Summary
Although oncogene-targeted therapy often elicits profound initial tumor responses in patients, responses are generally incomplete because some tumor cells survive initial therapy as residual disease that enables eventual acquired resistance. The mechanisms underlying tumor cell adaptation and survival during initial therapy are incompletely understood. Here, through the study of EGFR-mutant lung adenocarcinoma we show that NF-κB signaling is rapidly engaged upon initial EGFR inhibitor treatment to promote tumor cell survival and residual disease. EGFR oncogene inhibition induced an EGFR-TRAF2-RIP1-IKK complex that stimulated an NF-κB-mediated transcriptional survival program. The direct NF-κB inhibitor PBS-1086 suppressed this adaptive survival program and increased the magnitude and duration of initial EGFR inhibitor response in multiple NSCLC models, including a patient-derived xenograft. These findings unveil NF-κB activation as a critical adaptive survival mechanism engaged by EGFR oncogene inhibition and provide rationale for EGFR and NF-κB co-inhibition to eliminate residual disease and enhance patient responses.
Introduction
Epidermal growth factor receptor (EGFR)-mutant NSCLC is a paradigm-defining model of the success and limitations of targeted cancer therapy. Activating mutations in EGFR are present in approximately 10-35% of NSCLC patients (D'Angelo et al., 2011). Although the EGFR tyrosine kinase inhibitors (TKIs) erlotinib, gefitinib, and afatinib are approved as first-line therapy in advanced-stage EGFR-mutant NSCLC patients, resistance is a major challenge. Approximately 20-30% of patients exhibit innate resistance and fail to respond to initial treatment and 98% of patients who have an initial EGFR TKI response exhibit an incomplete response (Mok et al., 2009; Zhou et al., 2011). This incomplete therapy response results in residual disease that enables the emergence of acquired resistance in patients, often a lethal event. Although many mechanisms of either innate or acquired resistance have been deciphered (Bivona et al., 2011; Engelman et al., 2007; Ercan et al., 2012; Ng et al., 2012; Ohashi et al., 2013; Ohashi et al., 2012; Sequist et al., 2011; Takezawa et al., 2012; Turke et al., 2010; Yu et al., 2013; Zhang et al., 2012), the molecular basis of incomplete response and residual disease during initial EGFR TKI therapy is poorly understood. Filling this knowledge gap is essential to identify therapeutic strategies to combat tumor cell adaptation and survival during initial treatment and induce complete responses in patients.
Prior work uncovered a cancer cell population termed ‘drug tolerant persisters’ that withstood initial treatment via an IGF1R-mediated epigenetic program that could be pharmacologically reversed with chromatin-directed or IGF1R targeted therapy (Sharma et al., 2010). Subsequent clinical trials did not show a significant effect of either chromatin-directed or IGF1R targeted therapy on response to concurrent EGFR kinase inhibitor treatment in NSCLC patients (Goldberg et al., 2012; Ramalingam et al., 2011). Although this hypothesis remains promising, additional studies are required. Other work exploring initial response to targeted therapy in cancer cells showed that EGFR inhibition provokes STAT3 survival signaling (Lee et al., 2014). The precise molecular mechanism underlying this EGFR inhibitor-induced STAT3 signaling remains incompletely understood.
Here, we further investigated signaling events that occur in response to EGFR oncogene inhibition in NSCLC cells to enable their adaptation and survival during initial therapy and thereby promote residual disease. Although we previously found that NF-κB promotes innate EGFR TKI resistance (Bivona et al., 2011), herein we explored the distinct hypothesis that NF-κB activation might be triggered by initial EGFR TKI treatment as an adaptive event to promote NSCLC cell survival and residual disease, thus limiting EGFR inhibitor efficacy.
Results
EGFR oncogene inhibition triggers NF-κB activation in NSCLC models
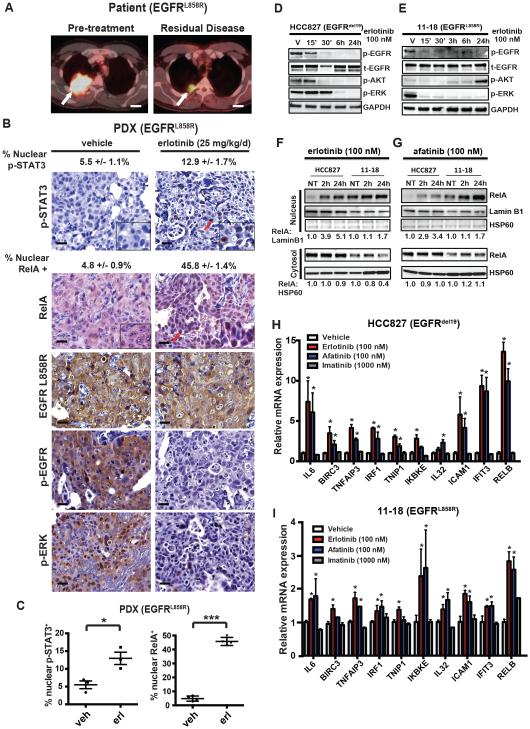
We explored whether NF-κB was activated in tumor cells obtained at the time of residual disease in the setting of an initial incomplete tumor response to EGFR TKI monotherapy. Although patient tumor specimens obtained at residual disease after an initial response to EGFR TKI monotherapy are rare, as surgical resection for metastatic disease is uncommon, we had the opportunity to generate and study a patient-derived tumor xenograft (PDX) obtained from a patient with oligometastatic EGFR-mutant NSCLC treated with erlotinib. This patient uncharacteristically underwent surgical resection of residual disease after an incomplete response to initial erlotinib therapy, which was discontinued prior to surgery (Figure 1A). The residual disease NSCLC specimen resected from this patient had the identical EGFR L858R mutation detected in the pre-treatment tumor by a clinical DNA sequencing assay and had no evidence of the EGFR T790M resistance mutation or other established oncogenic mutations by whole exome deep sequencing (mean coverage depth 100X, data not shown). Immunohistochemical (IHC) staining of the resected tumor confirmed expression of EGFR L858R, p-EGFR, and p-ERK in the tumor cells, indicating oncogenic EGFR signaling in the tumor (Figure S1A). The p-EGFR and p-ERK expression is consistent with the clinical course of the patient, as the patient was off of EGFR TKI at the time of surgery. We investigated NF-κB activation status, and that of STAT3, in the tumor using RelA and p-STAT3 antibodies in IHC studies in the resected tumor specimen. We found minimal RelA or p-STAT3 nuclear expression in the patient tumor specimen (Figure S1A), suggesting that these pathways were not significantly engaged in the absence of EGFR TKI in the patient (Figure S1A). We treated mice bearing this PDX tumor with erlotinib (or vehicle) and assessed the effects of treatment on signaling in the tumor after 48 hours. Tumors from vehicle treated mice were representative of the primary patient resected tumor specimen, exhibiting expression of EGFR L858R, p-EGFR, and p-ERK and minimal RelA or p-STAT3 nuclear expression (Figure 1B and S1A). Erlotinib treatment of these mice led to decreased levels of p-EGFR, p-ERK, and p-AKT (Figure 1B, and S1B). We observed a two-fold increase in nuclear p-STAT3 in PDX tumors treated with erlotinib (Figure 1B-C), consistent with the STAT3 activation observed in EGFR-mutant NSCLC cell lines treated with an EGFR TKI (Lee et al., 2014). Strikingly, we found that erlotinib treatment resulted in a 10-fold increase in nuclear RelA expression in the tumor cells that is indicative of NF-κB activation (O' Reilly et al., 2009) (Figure 1B-C). Thus, erlotinib treatment suppressed EGFR and canonical downstream MEK-ERK and AKT signaling while concurrently activating NF-κB and STAT3.
