SUMMARY
Lipids of the Escherichia coli membrane are mainly composed of 70–80% phosphatidylethanolamine (PE) and 20–25% phosphatidylglycerol (PG). Biochemical studies indicate that the depletion of PE causes inversion of the N-terminal helix bundle of the lactose permease (LacY), and helix VII becomes extramembraneous. Here we study this phenomenon using single-molecule force spectroscopy, which is sensitive to the structure of membrane proteins. In PE and PG at a ratio of 3:1, ~95% of the LacY molecules adopt a native structure. However, when PE is omitted and the membrane contains PG only, LacY almost equally populates a native and a perturbed conformation. The most drastic changes occur at helices VI and VII and the intervening loop. Since helix VII contains Asp237 and Asp240, zwitterionic PE may suppress electrostatic repulsion between LacY and PG in the PE:PG environment. Thus, PE promotes a native fold and prevents LacY from populating a functionally defective, non-native conformation.
Keywords: atomic force microscopy, single-molecule force spectroscopy, phospholipids, major facilitator superfamily, membrane transport proteins, dual topology, inverted topology
INTRODUCTION
The lactose permease of Escherichia coli (LacY) with 12 mostly irregular transmembrane α-helices organized into two pseudo-symmetrical 6-helix bundles connected by a relatively long cytoplasmic loop (Abramson et al., 2003; Chaptal et al., 2011; Guan et al., 2007; Kumar et al., 2014; Mirza et al., 2006) is a paradigm for the major facilitator superfamily (MFS) (Marger and Saier, 1993). In the native membrane, which is 70–80% zwitterionic phosphatidylethanolamine (PE) and 20–25% anionic phosphatidylglycerol (PG) plus cardiolipin, LacY adopts a native topology with the N- and C-termini on the cytoplasmic surface of the membrane (Bogdanov et al., 2008; Calamia and Manoil, 1990; Chen and Wilson, 1984; Foster et al., 1983; Seto-Young et al., 1985). However, in the absence of PE where 100% of the lipids are negatively charged, the N-terminal helix bundle in LacY becomes inverted relative to the C-terminal bundle, and transmembrane helix VII and the cytoplasmic loop VI/VII become accessible from the periplasmic side of the membrane (Bogdanov et al., 2008; Vitrac et al., 2013; Wang et al., 2002). Furthermore, the degree of inversion increases with a progressive decrease in PE level (Bogdanov and Dowhan, 2012; Vitrac et al., 2013), and the event is rapidly reversed upon introduction of PE after assembly of LacY in vivo (Bogdanov and Dowhan, 2012; Bogdanov et al., 2008; Vitrac et al., 2013). Both the dependence of topological orientation and the reversibility of transmembrane domain orientation with a change in lipid composition were fully replicated in an in vitro proteoliposome system (Bogdanov and Dowhan, 2012; Vitrac et al., 2013; Wang et al., 2002). Thus, membrane protein topology can be changed simply by changing membrane lipid composition confirming that topological interconversion is thermodynamically determined by the properties of the protein interacting with the lipid environment independent of other cellular factors (Bogdanov et al., 2014).
Two methods have been used thus far to document the phenomenon with LacY in vivo and in vitro (reviewed in (Bogdanov et al., 2014; Dowhan and Bogdanov, 2012). One method utilizes a site-specific monoclonal antibody to probe the conformation of periplasmic loop VII/VIII. Because the antibody recognizes this loop from the periplasmic side in PE-enriched membranes and no binding is observed when PE is replaced with PG, the results indicate that the structure of loop VII/VIII is perturbed in PG-rich membranes (Bogdanov et al., 1996; Vitrac et al., 2013). A second method utilizes Cys-scanning mutagenesis combined with site-directed alkylation (Bogdanov et al., 2010; Bogdanov et al., 2005) and similarly documents inversion of the N-terminal bundle (Bogdanov et al., 2002; Bogdanov et al., 2008; Vitrac et al., 2013; Wang et al., 2002). It is also notable that Cys replacements on the cytoplasmic side of LacY do not react with impermeant thiol reagents unless the membrane is disrupted (Bogdanov et al., 2002; Vitrac et al., 2013). Thus, it appears that the phenomenon is not reversible in the absence of PE enrichment.
