Abstract
Uncovering the relationships between peptide and protein sequences and binding properties is critical for successfully predicting, re-designing and inhibiting protein-protein interactions. Systematically collected data that link protein sequence to binding are valuable for elucidating determinants of protein interaction, but are rare in the literature because such data are experimentally difficult to generate. Here we describe SORTCERY, a high-throughput method that we have used to rank hundreds of yeast displayed peptides according to their affinities for a target interaction partner. The procedure involves fluorescence-activated cell sorting (FACS) of a library, deep sequencing of sorted pools, and downstream computational analysis. We have developed theoretical models and statistical tools that assist in planning these stages. We demonstrate SORTCERY’s utility by ranking 1026 BH3 peptides with respect to their affinities for the anti-apoptotic protein Bcl-xL. Our results are in striking agreement with measured affinities for 19 individual peptides with dissociation constants ranging from 0.1 to 60 nM. High-resolution ranking can be used to improve our understanding of sequence-function relationships, and to support the development of computational models for predicting and designing novel interactions.
Keywords: deep sequencing, yeast display, high-throughput assay, protein interaction, Bcl-2 family
Introduction
Understanding the relationships between protein sequences and their functions is a fundamental objective of protein science. Our ability to map these relationships has improved with advances in technology. Until recently, the ability to decode information from experiments that characterize protein function was limited by the need to clone and/or individually sequence every gene of interest at relatively low throughput. Next-generation sequencing has changed this, and a number of important publications describe techniques that combine phenotypic screening and deep sequencing to investigate how protein sequence influences structure, folding, binding or organism growth/fitness [1–10]. Araya and Fowler have written a good review of recent advances [11]. Generally, the experimental approach involves constructing a library of many different mutant variants of a protein of interest. The library is then screened/selected for some property or function. The retained library pool is sequenced, and features of sequences that are observed with high frequency are implicated as important for the relevant property. In this introduction, we discuss applications of this approach to the problem of determining protein interactions with a target.
Interaction systems that have been subjected to a screening-plus-sequencing approach include PDZ domain peptide ligands [4, 5], Pin WW domain peptide ligands [6], influenza haemaglutinnin inhibitors [7], LYN kinase interaction partners [8], computationally designed digoxigenin binders [9] and Bcl-2 type receptor/BH3 complexes [10]. Experiments varied in library size (~1,000 to ~ 600,000 members) as well as in the type of screening used to detect binding (phage display, yeast display, ribosome display, bacterial two-hybrid). These studies are exciting milestones that dramatically expand the amount of data available to describe protein interactions. Yet, it is important to consider what information the data from various interaction screens contain and how it can be used. A standard approach has been to quantify the enrichment of each sequence or point mutation among library members classified as binders, relative to the unselected library, and to use this as a proxy for affinity. This may be problematic, as it relies on adequate deep sequencing of the starting library and bias-free amplification of sequences throughout screening and sample preparation. In fact, Derda et al. found that the relative abundance of phage displayed peptides could be significantly skewed if phages were amplified after a selection step [12]. McLaughlin et al. have reported data that support an impressive correlation of enrichment scores with binding affinities [5], but the appropriateness and resolution of new methods for affinity determination is not well established.
Recently, Kinney et al. pioneered a detailed approach to the screen-and sequence scheme and applied it to measure protein-DNA interactions [13]. Adopting the expression level of GFP as an indicator of transcription factor binding strength, they employed fluorescence activated cell sorting (FACS) to sort a bacterial library of ~20000 mutant lacZ-promoters with different activities into pools and decoded these by deep sequencing. A maximum-likelihood computational routine transformed the sequencing data into a position specific scoring matrix that described the DNA-binding affinity of the transcription factor. In a similar approach, Sharon et al. monitored the affinity of transcription factors for hundreds of mutant yeast promoters that were coupled to YFP and derived a ranking of transcription factor activities [14].
Sharon et al. and Kinney et al. employed multi-bin sorts that increased the resolution of their experiments (i.e. the ability to distinguish between two different dissociation constants or equivalent measures of affinity) and permitted the analysis of frequency distributions rather than the more difficult to interpret enrichment values. However, issues remain to be addressed. First, only the expression of fluorescent protein was monitored in the protein-DNA binding studies, without accounting for variations in transcription factor levels that impact reporter gene expression. Prior work supports the importance of a correction. Liang et al. developed a two-color FACS screen for RNA gene regulatory devices [15]. One fluorescence signal reported the device activity, the other was a measure of basic transcription levels. This setup dramatically increased the resolution of the sorting scheme in comparison to a one-color strategy. Similarly, Dutta et al. gauged the stability of protein mutants by fragment reconstitution and yeast display [16]. They observed the expression and display of a mutant fragment with one fluorescence signal and the binding of a complementary fragment with another signal. Their findings suggested a correlation between the stabilities of the protein mutants and the ratio of the two fluorescence signals. Chao et al. showed qualitatively that a mixture of two yeast-displayed antibodies with very similar affinities for a target can be enriched for the stronger binder by FACS when expression levels are taken into account. Second, Kinney et al. [13] and Sharon et al. [14] considered averages of their detailed experimental information during computational analyses. They calculated position specific scoring matrices and mean expression values, respectively. Cooperative effects and signal variance may limit the accuracy of models derived with such assumptions.
High-throughput characterization of protein interactions will be most useful if it can deliver accurate estimates of affinity or affinity rankings. For example, such estimates could enable the construction of more accurate predictive models or could guide the refinement of protein designs [7]. We present a protocol that uses a rigorous sorting strategy in combination with downstream computational processing that returns a precise affinity ranking of individual sequences. Taking advantage of yeast-surface display, in which a signal resulting from a peptide binding to a protein can be normalized by the expression level of that peptide, we developed a theoretical framework to derive the expected signals for binders of different affinities. Experimental sorting using FACS, plus library sequencing, yielded coarse-grained signal distributions for ~1000 peptide-displaying clones in a single experiment. Computational processing generated a global ranking of peptide affinities, and our theoretical model allowed a detailed statistical analysis of sources of error in the final results. Because existing methods are already capable of discerning strong from weak and non-binders, we have focused on discriminating tight binders within a 500-fold range of affinities (0.1 nM-60 nM). Accurate data in this regime may aid in the design of very strong binders that can be important therapeutic and diagnostic agents [17–19]. We conducted our study using a small library of about 1000 yeast displayed BH3 peptides that bind to Bcl-xL, a key regulator of apoptosis. High-affinity binders of Bcl-xL are of great interest due to their potential for diagnosing or surmounting apoptotic blockades in numerous cancers [20–22].
Results
Our high-throughput method called SORTCERY analyzes the binding of yeast displayed peptide ligands to a target molecule and returns a ranking for the affinities of all considered ligands. The multi-step procedure involves sorting a yeast displayed library into several bins, deep-sequencing all bins and analyzing the resulting data (see figure 1). Our optimized sorting strategy is based on a theoretical model relating two fluorescence signals to the peptide-target dissociation constant. The model establishes how to set cell-sorting gates or bins for an optimal separation of ligands of varying affinities. The post-sequencing data analysis involves constructing frequency distributions over bins, and then exhaustively comparing pairs of distributions. Finally, the pairwise comparisons are combined into a global ranking of affinities. We have tested the performance of SORTCERY on a yeast display library and shown that we can produce very accurate affinity rankings of peptide ligands. More specifically, we have investigated the interaction between mutant Bim and Puma BH3 peptides and the anti-apoptotic receptor Bcl-xL. Bcl-xL plays a critical role controlling cell death by interacting with the helical Bcl-2 homology 3 (BH3) regions of many pro-apoptotic proteins. We monitored the interaction of yeast-surface displayed BH3 peptides with soluble Bcl-xL. Tags on the BH3 variants and the Bcl-xL protein were used to quantify expression and binding levels with fluorescently labeled antibodies, similar to Dutta et al. [23]. A more complete description of the experimental setup is given in the Methods section.