Figure 1. Adaptive NF-κB activation in response to EGFR oncogene inhibition in NSCLC.
(A) FDG-PET CT imaging of the patient’s thorax with metastatic EGFR-mutant NSCLC (indicated by the white arrow) prior to treatment with erlotinib, and post-erlotinib with residual disease present. Scale bar = 2 cm. (B) Representative IHC staining for the indicated proteins demonstrating p-STAT3 and RelA nuclear localization (red arrows), as well as EGFR L858R, p-EGFR, and p-ERK in tumors resected from PDX mice treated with vehicle or erlotinib (25 mg/kg/d) for 48 h. Scale bar = 20 microns. Percentage of cells positive for nuclear p-STAT3 and RelA (mean ± S.E.M.) are indicated. (C) Quantitation of nuclear p-STAT3 and nuclear RelA in EGFR L858R mutant PDX tumors treated with vehicle or erlotinib (mean ± S.E.M.). * p < 0.05, *** p < 0.001 as determined by two-tailed unpaired T-test. (D-E) HCC827 and 11-18 cells were treated with erlotinib (100 nM) for the indicated period of time and activity of the EGFR pathway was monitored by western blot. V = vehicle treated controls. GAPDH is used as a loading control. (F-G) HCC827 and 11-18 cells were treated with erlotinib (100 nM) or afatinib (100 nM) for the indicated periods of time and nuclear and cytosolic fractions were separated to show the translocation of the NF-κB subunit RelA to the nucleus. Lamin B1 is a marker of nuclear extraction. HSP60 is a marker of cytosolic extraction. (H-I) Quantitative real-time PCR (Q-PCR) analysis of indicated NF-κB target genes in (I) HCC827, and (H) 11-18 cells in response to vehicle, erlotinib, afatinib, or imatinib treatment for 12 hours (mean ± S.E.M.). * P < 0.05 compared to vehicle treated cells by Bonferroni’s multiple comparisons ANOVA test. See also Figure S1.
We used an established transgenic murine model of EGFR-mutant (EGFR L858R) NSCLC that recapitulates human EGFR-mutant NSCLC (Politi et al., 2006) to further explore the link between NF-κB activation and residual disease on EGFR TKI therapy. EGFR-mutant NSCLCs in these transgenic mice initially regress upon erlotinib treatment and then develop acquired erlotinib resistance after several months (Politi et al., 2009). This observation suggests that some tumor cells survive initial erlotinib exposure as residual disease that eventually grows to form a resistant tumor. To assess if NF-κB is activated in residual tumor cells that persist following initial erlotinib treatment, we treated EGFR L858R tumor bearing mice with erlotinib for 7 days and harvested lung tumors representing residual disease on EGFR TKI therapy to assess for evidence of NF-κB activation. Increased nuclear RelA expression that is indicative of increased NF-κB activity (O' Reilly et al., 2009) was observed in tumors harvested from mice treated with erlotinib (Figure S1C-E). These findings suggest that NF-κB activity is increased in tumor cells that persist through initial EGFR oncogene inhibition as residual disease in the in vivo and immunocompetent tumor microenvironment. The findings in both the PDX and transgenic EGFR-mutant NSCLC residual disease tumors suggest that NF-κB might be triggered by EGFR TKI treatment to promote tumor cell survival and residual disease.
We used human EGFR-mutant NSCLC cellular models to further investigate EGFR TKI-induced NF-κB activation. Using five different human EGFR-mutant NSCLC cell line models (HCC827 EGFR exon19 deletion, 11-18 EGFR L858R, H3255 EGFR L858R, PC9 exon19 deletion, H1975 EGFR L858R/T790M), we found that EGFR TKI treatment rapidly suppressed AKT and MAPK signaling (Figure 1D-E), as expected (Gong et al., 2007). In contrast, NF-κB was immediately hyperactivated upon erlotinib or afatinib treatment (Figure 1F-G and S1F-H) (Figure 1H-I) (Raskatov et al., 2012; Sun et al., 1994). This early activation of NF-κB was specific to EGFR oncogene inhibition, as the treatment of cells with the non-EGFR TKI imatinib did not impact transcription of NF-κB target genes (Figure 1H-I). H1975 NSCLC cells harboring the EGFR T790M erlotinib-resistance mutation exhibited increased RelA nuclear localization specifically upon treatment with the EGFR inhibitor afatinib that has activity against EGFR T790M, but not with erlotinib that does not (Figure S1H). Inhibition of the EML4-ALK oncogenic fusion protein with the ALK inhibitor crizotinib also failed to induce NF-κB activation in H3122 EML4-ALK positive NSCLC cells (Figure S1I). We found that NF-κB activation was present in the subpopulation of EGFR-mutant HCC827 NSCLC cells that persist during initial EGFR TKI therapy, representing residual disease (Figure S1J). These persistent tumor cells exhibited decreased p-EGFR, p-ERK, and p-AKT levels, indicating that canonical MEK-ERK and AKT signaling remained inhibited as NF-κB was specifically activated. The findings indicate that EGFR oncogene inhibition promotes rapid and adaptive hyperactivation of NF-κB in EGFR-mutant NSCLC and suggest that this heightened NF-κB signaling may limit initial EGFR TKI response.
EGFR oncogene inhibition induces TRAF2 ubiquitination and activation of an NF-κB-activating complex
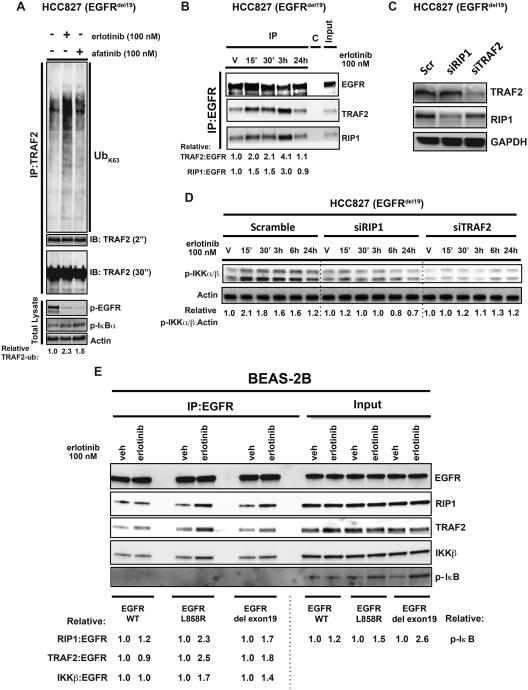
We next studied the mechanism by which NF-κB is activated by EGFR oncogene inhibition. Activation of canonical NF-κB signaling involves assembly of a TRAF2-RIP1 signaling complex via ubiquitination events initiated at an upstream receptor (Ea et al., 2006). TRAF2 ubiquitination and RIP1 association leads to IKK recruitment and activation, phosphorylation and degradation of IκB, and NF-κB (RelA) nuclear translocation (Hayden and Ghosh, 2008). We hypothesized that EGFR oncogene inhibition might promote immediate ubiquitination of proteins that drive NF-κB signaling. To identify ubiquitination events that might contribute to NF-κB activation in the context of EGFR inhibition, we performed a global enrichment of ubiquitinated proteins followed by mass spectrometry analysis on proteins harvested from HCC827 cells treated with vehicle or erlotinib. These studies revealed that the top-scoring ubiquitination event in erlotinib treated cells was increased ubiquitination of TRAF2 (Table 1), an established TRAF2 and NF-κB-activating event (Mahul-Mellier et al., 2012). We confirmed that treatment of HCC827 cells with either erlotinib or afatinib induced TRAF2 K63-linked ubiquitination (Figure 2A). We extended these findings to an additional EGFR-mutant NSCLC cell line, confirming that afatinib treatment induced TRAF2 K63-linked ubiquitination in H1975 cells (Figure S2A). These studies uncover early and dynamic regulation of ubiquitin-modifying events that may modulate response to EGFR targeted therapy in NSCLC.