Atomic force microscopy (AFM)-based single-molecule force spectroscopy (SMFS) localizes and quantifies intra- and intermolecular interactions that stabilize structural segments of membrane proteins (Bippes and Muller, 2011; Engel and Gaub, 2008; Kedrov et al., 2007; Muller et al., 2002). SMFS is applied to membrane proteins embedded in native or synthetic lipid membranes under physiologic conditions and has been useful for characterizing the stability of secondary structure segments of membrane proteins under a wide range of conditions (Bippes et al., 2009; Bippes et al., 2013; Ge et al., 2011; Janovjak et al., 2003; Kedrov et al., 2005; Park et al., 2007; Sapra et al., 2008; Sapra et al., 2006; Serdiuk et al., 2014; Zocher et al., 2012a; Zocher et al., 2012b). Thus, SMFS is particularly well suited to study conformational states of membrane proteins and the changes they undergo.
SMFS is applied here to characterize the influence of the lipid environment on the LacY structure. As shown previously (Serdiuk et al., 2014), LacY reconstituted into PE:PG (ratio 3:1) proteoliposomes predominantly exhibits a secondary structure consistent with the native conformer. However a very low percentage of conformers are now observed in which helices VI and VII, as well as the intervening loop are perturbed. When reconstituted into proteoliposomes composed of negatively charged PG only, the same two conformers are observed, but in about equal proportions. One conformer represents native LacY and the other exhibits a conformer that likely represents the perturbed N-terminal structure described previously (reviewed in (Bogdanov et al., 2014)). The SMFS results indicate that in the absence of zwitterionic PE, anionic PG changes the folding free energy landscape of LacY in such a manner that both native and perturbed conformers are equally favored. In the presence of PE, the free energy landscape promotes the native LacY fold and suppresses the perturbed structure confirming that lipid-dependent topogenesis is primarily a thermodynamically driven process, as suggested (Bogdanov and Dowhan, 2012; Dowhan, 2014; Vitrac et al., 2013).
RESULTS
Expression, Purification and Reconstitution of LacY in PE:PG and PG
WT LacY was expressed, purified by metal-affinity chromatography (Figure S1A) and reconstituted into 1-palmitoyl-2-oleoyl-sn-glycero-3- (PO) PE:POPG (3:1, mol:mol) proteoliposomes or into proteoliposomes composed of POPG only (Viitanen et al., 1986). In order to make LacY suitable for ready characterization by SMFS, the C-terminus was extended with a 36 amino acid (aa) long unstructured ‘polyGly’ polypeptide [GSM(G11)EAVEEAVEEA(G11)S] followed by a 8-aa-long His-Tag (His8-Tag), which has little effect on LacY transport activity (Serdiuk et al., 2014). Successful reconstitution of LacY in both types of proteoliposomes was confirmed by AFM imaging (Figures S1B and S1C).
LacY Populates Predominantly a Single State in PE:PG
To characterize the structural stability of LacY reconstituted in PE:PG proteoliposomes single LacY molecules were unfolded using SMFS. Therefore, we first localized proteoliposomes containing WT LacY by AFM imaging and then pressed the AFM stylus onto the membrane applying a force of ≈1 nN for 0.5–1 s (see Experimental Procedures). By this means, the AFM stylus was attached unspecifically to LacY (Bippes and Muller, 2011; Muller and Engel, 2007). The C-terminal end of WT LacY, which was extended by a 44 aa long unstructured polypeptide, increased the probability of unspecific attachment (Serdiuk et al., 2014). Withdrawal of the AFM stylus stressed the C-terminal polypeptide attached and mechanically unfolded single LacY molecules (Figure 1A). Force-distance (FD) curves recorded during this withdrawal of the AFM stylus exhibit sawtooth-like force peak patterns that describe the stepwise unfolding of single LacY molecules (Serdiuk et al., 2014)(Figures 1B and 1C).
