studies were limited either due to a small study population, or an inappropriate population stratification, or the lack of correction for multiple comparisons, or a low resolution typing technique [5,19]. Moreover, racial differences in the studies of association between the HLA patterns and the outcomes of HCV infection were also identified [18–21]. Nevertheless, there have been few studies focusing on the Chinese population, which is the largest HCV infected population in the world [22, 23].
In this study, we conducted a high resolution typing of the HLA class I (A and B) and II (DRB1) alleles among a cohort of Chinese voluntary blood donors that included 426 HCV-infected (HCV Ab+/RNA+) and 709 uninfected individuals (HCV Ab−/RNA−). The identified allelic and genotypic frequencies revealed an opposite association of HLA-B*15:01 and B*15:02 with HCV infection.
Materials and Methods
Subjects
Blood samples were collected from voluntary blood donors at Guangzhou Blood Center, China, during the period of July 2009 to November 2011. A total of 426 HCV-infected and 709 uninfected blood donors were recruited. Before blood donation, all individuals were informed to complete a Blood Donation Healthy Consulted Form for their participation in the study. Physicians ensured that individuals were personally interviewed to assure their complete understanding of the informed consent. All of the participants provided their verbal informed consent by telephone. This study strictly followed the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Medical Ethics Committee of Guangzhou Blood Center.
Pathogen Detection
The HCV, HBV, HIV and Treponemapallidum (TP) tests were performed for blood screening. Anti-HCV was assayed using two independent enzyme-linked immunosorbent (EIA) kits (Kehua Biotech Co. Ltd, Shanghai, China and Abbott HCV EIA 2.0, Abbott Laboratories, Illinois, USA) [24]. HCV RNA were detected using nucleic acid testing (NAT; the Procleix Ultrio Assay, Gen-Probe, San Diego, CA, USA) according to the providers’ guidance. HBV, HIV and TP were detected following the manufacturers’ protocols [25]. All the HCV infected donors were tested to be positive for both anti-HCV and HCV-RNA (HCV Ab+/RNA+), and negative for HIV antibodies, HBV surface antigen (HBsAg), TP antibodies, HBV and HIV nucleic acids, while the controls were found to be negative for all of these testing items.
HLA-A, B, and DRB1 typing
The HLA-A, B, and DRB1 alleles were assigned using the HLA sequencing based typing (HLA-SBT) by following the manufacturer’s protocols (AlleleSEQR HLA-A, AlleleSEQR HLA-B, AlleleSEQR HLA-DRB1, Atria Genetics, San Francisco, CA, USA). Briefly, the target genes were amplified in a mixture containing AmpliTaq Gold DNA polymerase. The amplicons were purified using ExoSAP-IT (Atria Genetics) and sequenced in both directions (ABI 3730 DNA Sequencer, Applied Biosystems, Foster City, CA, USA). The HLA alleles were assigned at a 4-digit level using the ASSIGN 3.5 software (Conexio Genomics, Applecross, Australia). Synonymous mutations were not recorded, while those samples with ambiguous results were applied to additional haplotype sequencing [26].
HCV genotyping
HCV genotypes were determined as previously described [25]. Some of the partial NS5B sequences of HCV have been previously reported [27]. In brief, the partial NS5B sequences of HCV were amplified using the Primer STAR kit (Takara, Dalian, China). The expected Amplicons were sequenced in both directions on an ABI Prism 3100 genetic analyzer (PE Applied Biosystems, FosterCity, CA, USA). The obtained sequences were aligned using the CLUSTAL_X program. Phylogenetic trees were estimated based on the maximum-likelihood method under the HKY+I+Γ6 substitution model using the MEGA5 software. Bootstrap resampling was performed in 1000 replicates.
Statistical analysis
The allelic and genotypic distribution at the HLA-A, B, and DRB1 loci and their association with chronic HCV infection among the Chinese voluntary blood donors were analyzed using Chi-square test with the SPSS 16.0 software. The strength of the associations was inferred by odds ratio (OR) with 95% confidence interval (95% CI). For multiple comparisons, false discovery rate (FDR) method (described by Benjamini and Hochberg) was used to calculate q values to control the false discovery rate. Statistically significant associations were indicated when q values were less than 0.1.
