Abstract
Innate immune responses depend on timely recognition of pathogenic or danger signals by multiple cell surface or cytoplasmic receptors and transmission of signals for proper counteractions through adaptor and effector molecules. At the forefront of innate immunity are four major signaling pathways, including those elicited by Toll-like receptors, RIG-I-like receptors, inflammasomes, or cGAS, each with its own cellular localization, ligand specificity, and signal relay mechanism. They collectively engage a number of overlapping signaling outcomes, such as NF-κB activation, interferon response, cytokine maturation, and cell death. Several proteins often assemble into a supramolecular complex to enable signal transduction and amplification. In this article, we review the recent progress in mechanistic delineation of proteins in these pathways, their structural features, modes of ligand recognition, conformational changes, and homo- and hetero-oligomeric interactions within the supramolecular complexes. Regardless of seemingly distinct interactions and mechanisms, the recurring themes appear to consist of autoinhibited resting-state receptors, ligand-induced conformational changes, and higher-order assemblies of activated receptors, adaptors, and signaling enzymes through conserved protein-protein interactions.
Keywords: Toll-like receptor, RIG-I-like receptor, inflammasome, cGAS, STING, death domain superfamily, higher-order assembly
INTRODUCTION
Innate immunity research has undergone a revolution since the first discovery, in the late 1990s, of a Toll-like receptor (TLR) that acts as a microbial sensor and as the immunologist’s dirty little secret for the induction of adaptive immunity (1–3). The innate immune system is composed of a large number of cell surface and cytosolic pattern-recognition receptors (PRRs) that are used for sensing conserved pathogen-associated molecular patterns (PAMPs) present on bacteria, viruses, and fungi and for detecting intrinsic danger-associated molecular patterns (DAMPs) elicited by cellular injury. In addition to TLRs, major PRR families include, but are not limited to, absent in melanoma 2 (AIM2)-like receptors (ALRs), nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), RIG-I-like receptors (RLRs), and the cGAS/STING system (Figure 1). Upon ligand binding, these receptors activate the NF-κB transcription factors for inflammatory responses, interferons (IFNs) for establishment of antiviral states, and caspases for cytokine maturation and cell death induction. Cytokine receptors in the interleukin-1 (IL-1) receptor (IL-1R) family signaling pathways are similar to those of TLRs. Collectively, these cellular responses provide the arsenal to fight infection and to restore homeostasis. Understanding the structure of these receptor signaling pathways is critical for revealing the molecular mechanisms that govern innate immunity. In this review, we focus on recent structural elucidations of receptor-proximal signaling components in these pathways.
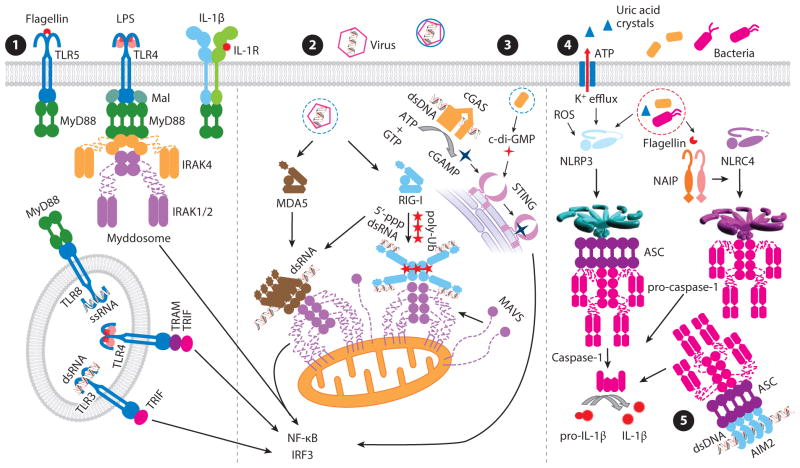
Figure 1.
Overview of TLR/IL-1R, RLR, cGAS, and inflammasome signaling pathways. The TLR/IL-1R signaling pathway ➊. TLRs are responsible for recognition of various pathogen-associated molecular patterns. IL-1R binds to IL-1β and associates with IL-1RAcP to form a ternary active complex. Both TLRs and IL-1Rs contain intracellular Toll-IL-1R domains, responsible for recruitment of downstream signaling molecules. Upon activation, IL-1Rs and all TLRs except TLR3 recruit MyD88 for signal transduction. In the case of TLR4, Mal is required for the recruitment of MyD88. Upon recruitment by TLRs/IL-1Rs, MyD88 further recruits IRAK4 and IRAK1/2 to form the Myddosome, through death domain interaction. IRAKs phosphorylate and activate downstream substrates, leading to the activation of NF-κB and IRF3. TLR3 recruits TRIF and activates TRIF-dependent signaling pathways. Endosomal TLR4 also activates TRIF-dependent signaling pathways through recruitment of TRAM and TRIF. The RLR signaling pathway ➋. RIG-I and MDA5 are responsible for detection of different types of viral RNAs. RIG-I recognizes viral RNA containing 5′-ppp, whereas MDA5 detects long viral dsRNA. Upon RNA stimulation, RIG-I forms a tetramer and nucleates MAVS filament formation in the presence of K63 polyubiquitin chains. MDA5 and RIG-I assemble into filamentous structures along long dsRNA and nucleate the filament assembly of downstream MAVS. MAVS filaments further activate downstream proteins, leading to the activation of IRF3 and induction of type I interferons. The cGAS/STING signaling pathway ➌. cGAS is a major cytosolic DNA sensor that synthesizes cGAMP upon activation by dsDNA. cGAMP acts as a second messenger to activate downstream STING. STING can also be directly activated by the bacterial second messenger c-di-GMP. Upon binding to stimulators, STING undergoes a conformational change and activates downstream signaling molecules. The inflammasome signaling pathways ➍➎. NLRP3 is activated by diverse stimuli, including pathogens, extracellular ATP, ROS, and phagocytosed particulates such as uric acid crystals. NAIPs recognize bacterial proteins and activate NLRC4. The AIM2 inflammasome is activated by cytosolic dsDNA released by viruses or bacteria. Upon activation, both NLRP3 and AIM2 nucleate filament formation of pro-caspase-1 through the adaptor protein ASC, whereas NLRC4 may directly nucleate pro-caspase-1 filament formation. The caspase domains of pro-caspase-1 are brought into proximity, leading to dimerization and activation of caspase-1. Active caspase-1 processes pro-IL-1β to generate mature IL-1β. Abbreviations: AIM2, absent in melanoma 2; ASC, apoptosis-associated speck-like protein containing a CARD; c-di-GMP, cyclic di-GMP; cGAMP, cyclic-GMP-AMP; cGAS, cGAMP synthase; IL-1RAcP, IL-1R accessory protein; IRAK, IL-1R-associated kinase; IRF, interferon regulatory factor; LPS, lipopolysaccharide; Mal, MyD88-adaptor-like; MAVS, mitochondrial antiviral-signaling protein; MDA5, melanoma differentiation-associated protein 5; NLR, NOD-like receptor; NLRC, NLR with N-terminal CARD; NLRP, NLR with N-terminal PYD; RLR, RIG-I-like receptor; ROS, reactive oxygen species; STING, stimulator of interferon genes; TLR, Toll-like receptor; TRAM, TRIF-related adaptor molecule; TRIF, TIR-domain-containing, adaptor-inducing interferon-β; Ub, ubiquitin.
THE TOLL-LIKE RECEPTOR/IL-1R SIGNALING PATHWAY
Background
TLRs and IL-1 receptors are classified into an evolutionarily conserved receptor superfamily, the TLR/IL-1R superfamily. Despite having distinct ectodomains, TLRs and IL-1Rs possess a common cytoplasmic signaling domain, the Toll-IL-1R (TIR) homology domain (4). The 10 TLRs that have been identified in humans recognize distinct families of PAMPs (5, 6). For example, TLR2 forms a heterodimer with either TLR1 or TLR6, each recognizing distinct bacterial lipoproteins. TLR4 in complex with MD2 is responsible for recognition of bacterial lipopolysaccharides (LPS), a major component of the outer membranes of gram-negative bacteria. TLR5 responds to a depolymerized form of bacterial flagellin (FliC). TLR3 senses viral dsRNA, whereas TLR7 and TLR8 are stimulated by ssRNA. TLR9 is responsive to unmethylated CpG islands of bacterial and viral DNA. TLR1-2 and 4–6 are cell surface receptors, whereas TLR3 and 7–9 reside on the surface of endosomes, which contain internalized viral and bacterial genomes.
The IL-1R family comprises receptors for proinflammatory cytokines such as IL-1β, whose production is activated through cooperation of a number of PRRs. Once secreted, IL-1β acts as a key cytokine that regulates innate and adaptive immunity (7). IL-1β forms a ternary signaling complex with its primary receptor and a receptor accessory protein. As the primary receptor of IL-1β, IL-1R type I (IL-1RI) directly binds to IL-1β. In contrast, IL-1R accessory protein (IL-1RAcP) does not interact with IL-1β directly, but its association with the IL-1β/IL-1RI binary complex is essential for the subsequent signal transduction. IL-1 receptor type II (IL-1RII), a decoy receptor of IL-1β, has an extracellular domain similar to IL-RI but lacks the intracellular TIR domain essential for signal transduction (8). Other IL-1R family members, such as the receptors for IL-18, IL-33, and IL-36, share similar signaling pathways.