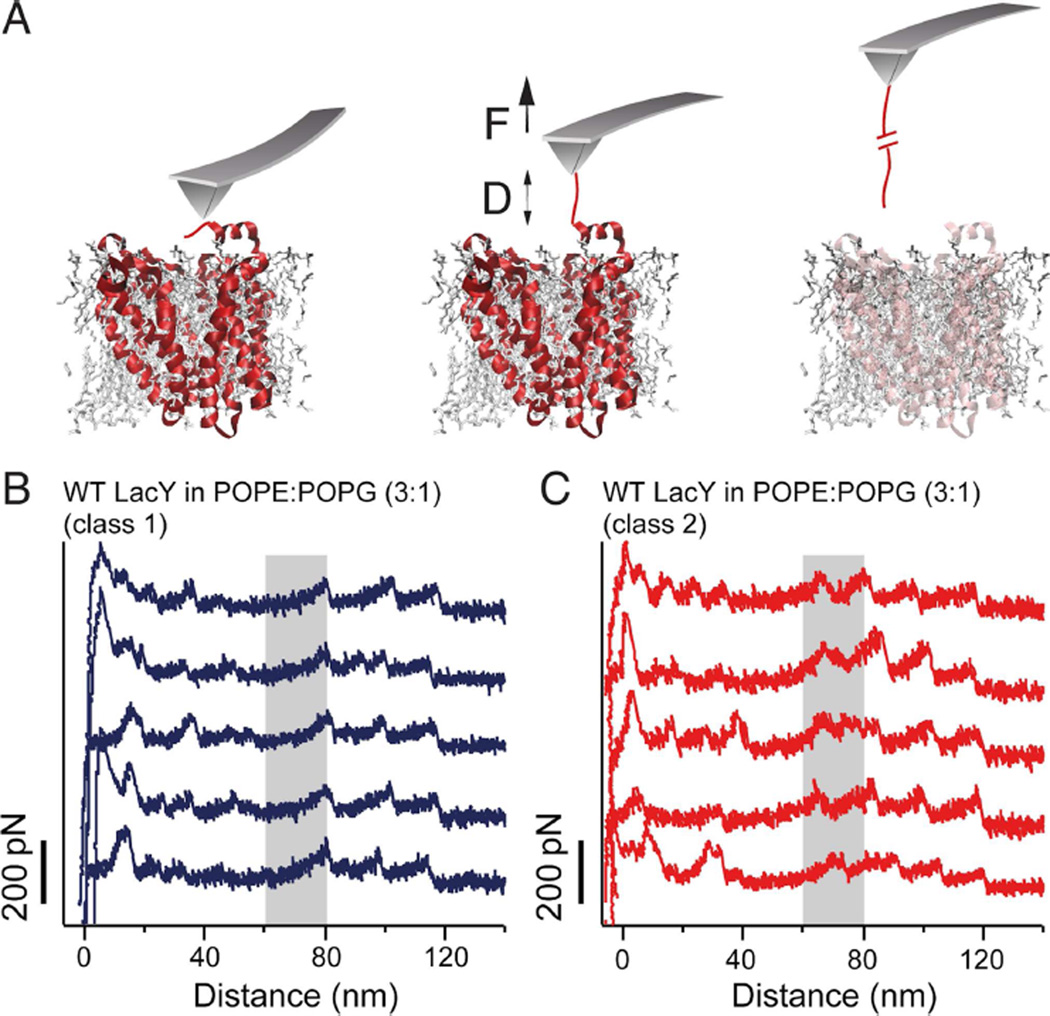
Figure 1. Unfolding of LacY using SMFS reveals two classes of FD curves.
(A) Schematic representation of SMFS with LacY in a phospholipid bilayer. Pushing the AFM stylus onto the protoliposomes unspecifically attaches the LacY terminal end to the stylus (left cartoon). Withdrawal of the cantilever increases the distance D between the AFM stylus and lipid membrane and applies a mechanical force F to the permease (middle cartoon). Further withdrawal of the AFM stylus induces the sequential unfolding of LacY (right cartoon). During this process, a force-distance (FD) curve is recorded. (B and C) Typical FD curves each one corresponding to the unfolding of a single wild-type (WT) LacY from POPE:POPG lipid (ratio 3:1) membranes. FD curves show two classes of force peak patterns, which differ in whether they have not (B) or have (C) an additional force peak at distances ranging from 60–80 nm (gray shaded area). To increase the probability of attaching the C-terminal end to the AFM stylus, the C-terminus of LacY was elongated by a 36 aa long unstructured polyGly polypeptide followed by a His8-tag. SMFS experiments were performed in 50 mM potassium phosphate (KPi), pH 7.2 and at 25°C. See also Figure S1.