Figure 1.
SORTCERY ranks yeast displayed peptide ligands according to their affinities for a target. A) The experimental setup comprises a yeast displayed peptide ligand and a soluble target molecule. The expression level of the ligand and the binding level of the target are monitored by fluorescence labeled antibodies that recognize respective tags. These two fluorescence signals enable two-color sorting by FACS according to the target affinities for the displayed ligands. The schematic depicts an example with twelve areas in signal space (FACS gates) that correspond to twelve different affinity ranges. B) A SORTCERY experiment consists of five steps. First, a yeast displayed library of peptides is sorted by FACS into a set of gates that correspond to different ranges of affinities. The schematic shows four clonal populations each displaying a peptide of different binding strength (green, red, brown, blue). Second, each sorted pool is deep sequenced. Third, frequency distributions over the FACS gates are constructed for each observed sequence. Fourth, pairs of distributions are compared to determine the probability that one peptide binds stronger than the other. Fifth, a global ranking is derived from the pairwise probabilities.
Theoretical model relating FACS profiles to binding
In this section we present a theoretical model that relates the dissociation constant of a protein-peptide complex to observed fluorescence signals for expression and binding in a flow cytometric measurement. We define a one-dimensional “axis of affinity” in two-dimensional fluorescence signal space that facilitates accurate discrimination between yeast displayed peptides of different affinities. Based on this concept, we present a strategy for sorting peptide ligands according to their affinities for a target. We also provide data for individual BH3 clones that support the predictions of our theory.
Results of the theoretical model
We derived our model with the following assumptions: First, binding occurs as a two-state process; i.e. yeast displayed ligands exist either in a heterodimer complex or in the unbound state, there are no intermediate stages. Second, the solution of target molecules is not depleted due to binding, i.e. the concentration of unbound target is approximately equal to the total target concentration. Note that yeast cells were incubated in sufficiently large volumes of Bcl-xL solution during experiments to ensure this condition was satisfied. Third, fluorescence signals are linearly related to the concentration of yeast-displayed ligand and bound target, respectively. Fourth, background fluorescence is small in comparison to binding and expression signals. This should generally hold true, since FACS machines record signals over several orders of magnitude. Fifth, the equilibrium between free and bound ligand is not disturbed before the measurement. Although we have derived the theoretical model under this assumption, SORTCERY may provide valuable data even when it is not strictly satisfied (see Expanded View).
Our derivation starts with four equations describing the mass action law for complex dissociation (equation 1), the conservation of ligand molecules (equation 2), and the relations between fluorescence signals and corresponding molecular concentrations (equations 3, 4).
| (1) |
| (2) |
| (3) |
| (4) |
Here [target], [ligand], [complex] and [ligand]total signify the concentrations of free target, free ligand, target-ligand complex and total ligand molecules, respectively. KD is the dissociation constant of the target-ligand complex. Fe is the fluorescence signal for ligand expression, Fb is the fluorescence signal for target binding. Fa signifies autofluorescence and fluorescence from non-specific binding. Combining equations 1 to 4, and considering the logarithmic form of the expression, because FACS data are generally displayed this way, yields:
| (5) |
The constant c arises from the proportionality constants ce and cb and depends on experimental parameters such as fluorescence yields and PMT voltages of the FACS machine. If the contribution of Fa to the binding signal is small eq. 5 simplifies to
| (6) |
This mathematical model predicts several features for FACS profiles of individual peptide ligands, some of which are illustrated in figure 2. First, at a given target concentration, the theoretical, idealized FACS profile (log(Fb) vs. log(Fe) signal curve) is a line with a slope of 1. Second, a change in affinity (KD) at constant target concentration shifts the line along a perpendicular axis; i.e. ligands of different affinities exhibit different y-intercepts, but the same slope. Third, a forbidden area exists where no well-behaved FACS profiles are observed. This area corresponds to binding of target in excess of displayed ligand. Fourth, at high target concentrations, FACS profiles converge at the boundary of the forbidden region, and the resolution between ligands with different affinities becomes poor. This trend is illustrated in figure 2C (for ligands with KD = 0.4 and 4 nM). We also note that, at too low a target concentration, the binding signals of FACS profiles fall below a threshold where equation 6 no longer applies (figure 2A for 40 and 400 nM ligands). Proper choice of target concentration is thus important for optimally resolving different KD intervals.
Figure 2.
Predicted relation between expression and binding fluorescence signals and dissociation constants (KD). Panels show the expected distribution of binders at target concentrations of A) 0.02 nM, B) 1 nM and C) 50 nM. The red lines correspond to binders with dissociation constants of 0.4 nM, 4 nM, 40 nM and 400 nM. The dark gray area in the upper left corner indicates a “forbidden region” that corresponds to binding of target molecules to the cell surface in excess of ligand expression. In the light gray area Fa contributes significantly to the binding signal and the linear assumption (eq. 6) does not hold. The orange arrow indicates an axis perpendicular to the red lines. Projection of the lines on this axis permits an estimate of affinities.
The fact that FACS profiles are predicted to be linear and exhibit parallel shifts to each other is of special interest. It permits the definition of an “axis of affinity”. This axis has the orientation of the orange arrows in figure 2. Projection of a theoretical FACS profile onto this axis results in a single point and such a “coordinate of affinity” is directly related to the binding strength of a ligand. Hence, different ligands can be directly compared in this one-dimensional coordinate system, rather than dealing with the more complicated two-dimensional system of fluorescence signals. The coordinate of affinity, a, can be expressed in terms of the binding and expression signals:
| (7) |
It is instructive to consider the dissociation constant as a function of the coordinate of affinity:
| (8) |
According to this expression, an infinitely tight binder (KD = 0) would have a coordinate of affinity of . Ligands with ever larger KDs are found at ever smaller values of a. Hence the relevant range of the axis of affinity is . The forbidden region comprises the interval . The free energy of dissociation, ΔG, is related to the coordinate of affinity via
| (9) |
There are two important regimes. When then . In this regime ΔG asymptotically goes to ∞ and even minute changes in a lead to large changes in ΔG (see supplemental figure 1). On the other hand, when then and
| (10) |
Note that it is possible to shift binding free energies into the better resolved linear region by adjusting the target concentration accordingly.
Testing the validity of the theoretical model
We tested three features of the theoretical model by recording FACS profiles for Bcl-xL binding of individual yeast displayed BH3 peptides. First, we considered the expected shape and orientation of FACS profiles. Experimental FACS profiles will not be infinitesimally narrow lines, due to measurement error. However, we expected elongated FACS profiles with a major principal component with a slope of 1. Second, as [target] → ∞, eq. 6 becomes log(Fb) = log(Fe)−log(c). Hence, at high Bcl-xL concentrations, binding will saturate and all BH3 clones should exhibit FACS profiles with very similar coordinates of affinity. Third, our model predicts the relationship between the dissociation constant and the coordinate of affinity. Rearrangement of equation 8 shows that log and a should be linearly correlated with a slope of .