Upon receptor activation, the TIR domains of TLRs and IL-1Rs are believed to oligomerize and recruit TIR-containing adaptors, such as MyD88, Mal, TRIF (TIR-domain-containing, adaptor-inducing interferon-β), and TRAM (TRIF-related adaptor molecule) (4). In addition to its TIR domain, MyD88 contains a death domain (DD) that interacts with IL-1R-associated kinases (IRAKs), including IRAK1, IRAK2, IRAK4, and IRAK-M, through DD-DD interactions to form the Myddosome (9, 10). Myddosome assembly leads to IRAK activation, stimulation of nondegradative polyubiquitination by TRAF6, and finally activation of transcription factors to execute the signaling pathway (11).
Structures of Ectodomains of Toll-Like Receptors in Complex with Ligands
The ectodomains of TLRs belong to the leucine-rich repeat (LRR) family, which is characterized by the presence of multiple LRR modules. Each LRR module is usually composed of ~20–30 amino acids, and typically in a β-turn-α structure (12). Each TLR ectodomain contains more than 10 LRR modules that are assembled into an α/β horseshoe fold with an inner surface of parallel β-sheet, an outer surface of α-helices or long loops, and a hydrophobic core between that is packed with many leucine residues. Crystal structures of most TLR ectodomains, alone or in complex with their ligands, are now available (13–20). These structures include those of TLR3, the TLR1/TLR2 complex, and the TLR2/TLR6 complex, which have been reviewed extensively (6, 21–23), and those in the TLR4, TLR5, and TLR8 systems, which were reported more recently (16, 18–20). The latter systems are taken as representative examples here to illustrate structures and mechanisms.
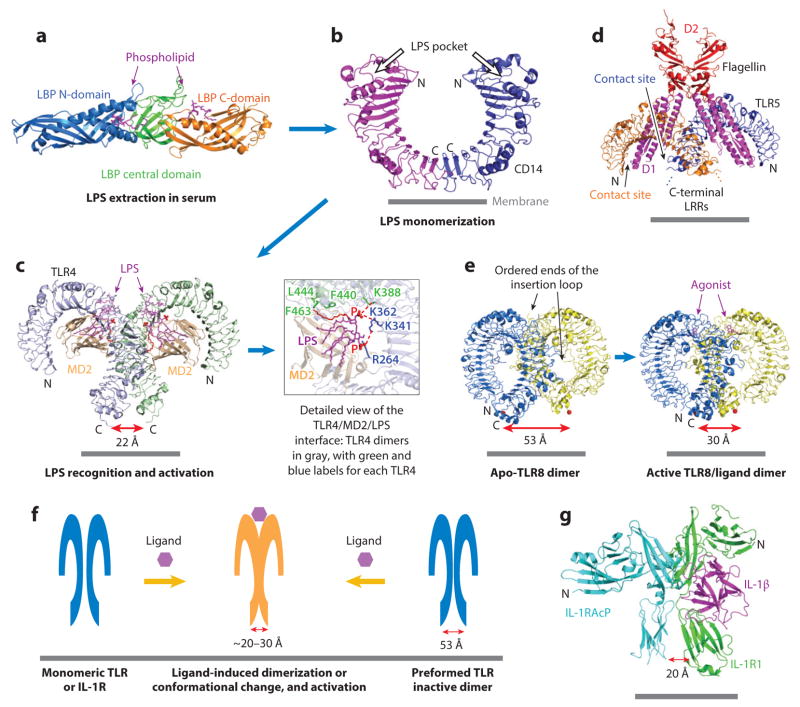
LPS is a major bacterial cell wall glycolipid consisting of a carbohydrate region and a lipid A region that is both hydrophobic and negatively charged (24). The sentinel recognition of LPS by TLR4 involves several steps (25, 26). First, LPS is extracted from bacterial membranes by the serum LPS-binding protein (LBP). Second, LPS is transferred from LBP to CD14, a glycosylphosphatidylinositol-linked protein on the cell surface or a secreted or shed soluble protein in the serum (26). Third, CD14 splits LPS aggregates into monomers and presents them to the TLR4/MD2 complex. The structure of LBP was determined recently (27), and crystal structures of all the proteins involved in these steps are available. LBP has an elongated structural architecture with topologically similar N-terminal and C-terminal barrel-shaped domains and a connecting central domain (Figure 2a). Both the N domain and C domain possess a hydrophobic pocket with bound lipid molecules. CD14 is a horseshoe-shaped LRR structure; it forms a dimer through its C-terminal LRR module (28) (Figure 2b). The LPS-binding pocket is located at each N-terminal end of the LRR horseshoe.
Figure 2.
The TLR/IL-1R superfamily: ligand recognition and receptor activation. (a) Cartoon representation of LBP. LBP is an elongated structure with three domains. The N-terminal and C-terminal domains of LBP bind phospholipids (purple). (b) CD14 forms a homodimer. (c) Left: cartoon representation of the dimeric TLR4/MD2/LPS ternary complex (TLR4: light blue and pale green; MD2: light orange; LPS: five acyl chains in magenta, the sixth acyl chain mediating TLR4 dimerization in red, and the two phosphate groups shown as red spheres). Right: zoomed-in view of the detailed interactions at one dimerization interface. (d) Overall structure of the dimeric TLR5/flagellin complex (flagellin: D1 in magenta and D2 in red; TLR5: orange and blue). D1 interacts with both TLR5 molecules to mediate dimerization. (e) Ribbon diagrams of preformed apo-TLR8 dimer and ligand-induced active TLR8 dimer structures. The measured distances between C termini (marked by red spheres) for inactive and active TLR8 dimers are indicated. (f) Model of TLR/IL-1R activation mechanism. IL-1Rs and some TLRs are monomeric and form dimers upon ligand binding. Other TLRs are preformed inactive dimers. In either case, ligand binding triggers the formation of a dimer in which the C termini preceding the transmembrane domain are brought into proximity to activate the receptor. (g) Cartoon representation of the IL-1β/IL-1RI/IL-1RAcP ternary complex structure. The measured distance between the IL-1RI C terminus and the IL-1RAcP C terminus is indicated. Abbreviations: IL-1RAcP, IL-1R accessory protein; LBP, LPS-binding protein; LPS, lipopolysaccharide; LRR, leucine-rich repeat; TLR, Toll-like receptor.
Unlike the LRR domain of CD14, the TLR4 LRR domain does not have an LPS-binding pocket. Instead, TLR4 constitutively associates with a soluble protein, MD2, that directly interacts with LPS. In the crystal structures of the monomeric TLR4/MD2/antagonist and dimeric TLR4/MD2/LPS complexes (16, 18), MD2 uses an edge of its β-sandwich fold to contact a concave surface at the N-terminal and central regions of the TLR4 LRR horseshoe (Figure 2c). MD2 harbors a large internal pocket between its opposing β-sheets that accepts the acyl chains of the LPS lipid A. Previous studies show that the total number of acyl chains is one of the most important factors governing the inflammatory activity of LPS (29, 30). The dimeric TLR4/MD2/LPS complex structure reveals that six acyl chains are optimal, because although five acyl chains are completely buried in the MD2 pocket, the sixth acyl chain is partially exposed to allow interaction with a hydrophobic patch on the surface of a neighboring TLR4 molecule for dimerization (Figure 2c). MD2 also undergoes localized structural changes to participate in hydrophilic interactions with the same region of TLR4. The two phosphate groups of lipid A form ionic interactions with a cluster of positively charged residues in MD2 and TLR4 to further stabilize the dimerization interface (18).
FliC is a major component of bacteria flagella, which are responsible for bacterial motility (31). It is a ligand for TLR5, the only protein-responsive TLR conserved in vertebrates from fish to humans (32). FliC is able to assemble into flagellar filaments, but TLR5 only responds to monomeric FliC. The crystal structure of TLR5 in complex with a truncated fragment of Salmonella FliC shows that the FliC D1 domain makes a major contribution to both binding and dimerization of TLR5 (19) (Figure 2d). One side of the long helices of D1 forms an extensive binding interface with the TLR5 surface near the N-terminal part of the LRR domain, while an adjacent side dimerizes with the neighboring TLR5 at more C-terminal LRRs. The D1 surfaces used for TLR5 interactions coincide with residues involved in conserved FliC oligomerization in the flagellar filament, demonstrating that TLR5 targets a common molecular pattern in FliC.
TLR7, TLR8, and TLR9 form a TLR subfamily featuring a relatively large ectodomain, localization to the endosomes, and sensing of pathogenic or self nucleic acids (33, 34). TLR8 is believed to respond to uridine- and guanosine-rich ssRNA (35, 36). As with other endocytic TLRs (37, 38), the crystal structure of TLR8 reveals an internal cleavage at an insertion loop in the ectodomain critical for endocytic TLR activation (20) (Figure 2e). Without the cleavage, the insertion loop would have occupied the dimerization interface. The long TLR8 LRR domain almost makes a full circle of the horseshoe, so that the N and the C termini are very close to each other. The ectodomain of TLR8, in contrast to most TLRs, is a constitutive dimer formed through extensive contacts between the sides of the LRR horseshoe. Crystal structures of TLR8 in complex with three agonistic chemical ligands show considerable reorganization of the TLR8 dimer because the ligands act to glue specific residues from both TLR8 monomers to stabilize a different dimer interface (20) (Figure 2e).