Figure 2. EGFR-TRAF2-NF-κB activating complex forms in response to EGFR oncogene inhibition in NSCLC.
(A) Activation of the NF-κB signalosome was demonstrated by an ubiquitination assay of the TRAF2 protein. HCC827 cells were transfected with HA-Ubiquitin K63 plasmid and treated with erlotinib (100 nM) or afatinib (100 nM) for 30 minutes. Subsequently, cell lysates were used for endogenous immunoprecipitation of TRAF2 to determine its ubiquitination status by Western blot. (B) Immunoprecipitation of endogenous EGFR from HCC827 cells treated with erlotinib at the indicated time points, showing the assembly of essential components of NF-κB signaling. (C-D) Western blot analysis of EGFR and NF-κB signaling in HCC827 cells treated with RIP1 and TRAF2 specific siRNAs ± erlotinib. (E) Co-immunoprecipitation of NF-κB signalosome with exogenously expressed mutant EGFR upon erlotinib (100 nM) treatment of BEAS-2B human bronchial epithelial cells. All results shown represent 3 independent experiments. See also Figure S2.
K63-linked TRAF2 ubiquitination is essential for RIP1 activation and subsequent activation of IKKβ/γ and NF-κB (Alvarez et al., 2010; Ea et al., 2006; Li et al., 2009; Mahul-Mellier et al., 2012). Overexpressed RIP1 can associate with EGFR in breast cancer and glioma cells (Habib et al., 2001; Puliyappadamba et al., 2013). Given that we uncovered increased TRAF2 ubiquitination upon EGFR oncogene inhibition, we hypothesized that EGFR TKI treatment might promote assembly and activation (via ubiquitination) of a signaling complex containing TRAF2, RIP1, and EGFR that activates IKK and downstream NF-κB signaling. Through co-immunoprecipitation experiments we found that EGFR associates with TRAF2 and RIP1 in HCC827 EGFR mutant cells (Figure 2B). The formation of this TRAF2-RIP1-EGFR complex was enhanced by EGFR TKI treatment (Figure 2B). Increased association of RIP1 and IKKβ with EGFR was observed upon either erlotinib or afatinib treatment in both HCC827 and 11-18 EGFR-mutant NSCLC cell lines (Figure S2B), but only upon treatment with the EGFR T790M inhibitor afatinib in erlotinib-resistant H1975 cells (Figure S2C). Attempts to detect this NF-κB activating complex in vivo in the PDX model were unsuccessful, perhaps because detecting protein-protein associations is more challenging in vivo where both tumor cells and contaminating normal cells are present and can obscure detection of relevant interactions occurring specifically in tumor cells. Hence, it remains unclear whether the complex we detected in patient-derived cell lines is present in vivo. Nevertheless, our collective findings indicate that formation of the NF-κB signaling complex is dynamically enhanced by EGFR oncogene inhibition in several human EGFR-mutant NSCLC in vitro models. Using genetic knockdown experiments in multiple NSCLC cell lines, we found that this NF-κB signaling complex was essential for EGFR TKI-induced NF-κB activation as RIP1 and TRAF2 were each required for maximal induction of p-IKKα/β and NF-κB target gene expression by EGFR TKI treatment (Figure 2C-D, and S2D-F). Lastly, using a genetically controlled system of primary immortalized human bronchial epithelial cells (BEAS-2B) (Lonardo et al., 2002), we further found that the presence of oncogenic EGFR was critical for formation of this NF-κB signaling complex upon EGFR TKI treatment. (Figure 2E).
Direct pharmacologic NF-κB inhibition overrides EGFR TKI-induced NF-κB activation and suppresses the emergence of EGFR TKI therapy resistance
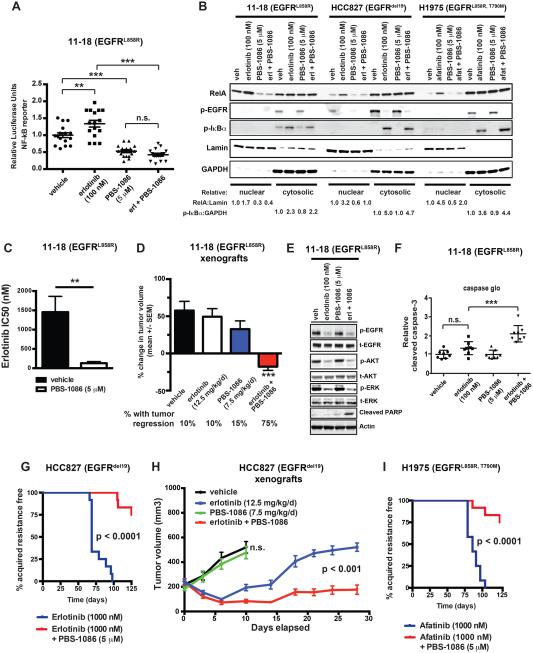
We next investigated the effects of pharmacologic inhibition of NF-κB in the context of EGFR TKI treatment. Our prior studies showed that NF-κB activation in EGFR-mutant NSCLC cells suppresses EGFR TKI-induced apoptosis, which can be reversed by genetic knockdown of RelA (Bivona et al., 2011). An obstacle to clinical translation is that pharmacologic inhibitors of NF-κB signaling clinically developed to date do not target NF-κB directly but rather do so indirectly by targeting either IKK or the proteasome. These indirect NF-κB inhibitors exhibit non-specific effects and limited therapeutic window in patients (Perkins, 2012). We took an alternative approach to credential a direct pharmacologic inhibitor of NF-κB for potential use in combination with an EGFR TKI in NSCLC patients. We tested PBS-1086 that acts as a specific inhibitor of Rel DNA binding (Figure S3A) (Fabre et al., 2012). We conducted several studies to validate PBS-1086 as a potent and selective NF-κB inhibitor. We profiled the effects of PBS-1086 on DNA binding by RelA (p65) and other common transcription factors in nuclear extracts using a transcription factor DNA binding assay (Renard et al., 2001) and found that PBS-1086 blocks RelA DNA binding (Figure S3B and Table S1). By gene set enrichment analysis (GSEA)(Mootha et al., 2003; Subramanian et al., 2005), we found PBS-1086 treatment specifically suppressed the expression of canonical NF-κB target genes and gene sets but did not modulate expression of genes regulated by other common transcription factors (Table S2). We verified that PBS-1086 inhibits NF-κB transcriptional activity in 11-18 cells using an established NF-κB activation reporter assay (Figure 3A) (Aoki and Kao, 1997). We next profiled the biochemical effects of PBS-1086, alone and combined with EGFR TKI, in EGFR-mutant NSCLC cell lines (Figure 3B). PBS-1086 treatment of myeloma cells leads to decreased levels of nuclear RelA (Fabre et al., 2012), as non-DNA bound RelA undergoes nuclear export (Harhaj and Sun, 1999). We observed that in each of three EGFR-mutant NSCLC cell lines, PBS-1086 treatment reduced nuclear RelA levels (Figure 3B). PBS-1086 treatment blunted the RelA nuclear localization induced by EGFR TKI treatment in each cell line (Figure 3B). EGFR TKI-induced phosphorylation of IκBα was not affected by PBS-1086 treatment, indicating that the drug inhibits NF-κB signaling at the level of Rel nuclear function and does not inhibit upstream components of the NF-κB pathway (Figure 3B). These data validate PBS-1086 as a potent and specific inhibitor of NF-κB transcriptional activity in human NSCLC cells.