Results
Characteristics of the studied donors
The general information of the studied donors was summarized in Table 1. They were all Chinese and predominantly of Han ethnicity. Of the 426 HCV-infected donors, 82.9% (353/426) were male and 17.1% (73/426) were female, while these percentages were 71.9% (510/709) and 28.1% (199/709), respectively, among the 709 controls. The male/female ratio was significantly higher in the former than in the latter group (P=2.95E-05), which verified previous findings that male are more susceptible to HCV infection than female [24,25]. No significant difference was identified in age between the HCV-infected and uninfected group (P=0.146).
Table 1.
General information about the studied blood donors.
| Demographic data | HCV-infected donors (%) | Uninfected donors (%) | P value |
|---|---|---|---|
| Total number | 426 | 709 | |
| Age (X̄ ±S) | 33.8±8.9 | 31.0±8.9 | 0.146 |
| Gender * | |||
| Male | 353 (82.9) | 510 (71.9) | 2.95E-05 |
| Female | 73 (17.1) | 199 (28.1) | |
| Ethnicity | |||
| Chinese Han | 418 (98.1) | 700 (98.7) | 0.414 |
| Others | 8 (1.9) | 9 (1.3) | |
Significantly different (P<0.05) between the two groups based on Chi-square test.
HLA alleles and chronic HCV infection
To reveal possible associations between the HLA polymorphism and HCV infection, alleles at the HLA-A, B, and DRB1 loci were sequenced. Of the 426 HCV-infected donors, reliable typing results were obtained for 413, 415, and 416 at the HLA-A, B, and DRB1 loci, respectively. Of the 709 control donors, reliable typing results were obtained for 658, 650, and 683. In total, 31 HLA-A, 64 HLA-B, and 54 HLA-DRB1 alleles were identified. Table 2 listed the alleles that had frequencies of >1% among the studied subjects. At the HLA-A locus, 14 alleles showed frequencies of >1% accounting for 97.9% of the total HLA-A alleles identified. At the HLA-B locus, 18 alleles showed frequencies of >1% accounting for 86.7% of the total HLA-B alleles sequenced. Similarly, at the DRB1 locus, 18 alleles showed the frequencies of >1% accounting for 93.5% of the total HLA-DRB1 alleles detected.
Table 2.
Frequencies of HLA alleles between the two cohorts of blood donors a
| Loci | 4-digit | HCV infected donors n (%) |
Uninfected donors n (%) |
P value | OR (95% CI) |
|---|---|---|---|---|---|
| HLA-A | 01:01 | 10 (1.2) | 19 (1.5) | 0.648 | 0.892(0.539,1.478) |
| 02:01 | 49 (6.1) | 0.648 | 0.334 | 0.894(0.709,1.129) | |
| 02:03 | 66 (8.2) | 120 (9.3) | 0.364 | 0.913(0.746,1.117) | |
| 02:06 | 23 (2.8) | 48 (3.7) | 0.276 | 0.835(0.594,1.174) | |
| 02:07 | 98(12.1) | 135 (10.5) | 0.247 | 1.103(0.938,1.296) | |
| 03:01 | 14 (1.7) | 11 (0.9) | 0.072 | 1.460(1.027,2.075) | |
| 11:01 | 244 (30.2) | 423 (32.8) | 0.199 | 0.926(0.822,1.043) | |
| 11:02 | 32 (4.0) | 34 (2.6) | 0.093 | 1.267(0.982,1.635) | |