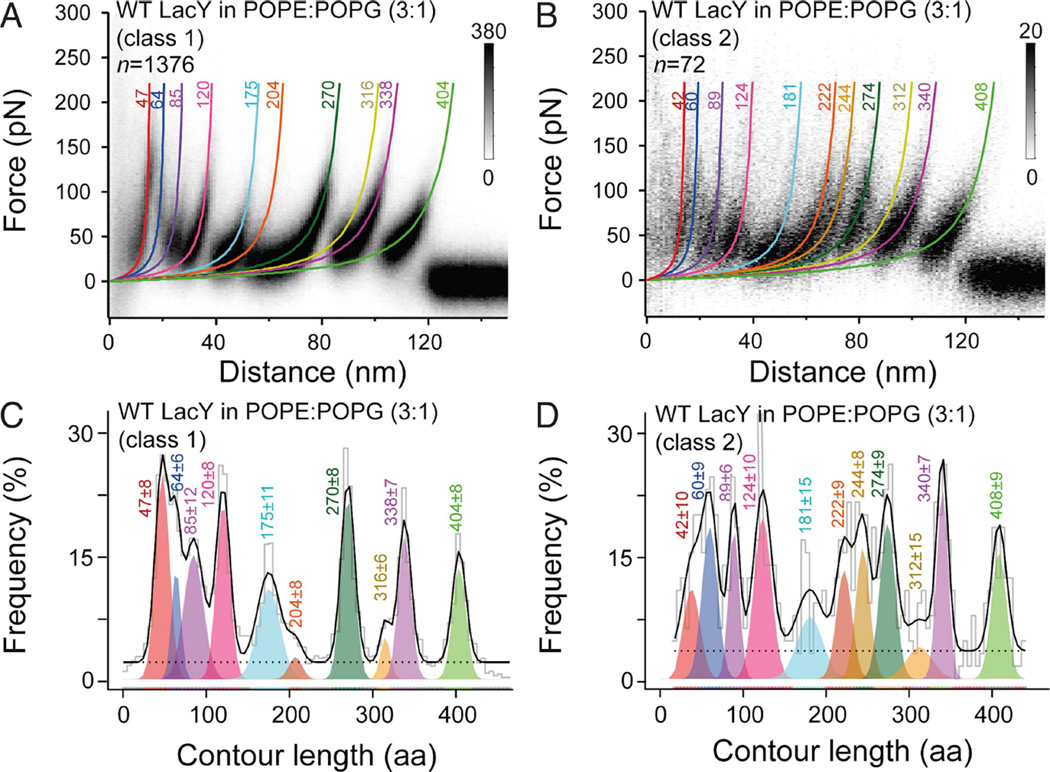
Although undetected initially, two classes of molecules are observed. Almost all the single-molecule FD curves show a force peak pattern similar to that observed upon unfolding of native LacY, as reported previously (Serdiuk et al., 2014). Therefore, we classified these FD curves as belonging to ‘class 1’. However, very few FD curves showed an additional force peak at distances between 60 and 80 nm. These FD curves were assigned to ‘class 2’. To obtain valid statistics, more than 1448 single-molecule FD curves were recorded from many (>20) different preparations and experiments (Figure 2). From these 1448 measurements, 1376 fall into class 1 (≈95%) and only 72 fall into class 2 (≈5%). The FD curves belonging to either one of the two classes were superimposed to highlight their common features (Figures 2A and 2B). Clearly, in PE:PG, WT LacY largely populates a single conformational state. To assign structural segments of LacY, every force peak in each FD curve (n=1448) was fit by the worm-like-chain (WLC) model to reveal the contour length (in aa) of the unfolded LacY polypeptide stretches (Figures 2A and 2B) (Bippes and Muller, 2011; Bustamante et al., 1994; Kedrov et al., 2007). From superimpositions of both classes of FD curves, histograms were made that show the mean contour lengths at which the force peaks reproducibly occur (Figures 2C and 2D). As shown previously (Serdiuk et al., 2014), class 1 FD curves exhibit 10 characteristic force peaks (Figure 2C). In contrast, the class 2 FD curves number 11 force peaks (Figure 2D). The class 2 FD curves exhibit an additional peak at ≈60–80 nm.