Figure 3. Pharmacologic direct NF-κB inhibition with PBS-1086 enhances EGFR TKI response and suppresses the emergence of acquired resistance in EGFR-mutant NSCLC models.
(A) NF-κB transcriptional activation activity in 11-18 cells treated as indicated and measured by luciferase reporter assay (mean ± S.E.M.). ** p < 0.01, *** p < 0.001 as determined by Bonferroni’s multiple comparisons ANOVA test. (B) Nuclear/cytoplasmic fractionation and western blot analysis of the indicated proteins in 11-18, HCC827, and H1975 cells treated as indicated. (C) Drug sensitivity as measured by half maximal inhibitory concentration (IC50) of erlotinib (mean ± S.E.M.) in 11-18 cells treated with vehicle or 5.0 micromolar PBS-1086. ** p < 0.01 as determined by two-tailed unpaired T-test. (D) Mean change in tumor volume (± S.E.M.) of 11-18 EGFR-mutant NSCLC tumor xenografts over a 10 day period after treatment of mice with the drugs indicated. A minimum of 10 tumors were evaluated per treatment group. The percentage of tumors with regression is shown under treatment cohort, revealing that combined erlotinib + PBS-1086 treatment (using the monotherapy dose of each drug in the combination) induces significantly more tumor regressions than monotherapy. *** p < 0.001 in comparison to each other treatment group by Bonferroni’s multiple comparisons ANOVA test. (E) Effect of combined erlotinib + PBS-1086 treatment (using the monotherapy dose of each drug in combination) on EGFR signaling components and apoptosis marker (cleaved PARP) in the erlotinib-resistant 11-18 cells. (F) Relative caspase activity in 11-18 cells upon treatment as indicated (mean ± S.E.M.), where the monotherapy dose of each drug was used in combination). *** p < 0.001 by Bonferroni’s multiple comparisons ANOVA test. (G) Comparison of time to the development of acquired resistance of HCC827 cells treated as indicated. P-values determined by logrank test comparing the median time to treatment resistance. (H) Growth of HCC827 tumor xenografts (mean ± S.E.M.) treated as indicated over a 28-day period. The monotherapy dose for each drug was also used in the combination. P-value determined by linear regression analysis. (I) Comparison of time to the development of acquired resistance of H1975 cells treated as indicated. P-values determined by logrank test comparing the median time to treatment resistance. See also Figure S3, Table S1, and Table S2.
We explored the impact of PBS-1086 treatment on EGFR TKI efficacy in EGFR-mutant NSCLC models. 11-18 cells are relatively less sensitive to EGFR TKI treatment (erlotinib IC50 ~1.5 μM) compared to HCC827 or H3255 cell lines (erlotinib IC50 ~10 nM and ~100 nM, respectively). 11-18 cells do not express resistance-conferring lesions associated with acquired resistance in patients and exhibit high basal NF-κB activity that is further enhanced by EGFR TKI treatment (Figure 1E-G, 3B). We used these cells as a model of NF-κB-mediated residual disease on EGFR TKI treatment and found that treatment of 11-18 cells or tumor xenografts with PBS-1086 enhanced erlotinib response (Figure 3C-D and Figure S3C). This enhanced response was associated with induction of apoptosis upon PBS-1086 and erlotinib polytherapy (Figure 3E-F). The effects of PBS-1086 treatment occurred predominantly through Rel (A, B) inhibition, as we observed minimal additive effect of PBS-1086 treatment on erlotinib sensitivity in 11-18 cells in which we genetically silenced RelA or RelB (Figure S3D). Exogenous expression of a constitutively active RelA rescued the effects of PBS-1086 treatment on erlotinib sensitivity (Figure S3D). These findings show the synthetic lethality of combined EGFR and NF-κB (Rel) inhibition (with PBS-1086), suggesting a polytherapy to eliminate residual disease.
We next explored the effects of pharmacologic NF-κB inhibition in EGFR-mutant cell lines that model the common clinical scenario in which a patient has a substantial, though incomplete, EGFR TKI response. We reasoned that HCC827 and H1975 cells, which are relatively sensitive to EGFR TKI monotherapy, would allow us to test if NF-κB inhibition with PBS-1086 might decrease the presence of residual disease and the emergence of acquired EGFR TKI resistance. We derived a panel of HCC827 sub-clones with acquired erlotinib resistance through prolonged, continuous erlotinib treatment and profiled NF-κB transcriptional activity (Aoki and Kao, 1997). We observed NF-κB hyperactivation in 50% (6 of 12) of the resistance sub-clones (Figure S3E). Erlotinib-resistant cells exhibited increased association of IKKβ with EGFR, indicating that these cells with acquired erlotinib resistance harbored increased formation of the NF-κB activating complex (Figure S3F). Erlotinib resistant sub-clones that exhibited NF-κB activation did not harbor the EGFR T790M resistance mutation or MET overexpression (data not shown). To test whether NF-κB activity was necessary for acquired EGFR TKI resistance, we treated the resistant sub-clones with PBS-1086 alone and in combination with erlotinib. Treatment with PBS-1086 and erlotinib reversed acquired erlotinib resistance specifically in the cellular models with NF-κB hyperactivation (Figure S3G). As these findings indicated that NF-κB activation may promote the survival of cells during initial EGFR TKI treatment that drive acquired resistance, we reasoned that initial treatment with PBS-1086 might suppress the development of acquired EGFR TKI resistance. PBS-1086 treatment in combination with erlotinib suppressed the emergence of acquired resistance in HCC827 cells both in vitro (Figure 3G) and in vivo (Figure 3H). These effects were phenocopied in H1975 cells with EGFR L858R/T790M, in which we similarly found that PBS-1086 treatment suppressed the emergence of acquired afatinib resistance in vitro (Figure 3I). These findings suggest that early activation of NF-κB is required for the survival of a subset of cells that subsequently emerge to promote acquired EGFR TKI resistance. Pharmacologic blockade of NF-κB with PBS-1086 may overcome and prevent the development of acquired resistance in EGFR-mutant NSCLC.