| 24:02 | 125 (15.5) | 187 (14.5) | 0.559 | 1.046(0.902,1.212) | |
| 26:01 | 14 (1.7) | 26 (2.0) | 0.639 | 0.906(0.592,1.386) | |
| 29:01 | 5 (0.6) | 26 (2.0) | 0.010 | 0.414(0.185,0.927) | |
| 30:01 | 27 (3.3) | 24 (1.9) | 0.033 | 1.385(1.063,1.805) | |
| 31:01 | 14 (1.7) | 9 (0.7) | 0.027 | 1.588(1.139,2.214) | |
| 33:03 | 88 (10.9) | 134 (10.4) | 0.731 | 1.031(0.868,1.224) | |
|
| |||||
| HLA-B b | 07:05 | 4 (0.6) | 30 (2.7) | 0.001 ** | 0.299(0.119,0.752) |
| 13:01 | 63 (8.8) | 105 (9.3) | 0.708 | 0.962(0.784,1.180) | |
| 13:02 | 25 (3.5) | 18 (1.6) | 0.009 * | 1.515(1.168,1.965) | |
| 15:01 | 21 (2.9) | 14 (1.2) | 0.010 * | 1.561(1.184,2.059) | |
| 15:02 | 61 (8.5) | 136 (12.0) | 0.016 * | 0.778(0.627,0.967) | |
| 27:04 | 9 (1.3) | 25 (2.2) | 0.135 | 0.677(0.386,1.190) | |
| 35:01 | 18 (2.5) | 16 (1.4) | 0.089 | 1.372(0.994,1.894) | |
| 38:02 | 35 (4.9) | 63 (5.6) | 0.514 | 0.915(0.697,1.202) | |
| 39:01 | 13 (1.8) | 14 (1.2) | 0.317 | 1.244(0.838,1.848) | |
| 40:01 | 135 (18.8) | 192 (17.0) | 0.318 | 1.078(0.933,1.245) | |
| 40:02 | 9 (1.3) | 21 (1.9) | 0.316 | 0.769(0.444,1.333) | |
| 46:01 | 131 (18.3) | 196 (17.4) | 0.618 | 1.038(0.897,1.203) | |
| 48:01 | 14 (2.0) | 18 (1.6) | 0.565 | 1.129(0.759,1.679) | |
| 51:01 | 34 (4.7) | 58 (5.1) | 0.704 | 0.949(0.722,1.247) | |
| 51:02 | 12 (1.7) | 20 (1.8) | 0.875 | 0.965(0.615,1.515) | |
| 54:01 | 27 (3.8) | 43 (3.8) | 0.962 | 0.993(0.734,1.342) | |
| 55:02 | 20 (2.8) | 37 (3.3) | 0.555 | 0.901(0.630,1.288) | |
| 58:01 | 86 (12.0) | 123 (10.9) | 0.467 | 1.068(0.898,1.269) | |
|
| |||||
| HLA-DRB1 | 03:01 | 64 (8.3) | 95 (7.4) | 0.472 | 1.077(0.883,1.313) |
| 04:03 | 19 (2.5) | 43 (3.4) | 0.251 | 0.810(0.555,1.184) | |
| 04:05 | 42 (5.4) | 68 (5.3) | 0.896 | 1.016(0.796,1.298) | |
| 04:06 | 14 (1.8) | 20 (1.6) | 0.664 | 1.097(0.731,1.646) | |
| 07:01 | 47 (6.1) | 58 (4.5) | 0.120 | 1.203(0.965,1.499) | |
| 08:03 | 42 (5.4) | 67 (5.2) | 0.836 | 1.026(0.804,1.310) | |
| 09:01 | 115 (14.9) | 184 (14.3) | 0.739 | 1.027(0.879,1.200) | |
| 10:01 | 9 (1.2) | 38 (3.0) | 0.008 | 0.504(0.279,0.908) | |
| 11:01 | 43 (5.6) | 82 (6.4) | 0.446 | 0.910(0.710,1.167) | |
| 12:01 | 20 (2.6) | 34 (2.7) | 0.931 | 0.985(0.692,1.401) | |
| 12:02 | 78 (10.1) | 162 (12.6) | 0.083 | 0.849(0.701,1.028) | |
| 13:02 | 22 (2.8) | 24 (1.9) | 0.147 | 1.280(0.942,1.740) | |
| 13:12 | 14 (1.8) | 13 (1.0) | 0.124 | 1.386(0.960,2.002) | |
| 14:01 | 47 (6.1) | 60 (4.7) | 0.165 | 1.179(0.945,1.472) | |
| 14:05 | 22 (2.8) | 28 (2.2) | 0.344 | 1.175(0.855,1.615) | |
| 15:01 | 98 (12.7) | 153 (11.9) | 0.614 | 1.044(0.885,1.232) | |
| 15:02 | 37 (4.8) | 50 (3.9) | 0.332 | 1.138(0.885,1.462) | |
| 16:02 | 40 (5.2) | 104 (8.1) | 0.012 | 0.725(0.553,0.949) | |
Alleles with frequencies higher than 1.0% were shown at the 4-digit level and analyzed.