We investigated whether FACS profiles of BH3 peptides exhibited the predicted characteristics. We found that FACS profile shapes were sensitive to growth conditions, but, when these were optimized, we could robustly obtain experimental data in good agreement with the theoretical predictions (see Methods). BH3-displaying clones that were incubated with either 1 nM or 500 nM Bcl-xL had elongated profiles (figures 3 A and B) and autofluorescence/non-specific binding was relatively low (compare the binding-signal for the non-expressing population, at lower left, to the expressing population). We recorded the FACS profiles for a number of clones in duplicate on twelve separate days, conducting a total of 24 measurements on each. At 1 nM Bcl-xL, all profiles exhibited a first principal component with a slope close to 1 (figure 3A). This was true even in cases where not all recorded signals were much stronger than the background fluorescence (lower right panel in figure 3A), which violates one of our assumptions in the derivation of equation 6. We observed the same pattern for other Bcl-2 family proteins binding to BH3 peptides (see supplemental figure 2), indicating the possibility of general applicability. We were also able to verify our second prediction: FACS profiles for clones of different affinities superimposed at 500 nM Bcl − xL (figure 3B). Our test of the third prediction involved a linear fit of log vs. a for 11 individual BH3 clones. KD values were determined by titrating Bcl-xL and monitoring the binding signal (see Methods). The linear fit yielded a slope of −1.19, with 95% confidence limits of [−1.41 , −0.96]. The R2 value was 0.94 (p-value < 10−6). When we constrained the slope to we obtained an R2 value of 0.93. Both constrained and unconstrained fits are shown in figure 3C. The data show a strong linear correlation, even though our estimate of the coordinates of affinity was relatively crude (see Methods). The expected slope of lies just beyond the 95% confidence limit of the best-fit slope. If this deviation did not arise from experimental noise, but reflects a true discrepancy between experiment and theory, then our model predicts a somewhat stronger change of KD with a than is actually the case and thus underestimates the maximum possible resolution. However, the good linear fit indicates that the overall relation between KD and a is well captured. In summary, all measurements of individual BH3 clones supported crucial features of the theoretical model.
Figure 3.
Experimental tests of model predictions. A) FACS profiles of four different BH3-displaying yeast clones. Dissociation constants (KD), mean slopes (ms) and corresponding standard deviations (stdv) of the first principal components from 24 independent measurements, determined using expressing cells with log(expression signal) > 3, are indicated in each panel. Cells with log(expression signal) < 3 are non-expressing. Red lines indicate the orientation of the first principal component of the expressing population. In the case of the weakest binder, a small set of points in the baseline was retained that biased the principal component towards smaller slopes. All clones were incubated with 1 nM Bcl-xL. B) FACS profiles of displayed BH3 peptides with different affinities converge under saturating conditions. The blue profile in the upper and lower panel is from the same measurement of a BH3-displaying clone with a dissociation constant of 10 nM. In the upper panel the red profile corresponds to a clone with a dissociation constant of 2.4 nM. In the lower panel the red profile is from a clone with a dissociation constant of 0.06 nM. All profiles are superimposed, as expected for a Bcl-xL concentration >> KD ([Bcl-xL] = 500 nM). C) Plot of log versus the coordinate of affinity for 11 different BH3 sequences. Data points indicate mean values of multiple experiments. We determined coordinates of affinity in duplicate and dissociation constants at least in duplicate. The red line indicates an unconstrained linear fit. The green line corresponds to a constrained linear fit with a slope of . D) Probability densities for individual BH3-displaying clones over the axis of affinity. Dissociation constants of the BH3 peptides are: black 0.06 nM, red 1 nM, green 10 nM, blue 60 nM. The vertical lines indicate the coordinates of affinity of the gate delimitations that run perpendicular to the axis of affinity.
Theory based strategy for sorting
Our theoretical model can be used to guide the selection of FACS gates for sorting displayed ligands according to their affinity for a target. The position, orientation and size of a gate are determined by the gate’s delimitations. Our model predicts that an optimal gate delimitation should include two boundaries perpendicular to the axis of affinity, so that the gate will exclusively collect ligands in a specific affinity regime (see figure 3D and red lines in supplemental figure 3).
The width of the gates along the axis of affinity impacts the resolution that is attainable. Our model makes predictions about the relation of dissociation free energy to the coordinate of affinity (see equation 9) and can thereby guide user choices. In this study, our emphasis was on discriminating ligands over a range of dissociation free energies. Because dissociation free energy is in first approximation a linear function of the coordinate of affinity (equation 10), we decided to use gates of equal width. The approximation only breaks down for extremely tight binders, and good resolution in this regime would require ever narrower gates (see supplemental figure 1). The cost to benefit ratio to attain better resolution for this small interval on the axis of affinity appears rather high.
A user should choose gates with the help of a few standards. Converting FACS profiles into densities over the axis of affinity (see Methods), the user can determine the interval on the axis he/she wants to operate on. For example, figure 3D shows four standards from our experiment that span affinities of ~ 0.1 nM to 60 nM and define the signal space of interest. The number of gates to be placed on this interval depends on the required resolution and feasibility in cost and time. A user can gauge the resolution by referring to the standards. Supplmenetal figure 4 shows probabilities to identify the stronger vs. weaker binder in pairwise comparisons of 11 standards in a three- vs. a twelve-gate setup. The twelve-gate setup shows significantly better performance, although the three-gate setup can also yield valuable information.
Post deep-sequencing data analysis Procedure for generating a global rank order of peptide ligand affinities
We developed a three-step computational protocol that derives a global affinity ranking of peptides by analyzing deep-sequencing data for samples from our sorting scheme. First, we constructed frequency distributions over the gates for each observed BH3 sequence, generating coarse-grained distributions over the axis of affinity. Second, for every pair of BH3 peptides, A and B, we calculated the probability that A is a better binder than B. In our scheme, this corresponds to the probability that A will be observed at a larger coordinate of affinity than B. To evaluate this, we computed the normalized frequency fhX with which a peptide X was observed in gate h. This frequency can be regarded as the empirical probability of observing peptide X in gate h. Then the probability that peptide A hits gate i and peptide B hits a gate that corresponds to smaller coordinates of affinity than i is:
| (11) |
Hence the probability that A is the stronger binder can be obtained by summing over all possible gates i:
| (12) |
We calculated all probabilities for all possible combinations of peptides and stronger binders using equation 12, i.e. p(A > B) and p(B > A) for all possible combinations of A and B. Third, we derived a global ranking of affinities from the calculated pairwise probabilities. The set of pairwise probabilities corresponds to a directed graph with weighted edges. Each vertex represents a BH3 peptide and the weight of an edge leading from vertex A to vertex B is the probability that peptide A binds with higher affinity than peptide B. The linear subgraph with the maximum product of edge weights that does not include any contradictory orderings of affinities provides the global ranking. In graph theoretical terms this solution would be equivalent to finding the minimum feedback arc set in a tournament. A simple yet effective approximate algorithm for this task has been described in [24].
Affinity ranking of 1026 BH3 peptides with SORTCERY
We used SORTCERY to sort and rank a library composed of Bim and Puma BH3 peptide mutants (see Methods). The library consisted of high-affinity ligands for Bcl-xL. The dynamic range of our experiment was about 500-fold, spanning a KD interval from ~0.1 nM to ~60 nM. Cell sorting was carried out according to the scheme described above and in the methods section. We deep-sequenced all sorted pools and analyzed the data for 1,026 unique sequences. We gauged the quality of the data according to copy numbers for each unique BH3 sequence and widths for the corresponding frequency distributions (see Methods). supplemental figure 5 shows how both measures were distributed in our data set, and supplemental figure 6 shows representative frequency distributions. About 75% of all sequences had a copy number > 1, 100 and a distribution width narrower than that shown in supplemental figure 6C. The final result of our data analysis was a global rank order of all 1026 observed BH3 sequences with respect to their binding strengths. We bootstrapped the deep-sequencing data and recalculated the global ranking to gauge uncertainties in our results. The average deviation of a BH3 sequence from its original rank was ≤ 23.5 in 95% of all bootstrap samples. This is a shift of < 2.3% of the total ranking.