A significant consequence of the reorganization is that the C termini in the agonist-bound TLR8 dimer are much closer to each other than those in the apo-TLR8 dimer, measured at ~30 Å in contrast to ~53 Å (Figure 2e). Because the C termini continue into the membrane to the cytosolic TIR domains, the closer proximity may facilitate oligomerization of the TIRs and subsequent adaptor recruitment for assembly of the signaling complex. Formation of an M-shaped double horseshoe may be a general feature of ligand-induced TLR activation in the ectodomain. In the TLR4/MD2/LPS complex structure, the C termini of the TLR4 dimer are also brought close, measured at a distance of ~22 Å (Figure 2c). Given that a TIR domain is ~30 Å in size (39), the juxtaposition of the transmembrane helices by the extracellular ligand interaction may position the intracellular domains optimally for interaction and signaling (Figure 2f).
Structures of Ectodomains of IL-1Rs in Complex with Ligands
Crystal structures of the IL-1β/IL-1RII/IL-1RAcP and IL-1β/IL-1RI/IL-1RAcP ternary complexes were published successively (40, 41). The overall architectures of the two ternary complexes are quite similar (Figure 2g). The ectodomains of IL-1RI, IL-1RII, and IL-1RAcP each comprise three immunoglobulin domains, each folded into a question mark–like structure. IL-1β is held in the center of the concave surface of IL-1RI or IL-1RII. IL-1RAcP associates with the IL-1β/IL-1RI and the IL-1β/IL-1RII binary complexes at the middle and C-terminal immunoglobulin domains. In the ternary complex, the juxtamembrane C termini of the two IL-1RI molecules are close to each other, measuring ~20 Å apart (41) (Figure 2g). Consequently, the intracellular TIR domains may be brought into proximity to facilitate oligomerization and recruitment of downstream signaling components, in a similar manner as TLR4 and TLR8 ectodomains (Figure 2f). Structural studies on IL-18 and IL-33 in complex with their receptors revealed similar structural architectures but with distinct determinants of specificity (42, 43).
Structural Implications on TIR-TIR Interactions
Despite extensive knowledge of the structural aspects of extracellular interactions, the mechanism by which TIR domains mediate intracellular signaling of TLR/IL-1R superfamily receptors remains much more enigmatic. Structures of many TIR domains from both receptors and adaptors are available (39, 44–48) and are generally similar to each other in that all of them include a central five-stranded β-sheet surrounded by five α-helices (Figure 3a). However, it has been extremely difficult to obtain stable TIR/TIR complexes in solution for structural studies.
Figure 3.
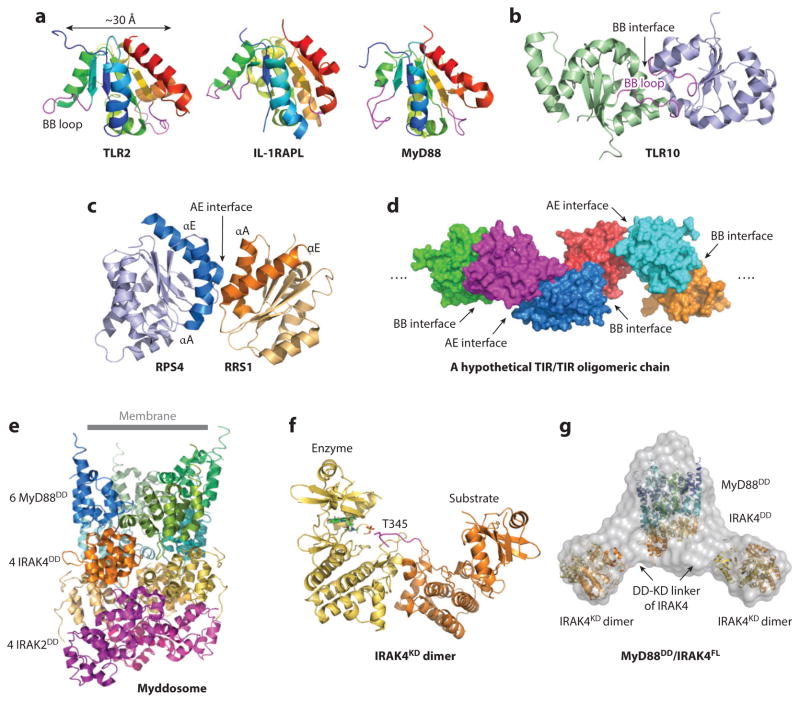
The TLR/IL-1R superfamily: intracellular signaling. (a) Cartoon representations of TIR domain structures. TIR domains of TLR2, IL-1RAPL, and MyD88 are aligned to the same orientation. Location of the BB loop is indicated. (b) Arrangement of TLR10 TIR homodimer structure using the BB interface. (c) Structure of the RPS4/RRS1 TIR domain heterodimer formed using the AE interface. The location of the dimerization interface is indicated. (d) Model of a hypothetical TIR/TIR oligomerization chain, based on iterative BB and AE interfaces. (e) Representation of the Myddosome DD complex structure. (f) Structure of the inactive IRAK4 KD dimer. (g) The SAXS/WAXS envelope of the MyD88DD/IRAK4FL complex fitted with one binary Myddosome DD complex and two IRAK4 KD dimers. Abbreviations: DD, death domain; FL, full length; IL-1RAPL, interleukin-1 receptor accessory protein-like; IRAK, IL-1R-associated kinase; KD, kinase domain; RPS, resistance to Pseudomonas syringae; RRS, resistance to Ralstonia solanacearum; TIR, Toll-IL-1R; TLR, Toll-like receptor.
Multiple clues appear to suggest the involvement of two interaction surfaces between TIR domains. First, the TLR10 TIR domain was crystallized as a symmetric homodimer with an extensive dimer interface, containing the BB loop (the loop between the βB-strand and the αB-helix), DD loop (the loop between the βD-strand and the αD-helix), and αC-helix (44), here denoted the BB interface after the BB loop (Figure 3b). Structure-based mutagenesis indicates that these interacting elements are likely part of the TIR-TIR interface for TLR activation (44). The BB loop region has also been identified as an important region for signaling (46). In addition, recent TRIF and TRAM TIR domain structures, mutagenesis, and yeast two-hybrid experiments further support that BB loops are essential for TIR-TIR homotypic interactions and downstream signal transduction (48). Second, the recently published structure of a heterodimeric TIR/TIR domain complex from the plant resistance proteins RPS4 and RRS1 includes a pseudosymmetric heterodimeric interface involving residues in the αA- and αE-helices, and EE loops of both TIR domains, here denoted the AE interface (49) (Figure 3c). Because the BB interface and the AE interface are mutually inclusive, we speculate that higher-order TIR/TIR complexes for TLR/IL-1R signaling may assemble under the cytoplasmic membrane, a two-dimensional surface that may greatly enhance protein-protein interactions (50). Building TIR-TIR interactions using iterative BB and AE interfaces leads to a potential TIR domain chain (Figure 3d), which may act as a platform for formation of downstream signaling complexes.
DD Interactions in the Myddosome Through Helical Symmetry
The oligomeric DD-complex structure of a Myddosome containing six MyD88 DDs, four IRAK4 DDs, and four IRAK2 DDs reveals the hierarchical assembly mechanism in TLR/IL-1R signal transduction (51). The DDs are arranged into a left-handed helical tower, with six MyD88 DDs forming the approximate top two helical turns, four IRAK4 DDs in the middle turn, and four IRAK2 DDs at the bottom turn (Figure 3e). Shape and charge complementarity between the top and bottom surfaces of each turn confers a sequential assembly order in the Myddosome. Upon recruitment to a TLR or an IL-1R, MyD88 may form an oligomeric platform through TIR-TIR interactions, which in turn promotes the assembly of the oligomeric Myddosome via DD-DD interactions. The structural evidence for oligomerization of the intracellular signaling complex, rather than dimerization, also suggests that dimeric TLR/ligand and IL-1R/ligand complexes may also further cluster on the cell surface to facilitate signaling.
DDs belong to the DD superfamily, which also includes caspase recruitment domain (CARD), pyrin domain (PYD), and death effector domain (9). Identification of the helical assembly mechanism in the Myddosome, as well as in the previous oligomeric DD complex structures (52, 53), marks a beginning for the realization that many more DD superfamily proteins form helical structures, including filaments to induce signaling. Indeed, as shown in the following sections, the CARDs and PYDs involved in RLR signaling and inflammasome formation assemble into helical oligomers or helical filaments. These structural insights provide the molecular basis for the involvement of higher-order signaling complexes in innate immunity.
The Allosteric Mechanism of IRAK4 Kinase Activation
A crucial activation event in the Myddosome is autophosphorylation of the IRAK4 kinase, which in turn leads to phosphorylation of other IRAKs to propagate the signal transduction process. Oligomerization-induced autophosphorylation of IRAK4 must execute through allosteric changes that are only possible at a high local concentration. The recent crystal structure of the IRAK4 kinase domain (KD) containing an active site mutation showed an asymmetric dimer structure caught in the act of trans-autophosphorylation (54). Despite being unphosphorylated, one IRAK4KD molecule (the “enzyme”) is in the active conformation, whereas the other IRAK4KD molecule (“substrate”) shows a protruded activation loop. The enzyme IRAK4 active site precisely accepts the cognate phosphorylation residue T345 from the substrate for trans-autophosphorylation (Figure 3f). Further biochemical and cellular studies show that the observed dimerization interface is vital for IRAK4 autophosphorylation and signal transduction (54). Small- and wide-angle X-ray scattering (SAXS/WAXS) of full-length (FL) IRAK4 in complex with MyD88DD reveals a central core with two flanking lobes, suggesting a MyD88/IRAK4 DD complex core at the center and two IRAK4KD dimers at the periphery (Figure 3g). IRAK4 exhibits a modest dimerization affinity in solution, and MyD88 enhances IRAK4KD dimerization and autophosphorylation by increasing its local concentration.