Alleles with statistical significance (q<0.1) were indicated in bold.
q < 0.1;
q <0.05
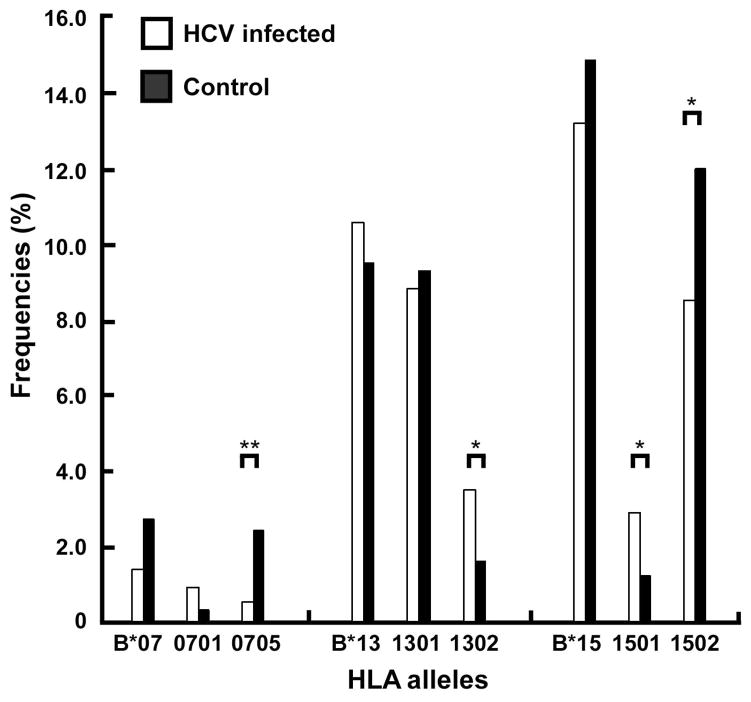
Next, we analyzed the allele frequencies at the 2-digit level. The allele frequencies at three loci, HLA-A, B, and DRB1, were not statistically different between the two study groups after adjusted for multiple variables (Table S1). However, when analyzed at the 4-digit level, four alleles at the HLA-B locus were found to be significantly different between the two groups (Table 2). B*13:02 (OR=1.515, P=0.009, q<0.1) and B*15:01 (OR=1.561, P=0.010, q<0.1) were significantly more often in the HCV-infected group than in the control group. Reversely, B*07:05 (OR=0.299, P=0.001, q<0.05) and B*15:02 (OR=0.778, P=0.016, q<0.1) were significantly more often in the control group than in the HCV-infected group (Table 2 and Figure 1).
Figure 1. Comparison of HLA alleles at the 2-digit and 4-digit levels in association with chronic HCV infection.
The HLA-B*07, B*13, B*15 alleles assigned at the 2-digit level were compared with their subordinates assigned at the 4-digit alleles. *q < 0.1; **q <0.05.
The analysis at the HLA-A and DRB1 loci also identified the alleles with marginally significant differences between the HCV-infected and uninfected groups. A*30:01 and A*33:03 (OR=1.385, P=0.033 and OR=1.588, P=0.027, respectively) appeared to be more often in the HCV-infected group, while A*29:01 (OR=0.414, P=0.010), DRB1*10:01 (OR=0.504, P=0.008), and DRB1*16:02 (OR=0.725, P=0.012) appeared to be more often among the control group (Table 2). However, they all failed to reach a significant level after an FDR adjustment (q>0.1).
Finally, a multivariate logistic regression analysis was used to estimate the independent risk factors for these alleles in association with HCV infection, adjusted for other possible modifiers or confounders such as age, gender, etc. Stepwise logistic regression analysis indicated that five variables were independently associated with HCV infection: ➀gender (OR = 0.532, P < 0.001, 95% CI [0.428–0.660]), ➁HLA-B*07:05 allele (OR = 0.204, P = 0.003, 95% CI [0.071–0.583]), ➂HLA-B*15:02 allele (OR = 0.690, P = 0.023, 95% CI [0.502–0.949]), ➃HLA-B1*13:02 allele (OR = 2.217, P = 0.012, 95% CI [1.193–4.120]), and ➄HLA-B*15:01 allele (OR =2.319, P = 0.017, 95% CI [1.165–4.620]) (Table 3).
Table 3.
Multiple Logistic regression analysis of variables associated with HCV infection
| Variable | OR (95%CI) | P value |
|---|---|---|
| Gender (female/male) | 0.532 (0.428–0.660) | < 0.001 |
| HLA-B *07:05 | 0.204 (0.071–0.583) | 0.030 |
| HLA-B *13:02 | 2.217 (1.193–4.120) | 0.012 |
| HLA-B *15:01 | 2.319 (1.165–4.620) | 0.017 |
| HLA-B *15:02 | 0.690 (0.502–0.949) | 0.023 |
P-values are less than 0.05. CI, confidence interval; OR, odds ratio.