We tested the accuracy of the global ranking for a subset of 19 BH3 peptides. These peptides were chosen to reflect the variety among sequences in the library as well as differences in the quality of the observed data (see table 1). We measured the dissociation constants of yeast displayed sequences by titrating cells with increasing concentrations of Bcl-xL and monitoring binding by flow cytometric analysis (supplemental figure 7). The relation between measured dissociation constants and our predicted ranking of affinities is shown in figure 4A. The predicted ranking shows outstanding agreement with individual measurements within experimental error (on KD values) and 95% bootstrap confidence intervals (on ranks).
Table 1.
BH3 sequences chosen from the global ranking for individual measurements of dissociation constants by titration of yeast clones
| variable region | sequence context a) |
reindexed ranking (original rank index) |
dissociation constant ± std dev (nM) b) |
observed copy number |
width parameter c) |
|---|---|---|---|---|---|
| VGAQLRRIADDV | P | 1 (2) | 0.26 ± 0.06 | 1468 | 0.47 |
| VGAQFRRIADDI | P | 2 (3) | 0.35 ± 0.09 (0.07 ± 0.1) | 1324 | 0.30 |
| VAQELKRIGDEF | B | 3 (10) | 0.18 ± 0.05 (0.1 ± 0.3) | 1452 | 0.40 |
| VAQELRRYGDEY | B | 4 (35) | 2.4 ± 0.3 | 2245 | 0.61 |
| IGAQFGRFADDF | P | 5 (41) | 1.3 ± 0.3 | 773 | 0.72 |
| FGAQLNRIAEDF | P | 6 (136) | 2.1 ± 0.2 | 6761 | 0.52 |
| IGAQLRRMADDV | P | 7 (186) | 0.59 ± 0.09 | 1216 | 0.28 |
| AAQELRRYGDEY | B | 8 (250) | 4 ± 2 | 3034 | 0.57 |
| VAQELRRIGDEV | B | 9 (272) | 3.3 ± 0.6 | 3921 | 0.35 |
| YGAQLDRYAQDF | P | 10 (360) | 3.9 ± 0.9 | 1025 | 0.47 |
| NGAQLKRIADDY | P | 11 (427) | 5 ± 2 (7 ± 1) | 1232 | 0.57 |
| AGAQLHRFADDY | P | 12 (546) | 30 ± 9 | 949 | 0.56 |
| YAQELQRYGDET | B | 13 (565) | 5.2 ± 0.5 | 3172 | 0.54 |
| FGAQLGRVASDF | P | 14 (626) | 9 ± 2 | 2241 | 0.53 |
| DAQELKRFGDEF | B | 15 (741) | 11 ± 1 (13 ± 2) | 1384 | 0.45 |
| YAQEIGRNGDEF | B | 16 (797) | 11 ± 3 | 3282 | 0.45 |
| VGAQFHRFANDF | P | 17 (878) | 16 ± 4 | 26372 | 0.42 |
| AAQELQRNGDEY | B | 18 (940) | 40 ± 14 (31 ± 6) | 12302 | 0.54 |
| IGAQLDRMADDL | P | 19 (989) | 60 ± 15 (290 ± 60) | 4215 | 0.58 |
“B” indicates Bim, “P” indicates Puma
Errors are the standard deviation of KD values from four independent measurements. Values without parentheses indicate KD values from titration of yeast clones. Dissociation constants from fluorescence polarization experiments are given in parentheses, where available.
see Methods for explanation
Figure 4.
Ranking by SORTCERY agrees well with ranking based on individual measurements. In both panels global SORTCERY ranks were re-indexed from 1 to 19. A) Dissociation constants versus corresponding predicted rankings for 19 out of 1026 BH3 sequences. Error bars for the rankings indicate the ranks that each BH3 peptide assumed within 95% confidence during the bootstrap runs. Error bars for the KD values are the standard deviation over four independent measurements for each BH3 peptide. B) Comparison of rankings obtained from individual titrations with the rankings obtained from SORTCERY.
To judge the reproducibility of our protocol, we repeated the SORTCERY experiment (including sorting, deep sequencing and data analysis) and re-determined the ranking for the 19 clones. Because the clone with rank 12 in the original experiment (table 1, figure 4) showed very low deep sequencing coverage in the repeat experiment, we relaxed our quality criteria for coverage so that this clone could be included in the data analysis (see Methods). This led to a data set of 5518 unique sequences. Despite the larger number of sequences, and possibly worse statistics due to the low copy number of many sequences, SORTCERY again ranked the 19 chosen clones in a very similar fashion (supplemental figure 8). We also compared the ranking order of all sequences common to the original and the repeat data sets. These 973 sequences were ranked very similarly in both analyses (supplemental figure 9): the average difference in ranks was ~69, or ~7% of the size of the total subset.
In both data sets we observed the largest bootstrap uncertainties in ranks for strong binders. These uncertainties can be explained by sampling noise during cell sorting (see Error Analysis in the Expanded View). In fact, we observed uncertainties exactly where our theoretical model predicted resolution to decline (supplemental figure 1). The least accurately ranked sequences in the original data set (at ranks 7 and 12 in figure 4A) were associated with no bootstrap uncertainty in rank. The deviation from their “true” rank, therefore, must have originated from bias rather than sampling noise. The least accurately ranked sequence in the repeat SORTCERY experiment (rank 10 in the repeat; rank 12 in the original; rank based on KD was 17) showed only small bootstrap error, indicating that in this data set ranking of this sequence also was biased.
In our error analysis in the Expanded View, we show that bias leading to ranking errors most likely originated from the deep sequencing step. Quantitative deep sequencing analyses are known to be affected by bias [25, 26]. Our overall ranking was not dramatically affected by deep sequencing bias, but a correction algorithm for peptide libraries could be beneficial. Note that both outlier sequences in the original data set exhibited a low-value quality parameter (rank 7 had a particularly broad distribution and rank 12 a low copy number, see table 1). The quality parameters could serve as a warning of possible bias.
Yeast display experiments are often performed to identify peptide ligands for applications in solution. To check whether the SORTCERY ranking for displayed ligands accurately reflected the affinities of soluble peptides, we chose 6 of the 19 investigated BH3 peptides with KD values of different orders of magnitude. We measured the affinities for 28-residue peptides binding to Bcl-xL by competitive fluorescence polarization experiments. Affinity rank orders for these six peptides were identical as assessed by yeast display and fluorescence polarization, and measured KD values agreed within the margins of error in 4 out of 6 examples (supplemental figures 10, 11 and table 1).