THE RIG-I-LIKE RECEPTOR SIGNALING PATHWAY
Background
RLRs, which include RIG-I, MDA5 (melanoma differentiation-associated protein 5), and LGP2, detect viral RNA molecules in the cytoplasm to trigger innate immunity and inflammation (55). In cells, RIG-I is essential for antiviral immunity against RNA viruses including paramyxoviruses, influenza virus, and Japanese encephalitis virus, whereas MDA5 is critical for immunity against picornaviruses (56). In vitro, RIG-I can recognize ssRNA with 5′-triphosphate (5′-ppp), a typical feature of replicated viral RNA, as well as blunt-ended dsRNA with or without a 5′-ppp (56, 57). In contrast, MDA5 does not distinguish 5′-ppp in RNA but can recognize long dsRNA such as synthetic poly(I:C) (56). Upon RNA sensing, activated RIG-I and MDA5 engage the innate immune response by activating the downstream mitochondrial adaptor protein MAVS (mitochondrial antiviral-signaling protein; also known as IPS-1/VISA/Cardiff) (58–61). MAVS recruits TRAF proteins, which in turn activates IKK and TANK-binding kinase 1 (TBK1) to result in NF-κB activation and IFN responses.
Crystal Structure of Full-Length RIG-I in an Autoinhibited State
RLRs contain an N-terminal tandem CARD (2CARD), a central DECH-box RNA helicase domain, and a C-terminal domain (CTD; also referred to as the repressor domain, RD). The helicase domain is closely related to DEAD-box helicases and comprises N- and C-terminal RecA-like domains Hel1 and Hel2. Crystal structures of RIG-I show that an insertion domain, denoted Hel2i, forms within the primary sequence of the Hel2 domain and that a V-shaped helical structure, denoted the pincer domain, bridges Hel2 back to Hel1 (62–66) (Figure 4a). The apo-full-length RIG-I structure provides a glimpse of an autoinhibited state in which the ATPase core is partially ordered while the CTD is invisible (62). The CARD1 and CARD2 in 2CARD are rigidly associated with each other. Importantly, the Hel2i domain sequesters the 2CARD by direct, extensive interaction with CARD2 (Figure 4a), preventing self-association of 2CARD and, thus, activation of the downstream pathway in the absence of RNA binding (62). Conversely, the 2CARD/Hel2i interaction impedes Hel2i interaction with RNA, creating a competition between ligand and 2CARD for Hel2i that contributes to ligand-mediated regulation of RIG-I. Thus, full-length RIG-I exhibits greater RNA-dependent ATPase activity in the presence of the tighter binder 5′-ppp-dsRNA, whereas RIG-I lacking the 2CARD is indiscriminately active with either 5′-ppp-or 5′-OH-dsRNA (67).
Figure 4.
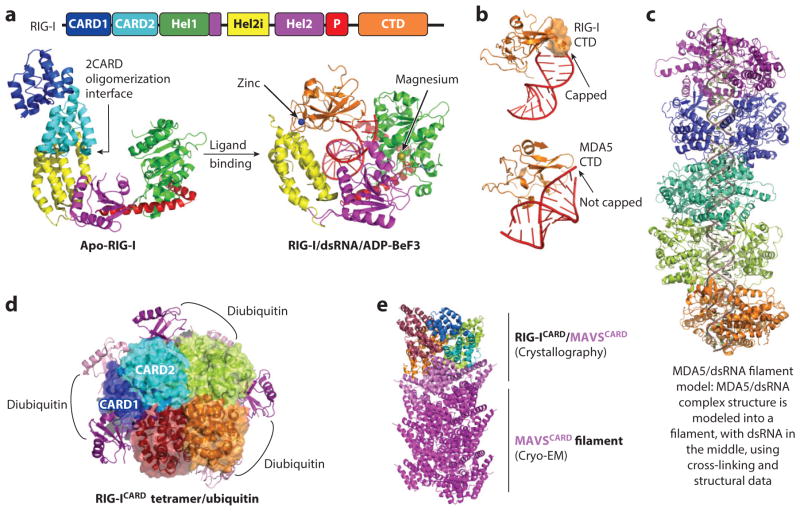
The RLR family: ligand recognition and receptor activation. (a) Structures of RIG-I in the ligand-free state (left) and the RNA-bound and transition state ATP analog–bound state (right). Zinc and magnesium ions are shown as blue and yellow spheres, respectively. (b) The flexible CTD loop (residues 846–859, orange surface) of RIG-I interacts with and caps the 5′-ppp dsRNA end, whereas the equivalent loop in MDA5 is disordered. (c) A model of the MDA5/dsRNA filament based on the crystal structure of a ΔCARD MDA5/dsRNA complex. (d) Structure of the RIG-I 2CARD/diubiquitin complex diubiquitin: magenta and pink; 2CARDs: dark blue and cyan for CARD1 and CARD2 of the first 2CARD molecule and lime green, orange, and dark red for the other 2CARD molecules). (e) A model depicting the nucleation of MAVSCARD by RIG-ICARD based on both crystallographic and cryo-EM data. Abbreviations: CARD, caspase recruitment domain; CTD, C-terminal domain; EM, electron microscopy; MAVS, mitochondrial antiviral-signaling protein; MDA5, Melanoma differentiation-associated protein 5; P, V-type pincer domain P; RLR, RIG-I-like receptor.
Crystal Structures of RIG-I-Like Receptor/RNA Complexes
Crystal structures of multiple RIG-I/dsRNA complexes reveal significant conformational changes upon dsRNA binding (62–65, 68). In RIG-I/dsRNA complexes in the absence of nucleotide or in the presence of ADP, Hel1, Hel2, Hel2i, and CTD encircle the A-form dsRNA to contact both strands of the duplex, with ordering of CTD and compaction of Hel1, Hel2, and Hel2i closer to one another relative to their positions in the apo-RIG-I. There are abundant contacts between RIG-I and 2′-hydroxyl groups in the RNA ribose, suggesting that the mechanism of specific recognition of dsRNA, but not dsDNA, relies on both shape sensing of the A-form duplex and chemical sensing of the RNA backbone. Grafting the observed Hel2i-2CARD interaction in apo-RIG-I to these structures shows that 2CARD begins to clash with the ordered CTD, but the structural overlap between 2CARD and CTD is still quite minimal, suggesting that dsRNA binding is not sufficient to overcome 2CARD autoinhibition.
Much more dramatic conformational changes are observed upon cooperative dsRNA and ATP binding, as shown in RIG-I/dsRNA complexes in the presence of a transition-state ATP analog such as ADP-BeF3 or ADP-AlF3 (Figure 4a). All domains move relative to each other, with close apposition of the two RecA domains Hel1 and Hel2 to form the ATP-binding site, as well as ordering of many previously disordered loops. These movements result in further closure of the domains around the dsRNA. Grafting the observed Hel2i-2CARD interaction in apo-RIG-I to the RIG-I/dsRNA/ADP-BeF3 structure reveals that the 2CARD, if in the Hel2i-bound position, would have been completely clashed with the CTD and dsRNA, suggesting that cooperative dsRNA and ATP binding disengages 2CARD from the rest of the RIG-I domains to allow signal transduction.
The crystal structure of MDA5 in complex with dsRNA and an ATP analog is a closed conformation, similar to the RIG-I/dsRNA complexes in the presence of transition-state ATP analogs (69). However, despite having the same zinc-containing β-sheet fold, the CTD in MDA5 is quite different from that in RIG-I in relative orientation to the helicase domain and in structure (Figure 4b). In RIG-I, the CTD caps one end of the dsRNA in the same manner the isolated CTD of RIG-I interacts with 5′-ppp-RNA duplex using a lysine-rich basic cleft, providing a structural explanation for the preference of RIG-I for 5′-ppp RNA substrates (68, 70, 71). In MDA5, the CTD moves away from the dsRNA end, allowing binding to the internal duplex stem rather than the end. In addition, the major capping loop in RIG-I is disordered in the corresponding region of MDA5; if ordered as in RIG-I, the loop would have occupied a similar position as the bound dsRNA. In an NMR structure of isolated MDA5 CTD, this loop adopts a conformation similar to that of RIG-I (72), suggesting that its displacement is required for dsRNA binding (69). An independent structure of the MDA5 helicase domain in complex with the paramyxovirus V protein shows that MDA5 is partially unfolded by a β-hairpin motif of the V protein (73).
RIG-I-Like Receptor Filament Formation Along dsRNA
Imaging by electron microscopy (EM) has revealed that MDA5 stacks along dsRNA to form filaments (74, 75), suggesting a mode of recognition for long dsRNA. A model of the MDA5/dsRNA filament structure with head-to-tail arrangement was built based on the crystal structure of the 1:1 MDA5/dsRNA complex and structurally designed cross-linking experiments (69) (Figure 4c). A similar model was reached independently using an ~22-Å resolution EM density from helical reconstruction (76). In the presence of ATP, RIG-I also assembles into filaments on dsRNA (77). It appears that ATP hydrolysis allows RIG-I to translocate from the initial binding site on dsRNA ends to dsRNA stems to promote wrapping along dsRNA (77). The observed conformation changes between the open and the closed states of RIG-I upon dsRNA and ATP binding may power the motion for the translocation (78).
The RLR family member LGP2 lacks the N-terminal 2CARD and was proposed to act as an inhibitor of RIG-I and MDA5 signaling. However, recent studies show that LGP2 assists, rather than inhibits, MDA5/dsRNA interactions and enhances MDA5-mediated antiviral signaling (79). In particular, LGP2 possesses a much faster association rate with dsRNA than MDA5 does and acts as seeds to increase the rate of formation of MDA5/dsRNA filaments. MDA5 and RIG-I filament formation on dsRNA brings its 2CARD into proximity to nucleate the MAVS CARD filament to mediate downstream signaling without assistance from polyubiquitination (74, 75, 77).