Figure 2. LacY reconstituted in POPE:POPG (ratio 3:1) membranes shows one major and one minor force peak pattern.
(A and B) Density plots of superimposed FD curves classified into the two distinct force peak patterns observed upon unfolding of WT LacY in PE:PG lipid membranes. WLC fits corresponding to the mean contour lengths of each force peak are represented by colored curves. n gives the number of classified superimposed and analyzed FD curves.
(C and D) Contour length histogram of force peaks detected in class 1 (C) and class 2 (D) FD curves. Histograms were fitted with a Gaussian mixture model (Kawamura et al., 2013). Each colored Gaussian distribution gives the mean contour length of a force peak class. Ten force peak classes were found for the class 1 FD curves (A and C). Eleven force peak classes were found for the class 2 FD curves (B and D). The black solid line shows the sum of weighted contour lengths for all force peak classes. The dashed line represents the uniform baseline noise. Colored numbers at each WLC curve and histogram peak represent the mean contour length in aa ± SD.
LacY Populates Two States Equally In PG
SMFS of WT LacY reconstituted in PG membranes was performed in the same manner as WT LacY reconstituted in PE:PG membranes. More than 612 single LacY molecules were unfolded and recorded, from which 285 curves (47%) belong to the class 1 FD curves and 327 (53%) to the class 2 FD curves (Figures 3A and 3B). Although superimposition of FD curves from each class reveals a high degree of reproducibility (Figures 3A and 3B), the two patterns may have originated conceivably from unfolding LacY either from the C- or the N-terminus. In order to assign from which end the FD curves were recorded, curves from WT LacY with only a 6-His-tag on the C-terminus were compared to those from WT LacY with the 44-residue extension (Figure S2 and S3). The FD curves for the two constructs reconstituted into PG membranes exhibit the same unfolding patterns except that curves recorded from polyGly WT LacY are shifted to longer distances from the AFM stylus. The longer distance of 7–10 nm corresponds to a polypeptide length of ≈20–27 aa, which is due to the longer C-terminal extension. Therefore, it is apparent that both sets of FD curves were recorded from the C-termini. In addition, the initial force peaks at short distances from the stylus (0–15 nm) are masked by noise in the FD spectra of LacY with the short extension, but clearly observed in the construct with the long polyGly extension (Figures S2 and S3). Thus, it is clear that class 1 and class 2 FD curves describe individual LacY molecules in two different conformational states.
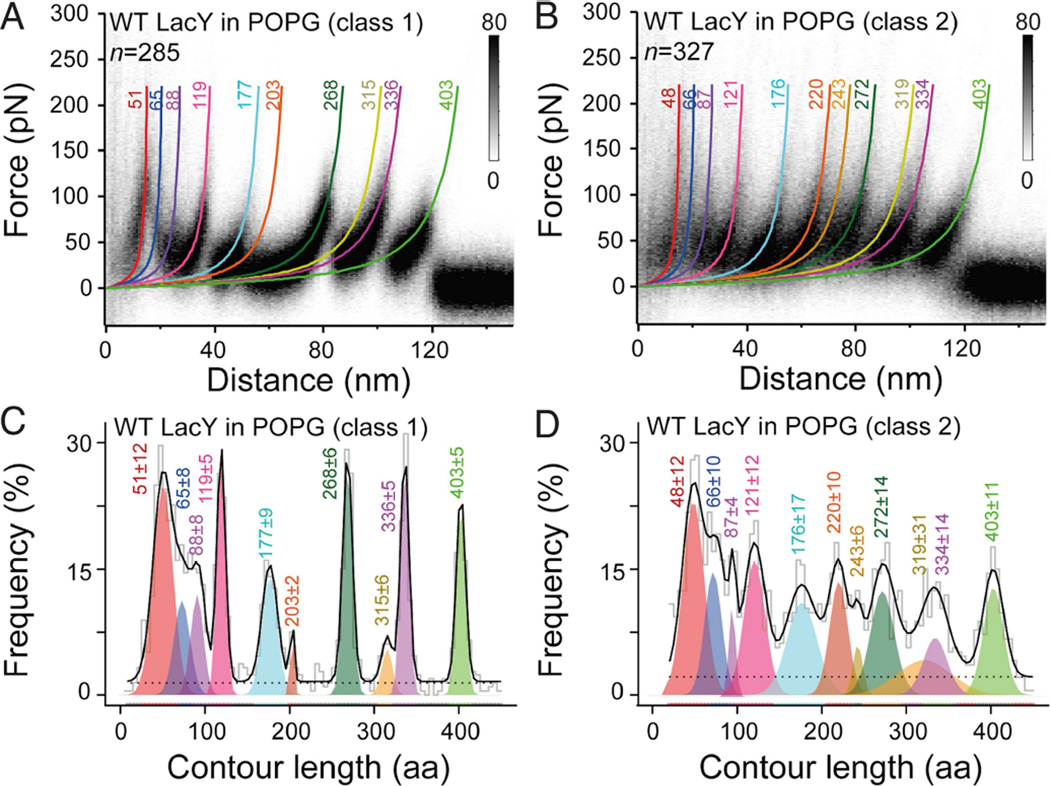
Figure 3. LacY reconstituted in POPG membranes shows two distinct unfolding patterns.