Sequence features in the sorted affinity regimes
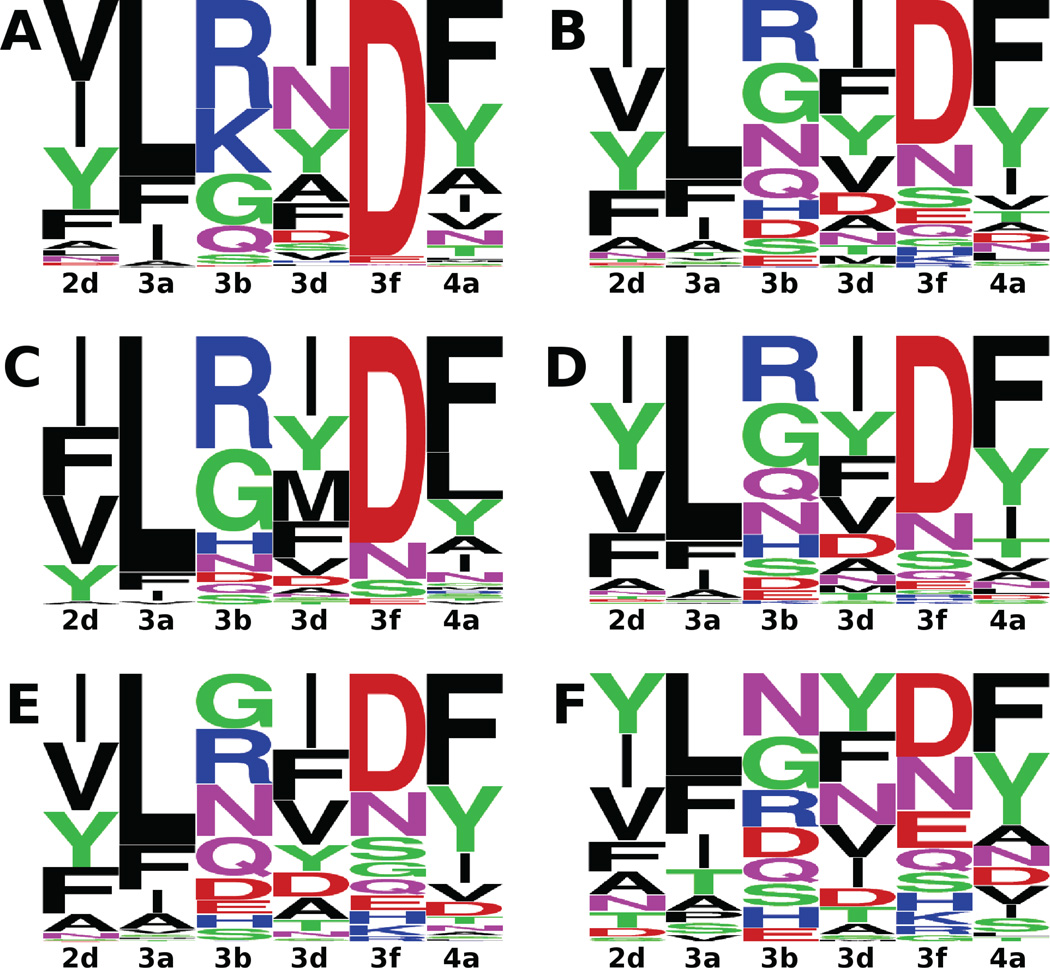
SORTCERY yields detailed, high resolution information about peptide binding that can potentially be linked to sequence features. We first created sequence logos from all the unique Bim and Puma sequences in our ranking (see figure 5). The 310 observed Bim mutant sequences show a highly conserved aspartate in position 3f, consistent with strong conservation of this residue in natural and engineered BH3 peptides. Among the 716 Puma mutant sequences, however, we detected significant sequence variation at position 3f. We examined Puma sequence features in different affinity regimes, dividing our data into four bins: peptides between ranks 1 and 185 (KD values ≲ 1 nM), peptides between ranks 186 and 594 (1 nM ≲ KD ≲ 10 nM), peptides between ranks 595 and 908 (10 nM ≲ KD ≲ 40 nM) and peptides between ranks 909 and 1026 (KD ≳ 40 nM). Sequence logos for all four bins are shown in figure 5 and show that Asp at 3f is highly conserved among very strong binders (KD ≲ 1 nM), with only three other possible alternatives. In lower affinity bins, sequence variety increases at 3f. A similar trend exists for Leu in position 3a. We observed more subtle changes with KD in the residue composition of the other four positions.
Figure 5.
Sequence logos for different subsets of the observed sequences in the SORTCERY experiment. A) and B), Bim and Puma variant sequences. C), D), E) and F), sequence logos for Puma variant sequences in different KD regimes. C) KD ≤ 1 nM, D) 1 nM < KD < 10 nM, E) 10 nM ≤ KD ≤ 40 nM, F) KD > 40 nM.
Comparison to one-gate experiment
Engineering binding peptides using standard library selection methods involves enrichment steps. Deep sequencing data from an enriched pool can be analyzed by calculating enrichment values for each unique sequence [6, 8], or positional enrichment values [5–9]. PSSMs built from residue frequencies in enriched populations have also been employed [10]. All of these scores have been assumed to reflect affinities. For comparison to other methods, we carried out a series of single-gate FACS experiments, mimicking common yeast-display screens [23, 27]. We chose either the highest-affinity gate or combined two to twelve of the highest-affinity gates from our SORTCERY scheme (supplemental figure 3). We deep sequenced each sorted pool and determined scores for the 19 experimentally characterized clones in table 1 according to sequence enrichment, positional enrichment, or a PSSM (see Methods). We then ranked the 19 clones and compared the ranking to the ranking based on directly measured affinities (in analogy to our comparison of the SORTCERY ranking to the experimental ranking in figure 4B). Plots of standard deviations between these two rankings vs. the chosen number of combined gates are shown in figures 6A, C and E. For all three scoring methods, the number of chosen gates significantly influenced the quality of the derived ranking, indicating that a successful experiment may require prior information on how to set the gate. Interestingly, the PSSM method exhibited by far the best ranking and the least variation with the number of chosen gates. Yet, even the lowest observed standard deviation (4.2) was two-fold higher than the standard deviation observed for SORTCERY (1.9). We investigated the performance for the best gating choice more closely for each scoring method. Figure 6 B, D and F shows plots of the experimentally determined KD values vs. the ranking indices that resulted from each scoring method. We observed a clear trend with the PSSM method, especially when Bim and Puma variants were considered separately. Nevertheless, SORTCERY’s accuracy is much higher. There are several examples where the PSSM approach cannot resolve KD values with an order of magnitude difference (figure 6F). The other two scoring methods were less successful at predicting relative affinities.
Figure 6.
Comparison of the ranking from individually measured KD values vs. rankings from different one-gate sorting schemes and analyses. A), C) and E) Standard deviations between the two types of rankings for different numbers of combined highest-affinty gates in the one-gate scheme. The standard deviation of the SORTCERY ranking from the ranking by individually determined KD values was 1.9. B), D) and F) Comparison of experimental KD values to ranks from the one-gate sorting methods. Black data points in panels B, D and F indicate Puma variants, gray data points indicate Bim variants. Error bars on the rankings indicate the ranks that each BH3 peptide assumed within a 95% confidence interval during a bootstrap analysis. Error bars for the KD values indicate the standard deviation over four independent measurements. A–B) Ranking by enrichment values using the 5 highest-affinity gates for data in panel B. C–D) Ranking by positional enrichment using the 4 highest-affinity gates for data in panel D. E–F) Ranking by PSSM scores using the 10 highest-affinity gates for data in panel F.
Discussion
Biophysical characterization of the binding of proteins and their mutational variants is typically conducted using low-throughput one-at-a-time analyses. Higher-throughput studies can provide qualitative information, and methods for obtaining higher resolution are being explored [11, 13, 14]. In this study we ranked 1026 BH3 sequences based on affinity for Bcl − xL over a dynamic range of dissociation constants from ~0.1 nM to ~60 nM in a single experiment. We gauged the effect of combinations of mutations in different sequence contexts (Bim and Puma based peptides), demonstrating that SORTCERY can accurately operate in a diverse sequence space. Indeed, SORTCERY’s resolution in this application was equivalent to the resolution obtained by individual measurements, and could in many cases distinguish two- to three-fold differences in the dissociation constants of peptide ligands. Individual measurements for a few members of the ranking can therefore provide very accurate estimates of the dissociation constants of all observed sequences. Alternative analyses applied to this problem showed relatively poor performance on the challenging ranking task.