IL6 is a critical NF-κB target gene induced by EGFR TKI treatment that underlies NF-κB-mediated survival during EGFR TKI therapy
We explored the downstream targets through which activated NF-κB suppresses apoptosis and promotes survival during EGFR TKI therapy. As the NF-κB target gene repertoire is context-specific, we used an unbiased approach to define by whole transcriptome sequencing (RNA-seq) a transcriptional profile of NF-κB activation in EGFR-mutant NSCLC cells (GEO:GSE65420). We genetically inhibited NF-κB signaling through expression of a non-phosphorylatable form of IκB (IκBSR) (Jiang and Clemens, 2006). Conversely, we genetically activated NF-κB signaling through expression of a constitutively active form of RelA (RelA S536E) (Hu et al., 2004), which promoted erlotinib resistance (Figure S3D). Using these tools, we defined a set of NF-κB target genes that through differential expression analysis of the transcriptome data were both activated by the expression of RelA S536E and suppressed by IκBSR expression, and that were established direct transcriptional targets of NF-κB in the ENCODE dataset (Bernstein et al., 2012) (Figure S4A). We then determined which of these NF-κB target genes were differentially regulated by erlotinib, PBS-1086, or the combination. We established the genetic signature of adaptive NF-κB activation induced upon EGFR oncogene inhibition by identifying the genes whose expression was increased by erlotinib treatment and also decreased by PBS-1086 in the presence of erlotinib. This signature of EGFR TKI-induced NF-κB output consisted of 36 NF-κB target genes, including established regulators of NF-κB signaling and cell survival such as TNFAIP3, BIRC3, and IL6 (Figure 4A and S4A). We confirmed that the expression of a subset of the genes in this signature was increased upon treatment with multiple EGFR TKIs and decreased with PBS-1086 in multiple EGFR-mutant NSCLC cell lines (Figure S4B-C). These data uncover the NF-κB-regulated genetic signature and components of the adaptive response to EGFR oncogene inhibition in NSCLC cells.
Figure 4. An NF-κB transcriptional survival program triggered by EGFR oncogene inhibition promotes tumor cell survival and resistance.
(A) Supervised-hierarchical clustering of NF-κB regulated gene expression changes in 11-18 cells treated as indicated based upon RNA-sequencing analysis. The analysis revealed a genetic signature of the NF-κB medicated adaptive response to EGFR TKI treatment that consisted of a core set of NF-κB target genes induced by erlotinib and, in turn, suppressed by NF-κB inhibition. (B) RNA-sequencing analysis of 11-18 cells demonstrating induction or inhibition of IL6 expression by pharmacologic and genetic manipulation (mean ± S.E.M.). *** p < 0.001 by Bonferroni’s multiple comparisons ANOVA test. (C) ELISA demonstrating the effect of the indicated drug treatments on soluble IL6 protein expression in 11-18 cells treated as indicated (mean ± S.E.M.). * P < 0.05 by Bonferroni’s multiple comparisons ANOVA test. The monotherapy dose for each drug was also used in the combination. (D) Drug sensitivity as measured by half maximal inhibitory concentration (IC50) of erlotinib (mean ± S.E.M.) in 11-18 cells transfected with empty vector (EV) or an IL6 expressing construct (IL6) and treated with the indicated drug combinations. The PBS-1086 + Ruxolitinib combination condition used the indicated monotherapy dose for each drug. * p < 0.05, ** p < 0.01 as determined by Bonferroni’s multiple comparisons ANOVA test. (E) Relative mRNA expression of IL6 as determined by Q-PCR of 11-18 cells transfected with control siRNA, IL6 siRNA, empty vector (EV), or IL6 overexpression construct (IL6) treated with PBS-1086 (mean ± S.E.M.). ** p < 0.01, *** p < 0.001 as determined by two-tailed unpaired test. (F) Drug sensitivity as measured by half maximal inhibitory concentration (IC50) of erlotinib (mean ± S.E.M.) in 11-18 cells treated with IL6 specific or control siRNAs. *** p < 0.001 as determined by two-tailed unpaired test. (G) Western blots showing the effects of the indicated drug treatments on STAT3 activation (phosphorylation) in stably transfected 11-18 (EV) and 11-18 (IL6) cell lines. See also Figure S4.
We next investigated if individual components of the NF-κB output signature promoted EGFR TKI resistance. IL6 was a component of this EGFR TKI-induced, adaptive NF-κB-mediated transcriptional program. As a secreted factor, IL6 could act in an autocrine/paracrine manner to promote survival of a subset of tumor cells in the bulk population during initial therapy. IL6 was consistently and significantly increased by EGFR TKI treatment in two distinct EGFR-mutant NSCLC model (Figure 1H-I), and was recently implicated as a mediator of EGFR TKI-induced STAT3 activation (Lee et al., 2014). We confirmed that EGFR TKI treatment increased IL6 mRNA and secreted IL6 protein levels in 11-18 cells (Figure 4B,C). This induction of IL6 was suppressed by PBS-1086 (Figure 4B, 4C and S4B-C), suggesting that NF-κB promotes IL6 expression induced by EGFR inhibitor treatment. We found that exogenous expression of IL6 rescued the effects of PBS-1086 treatment on erlotinib sensitivity in 11-18 cells (Figure 4D-E), while genetic silencing of IL6 was sufficient to sensitize 11-18 cells to erlotinib treatment (Figure 4F). These data reveal that IL6 may operate downstream of NF-κB in the adaptive response to EGFR oncogene inhibition in NSCLC.
As IL6 promotes JAK2/STAT3 activation (Berishaj et al., 2007), we hypothesized that NF-κB may be required for EGFR TKI-induced STAT3 activation. We found that PBS-1086 inhibited erlotinib-induced STAT3 activation in three EGFR-mutant NSCLC cell lines (Figure 4G and S4D-E), as well as in HCC827 tumor xenografts (Figure S4F-G). We tested if direct JAK2 inhibition with a specific inhibitor, ruxolitinib (Quintas-Cardama et al., 2010; Tefferi et al., 2011), could reverse the pro-survival effects of IL6 induced by EGFR TKI treatment. Co-treatment with erlotinib and ruxolitinib overcame IL6-mediated erlotinib resistance in 11-18 cells (Figure 4D,E,G). IL6 functioned downstream of NF-κB, as we observed that PBS-1086 treatment suppressed the induction of p-STAT3 by EGFR TKI treatment in the absence but not presence of exogenous IL6 (Figure 4G). Inhibition of IL6 signaling by JAK2 inhibitor treatment was required to suppress p-STAT3 in the presence of exogenous IL6 (Figure 4G). The data show that EGFR TKI treatment induces adaptive NF-κB activation that drives a transcriptional program that includes upregulation of IL6-STAT3 signaling. Although IL6-STAT3 signaling has been implicated in survival and resistance in some cancers, including EGFR-mutant NSCLC, our work reveals that this pathway functions downstream of NF-κB during the early adaptive response to EGFR oncogene inhibition in NSCLC (Bromberg et al., 1999; Gao et al., 2007; Kim et al., 2012; Lee et al., 2014; Timofeeva et al., 2013).