Structural Basis for Ubiquitin Recognition by RIG-I and MAVS Filament Formation
RIG-I 2CARD has been shown to directly interact with K63-linked polyubiquitin chains, an interaction shown to play an important role in RIG-I activation (80, 81). One possibility is that polyubiquitin interaction promotes RIG-I 2CARD oligomerization upon its release from the autoinhibited state by RNA and ATP binding. The recently published crystal structure of a RIG-I 2CARD/diubiquitin complex reveals that 2CARD assembles into a helical tetramer (82) (Figure 4d) that is similar in architecture to DD oligomers (9). Diubiquitin is bound along the outer rim of the 2CARD helical tetramer, connecting neighboring subunits and stabilizing tetramerization (82) and providing an elegant mechanism for polyubiquitin-enhanced RIG-I oligomerization and signaling.
RIG-I downstream signaling requires CARD-CARD interactions between RIG-I 2CARD and MAVS CARD, and the subsequent filament formation of MAVS CARD. Cryo-EM structure at 3.6-Å resolution shows that the MAVS CARD has a helical assembly that exhibits compatible symmetry and interfaces with the diubiquitin-stabilized RIG-I 2CARD tetramer (83) (Figure 4e). Mutational engineering was used to limit MAVS filament formation that enabled the formation and crystallization of a RIG-I 2CARD/MAVS CARD tetrameric complex. In this structure, MAVS CARDs stack against the CARD2 of the RIG-I 2CARD, and RIG-I 2CARD tetramer is arranged in the same way as in the RIG-I 2CARD/diubiquitin complex. Thus, the complex captures RIG-I 2CARD in the act of nucleating a MAVS CARD filament (83) (Figure 4e).
THE AIM2-LIKE RECEPTOR AND NOD-LIKE RECEPTOR INFLAMMASOMES
Background
ALR and NLR family members are best known for their ability to form inflammasomes, supramolecular complexes for activation of inflammatory caspases, such as caspase-1 (84, 85). AIM2 and IFI16 are ALRs with an N-terminal effector PYD and one or two C-terminal hemopoietic expression, interferon-inducibility, nuclear localization (HIN) domains for sensing cytosolic microbial dsDNAs (86–89). Upon activation, the PYDs of AIM2 and IFI16 engage apoptosis-associated speck-like protein containing a CARD (ASC) via PYD-PYD interactions. The CARD of ASC also recruits and activates caspase-1 through CARD-CARD interactions. Activated caspase-1 converts cytokines IL-1β and IL-18 to their mature forms by proteolysis, and/or to induce caspase-1-dependent pyroptotic cell death. The mouse p202 protein, which contains two HIN domains without PYD and directly associates with microbial dsDNA, inhibits AIM2 inflammasome formation (86).
NLR family members share the domain architecture of a central NOD and a C-terminal LRR domain. Depending on the N-terminal effector domain, the NLR family can be further divided into a number of subfamilies, such as NLRPs, those with N-terminal PYDs; NLRCs, those with N-terminal CARDs; and NLRBs (or NAIPs), those with N-terminal baculoviral IAP-repeat domains (90). Like PYD-containing ALRs, NLRPs can potentially use ASC to bridge caspase-1 recruitment and activation. NLRCs may directly engage caspase-1 via CARD-CARD interactions. For most NLRs—including the extensively studied NLRP3, which responds to a wide spectrum of stimuli, such as extracellular ATP, bacterial toxins, and uric acid crystals—the direct activating ligands are unknown. One exception is the NAIP/NLRC4 system, in which NAIPs directly bind FliC and other bacterial proteins, and NLRC4 acts as the adaptor to connect to caspase-1 activation (91, 92). ALR and NLR inflammasomes are tightly regulated, and unchecked activation often leads to autoinflammatory diseases, including hereditary fever syndromes, gout, psoriasis, lupus, and inflammatory bowel diseases such as ulcerative colitis and Crohn’s disease (84, 85, 93).
Crystal Structures of AIM2-Like Receptor/dsDNA Complexes and dsDNA-Mediated Filament Formation
The DNA-sensing HIN domains contain ~200 amino acids and are predicted to consist of tandem oligonucleotide/oligosaccharide-binding (OB) folds. Structural studies of AIM2, IFI16, and p202 show that the two OB folds are rigidly linked via a connecting α-helix and almost orthogonal to each other (94–97) (Figure 5a). AIM2 and IFI16 HIN domains interact with classical B-form dsDNA in a similar manner, using mostly electrostatic contacts between the highly charged surface of HIN domains and backbone phosphates of dsDNA (94, 96). Few base interactions are involved, consistent with sequence-independent recognition.
Figure 5.
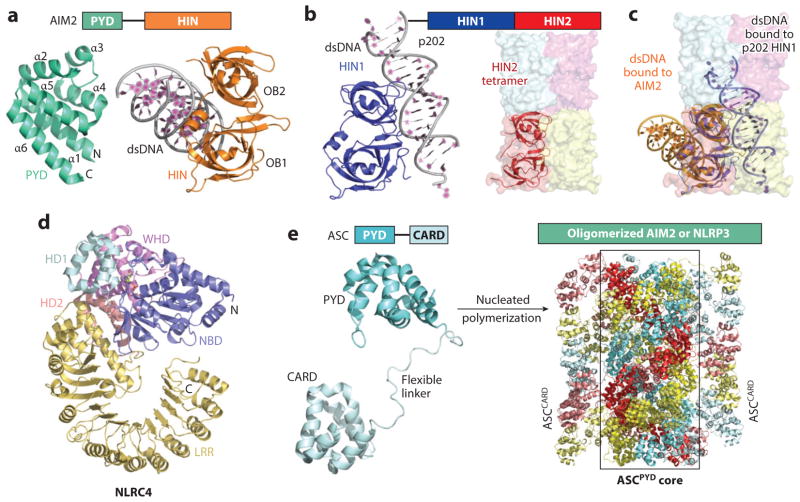
The AIM2-like receptor and NLR families: autoinhibition, ligand binding, and filament assembly. (a) Structures of AIM2 PYD and the HIN/dsDNA complex. (b) A ribbon diagram of p202 HIN1 in complex with DNA (left) and a surface diagram of HIN2 tetramer (right). One HIN2 molecule is also shown in a ribbon representation. (c) Superposition of AIM2 HIN, p202 HIN1, and p202 HIN2. AIM2 HIN and p202 HIN1 use opposite surfaces for dsDNA binding. The tetramerization interface of p202 HIN2 overlaps with the dsDNA binding site for p202 HIN1. (d) Structure of ΔCARD NLRC4. (e) Left: NMR structure of full-length ASC. Right: Cryo-EM structure of the ASC PYD filament in the center, with ASC CARD at the periphery modeled based on the structure of full-length ASC. The ASC filament formed upon nucleation by oligomerized AIM2 or NLRP3 is a three-start helical assembly, shown in red, cyan, and yellow for the PYDs in each of the helical strands and in faded red, faded cyan, and faded yellow for the corresponding CARDs. Abbreviations: AIM2, absent in melanoma 2; ASC, apoptosis-associated speck-like protein containing a CARD; CARD, caspase recruitment domain; HIN, hemopoietic expression, interferon-inducibility, nuclear localization; LRR, leucine-rich repeat; NBD, nucleotide-binding domain; NLR, NOD-like receptor; NLRC, NLR with N-terminal CARD; NLRP, NLR with N-terminal PYD; OB, oligonucleotide/oligosaccharide-binding; PYD, pyrin domain; WHD, winged-helix domain.
Unexpectedly, p202 HIN1 binds to dsDNA using an almost opposite surface (95, 96) (Figure 5b,c) that is, however, equivalent to the surface used by other OB-fold proteins in recognition of ssDNA (98). The p202 HIN2 domain completely loses the DNA-binding feature and instead assembles into a tetramer, which is a tail-to-tail dimer (OB2 to OB2) of an intimate lateral dimer (OB1-OB2 to OB1-OB2) (95) (Figure 5b,c). Therefore, full-length p202 exists as a tetramer with a HIN2 core and four flexibly appended HIN1 domains (95). The p202 HIN2 interacts specifically with the AIM2 HIN, which contributes to p202’s interference of AIM2, but not IFI16, inflammasome formation (95).
Full-length IFI16 is shown to cooperatively assemble onto dsDNA in a length-dependent manner to form filaments (99). Although the HIN domains engage dsDNA, these domains alone interact with dsDNA weakly without filament formation. It is the non-DNA-binding N-terminal PYD that enhances DNA binding affinity and promotes the cooperative filament assembly (99). This suggests that PYDs from multiple IFI16 molecules bound to the same dsDNA further cluster together to provide the cooperativity for filament formation. For AIM2, the PYD structure is a six-helix bundle that is characteristic of DD superfamily domains (100, 101) (Figure 5a), and the PYD clustering equals assembly into a different type of filament using the three conserved asymmetric interactions observed in the DD superfamily (101). We propose that dsDNA filaments and PYD filaments are formed concurrently, leading to a central dsDNA filament decorated by short PYD filaments along its length. These AIM2PYD or IFI16PYD filaments provide the platforms for ASC recruitment and inflammasome assembly.