Mutational analysis is a critical tool for understanding and re-engineering protein interactions. Over the past several decades, landmark alanine-scanning studies have led to an appreciation of the important roles of hot-spot residues in protein interfaces [28], and other mutational studies have provided insights into interfacial sequence tolerance, structural modularity, and positional site independence vs. cooperativity in binding [29, 30]. Such studies can now be scaled up dramatically using SORTCERY, without significant loss of resolution. The potential to probe combinations of mutations is particularly exciting in this respect. In the realm of engineering, introducing mutations to modulate stability, affinity or binding specificity is critical for industrial efforts in antibody design and affinity maturation, and is important for the exploration of alternative scaffolds. The use of SORTCERY to categorize large sets of mutations, and combinations of mutations, can accelerate designs efforts. Finally, we are especially excited about the potential of SORTCERY to provide semi-quantitative data for computational model building. Many groups have developed computational methods to predict the influence of mutations on protein binding [31]. But testing these methods relies on the availability of large experimental data sets, which so far are scarce. The data we report here provide a challenging prediction benchmark, and SORTCERY can generate many other tests of this variety. Data from SORTCERY experiments can also be incorporated directly into models, similar to the way experimental binding data have been used to build models of coiled-coil interactions and PDZ-ligand binding in the past [32, 33].
It should be possible to expand SORTCERY’s potential in several ways. For example, our theory predicts that changing the target molecule concentration will make it possible to collect data from different KD regimes. A combination of rankings could then yield information over a larger dynamic range with high resolution. Processing more cells in a SORTCERY experiment would make it possible to rank larger libraries. Finally, SORTCERY may have potential to identify highly specific binders. This could be achieved by creating several rankings for one library, each corresponding to the binding of a different target molecule.
Systems with high dissociation rates may pose a challenge for SORTCERY. Our experimental protocol was optimized to minimize target dissociation before sorting (see Methods). But very fast off-rates will lead to shifts on the axis of affinity. In such scenarios SORTCERY could still be a valuable procedure if dissociation rates correlate sufficiently with equilibrium constants. This would lead to consistent shifts in the binding signal and consistent shifts on the axis of affinity (as described in the supplemental material). Fast off-rates uncorrelated with equilibrium constants could cause SORTCERY to fail, as ligands with similar affinities would be assigned very different coordinates of affinity.
Although our study focused on the affinities of yeast displayed peptide ligands, SORTCERY’s potential applicability is more general. E.g. Chao et al. observed FACS profiles of yeast displayed antibodies that could be described by our theory and demonstrated that expression levels matter for optimal enrichment of high affinity antibodies by FACS [34]. SORTCERY could be a powerful tool to gather data for antibody design and engineering. Furthermore, in vivo gene regulation could be monitored with similar models as used here for binding profiles. In fact, Liang et al. reported a two-color screening method for RNA regulatory devices that exploits the linear relationship between a fluorescence signal for gene expression and a fluorescence signal for baseline transcription [15]. Liang et al. found this relationship empirically, without the formulation of any theoretical foundation. However, a model similar to the one derived here would describe their experiment. A variant of our model could also help gauge the stability of protein mutants. Dutta et al. developed a method that probes protein stability by fragment reconstitution and yeast display [16]. They distinguished between different stabilities with empirically determined, diagonally shaped gates similar to those observed in this study. SORTCERY-style procedures could, therefore, significantly increase our knowledge of the sequence-function relationships of proteins, peptides, RNA and DNA. This treasure trove of data may ultimately lead to better computational models and advances in protein design, biotechnology, synthetic biology and pharmaceutical applications.
Materials and methods
Yeast display setup
The yeast-surface display experiment was similar to that described by Dutta et al. and used many of the same reagents [23]. Briefly, we displayed BH3-peptides fused to the C-terminus of the Aga2 yeast cell-surface protein. The construct included HA and FLAG tags N-and C-terminal to the BH3 peptide, respectively. All BH3 peptides were variants of either the Bim or Puma human BH3 sequences. The Bim wild-type sequence consisted of the 31 residues
RPEIWIAQELRRIGDEFNAYYARRVFLNNYQ
and the Puma wild-type sequence comprised the 33 residues
GEEEQWAREIGAQLRRMADDLNAQYERRRQEEQ.
Mutations were introduced in positions 2d, 3a, 3b, 3d, 3f and 4a of the BH3 helix, which are indicated in bold text above (consult [23] for notation). In all experiments, we incubated cells with soluble Myc-tagged Bcl-xL in BSS (50 mM Tris, 100 mM NaCl, pH 8.0, plus 1 mg/ml BSA to block Bcl-xL from non-specific binding) at room temperature for 2 h (2·106 cells in 1.4 ml volume to meet the assumptions of the theoretical model). Cells were washed twice with BSS before the application of antibodies. We incubated the cells with a mixture of primary mouse anti-HA antibodies to detect peptide expression and primary rabbit anti-Myc antibodies to detect Bcl − xL binding (Roche, catalog # 11583816001 and Sigma, catalog # P9537). The incubation was carried out for 15 min at 4 °C (to minimize dissociation), applying 10 µl of a 1:100 dilution per 106 cells. After a second wash step, we incubated the cells with a secondary APC-labeled anti-mouse and a secondary PE-labeled anti-rabbit antibody (BD Biosciences, catalog # 550826 and Sigma, catalog # P9537). Dilutions were 1:40 for the anti-mouse and 1:100 for the anti-rabbit antibody. All other incubation parameters were the same as for the incubation with the primary antibodies. No significant dissociation was observed once sample preparation was complete. FACS profiles remained the same even when samples were remeasured after several hours. We suspect that the antibodies crosslink bound target across the cell surface and thus prevent further dissociation.
Cell growth was rigorously monitored. First, we grew cells in selective media containing glucose (SD+CAA) for 8h at 30 °C from a starting OD600nm of 0.05 and then diluted to an OD600nm of 0.005 and grew to an OD600nm of 0.1 to 0.4. Induction of BH3 expression was carried out in selective media with galactose (SG+CAA) at 30°C from a starting OD600nm of 0.025 until an OD600nm of 0.2 to 0.5 was reached.
Testing of model predictions
We collected signals from about 10000 cells for each FACS profile. To determine coordinates of affinity for individual BH3 peptides, we transformed FACS profiles into two-dimensional probability densities with an R-based kernel density routine [35] (Hpi routine with options pilot = “samse“ and pre = “sphere“). Integration perpendicular to the axis of affinity gave probability densities over this axis for each BH3 peptide. We considered the position of the global maximum of density as the corresponding peptide’s coordinate of affinity. Although densities are not fully symmetric and may show local maxima (see figure 3D), we considered this a good approximate measure. We subsequently applied a linear fit to the data set of pairs. KD values for individual clones were determined as described in [23]. The fit involved the minimization of absolute residuals rather than the more commonly used square residuals. Least-square residuals may result in a somewhat better R2 goodness of fit value, but does this by favoring extreme points. Apart from checking for linear correlation, we also desired a robust estimate of the slope. Hence, least absolute residuals were the better choice. Plots of experimental single clone FACS profiles were generated with the help of the Matlab routine dscatter as described in [36].