Direct pharmacologic inhibition of NF-κB with PBS-1086 enhances response to EGFR TKI treatment and suppresses residual disease in vivo
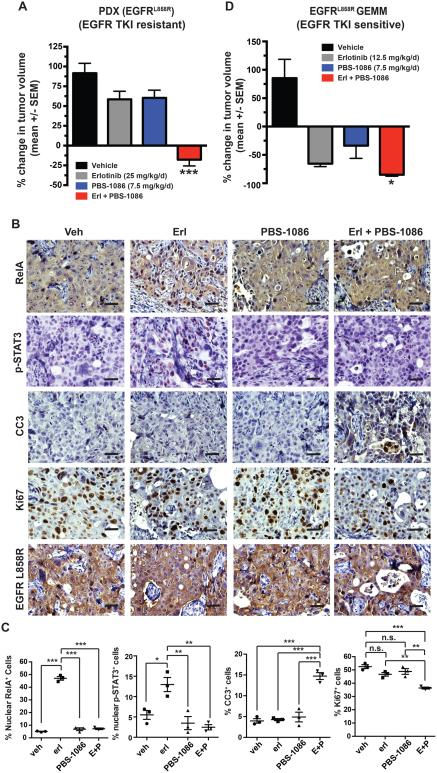
We used the EGFR-mutant PDX tumor model we generated and in which we observed induction of NF-κB activity upon EGFR TKI treatment to assess whether combined treatment with PBS-1086 and erlotinib was more effective than EGFR TKI monotherapy. As these tumors were derived from a patient with residual disease following erlotinib treatment (Figure 1A), they did not respond to erlotinib monotherapy (Figure 5A, S5A and Table S3). In contrast, treatment of mice with PBS-1086 combined with erlotinib promoted significant tumor responses, indicating that PBS-1086 could overcome NF-κB-mediated survival of the tumor cells in this residual disease NSCLC specimen (Figure 5A, S5A and Table S3).
Figure 5. Direct pharmacologic inhibition of NF-κB with PBS-1086 effectively targets residual disease to enhance response to EGFR TKI treatment in vivo.
(A) Mean change (± S.E.M.) in tumor volume of PDX treated with the indicated drugs for 10 days. *** p < 0.001 compared to all other groups by Bonferroni’s multiple comparisons ANOVA test. The monotherapy dose for each drug was also used in the combination. (B-C) Representative IHC staining and quantitation (mean ± S.E.M.) for RelA, p-STAT3, cleaved-caspase 3 (CC3), Ki67, or EGFR-L858R performed on PDX tumors treated as indicated for 48 hours. Scale bar = 20 microns. ** p < 0.01, *** p < 0.001 as determined by Bonferroni’s multiple comparisons ANOVA test. (D) Mean change in tumor volume (± S.E.M.) as determined by magnetic resonance imaging (MRI) of CC10-rtTA; TetO-EGFRL858R transgenic mice treated with doxycycline for 10 weeks (d0) followed by treatment with the indicated drugs for 7 days (d7). * p < 0.05 in comparison to erlotinib monotherapy treated mice by two-tailed unpaired T-test. The monotherapy dose for each drug was also used in the combination. See also Figure S5 and Table S3.
Erlotinib monotherapy induced RelA nuclear translocation and transcriptional upregulation of NF-κB target genes (Figure 1B, 5B-C and S5B). IL6 was most significantly increased with erlotinib treatment (Figure S5B), consistent with our in vitro findings (Figure 4). Co-treatment with PBS-1086 and erlotinib reversed the RelA nuclear translocation and induction of NF-κB target gene expression (Figure 5B-C and S5B), consistent with the tumor response to this combination therapy. This improved tumor and signaling response was associated with increased tumor cell apoptosis and decreased proliferation (Figure 5B-C). The data demonstrate the potential for combined PBS-1086 and EGFR TKI treatment to overcome residual disease.
We also studied the EGFR-mutant transgenic murine model that represents the common clinical scenario in which a patient has a profound initial response to EGFR TKI therapy. We found that these mice often exhibit a partial response to EGFR TKI monotherapy, as do most patients, and that some tumor cells persisting during this incomplete response and residual disease had increased nuclear RelA (Figure S1). We treated these mice with erlotinib and PBS-1086 and compared the response to treatment with erlotinib or PBS-1086 monotherapy. Treatment of mice with the combination of erlotinib and PBS-1086 reversed the effects of erlotinib on RelA nuclear localization and increased tumor response (Figure 5D and S5C-E). These data suggest that PBS-1086 treatment suppressed EGFR-mutant tumor cell persistence during initial EGFR TKI therapy within the native tumor microenvironment and intact host immune system. Animals treated with PBS-1086 either alone or combined with erlotinib exhibited no significant toxicity at the efficacious doses we tested (data not shown). These in vivo data provide further evidence that adaptive NF-κB activation in tumor cells exposed to an EGFR TKI enables incomplete response and residual disease that can be overcome by NF-κB inhibition with PBS-1086, leading to improved EGFR TKI response.
Discussion
We provide insight into the adaptive signaling events that occur in response to targeted cancer therapy to enable tumor cells to survive initial oncogene-targeted therapy. NF-κB hyperactivation in response to EGFR oncogene inhibition may counteract the effects of oncogene withdrawal in the tumor cell population, enabling tumor cell persistence manifesting as an incomplete tumor response and residual disease. This surviving tumor cell population with NF-κB hyperactivation may ultimately promote acquired resistance, potentially in conjunction with additional resistance-conferring alterations such as EGFR T790M or MET amplification.
Our findings have important implications for overcoming EGFR TKI resistance in NSCLC patients, offering an alternative approach to combat the emergence of resistance. Most clinical strategies aimed at overcoming EGFR TKI resistance in NSCLC have focused on counteracting the effects of individual genetic alterations present in drug-resistant tumors in patients who have already developed resistance (Koeppen et al., 2014; Miller et al., 2012; Solca et al., 2012; Walter et al., 2013). Our findings suggest that even when individual resistance lesions such as EGFR T790M are inhibited, adaptive activation of NF-κB may still drive tumor cell survival. This adaptive NF-κB activation and consequent tumor cell survival can be suppressed by NF-κB inhibition. Our findings provide mechanistic rationale for upfront combination therapy with PBS-1086 and an approved or emerging EGFR TKI to minimize or eliminate residual disease and thereby enhance response magnitude and duration in patients.
Experimental Procedures
Cell Lines and Assays
Cell lines were acquired from ATCC and as previously described (Bivona et al., 2011; Zhang et al., 2012). All drugs were purchased from Selleck Chemicals (Houston, TX). PBS-1086 was provided by rel-MD (Baltimore, MD). 72-hour viability assays were performed as described (Zhang et al., 2012). Each assay consisted of six replicate wells and was repeated at least twice in independent experiments.