Crystal Structure of NLRC4 in an Autoinhibited Conformation
NLRC4 is an adaptor in NAIP/NLRC4/caspase-1 inflammasomes. The crystal structure of ΔCARD of mouse NLRC4 reveals a monomeric, autoinhibited conformation in which the NOD is not capable of oligomerization (102). Structurally, the central NOD comprises the nucleotide-binding domain (NBD), the helical domain HD1, and the winged-helix domain (WHD). NLRC4 also contains a second helical domain (HD2) that connects NBD to the C-terminal LRRs (Figure 5d). The C-terminal LRR domain prevents NLRC4 from oligomerizing because its footprint on the NBD overlaps with the oligomerization interface. An ADP molecule copurified with NLRC4 from insect cells occupies the nucleotide-binding pocket and locks NLRC4 in the closed inactive conformation by holding the WHD. Removal of the LRR domain causes spontaneous oligomerization and activation of NLRC4, bypassing the requirement of NAIPs and bacterial ligands. We propose that NLRPs also exert autoinhibition using a similar mechanism as NLRC4. EM studies provide a first glimpse of oligomerized NLRC4, NLRP3, and the FliC-induced NAIP5/NLRC4 complex, in which NLRC4 and NLRP3 alone form rodlike, layered structures that may be helical stacks of AAA+ ATPase-like lock washers (103, 104) and the NAIP5/NLRC4 complex shows double disks of 11- or 12-fold symmetry (103).
Cryo-EM Structure of the ASCPYD Filament
Recent studies using fluorescence polarization to monitor filament formation show that assembly of ALR and NLRP inflammasomes proceeds through two successive steps of nucleation-polymerization (104). First, a substoichiometric amount of activated AIM2 or NLRP3 robustly nucleates ASC filaments through PYD-PYD interactions. Second, a substoichiometric amount of ASC that has been polymerized by the PYD filaments forms ASCCARD platforms to nucleate caspase-1 filaments through CARD-CARD interactions. The reconstituted ternary AIM2/ASC/caspase-1 inflammasome shows star-shaped structures in which multiple filaments protrude radially from a single central hub (104). In vitro, these star-shaped structures can further aggregate over time to form filamentous spheres from hundreds of nanometers to microns in size, similar to the single ASC punctum formed in cells upon AIM2 and NLRP3 inflammasome activation (88, 105).
The self-aggregation tendencies of ASC made it difficult to study ASC assembly mechanisms structurally. The monomeric structures of ASCFL and ASCPYD have been solved using NMR at highly acidic conditions that abolished the self-association (106, 107) (Figure 5e). Recent cryo-EM of ASCPYD filaments, at 3.8-Å resolution, has revealed that ASCPYD subunits pack densely in a spiral to create a cylinder-like structure. Superimposing the ASCFL structure onto the ASCPYD subunit in the filament creates a double-ring structure in which the inner ring is the core ASCPYD filament and the outer ring represents flexibly attached ASCCARD domains (Figure 5e). The latter can further oligomerize to nucleate caspase-1 filaments.
The ASCPYD filament structure reveals in molecular detail the three types of asymmetric interfaces in PYD-PYD interactions that are similar to the conserved interfaces in DD interactions (9). Structure-based mutagenesis confirms the importance of these ASC interfacial residues in filament formation in vitro and in ASC-induced IL-1β processing. These data also confirm previous extensive mutagenesis studies on the requirement for ASCPYD filament formation in cells (108). Presuming that the nucleators of ASCPYD, such as AIM2PYD and NLRP3PYD, adopt the same helical assembly, AIM2 and NLRP3 residues important for ASC interaction have been identified. Mutating these residues compromises the ability of AIM2 and NLRP3 to nucleate ASCPYD filaments, confirming the validity of the predicted interactions.
THE cGAS/STING SIGNALING PATHWAY
Background
Nucleic acids are a major class of PAMPs. Presence of microbial-derived dsDNA elicits potent immune and inflammatory responses. Nuclear or mitochondrial DNA wrongly placed in the cytosol induces similar responses, often underlying the mechanisms of autoimmune diseases. Stimulator of interferon genes (STING) (also named MITA, MPYS, and TMEM173) is an endoplasmic reticulum–localized transmembrane protein found to be essential in cytosolic DNA–induced innate immunity (109–113). It responds to transfected, bacterial, or viral genomic DNA, leading to activation of transcription factors in the IFN regulatory factor (IRF) and NF-κB families via TBK1 and IKK kinases, respectively.
Unique conserved nucleic acids known as cyclic dinucleotides (CDNs) [such as cyclic di-GMP (c-di-GMP), important for signaling in bacteria] have been found to act directly on STING to elicit the IFN response (114). The search for dsDNA sensors upstream of STING has recently led to the identification of a dsDNA-activated mammalian cyclase known as cyclic-GMP-AMP (cGAMP) synthase (cGAS), which converts GTP and ATP into cGAMP (115, 116). Further studies soon discovered that the major product of cGAS is not a canonical CDN (as in c-di-GMP) that contains only the 3′,5′-cyclic phosphodiester bond (3′,5′-cGAMP), but rather a noncanonical CDN, cyclic-[G(2′,5′)pA(3′,5′)p] (2′,5′-cGAMP), that serves as the endogenous ligand of STING (117–119). Collectively, these studies unexpectedly showed that CDNs represent the common element in sensing bacteria and dsDNA by the STING pathway.
Crystal Structures of cGAS and cGAS/dsDNA Complexes
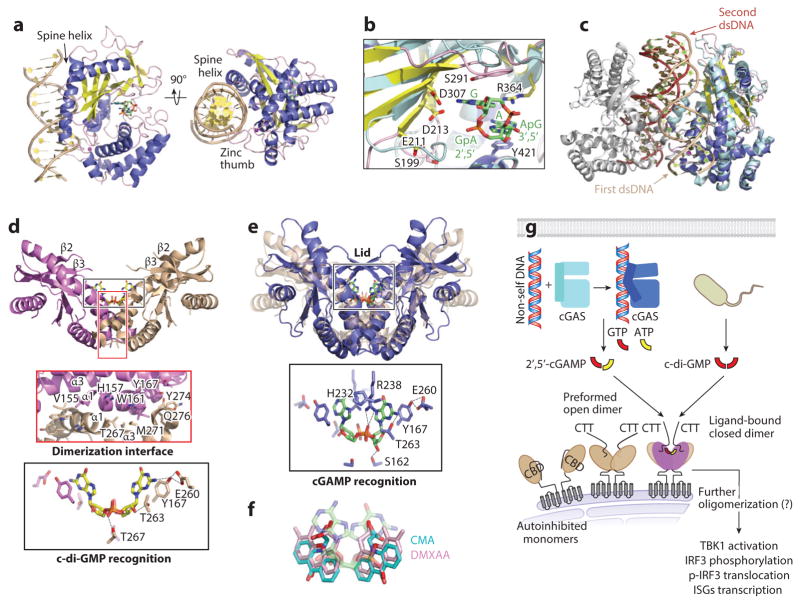
Crystal structures of cGAS and its complexes with dsDNA and/or various nucleotides (117, 120–124) reveal that the catalytic domain of cGAS is a bilobal structure belonging to the nucleotidyl transferase (NTase) superfamily with a mixed α+β fold (Figure 6a). The active site is located in the central twisted β-sheet, and dsDNA binds to the opposite side of the protein. A long helix (named the spine helix) on one end and a highly conserved zinc thumb on the other end firmly grip the dsDNA at the major groove (Figure 6a). cGAS mainly contacts the sugar-phosphate backbone of dsDNA, consistent with its sequence-independent recognition.
Figure 6.
The cGAS/STING pathway. (a) Structure of mouse cGAS with bound dsDNA and 2′,5′-cGAMP in two orthogonal orientations. cGAS is colored according to secondary structures (α-helices in blue-gray, β-strands in yellow, and loops in light pink; dsDNA: tan; zinc ion: purple sphere; 2′,5′-cGAMP: stick model with carbon in green, nitrogen in blue, oxygen in red, and phosphorus in orange). The spine helix and the zinc thumb clamp dsDNA from two sides. (b) Conformational changes at cGAS active site upon dsDNA binding. cGAS in complex with dsDNA and cGAMP, colored as in panel a, is superimposed on apo-cGAS in light blue. Selective side chains important for catalysis and cGAMP binding are depicted with sticks. (c) Structure of the 2:2 dimer complex of cGAS and dsDNA. One monomer is colored as in panel a and superimposed with apo-cGAS in pale cyan. The other monomer is gray. Tan and red arrows denote the first and second dsDNA molecules relative to the superimposed cGAS monomer. (d) Structure of human STINGH232 in complex with c-di-GMP (=), detailed STING dimer interface (middle), and detailed interactions with c-di-GMP (bottom), for which main chains are omitted and only one side is labeled (STING: purple and tan). (e) Structure of human STINGH232 (blue) in complex with cGAMP (green) superimposed on STINGH232 in complex with c-di-GMP (transparent tan) (top), and detailed contacts between STING and cGAMP (bottom). (f) Superimposed, mSting-bound conformations of CMA (cyan) and DMXAA (pink), with cGAMP in the background (transparent green). (g) Schematic diagram for the cGAS/STING pathway. cGAS-synthesized 2′,5′-cGAMP and bacteria-derived c-di-GMP can both activate STING. Activation of STING may require dimerization of monomeric STING molecules, closure of preformed STING dimer, and/or CTT displacement from STING CBD. Abbreviations: CBD, cyclic dinucleotide–binding domain; c-di-GMP, cyclic di-GMP; cGAMP, cyclic-GMP-AMP; cGAS, cGAMP synthase; CTT, C-terminal tail; IRF, interferon regulatory factor; ISG, interferon-stimulated gene; STING, stimulator of interferon genes.