HLA genotypes and chronic HCV infection
A potential association between the HLA genotypes and HCV infection was also assessed. Different HLA-A genotypes showed varied frequencies between the two study groups (χ2=41.565, P=0.007) (Table 4), while this was not the case for the HLA-B and DRB1 alleles (χ2=27.537, P=0.154, and χ2=31.086, P=0.411, respectively) (Table S2 and S3). Both A*02:07/A*02:07 and A*11:01/A*11:02 were significantly more frequent in the HCV-infected than in the control group (OR=1.831, P=0.021, and OR=1.824, P=0.011, respectively). A*02:07/A*11:01 was significantly more frequent in the control than in the HCV-infected group (OR=0.612, P=0.011) (Table 4). However, after an adjustment for multiple variables, this significance was not remained.
Table 4.
Frequencies of HLA-A genotype in the HCV-infected and control donors a
| HLA-A genotype | HCV infected donors (%) | Uninfected donors (%) | P value | OR (95% CI) |
|---|---|---|---|---|
| A*02:01/A*02:03 | 3 (1.0) | 9 (1.9) | 0.339 | 0.649 (0.243,1.736) |
| A*02:01/A*02:07 | 8 (1.5) | 4 (0.8) | 0.083 | 1.760 (1.168,2.652) |
| A*02:01/A*11:01 | 11 (3.6) | 24 (4.9) | 0.391 | 0.813 (0.494,1.338) |
| A*02:01/A*24:02 | 6 (2.0) | 11 (2.3) | 0.795 | 0.919 (0.480,1.761) |
| A*02:01/A*33:03 | 6 (2.0) | 8 (1.6) | 0.725 | 1.121 (0.608,2.065) |
| A*02:03/A*02:07 | 12(4.0) | 10 (2.1) | 0.112 | 1.441 (0.973,2.132) |
| A*02:03/A*11:01 | 20 (6.6) | 45 (9.3) | 0.191 | 0.789 (0.542,1.149) |
| A*02:03/A*24:02 | 11 (3.6) | 11 (2.3) | 0.253 | 1.316 (0.858,2.018) |
| A*02:03/A*33:03 | 3 (1.0) | 12 (2.5) | 0.141 | 0.517 (0.187,1.428) |
| A*02:06/A*11:01 | 8 (2.6) | 18 (3.7) | 0.42 | 0.797 (0.445,1.429) |
| A*02:07/A*02:07 | 9 (3.0) | 4 (0.8) | 0.021* | 1.831 (1.260,2.660) |
| A*02:07/A*11:01 | 17 (5.6) | 53 (10.9) | 0.011* | 0.612 (0.401,0.934) |
| A*02:07/A*24:02 | 15 (5.0) | 27 (5.6) | 0.721 | 0.928 (0.613,1.407) |
| A*02:07/A*33:03 | 14 (4.6) | 13 (2.7) | 0.141 | 1.370 (0.942,1.993) |
| A*11:01/A*11:01 | 44 (14.6) | 62 (12.8) | 0.469 | 1.097 (0.858,1.403) |
| A*11:01/A*11:02 | 11 (3.6) | 5 (1.0) | 0.011* | 1.824 (1.295,2.569) |
| A*11:01/A*24:02 | 32 (10.6) | 68 (14.0) | 0.164 | 0.815 (0.604,1.101) |
| A*11:01/A*26:01 | 4 (1.3) | 11 (2.3) | 0.348 | 0.692 (0.297,1.609) |
| A*11:01/A*30:01 | 9 (3.0) | 7 (1.4) | 0.136 | 1.482 (0.953,2.305) |
| A*11:01/A*33:03 | 27 (8.9) | 48 (9.9) | 0.863 | 0.933 (0.681,1.280) |
| A*24:02/A*24:02 | 11 (3.6) | 12 (2.5) | 0.341 | 1.257 (0.813,1.945) |
| A*24:02/A*33:03 | 16 (5.3) | 16 (3.3) | 0.165 | 1.322 (0.924,1.891) |
| A*33:03/A*33:03 | 5 (1.7) | 8 (1.6) | 0.992 | 1.004 (0.502,2.008) |
Genotypes with frequencies of <1% were not shown.