Animal Studies
11-18 tumor xenografts were generated as previously described (Bivona et al., 2011). For GEMM (Genetically-engineered mouse model) studies, male and female mice of the desired genotype were treated with doxycycline at 6-8 weeks of age, and tumor-bearing mice were treated with the indicated compounds at 16-18 weeks of age. Mice were genotyped and treated with doxycycline to induce lung adenocarcinoma formation as previously described (Politi et al., 2006). Mice were randomized to receive vehicle, erlotinib, PBS-1086, or erlotinib + PBS-1086 (minimum of 3 mice per treatment group) daily for 7 days by i.p. injection. For generation of PDX, informed consent was obtained from the patient as per an open IRB-approved protocol. Tumors were allowed to grow until they reached a minimum volume of 200 mm3 at which point mice were treated with combinations of erlotinib and/or PBS-1086 as described above.
Protein Studies
For western blotting, cells were scraped and lysed in lysis buffer (50 mM Tris·HCl pH 8.0, 150 mM sodium chloride, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X 100, 5mM EDTA containing protease and phosphatase inhibitors (Roche Diagnostics. Indianapolis, IN). All Western blot results shown represent 3 independent experiments. Where indicated, nuclear and cytosolic fractionation was performed as previously described (Fabre et al., 2012). For immunoprecipitations, cell lysates were incubated overnight at 4°C with the antibodies of interest. Immune complexes were precipitated with Protein-G and the beads were washed with RIPA buffer. Immunoprecipitated proteins were separated in a 4-15% SDS-PAGE and transferred onto a nitrocellulose membrane (Bio-Rad). Membranes were blocked with 5% FBS in TBS containing 0.1% Tween and incubated with the appropriate antibodies. Raw mass spectrometry data were analyzed using the MaxQuant software package (version 1.3.0.5)(Cox and Mann, 2008). Data were matched to the SwissProt human reference protein database. Data were searched against a concatenated database containing all sequences in both forward and reverse directions with reverse hits indicating the false discovery rate of identifications. The data were filtered to obtain a peptide, protein, and site-level false discovery rate of 0.01. The minimum peptide length was 7 amino acids. Results were matched between runs with a time window of 2 minutes for technical duplicates. A Z-score was calculated to assess the statistical probability of an increase in ubiquitination of a given peptide in the erlotinib treated samples compared to the DMSO (vehicle) treated samples, and a decrease in ubiquitination in the DMSO treated samples compared to the erlotinib treated samples. Only peptides with a Z-score > 0.5 in both directions were considered significant.
RNA Analysis
RNA-seq was performed in triplicate for each treatment condition on the Illumina Hi-Seq 2000 using paired-end 100bp reads as described (Li and Dewey, 2011; Lin et al., 2014). Differential expression analysis between sets of conditions was performed using DESeq(Anders and Huber, 2010) and as described(Lin et al., 2014). Quantitative PCR (Q-PCR) was performed on the QuantStudioTM 12K Flex Real-Time QPCR System using Taqman probes (Applied Biosystems, Life Technologies) and analysed as described (Lin et al., 2014). RNA-seq data has been deposited in the NCBI GEO database, accession number GSE65420.
Supplementary Material
Table 1.
Ubiquitination events increased by erlotinib treatment in human EGFR-mutant NSCLC cells.
| Peptide | Protein | Description | Z-Score ErI/DMSO |
|---|---|---|---|
| FQDHVKTCGK(gl) | Q12933 | TRAF2 (TNF receptor-associated factor 2) | 1.94 |
| SKHSEDVNVK(gl)VSNAQSVTSER | 060669 | M0T2 (Monocarboxylate transporter 2) | 1.31 |
| WGK(gl)QDGGEGHVGTVR | Q86YT6 | MIB1 (E3 ubiquitin-protein ligase MIB1) | 1.22 |
| FLLGYFPWDSTK(gl)EER | Q8TC07 | TBC15 (TBC1 domainfamily member 15) | 1.06 |
| HIYYITGETK(gl)DQVANSAFVER | P07900 | HS90A (Heat shock protein HSP 90-alpha) | 1.05 |
| SDALETLGFLNHYQMK(gl)NPNGPYPYTLK | P14866 | HNRPL (Heterogeneous nuclear ribonucleoprotein L) | 1.04 |
| WLTLSEVMKLLK(gl) | Q8IUC4 | RHPN2 (Rhophilin-2 |Homo sapiens) | 0.99 |
| SKASLEK(gl)AGK | P53985 | MOT1 (Monocarboxylate transporter 1) | 0.97 |
| YALTGDEVK(gl)K | P62701 | RS4X (40S ribosomal protein S4, X isoform) | 0.90 |
| ERPPNPIEFLASYLLK(gl)NK | Q9C005 | DPY30 (Protein dpy-30 homolog) | 0.86 |
| SIQLDGLVWGASK(gl)LVPVGYGIR | P29692 | EF1D (Elongation factor 1-delta) | 0.78 |
| QLMTLENK(gl)LK | Q7L7X3 | TA0K1 (Serine/threonine-protein kinase TA01) | 0.75 |
| TYVDPHTYEDPNQAVLK(gl)FTTEIHPSCVTR | P29317 | EPHA2( Ephrin type-A receptor 2) | 0.61 |
Acknowledgments
The authors acknowledge funding support from the following sources: Bonnie J. Addario Lung Cancer Foundation, NIH T32 2T32HL007185-36 (to C.M.B.); NIH Director’s New Innovator Award, NIH R01 CA169338, NIH K08 CA154787, Howard Hughes Medical Institute, Doris Duke Charitable Foundation, American Lung Association, Sidney Kimmel Foundation for Cancer Research, Searle Scholars Program, California Institute for Quantitative Biosciences (to T.G.B), NIH R01 GM107671 (to S.B. and N.K.), and the Li-Ka Shing Foundation (to S.B, N.K., and T.G.B). The authors thank Katerina Politi and Harold E. Varmus for generously providing CC10-rtTA and TetO-EGFRL858R transgenic mice.