Dramatic conformational changes are observed upon triggering by dsDNA interaction. In particular, the single spine helix breaks into two helices and shifts in position, causing lobe closure and rearrangement of the active site to turn on cGAS (Figure 6a–c). In contrast, binding of various nucleotides does not substantially alter the overall structure and the active site geometry. The overall structure and conformational changes are strikingly similar between cGAS and 2′-5′-oligoadenylate synthetase 1 (OAS1), a member of the NTase superfamily and a dsRNA immune sensor that synthesizes 2′,5′-oligoadenylates (2-5As) in the presence of dsRNA (125). 2-5As dimerize and activate RNase L, an effector of the IFN response that cleaves intracellular RNA to suppress translation (126, 127).
Most recent reports of structures of cGAS/dsDNA complexes unveil hitherto unnoticed 2:2 dimers (123, 124) that are also present in all previously reported crystal lattices. In the dimer, a second dsDNA, while clamped by the spine helix and zinc thumb of a second cGAS, interacts with the first cGAS at a location roughly 90° from the first binding site, likely assisting the conformational changes required for activation by pushing a loop connected to the central β-sheet (Figure 6c). Protein-protein contacts in the dimer, involving the zinc thumb region, may further aid a cooperative assembly of the 2:2 dimer. Structure-based mutations on the second site compromise cGAS activity in vitro and IFN signaling in cells (123, 124).
Given that there is only one active site in cGAS, it is intriguing how 2′,5′-cGAMP, which contains two phosphodiester bonds, is formed. Crystal structures of cGAS in complex with dsDNA and different nucleotides captured various stages in the catalytic landscape, resulting in the deduction that 2′,5′-cGAMP is synthesized in two steps (117, 119). First, a reaction occurs between the 2′-OH of GTP and the α-phosphate of ATP, forming the dinucleotide pppG(2′,5′)pA. Second, the intermediate dinucleotide undergoes a flipover, placing the 3′-OH of pA closer to the active site to facilitate the formation of the 3′-5′ phosphodiester bond to complete cyclization. The product 2′,5′-cGAMP possibly undergoes another flipover before being released (117, 123).
Crystal Structures of STING in Complex with CDNs
Previous sequence analyses predicted that STING possesses N-terminal transmembrane helices, a soluble CDN-binding domain (CBD) that senses the environment in the cytoplasm, and a C-terminal tail (CTT). A series of crystal structures of human STING CBD, alone and in complex with c-di-GMP, reveal a V-shaped dimer (128–132) (Figure 6d, top). Each protomer assumes an α+β fold with a five-strand β-sheet sandwiched between two sets of α-helices. The dimer interface is formed mainly by the longest helix, α1 (previously mistaken for a transmembrane helix); and α3 via hydrophobic interactions (Figure 6d, middle). The golden ticket mutation I199N in mouse Sting (mSting; equivalent to I200N in human STING), which causes failure in c-di-GMP-induced IFN response, renders STING unfolded (129). STING specifically recognizes the intrinsically symmetric c-di-GMP using hydrogen bonds to the ribose and a phosphoryl oxygen, and a stacking interaction with the guanine base (Figure 6d, bottom). Of note, the stacking residue Y167 itself is held in position by a hydrogen bond with E260.
Recent studies reveal that the human STING reference sequence contains a histidine residue at position 232 (STINGH232), whereas the STING sequence from a macrophage cell line highly responsive to CDNs contains R232 (STINGR232) (118). Functionally, human STINGR232 and the corresponding mStingR231 are more responsive to c-di-GMP than STINGH232 or mStingA231 (118). In contrast, both STINGH232 and STINGR232 respond robustly to cGAS expression and to 2′,5′-cGAMP (118). It was first proposed that an affinity difference might explain the higher stimulatory activity of 2′,5′-cGAMP compared with c-di-GMP (123, 133). However, a more thorough binding study showed that 2′,5′-cGAMP only binds STINGR232 with greater affinity while it binds STINGH232 with μM affinity similar to the interactions between c-di-GMP and both STING alleles (134).
A structural observation on STING dimer conformation upon binding to CDNs may offer an alternative, potentially plausible mechanism for STING activation. All human STING structures in the absence of CDN binding show an open dimer structure in which the span of the V shape is quite wide and the region between β2 and β3 is disordered (Figure 6d). When in complex with either STINGH232 or STINGR232, cGAMP indiscriminately induced a closed conformation of the STING dimer in which the V shape is narrower and the disordered β2-β3 loop is rearranged into a long β-hairpin to form the lid above the bound cyclic nucleotide (133, 134) (Figure 6e). Although R232 does directly interact with 2′,5′-cGAMP, H232 points away from 2′,5′-cGAMP (Figure 6e), indicating that STING conformation upon 2′,5′-cGAMP binding is not influenced by the residue type at position 232. For c-di-GMP binding, all three STINGH232 structures (128–130) remain in an open conformation. However, only one of the two STINGR232 structures shows V dimer closure when bound to c-di-GMP (132), and mStingR231 structures are all in the closed conformation, regardless of presence or absence of ligands (134, 135), indicating that although the correlation between activation and closed STING conformation is true in many cases, it is not absolute.
Interestingly, mSting is found to be the target of activation for certain flavonoid compounds, including CMA, FAA, and DMXAA, that showed impressive antiviral or antitumor activity in mice but not in humans (136, 137). Crystallographic studies show that two CMA or DMXAA molecules bind to one mSting dimer, mimicking CDN binding and conforming to the twofold symmetry (134, 136). The two planar molecules occupy the same CDN-binding pocket but sit somewhat deeper in the pocket (Figure 6f). A critical residue in mSting that confers binding reactivity to these compounds has been identified, and a mutation to this residue in human STING renders it responsive to DMXAA (134). Collectively, the recent structural studies demonstrate the mode of CDN binding and provide clues to the mechanism of its activation. Although many questions remain, induction of a closed conformation may be a critical factor in STING activation, which likely induces further clustering of STING through its CTT and activation of the IFN pathway (Figure 6g).
SUMMARY AND FUTURE PERSPECTIVES
Structural studies of innate immune signaling pathways have provided fundamental biological insights into their activation and regulation processes, with several emerging themes that especially suit the functions of the immune system. First, the sensor proteins in these pathways are often autoinhibited through intramolecular interactions to avoid accidental firing. Ligand binding then induces signaling-competent states to initiate the pathways. Second, higher-order signaling complexes are formed to execute signal transduction and signal amplification. Formation of filaments on long nucleic acid chains helps to bring the sensor proteins into proximity to facilitate oligomerization. The effector domains, which are often in the DD superfamily, then mediate oligomerization, recruitment, and additional filament formation using helical symmetry. These higher-order structures provide a common molecular scaffold that in turn promotes proximity-induced allosteric activation of signaling enzymes. Third, although not elaborated in this review, these supramolecular complexes may pose difficulty in disassembly and degradation and would therefore require the elicitation of autophagy as a regulatory mechanism (138, 139). Finally, recent studies suggest that DD superfamily filaments may possess prion-like activities, and such signaling complexes released to the extracellular space may act to mediate intercellular signaling (140–143). Altogether, these structural studies offer novel insights for understanding innate immunity on new mechanistic levels (144).
Acknowledgments
We apologize for incomplete coverage due to the space limitations and the vast data in the highly active field of innate immunity. The work was supported by NIH (R01 AI050872, AI089882 and AI045937 to HW and K99 AI108793 to QY) and National Natural Science Foundation of China (Project 31470724 to JL).
Contributor Information
Jixi Li, Email: lijixi@fudan.edu.cn.
Hao Wu, Email: hao.wu@childrens.harvard.edu.