P<0.05
HLA alleles and HCV genotypes
To explore possible association between the HLA polymorphism and HCV genotypes, the alleles at the A, B, and DRB1 loci were further correlated with the 166 and 136 isolates of HCV, which were determined from the HCV-infected donors and classified into subtypes 1b and 6a (Figure S1), respectively. However, no significant differences were identified in this analysis (Table S4).
DISCUSSION
In this study, we examined the association between the HLA alleles and HCV infection among a cohort of voluntary blood donors. To our knowledge, this represents the first study of such in China. Among the studied donors, all of those infected with HCV were asymptomatic, treatment naive, and are therefore ideal to display the natural outcomes of HCV infection for representing the general population in the country where the HCV prevalence is above the global average.
Our results revealed that, at different levels, four HLA alleles B*07:05, B*13:02, B*15:01, and B*15:02 were associated with HCV infection. Although such a relationship was not reported before, previous studies have shown the four alleles in association with other viral infections. Among Chinese patients, B*07 might protect hosts from HBV infection, B*15 was identified in association with the HBV-C2 clearance [12], while B*13 was detected more often in the HIV-1-infected individuals with unusually low viral loads [15]. These findings, together with that from the present study, may indicate the varied antigen-binding properties of the polymorphic HLA alleles and their different roles played in the host anti-viral immune responses.
The association between HCV infection and the four alleles B*07:05, B*13:02, B*15:01, and B*15:02 may be simply attributed to the differences in certain ethnic factors. Previously, most of the studies on this regard were performed on Caucasians, Hispanics and Africans [10,16–18], showing different results. For example, conflicting associations with HCV infection have been observed for the A*02 and DRB1*12 alleles when both Caucasians and non-Caucasians were compared [19]. Thio and his colleagues have identified that between the white and black people, the specific HLA class II alleles including the DRB1*01 are differently associated with the HCV infection outcomes [18]. With reference to the two studies described above, the present study showed distinct patterns of the HLA polymorphism in association with the HCV infection among a sunset of the Chinese blood donors.
To assess the association between the HLA polymorphism and HCV infection, we applied a high-resolution genotyping method, which can provide more robust information than using a low resolution approach [5,16]. Although no statistical differences were found in the frequency of alleles at the 2-digit level between the HCV-infected and uninfected groups, at the 4-digit level four HLA-B alleles showed significantly different frequencies between these two groups. B*15:01 and B*15:02 showed opposite associations with the former appearing as a risk allele while the latter a possibly favoring allele (Fig. 1). A similar situation was also observed between the B*07:01 and B*07:05 alleles, although the former failed to reach a statistical threshold likely due to its very low frequency of <1%. In a previous study on HIV-1 infection, opposing effects have been also observed for B*58:01 and B*58:02 that may control the level of viremia [28]. To our knowledge, the opposing effects identified in this study on HCV infection could represent the first such report hitherto. We hypothesize that the differences in the amino acids encoded by these polymorphic alleles (B*15:01/B*15:02, and B*07:01/B*07:05) may have affected the HLA properties in binding antigens, either structurally or functionally or both, and this could have contributed to the different outcomes of HCV infection. However, because the underlying molecular mechanism is not clear, further studies are therefore required. Such studies will not only lead to better understanding of the host-HCV interactions, they will also help in developing new strategies for the ultimate HCV eradication.
In this study, we also examined the association between the HLA alleles and HCV genotypes. HCV is known to be genetically diverse with seven genotypes and more than 80 subtypes that are now classified [29]. Such a high degree of genetic heterogeneity may have caused variable immune responses of the hosts that are determined by different patterns of the HLA alleles. A previous study has identified different distributions of the HLA class I alleles between infections with HCV subtypes 1a and 1b [30]. The HLA-restricted variations between HCV infections with subtypes 1a and 3a have been also indicated [31]. Recently, we have demonstrated that the infection with HCV subtype 6a is increasingly prevalent among patients and blood donors in Guangdong province, China, and this has made 6a and 1b as the two major HCV subtypes detected in that geographic region [25,27]. However, there remains a knowledge gap about the association between the HLA polymorphism and HCV genotypes in the general population in the country. In this study, we tried to address this issue but failed to obtain significant findings, likely for two reasons. First, some of the HLA alleles were undetected in this study possibly due to their very low frequencies among the asymptomatic blood donors we recruited, while these alleles may be more common among the symptomatic patients infected with more virulent strains of HCV. Second, possible associations between the HLA alleles and HCV amino acid variations were not inspected, while their interactions are deemed to play important roles in predicting the outcomes of HCV infection [16,32,33]. Future studies are needed to examine these possibilities.