K.B., T. F., and J.M. are employees of Rel-MD, Inc. T.G.B. has received compensation as a consultant for Driver Group and Novartis and is a recipient of a research grant from Servier.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Kao PN. Cyclosporin A-sensitive calcium signaling represses NFkappaB activation in human bronchial epithelial cells and enhances NFkappaB activation in Jurkat T-cells. Biochem Biophys Res Commun. 1997;234:424–431. doi: 10.1006/bbrc.1997.6658. [DOI] [PubMed] [Google Scholar]
- Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast cancer research : BCR. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, Moonsamy P, Dahlman K, Miller VA, Costa C, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- D'Angelo SP, Pietanza MC, Johnson ML, Riely GJ, Miller VA, Sima CS, Zakowski MF, Rusch VW, Ladanyi M, Kris MG. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol. 2011;29:2066–2070. doi: 10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J, Butaney M, Shimamura T, Sholl L, Ivanova EV, et al. Reactivation of ERK Signaling Causes Resistance to EGFR Kinase Inhibitors. Cancer Discov. 2012;2:934–947. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre C, Mimura N, Bobb K, Kong SY, Gorgun G, Cirstea D, Hu Y, Minami J, Ohguchi H, Zhang J, et al. Dual inhibition of canonical and non-canonical NF-kappaB pathways demonstrates significant anti-tumor activities in multiple myeloma. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg SB, Supko JG, Neal JW, Muzikansky A, Digumarthy S, Fidias P, Temel JS, Heist RS, Shaw AT, McCarthy PO, et al. A phase I study of erlotinib and hydroxychloroquine in advanced non-small-cell lung cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2012;7:1602–1608. doi: 10.1097/JTO.0b013e318262de4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib AA, Chatterjee S, Park SK, Ratan RR, Lefebvre S, Vartanian T. The epidermal growth factor receptor engages receptor interacting protein and nuclear factor-kappa B (NF-kappa B)-inducing kinase to activate NF-kappa B. Identification of a novel receptor-tyrosine kinase signalosome. J Biol Chem. 2001;276:8865–8874. doi: 10.1074/jbc.M008458200. [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Sun SC. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Molecular and cellular biology. 1999;19:7088–7095. doi: 10.1128/mcb.19.10.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hu J, Nakano H, Sakurai H, Colburn NH. Insufficient p65 phosphorylation at S536 specifically contributes to the lack of NF-kappaB activation and transformation in resistant JB6 cells. Carcinogenesis. 2004;25:1991–2003. doi: 10.1093/carcin/bgh198. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Clemens PR. Cellular caspase-8-like inhibitory protein (cFLIP) prevents inhibition of muscle cell differentiation induced by cancer cells. FASEB J. 2006;20:2570–2572. doi: 10.1096/fj.06-6347fje. [DOI] [PubMed] [Google Scholar]
- Kim SM, Kwon OJ, Hong YK, Kim JH, Solca F, Ha SJ, Soo RA, Christensen JG, Lee JH, Cho BC. Activation of IL-6R/JAK1/STAT3 Signaling Induces De Novo Resistance to Irreversible EGFR Inhibitors in Non-Small Cell Lung Cancer with T790M Resistance Mutation. Mol Cancer Ther. 2012;11:2254–2264. doi: 10.1158/1535-7163.MCT-12-0311. [DOI] [PubMed] [Google Scholar]
- Koeppen H, Yu W, Zha J, Pandita A, Penuel E, Rangell L, Raja R, Mohan S, Patel R, Desai R, et al. Biomarker Analyses from a Placebo-Controlled Phase II Study Evaluating Erlotinib {+/−} Onartuzumab in Advanced Non-Small-Cell Lung Cancer: MET Expression Levels Are Predictive of Patient Benefit. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Dorf ME. PKC phosphorylation of TRAF2 mediates IKKalpha/beta recruitment and K63-linked polyubiquitination. Mol Cell. 2009;33:30–42. doi: 10.1016/j.molcel.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Asthana S, Chan E, Bandyopadhyay S, Martins MM, Olivas V, Yan JJ, Pham L, Wang MM, Bollag G, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E748–757. doi: 10.1073/pnas.1320956111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardo F, Dragnev KH, Freemantle SJ, Ma Y, Memoli N, Sekula D, Knauth EA, Beebe JS, Dmitrovsky E. Evidence for the epidermal growth factor receptor as a target for lung cancer prevention. Clin Cancer Res. 2002;8:54–60. [PubMed] [Google Scholar]
- Mahul-Mellier AL, Pazarentzos E, Datler C, Iwasawa R, AbuAli G, Lin B, Grimm S. Deubiquitinating protease USP2a targets RIP1 and TRAF2 to mediate cell death by TNF. Cell Death Differ. 2012;19:891–899. doi: 10.1038/cdd.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, Zhou C, Su WC, Wang M, Sun Y, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528–538. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, Ariyaratne PN, Takahashi N, Sawada K, Fei Y, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- O' Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, Haynes NM, Tarlinton DM, Zhang JG, Belz GT, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature. 2009;461:659–663. doi: 10.1038/nature08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, Pan Y, Wang L, de Stanchina E, Shien K, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- Politi K, Fan PD, Shen R, Zakowski M, Varmus H. Erlotinib resistance in mouse models of epidermal growth factor receptor-induced lung adenocarcinoma. Dis Model Mech. 2009;3:111–119. doi: 10.1242/dmm.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puliyappadamba VT, Chakraborty S, Chauncey SS, Li L, Hatanpaa KJ, Mickey B, Noorani S, Shu HK, Burma S, Boothman DA, et al. Opposing effect of EGFRWT on EGFRvIII-mediated NF-kappaB activation with RIP1 as a cell death switch. Cell reports. 2013;4:764–775. doi: 10.1016/j.celrep.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A, Vaddi K, Liu P, Manshouri T, Li J, Scherle PA, Caulder E, Wen X, Li Y, Waeltz P, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood. 2010;115:3109–3117. doi: 10.1182/blood-2009-04-214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam SS, Spigel DR, Chen D, Steins MB, Engelman JA, Schneider CP, Novello S, Eberhardt WE, Crino L, Habben K, et al. Randomized phase II study of erlotinib in combination with placebo or R1507, a monoclonal antibody to insulin-like growth factor-1 receptor, for advanced-stage non-small-cell lung cancer. J Clin Oncol. 2011;29:4574–4580. doi: 10.1200/JCO.2011.36.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskatov JA, Meier JL, Puckett JW, Yang F, Ramakrishnan P, Dervan PB. Modulation of NF-kappaB-dependent gene transcription using programmable DNA minor groove binders. Proc Natl Acad Sci U S A. 2012;109:1023–1028. doi: 10.1073/pnas.1118506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard P, Ernest I, Houbion A, Art M, Le Calvez H, Raes M, Remacle J. Development of a sensitive multi-well colorimetric assay for active NFkappaB. Nucleic acids research. 2001;29:E21. doi: 10.1093/nar/29.4.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002003. 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, Kraemer O, Himmelsbach F, Haaksma E, Adolf GR. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. The Journal of pharmacology and experimental therapeutics. 2012;343:342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Beraud C, Ballard DW, Greene WC. Autoregulation of the NF-kappa B transactivator RelA (p65) by multiple cytoplasmic inhibitors containing ankyrin motifs. Proc Natl Acad Sci U S A. 1994;91:1346–1350. doi: 10.1073/pnas.91.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, Ohashi K, Janjigian YY, Spitzler PJ, Melnick MA, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Litzow MR, Pardanani A. Long-term outcome of treatment with ruxolitinib in myelofibrosis. N Engl J Med. 2011;365:1455–1457. doi: 10.1056/NEJMc1109555. [DOI] [PubMed] [Google Scholar]
- Timofeeva OA, Tarasova NI, Zhang X, Chasovskikh S, Cheema AK, Wang H, Brown ML, Dritschilo A. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc Natl Acad Sci U S A. 2013;110:1267–1272. doi: 10.1073/pnas.1211805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov. 2013;3:1404–1415. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, Riely GJ. Analysis of Mechanisms of Acquired Resistance to EGFR TKI therapy in 155 patients with EGFR-mutant Lung Cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lee JC, Lin L, Olivas V, Au V, Laframboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.