LITERATURE CITED
- 1.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–97. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 2.Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, et al. Genetic and physical mapping of the Lps locus: identification of the Toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998;24:340–55. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Part 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 5.O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol Rev. 2008;226:10–18. doi: 10.1111/j.1600-065X.2008.00701.x. [DOI] [PubMed] [Google Scholar]
- 6.Song DH, Lee JO. Sensing of microbial molecular patterns by Toll-like receptors. Immunol Rev. 2012;250:216–29. doi: 10.1111/j.1600-065X.2012.01167.x. [DOI] [PubMed] [Google Scholar]
- 7.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 8.Lang D, Knop J, Wesche H, Raffetseder U, Kurrle R, et al. The type II IL-1 receptor interacts with the IL-1 receptor accessory protein: a novel mechanism of regulation of IL-1 responsiveness. J Immunol. 1998;161:6871–77. [PubMed] [Google Scholar]
- 9.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–47. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napetschnig J, Wu H. Molecular basis of NF-κB signaling. Annu Rev Biophys. 2013;42:443–68. doi: 10.1146/annurev-biophys-083012-130338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor–interleukin-1 receptor superfamily. Sci Signal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–32. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 13.Bell JK, Botos I, Hall PR, Askins J, Shiloach J, et al. The molecular structure of the TLR3 extracellular domain. J Endotoxin Res. 2006;12:375–78. doi: 10.1179/096805106X118780. [DOI] [PubMed] [Google Scholar]
- 14.Choe J, Kelker MS, Wilson IA. Crystal structure of human Toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–85. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 15.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Kim HM, Park BS, Kim JI, Kim SE, Lee J, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–17. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, et al. Structural basis of Toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–81. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–95. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 19.Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–64. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science. 2013;339:1426–29. doi: 10.1126/science.1229159. [DOI] [PubMed] [Google Scholar]
- 21.Jin MS, Lee JO. Structures of TLR-ligand complexes. Curr Opin Immunol. 2008;20:414–19. doi: 10.1016/j.coi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Jin MS, Lee JO. Structures of the Toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–91. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Kang JY, Lee JO. Structural biology of the Toll-like receptor family. Annu Rev Biochem. 2011;80:917–41. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 24.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulero JJ, Boyle BJ, Bradley S, Bright JM, Nelken ST, et al. Three new human members of the lipid transfer/lipopolysaccharide binding protein family (LT/LBP) Immunogenetics. 2002;54:293–300. doi: 10.1007/s00251-002-0467-3. [DOI] [PubMed] [Google Scholar]
- 26.Tobias PS, Soldau K, Gegner JA, Mintz D, Ulevitch RJ. Lipopolysaccharide binding protein-mediated complexation of lipopolysaccharide with soluble CD14. J Biol Chem. 1995;270:10482–88. doi: 10.1074/jbc.270.18.10482. [DOI] [PubMed] [Google Scholar]
- 27.Eckert JK, Kim YJ, Kim JI, Gurtler K, Oh DY, et al. The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity. Immunity. 2013;39:647–60. doi: 10.1016/j.immuni.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, et al. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J Biol Chem. 2005;280:11347–51. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]
- 29.Erridge C, Bennett-Guerrero E, Poxton IR. Structure and function of lipopolysaccharides. Microbes Infect. 2002;4:837–51. doi: 10.1016/s1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- 30.Rietschel ET, Kirikae T, Schade FU, Ulmer AJ, Holst O, et al. The chemical structure of bacterial endotoxin in relation to bioactivity. Immunobiology. 1993;187:169–90. doi: 10.1016/S0171-2985(11)80338-4. [DOI] [PubMed] [Google Scholar]
- 31.Kojima S, Blair DF. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 33.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human Toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11:372–78. [PubMed] [Google Scholar]
- 34.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, et al. The evolution of vertebrate Toll-like receptors. PNAS. 2005;102:9577–82. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 36.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, et al. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science. 2004;303:1526–29. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 37.Asagiri M, Hirai T, Kunigami T, Kamano S, Gober HJ, et al. Cathepsin K-dependent Toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319:624–27. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- 38.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–48. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–15. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Zhang S, Li L, Liu X, Mei K, Wang X. Structural insights into the assembly and activation of IL-1β with its receptors. Nat Immunol. 2010;11:905–11. doi: 10.1038/ni.1925. [DOI] [PubMed] [Google Scholar]
- 41.Thomas C, Bazan JF, Garcia KC. Structure of the activating IL-1 receptor signaling complex. Nat Struct Mol Biol. 2012;19:455–57. doi: 10.1038/nsmb.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutsumi N, Kimura T, Arita K, Ariyoshi M, Ohnishi H, et al. The structural basis for receptor recognition of human interleukin-18. Nat Commun. 2014;5:5340. doi: 10.1038/ncomms6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Hammel M, He Y, Tainer JA, Jeng U-S, et al. Structural insights into the interaction of IL-33 with its receptors. PNAS. 2013;110:14918–23. doi: 10.1073/pnas.1308651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyman T, Stenmark P, Flodin S, Johansson I, Hammarstrom M, Nordlund P. The crystal structure of the human Toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283:11861–65. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 45.Khan JA, Brint EK, O’Neill LA, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem. 2004;279:31664–70. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- 46.Ohnishi H, Tochio H, Kato Z, Orii KE, Li A, et al. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. PNAS. 2009;106:10260–65. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valkov E, Stamp A, Dimaio F, Baker D, Verstak B, et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. PNAS. 2011;108:14879–84. doi: 10.1073/pnas.1104780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enokizono Y, Kumeta H, Funami K, Horiuchi M, Sarmiento J, et al. Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling. PNAS. 2013;110:19908–13. doi: 10.1073/pnas.1222811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–13. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–90. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park HH, Logette E, Rauser S, Cuenin S, Walz T, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–46. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat Struct Mol Biol. 2010;17:1324–29. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrao R, Zhou H, Shan Y, Li Q, Shaw DE, et al. IRAK4 dimerization and trans-autophosphorylation are induced by Myddosome assembly. Mol Cell. 2014;55:891–903. doi: 10.1016/j.molcel.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–92. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 57.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–74. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell. 2005;122:669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 59.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 60.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 61.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, et al. IPS-1, an adaptor triggering RIG-I-and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–88. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 62.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–35. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 63.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–27. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–22. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kohlway A, Luo D, Rawling DC, Ding SC, Pyle AM. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–79. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Civril F, Bennett M, Moldt M, Deimling T, Witte G, et al. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–34. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–79. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 68.Luo D, Kohlway A, Vela A, Pyle AM. Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure. 2012;20:1983–88. doi: 10.1016/j.str.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu B, Peisley A, Richards C, Yao H, Zeng X, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–89. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–87. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–43. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–74. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, et al. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–93. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- 74.Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. PNAS. 2011;108:21010–15. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–26. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. PNAS. 2012;109:18437–41. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–83. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 78.Rawling DC, Pyle AM. Parts, assembly and operation of the RIG-I family of motors. Curr Opin Struct Biol. 2014;25:25–33. doi: 10.1016/j.sbi.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55:771–81. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeng W, Sun L, Jiang X, Chen X, Hou F, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–30. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gack MU, Shin YC, Joo CH, Urano T, Liang C, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 82.Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–14. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, et al. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–23. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–42. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–22. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 86.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 87.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–18. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 90.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–87. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vavere AL, Simon GG, George RT, Rochitte CE, Arai AE, et al. Diagnostic performance of combined noninvasive coronary angiography and myocardial perfusion imaging using 320 row detector computed tomography: design and implementation of the CORE320 multicenter, multinational diagnostic study. J Cardiovasc Comput Tomogr. 2011;5:370–81. doi: 10.1016/j.jcct.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 93.Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythe-matosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jin T, Perry A, Jiang J, Smith P, Curry JA, et al. Structures of the HIN domain: DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–71. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013;4:327–39. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ru H, Ni X, Zhao L, Crowley C, Ding W, et al. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 2013;23:855–58. doi: 10.1038/cr.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sung MW, Watts T, Li P. Crystallographic characterization of mouse AIM2 HIN-200 domain bound to a 15 bp and an 18 bp double-stranded DNA. Acta Crystallogr Sect F. 2012;68:1081–84. doi: 10.1107/S174430911203103X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 99.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. PNAS. 2014;111:E62–71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288:13225–35. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu A, Kabaleeswaran V, Fu T, Magupalli VG, Wu H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J Mol Biol. 2014;426:1420–27. doi: 10.1016/j.jmb.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu Z, Yan C, Liu P, Huang Z, Ma R, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–75. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 103.Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk TH, Huizinga EG. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J Biol Chem. 2012;287:38460–72. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu J, Fernandes-Alnemri T, Alnemri ES. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J Clin Immunol. 2010;30:693–702. doi: 10.1007/s10875-010-9425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J Biol Chem. 2009;284:32932–41. doi: 10.1074/jbc.M109.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E, Otting G. The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol. 2003;332:1155–63. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 108.Moriya M, Taniguchi S, Wu P, Liepinsh E, Otting G, Sagara J. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry. 2005;44:575–83. doi: 10.1021/bi048374i. [DOI] [PubMed] [Google Scholar]
- 109.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–78. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–92. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhong B, Yang Y, Li S, Wang YY, Li Y, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–50. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 112.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–26. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun W, Li Y, Chen L, Chen H, You F, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. PNAS. 2009;106:8653–58. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–18. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu J, Sun L, Chen X, Du F, Shi H, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–30. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–61. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–84. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–68. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–37. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kato K, Ishii R, Goto E, Ishitani R, Tokunaga F, Nureki O. Structural and functional analyses of DNA-sensing and immune activation by human cGAS. PLOS ONE. 2013;8:e76983. doi: 10.1371/journal.pone.0076983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li X, Shu C, Yi G, Chaton CT, Shelton CL, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–31. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang X, Wu J, Du F, Xu H, Sun L, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–30. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. PNAS. 2013;110:1652–57. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Han Y, Donovan J, Rath S, Whitney G, Chitrakar A, Korennykh A. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343:1244–48. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang H, Zeqiraj E, Dong B, Jha BK, Duffy NM, et al. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol Cell. 2014;53:221–34. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ouyang S, Song X, Wang Y, Ru H, Shaw N, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–86. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–45. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–24. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shang G, Zhu D, Li N, Zhang J, Zhu C, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–27. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 132.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–30. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 133.Zhang X, Shi H, Wu J, Zhang X, Sun L, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–35. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao P, Ascano M, Zillinger T, Wang W, Dai P, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–62. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chin KH, Tu ZL, Su YC, Yu YJ, Chen HC, et al. Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING. Acta Crystallogr D. 2013;69:352–66. doi: 10.1107/S0907444912047269. [DOI] [PubMed] [Google Scholar]
- 136.Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–50. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim S, Li L, Maliga Z, Yin Q, Wu H, Mitchison TJ. Anticancer flavonoids are mouse-selective STING agonists. ACS Chem Biol. 2013;8:1396–401. doi: 10.1021/cb400264n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36:947–58. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–68. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 140.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cai X, Chen J, Xu H, Liu S, Jiang QX, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–22. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Baroja-Mazo A, Martin-Sanchez F, Gomez AI, Martinez CM, Amores-Iniesta J, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15:738–48. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 143.Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat Immunol. 2014;15:727–37. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–92. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]