It should be noted that if more information can be obtained from those donors who have spontaneously cleared the HCV infection, namely they were HCV-Ab+/HCV RNA− when analyzed, in comparison with those who are having existing HCV infection, more robust conclusion would have been made that is useful for predicting the outcomes of the natural HCV infection. Based on the results from the present study, it may be hypothesized that the described patterns of HLA alleles somehow interact with certain ethnic factors in determining the host susceptibility to persistent HCV infection and this is more prominent among a subset of Chinese population. Unfortunately, due to the lack of such cases who had subsequently cleared the HCV infection during the period of prospective observation, we were unable to test this hypothesis in detail. Nevertheless, for this purpose we are now recruiting more blood donors with natural HCV infection. It is our expectation that such a wide-organized prospective study will greatly facilitate the exploration of the roles played by the described patterns of HLA alleles in association with spontaneous HCV clearance in the Chinese population.
In conclusion, we identified the possible association of four HLA alleles, B*07:05, B*13:02, B*15:01, and B*15:02, with HCV infection among a cohort of voluntary blood donors who may ideally represent the general population in China, among whom B*15:01 and B*15:02 appeared to play opposing roles. This finding not only support the premise that the HLA polymorphic patterns are ethnically specific, it also partially fills in a knowledge gap about of the association between the HLA alleles and HCV infection in the Chinese population.
Supplementary Material
Acknowledgments
This work was supported by the Key Medical Disciplines and Specialties’ Program of Guangzhou, a grant from the National Natural Science Foundation of China (No.81273145), a grant from The 12th Five-year National Science & Technology Major Project (2012 ZX10004702), and a grant from NIAID/NIH (5 R01 AI080734-03A). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hepatitis C--global prevalence (update) Wkly Epidemiol Rec. 1999;74:425–427. [PubMed] [Google Scholar]
- 2.El-Shabrawi MH, Kamal NM. Burden of pediatric hepatitis C. World J Gastroenterol. 2013;19:7880–7888. doi: 10.3748/wjg.v19.i44.7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chevaliez S, Pawlotsky JM. Hepatitis C virus: virology, diagnosis and management of antiviral therapy. World J Gastroenterol. 2007;13:2461–2466. doi: 10.3748/wjg.v13.i17.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 5.Yee LJ. Host genetic determinants in hepatitis C virus infection. Genes Immun. 2004;5:237–245. doi: 10.1038/sj.gene.6364090. [DOI] [PubMed] [Google Scholar]
- 6.Lechner F, Gruener NH, Urbani S, Uggeri J, Santantonio T, Kammer AR, Cerny A, Phillips R, Ferrari C, Pape GR, Klenerman P. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol. 2000;30:2479–2487. doi: 10.1002/1521-4141(200009)30:9<2479::AID-IMMU2479>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 7.Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker BD. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klenerman P, Hill A. T cells and viral persistence: lessons from diverse infections. Nat Immunol. 2005;6:873–879. doi: 10.1038/ni1241. [DOI] [PubMed] [Google Scholar]
- 10.Hraber P, Kuiken C, Yusim K. Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology. 2007;46:1713–1721. doi: 10.1002/hep.21889. [DOI] [PubMed] [Google Scholar]
- 11.Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity (Edinb) 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Liu W, Wang H, Jin X, Fang S, Shi Y, Liu Z, Zhang S, Yang S. The influence of HLA alleles and HBV subgenotyes on the outcomes of HBV infections in Northeast China. Virus Res. 2012;163:328–333. doi: 10.1016/j.virusres.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Roe DL, Lewis RE, Cruse JM. Association of HLA-DQ and -DR alleles with protection from or infection with HIV-1. Exp Mol Pathol. 2000;68:21–28. doi: 10.1006/exmp.1999.2287. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Kaul R, Kaul A, Khan K. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J Gastroenterol. 2007;13:1770–1787. doi: 10.3748/wjg.v13.i12.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G, Allen S, Musonda R, Kaslow RA. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuniholm MH, Kovacs A, Gao X, Xue X, Marti D, Thio CL, Peters MG, Terrault NA, Greenblatt RM, Goedert JJ, Cohen MH, Minkoff H, Gange SJ, Anastos K, Fazzari M, Harris TG, Young MA, Strickler HD, Carrington M. Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology. 2010;51:1514–1522. doi: 10.1002/hep.23515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, O’Brien SJ, Karacki P, Astemborski J, Carrington M, Thomas DL. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thio CL, Thomas DL, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, O’Brien SJ, Karacki P, Marti D, Astemborski J, Carrington M. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- 19.Wang JH, Zheng X, Ke X, Dorak MT, Shen J, Boodram B, O’Gorman M, Beaman K, Cotler SJ, Hershow R, Rong L. Ethnic and geographical differences in HLA associations with the outcome of hepatitis C virus infection. Virol J. 2009;6:46. doi: 10.1186/1743-422X-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarmiento OL, Ford CL, Newbern EC, Miller WC, Poole C, Kaufman JS. The importance of assessing effect modification when asserting racial differences in associations between human leukocyte antigen class II alleles and hepatitis C virus outcomes. J Infect Dis. 2002;185:266–268. doi: 10.1086/338198. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y, Cao D, Li Y, Kulski JK, Shi L, Jiang H, Ma Q, Yu J, Zhou J, Yao Y, Shi L. Distribution of HLA-A, -B, and -C Alleles and HLA/KIR Combinations in Han Population in China. J Immunol Res. 2014;2014:565296. doi: 10.1155/2014/565296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu RB, Hong X, Ding WL, Tan YF, Zhang YX, Sun NX, Wu GL, Zhan SW, Ge DF. The association between the genetic polymorphism of HLA-DQA1, DQB1, and DRB1 and serum alanine aminotransferase levels in chronic hepatitis C in the Chinese population. J Gastroenterol Hepatol. 2008;23(9):1394–402. doi: 10.1111/j.1440-1746.2007.05215.x. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y, Xia W, Wang Y, Tian L, Pybus OG, Lu L, Nelson K. The seroprevalence of hepatitis C virus (HCV) among 559,890 first-time volunteer blood donors in China reflects regional heterogeneity in HCV prevalence and changes in blood donor recruitment models. Transfusion. 2010;50:1505–1511. doi: 10.1111/j.1537-2995.2010.02616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y, Wang Y, Xia W, Pybus OG, Qin W, Lu L, Nelson K. New trends of HCV infection in China revealed by genetic analysis of viral sequences determined from first-time volunteer blood donors. J Viral Hepat. 2010;18:42–52. doi: 10.1111/j.1365-2893.2010.01280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Z, Wang D, Xu Y, Gao S, Zhou H, Yu Q, Yang B. HLA-C polymorphisms and PCR dropout in exons 2 and 3 of the Cw*0706 allele in sequence-based typing for unrelated Chinese marrow donors. Hum Immunol. 2010;71:577–581. doi: 10.1016/j.humimm.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Rong X, Lu L, Wang J, Xiong H, Huang J, Chen J, Huang K, Xu R, Wang M, Zhang X, Guo T, Liu Y, Gao G, Fu Y, Nelson KE. Correlation of Viral Loads with HCV Genotypes: Higher Levels of Virus Were Revealed among Blood Donors Infected with 6a Strains. PLoS One. 2012;7:e52467. doi: 10.1371/journal.pone.0052467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 29.Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 30.Lange CM, Roomp K, Dragan A, Nattermann J, Michalk M, Spengler U, Weich V, Lengauer T, Zeuzem S, Berg T, Sarrazin C. HLA class I allele associations with HCV genetic variants in patients with chronic HCV genotypes 1a or 1b infection. J Hepatol. 2010;53:1022–1028. doi: 10.1016/j.jhep.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Rauch A, James I, Pfafferott K, Nolan D, Klenerman P, Cheng W, Mollison L, McCaughan G, Shackel N, Jeffrey GP, Baker R, Freitas E, Humphreys I, Furrer H, Gunthard HF, Hirschel B, Mallal S, John M, Lucas M, Barnes E, Gaudieri S. Divergent adaptation of hepatitis C virus genotypes 1 and 3 to human leukocyte antigen-restricted immune pressure. Hepatology. 2009;50:1017–1029. doi: 10.1002/hep.23101. [DOI] [PubMed] [Google Scholar]
- 32.McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, Walsh A, Hegarty J, Lawlor E, Kelleher D. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 33.Rauch A, Gaudieri S, Thio C, Bochud PY. Host genetic determinants of spontaneous hepatitis C clearance. Pharmacogenomics. 2009;10:1819–1837. doi: 10.2217/pgs.09.121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.