Gate setting
Gate delimitations perpendicular to the axis of affinity were chosen guided by the model theory, as discussed in the text (red lines in supplemental figure 3). To define other borders, we considered the FACS profiles from expressing but non-binding cells, which are not described by our theory but experimentally partially overlap the binding population. We defined gate borders in the low fluorescence region such that non- and weak binders could enter low affinity gates only, and non-expressing cells were excluded (green lines in supplemental figure 3). In the high fluorescence region, we excluded cells with maximum fluorescence/maximum channel numbers because the affinities of their displayed ligands could not be determined (blue lines in supplemental figure 3). Using experimentally recorded probability densities of individual BH3 peptides over the axis of affinity (e.g. profiles in figure 3A) we determined that 12 gates with the width and placement shown in supplemental figure 3 should give high resolution.
Sorting and sequencing a high-affinity BH3 library
We constructed a library of strong BH3 binders from existing Bim- and Puma- libraries in our laboratory. The Bim library is described in [23]. The Puma library contained the same variable regions and the same sequence variety. We started with a Bim library pool that had previously been sorted four times for binding to 1 µM Bcl-xL and a Puma library pool that had previously been sorted three times for binding to 1 µM Bcl-xL. Both library pools were sorted at 1 nM Bcl −xL according to our twelve-gate scheme. Equal numbers of cells from all gates and pools were combined (apart from samples of the three highest affinity gates, which were added at half the number of cells only due to a much lower sorting yield). About 10,000 cells from this mixture were grown to comprise the high-affinity library.
We sorted > 100,000 cells from the high-affinity library into each of the 12 established gates with a BD FACS Aria cell sorter and the BD FACS Diva software. The FACS Aria can sort four samples at the same time and we sorted into gates in ascending order. Cells were subsequently prepared for multiplexed deep-sequencing. The protocol we followed has been described in great detail in [37]. We assigned two different five basepair barcodes to each gate and performed a paired-end deep sequencing run on an Illumina GaII with forward and reverse reads yielding 80 base long partially overlapping sequences. For the one gate experiment we sorted between 200,000 and 1,000,000 cells into each combination of gates.
Data analysis
For subsequent analysis, we only considered those deep sequencing reads with a Phred confidence probability of > 0.95 for the combined variable positions in the sequence. 8720221 sequences satisfied this constraint. We proceeded to analyze all unique BH3 sequences for which we observed at least 750 copies. Copy numbers varied widely, indicating that although all BH3 clones likely were strong binders, their enrichment did not depend on their affinity for Bcl −xL alone (see supplemental figure 5A). After applying these criteria, we retained data for 1026 BH3 clones.
We constructed a frequency distribution over the 12 sorting gates for each BH3 clone from the deep-sequencing information. Because the FACS Aria cell sorter can only sort 4 samples at a time, we had to sample sets of gates sequentially. Therefore, counting the copy numbers of a specific BH3 sequence in each deep sequencing sample will not result in the frequency distribution of this sequence over the 12 gates. This distribution may be determined as follows. The relative frequency for BH3 clone j to hit gate i, fij, is
| (13) |
Here aij is the fraction of the sequence of BH3 clone j in the deep sequencing sample corresponding to gate i. zy is the relative frequency with which cells of the library would hit gate y if all gates could be sorted into at the same time. This information can be recorded during a FACS experiment without having to sort into every individual gate at the same time.
Despite our conservative gating scheme (see supplemental figure 3), preliminary experiments indicated a small probability for weak binders to enter high affinity gates. This could potentially lead to a diminished resolution in the final affinity ranking. We therefore set all frequencies < 0.05 in all distributions to 0 and renormalized. The quality of the final distributions was determined by their distance from the simplex center (see below and supplemental figure 5B). We subsequently used equation 12 to calculate probabilities for the stronger binder of all possible pairs of BH3 peptides and constructed a graph as described in the main text. We then employed a computational routine to determine the global affinity ranking for all peptides. The routine generated 1,000 candidate rankings with the algorithm described in [24] and retained the highest-scoring ranking (maximum product of edge weights of corresponding sub-graph) for further improvement. The improvement involved successive attempts to insert each individual peptide into each possible position in the retained ranking. We ran through this procedure 2,000 times, alternately starting with the highest and the lowest ranking peptide and proceeding to the lowest and highest ranking peptide, respectively. Finally, we conducted 10,000 Monte-Carlo steps. In each step an attempt was made to exchange the ranks of two different peptides. To determine the uncertainty in our results, we drew bootstrap samples from the high quality sequences in each deep sequencing sample and reran the whole subsequent analysis. Overall, we performed 10,000 such bootstrap repetitions resulting in the error bars in figure 4A.
Repeat Experiment
Library sorting and sequencing were performed as in the first experiment. In our repeat experiment, we obtained 105454910 high-quality reads from a HiSeq deep sequencing run. Most of the 19 individual test sequences from the original experiment had reasonably high copy numbers in this new data set. However, the sequence with original rank 12 (table 1, figure 4) had 467 reads. Although this is 4.4 · 10−6 of the total number of reads (corresponding to less than 10 sorted cells), and thus may contain insufficient statistical information, we decided to lower our cut-off on copy number to 467 so that this sequence could be included. A total of 5518 sequences met this criterion and were processed to generate a final ranking.
Analysis of one-gate data
Sorted pools from the one-gate experiment were deep sequenced and the observed sequences were scored in three different ways. First, we calculated enrichment values for each sequence, . Here f and fn signify the relative frequency in the sorted and naive library pools, respectively. Second, we determined a positional enrichment value ∏i, with index i running over all positions of the sequence and fi and fni indicating the relative frequency of the residue in position i in the sorted and naive library pool. Third, we built a PSSM from the sorted pool where the score is given by ∏i fi. We drew 10,000 bootstrap samples from the deep sequencing data on the sorted pools and the naive library and calculated the aforementioned scores each time to determine uncertainties in the rankings shown in figures 6 B, D and F.
Gauging the widths of frequency distributions
A parameter that expresses the spread of a distribution over the FACS gates can be very helpful when gauging the distribution’s quality or simulating a SORTCERY experiment (see Error Analysis in Supplemental Material). We noted that each normalized distribution over the 12 gates can be considered as a point in 12-dimensional space. Due to the normalization, points will be constrained to a so-called 11-simplex, a specific hypervolume in 12-dimensional space. The worst-case scenario (uniform distribution) corresponds to the center of this simplex. Thus the more distant a point lies from the simplex center the less uniform its corresponding distribution and the more accurate the resulting ranking. Although our metric assigns equal values to a distribution with two neighboring peaks and a distribution with two widely separated peaks, we observed that most experimental distributions exhibited one large cluster of frequencies only (see supplemental figure 6 for representative data). Hence, our metric is a good gauge for quality in practice and we generally refer to it a ”distribution width“ in the main text.
Titration of individual clones
We picked the 19 sequences listed in table 1 from the global ranking of 1026 BH3-peptides and cloned them into our yeast-display vector. We titrated each individual clone with Bcl − xL and recorded binding signals with a flow cytometry analyzer in four independent experiments. The KD values of the titration curves were determined as described in [23]. Example curves are shown in supplemental figure 7.
We also synthesized 6 of the 19 sequences as 28-residue long soluble peptides, corresponding to the region
RPEIWIAQELRRIGDEFNAYYARRVFLN
in the Bim context and the region
EEQWAREIGAQLRRMADDLNAQYERRRQ
in the Puma context. N- and C-termini were acetylated and amidated, respectively. We conducted four independent competition fluorescence polarization experiments with each of these peptides. Peptides were incubated with 50 nM Bcl −xL in the presence of 10 nM fluorescein labeled competitor with either 6 nM or 40 nM affinity for Bcl − xL (sequences of competitors were
SIRPKAQELRHLADQFSAEIARR
and
RSMVFARHLREVGDEFRSRHLNS
, respectively). The full experimental protocol has been described in [23]. We determined KD values by fitting the data to the model described in [38].
Supplementary Material
Highlights.
-
-
relating sequence to binding function is a fundamental goal in proteomics
-
-
SORTCERY determines binding semi-quantitatively for large peptide libraries
-
-
the method combines cell sorting, deep-sequencing and computational post-analysis
-
-
1000 peptide ligands of Bcl-xL were ranked with high accuracy using SORTCERY
-
-
this approach has high potential to provide binding data for many protein families
Acknowledgements
The authors thank Christos Kougentakis for help with the experiments. The authors express their gratitude to the Swanson Biotechnology Center Flow Cytometry Facility and the MIT BioMicro Center for technical support. This study was also funded by NIH award GM096466 to AK and grant no. RE 3111/1-1 of the German Merit Foundation to LR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lothar “Luther” Reich, Email: kuranes@mit.edu.
Sanjib Dutta, Email: sdutta@mit.edu.
Amy E Keating, Email: keating@mit.edu.
References
- 1.Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. Proc Natl Acad Sci USA. 2011;108:78967901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeKosky BJ, G C Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynolds KA, McLaughlin RN, Ranganathan R. Hot Spots for Allosteric Regulation on Protein Surfaces. Cell. 2011;147:15641575. doi: 10.1016/j.cell.2011.10.049. D Gfeller and Z Kan and S Seshagiri and P M Kim and G D Bader and S S Sidhu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst A, Gfeller D, Kan Z, Seshagiri S, Kim PM, Baderet GD, et al. Coevolution of PDZ domain-ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol Biosyst. 2010;6:1782–1790. doi: 10.1039/c0mb00061b. [DOI] [PubMed] [Google Scholar]
- 5.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J, Larman HB, Gao G, Somwar R, Zijuan Zhang Z, Lasersonet U, et al. Protein interaction discovery using parallel analysis of translated ORFs (PLATO) Nat Biotechnol. 2013;31:331–333. doi: 10.1038/nbt.2539. S D Khare and J Dou and L Doyle and J W Nelson and A Schena. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–218. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeBartolo J, Dutta S, Reich L, Keating AE. Predictive Bcl-2 Family Binding Models Rooted in Experiment or Structure. J Mol Biol. 2012;422:124–144. doi: 10.1016/j.jmb.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araya CL, Fowler DM. Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol. 2011;29:435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derda R, Tang SKY, Li SC, Ng S, Matochko W, Jafari MR. Diversity of Phage-Displayed Libraries of Peptides during Panning and Amplification. Molecules. 2011;16:1776–1803. doi: 10.3390/molecules16021776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinney JB, Murugana A, Callan CG, Jr, Cox EC. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci USA. 2010;107:9158–9163. doi: 10.1073/pnas.1004290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang JC, Chang AL, Kennedy AB, Smolke CD. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity. Nucleic Acids Res. 2012;40:138–142. doi: 10.1093/nar/gks636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutta S, Koide A, Koide S. High-throughput Analysis of the Protein Sequence Stability Landscape using a Quantitative Yeast Surface Two-hybrid System and Fragment Reconstitution. J Mol Biol. 2008;382:721–733. doi: 10.1016/j.jmb.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu L, Wang H, Wang L, Wang Y, Jiang K, Li C, et al. High-affinity peptide against MT1-MMP for in vivo tumor imaging. J Control Release. 2011;150:248–255. doi: 10.1016/j.jconrel.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Luo J, Li Y, Henderson PT, Wang Y, Wachsmann-Hogiu S, et al. Characterization of high-affinity peptides and their feasibility for use in nanotherapeutics targeting leukemia stem cells. Nanomed-Nanotechnol. 2012;8:1116–1124. doi: 10.1016/j.nano.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrivastava A, Wronski MA, Sato AK, Dransfield DT, Sexton D, Bogdan N, et al. A distinct strategy to generate high-affinity peptide binders to receptor tyrosine kinases. Protein Eng Des Sel. 2005;18:417424. doi: 10.1093/protein/gzi049. [DOI] [PubMed] [Google Scholar]
- 20.Pierceall WE, Kornblau SM, Carlson NE, Huang X, Blake N, Lena R, et al. BH3 Profiling Discriminates Response to Cytarabine-based Treatment of Acute Myeloid Leukemia. Mol Cancer Ther. 2013 doi: 10.1158/1535-7163.MCT-13-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBelle JL, Katz SG, Bird GH, Gavathiotis E, Stewart ML, Lawrence C, et al. A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest. 2012;122:20182031. doi: 10.1172/JCI46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Gaizo Moore V, Letai A. BH3 profiling–measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer Lett. 2013;332:202–205. doi: 10.1016/j.canlet.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta S, Gulla S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 Binding Specificity for Mcl-1 versus Bcl-xL. J Mol Biol. 2010;398:747–762. doi: 10.1016/j.jmb.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ailon N, Charikar M, Newman A. Aggregating Inconsistent Information: Ranking and Clustering. J ACM. 2008;55 article 23. [Google Scholar]
- 25.Ming-Sin C, Down TA, Latorre I, Ahringer J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res. 2011;39:e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toedling J, Servant N, Ciaudo C, Farinelli L, Voinnet O, Heard E. Deep-Sequencing Protocols Influence the Results Obtained in Small-RNA Sequencing. PLOS ONE. 2012;7:e32724. doi: 10.1371/journal.pone.0032724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. PNAS. 2007;104:66326637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;5196:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 29.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. PNAS. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal G, Kouadio JL, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for proteinprotein interactions by rapid combinatorial scanning. J Biol Chem. 2006;281:22378–22385. doi: 10.1074/jbc.M603826200. [DOI] [PubMed] [Google Scholar]
- 31.Moretti S, Fleishman J, Agius R, Torchala M, Bates PA, Kastritis PL, et al. Community-wide evaluation of methods for predicting the effect of mutations on protein-protein interactions. Proteins. 2013;81:1980–1987. doi: 10.1002/prot.24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. JMB. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 33.Chen JR, Chang BH, Allen JE, Stiffler MA, MacBeath G. Predicting PDZ domain-peptide interactions from primary sequences. Nat Biotechnol. 2008;26:1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, K D Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Prot. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 35.Duong T. ks: Kernel Density Estimation and Kernel Discriminant Analysis for Multivariate Data in R. J Stat Softw. 2007;21:1–16. [Google Scholar]
- 36.Eilers PHC, Goeman JJ. Enhancing scatter plots with smoothed densities. Bioinformatics. 2004;20:623–628. doi: 10.1093/bioinformatics/btg454. [DOI] [PubMed] [Google Scholar]
- 37.Hietpas R, Roscoe B, Jiang L, Bolon DNA. Fitness analyses of all possible point mutations for regions of genes in yeast. Nat Protoc. 2012;7:1382–1396. doi: 10.1038/nprot.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mocz G, Helms MK, Jameson DM, Gibbons IR. Probing the Nucleotide Binding Sites of Axonemal Dynein with the Fluorescent Nucleotide Analogue 2(3)-O-(-N-Methylanthraniloyl)-adenosine 5-Triphosphate. Biochemistry. 1998;37:9862–9869. doi: 10.1021/bi9730184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.