(A and B) Density plots of superimposed FD curves reveal two distinct force peak patterns matching either class 1 (A) or class 2 (B) FD curves. FD curves were recorded upon unfolding of WT LacY in PG membranes. WLC fits corresponding to the mean contour lengths of each force peak are represented by colored curves. n gives the number of classified, superimposed and analyzed FD curves.
(C and D) Contour length histogram of force peaks detected in class 1 (C) and class 2 (D) FD curves. Histograms were fitted with a Gaussian mixture model (Kawamura et al., 2013). Each colored Gaussian distribution gives the mean contour length of a force peak class. Ten force peak classes were found for the class 1 FD curves (A and C). Eleven force peak classes were found for the class 2 FD curves (B and D). The black solid line shows the sum of weighted contour lengths for all force peak classes. The dashed line represents the uniform baseline noise. Colored numbers at each WLC curve and histogram peak represent the mean contour length in aa ± SD. See also Figures S2 and S3.
After having shown that in PG membranes LacY equally populates two states, histograms showing the contour lengths at which the force peaks reproducibly occurred were generated for both classes (Figures 3C and 3D). To determine the mean contour lengths, force peaks from each histogram were fit by a Gaussian mixture model (Kawamura et al., 2013). For class 1 FD curves, 10 characteristic force peaks are detected that occur with a mean amino acid content of 51±12, 65±8, 88±8, 119±5, 177±9, 203±2, 268±6, 315±6, 336±5, and 403±5 residues (mean ± SD, Figure 3C), as described previously (Serdiuk et al., 2014). In contrast, class 2 FD curves exhibit 11 characteristic force peaks at mean contour lengths of 48±12, 66±10, 87±4, 121±12, 176±17, 220±10, 243±6, 272±14, 319±31, 334±14, and 403±11 aa (Figure 3D).
Since the 10 force peaks detected for class 1 FD curves recorded from WT LacY in PG membranes are at the same contour lengths as observed for the class 1 FD curves recorded from PE:PG membranes (Figure 2C), the class 1 FD curves in either PE:PG or PG represent only native WT LacY. For the class 2 FD curves, the Gaussian mixture model exhibits 11 force peaks the contour lengths of which match the force peak contour lengths observed for the minor fraction of class 2 curves in PE:PG plus the additional peak at ≈60–80 nm. Because the structural segments detected for a membrane protein by SMFS are sensitive to conformational state (Bippes and Muller, 2011; Engel and Gaub, 2008), it seems clear that class 1 and class 2 FD curves describe individual LacY molecules in two different conformations. Because the ratio of class 1 to class 2 FD curves is ≈1:1 (Figure 3), WT LacY reconstituted in PG populates the two conformational states almost equally.
Mapping the structural segments of FD curves
Each force peak class (Figures 2 and 3) reflects the unfolding of a single stable segment of LacY. The first force peak in an FD spectrum describes unfolding of the first stable segment and localizes the beginning of that segment. The next force peak indicates the end of the previous stable segment and the beginning of the next (Bippes and Muller, 2011; Engel and Gaub, 2008). Following this scheme, the mean contour lengths of the unfolding force peaks of class 1 FD curves were used to determine the beginning and end of each structural segment in the secondary structure model based upon the X-ray crystal structure (Figure 4A) (Abramson et al., 2003).
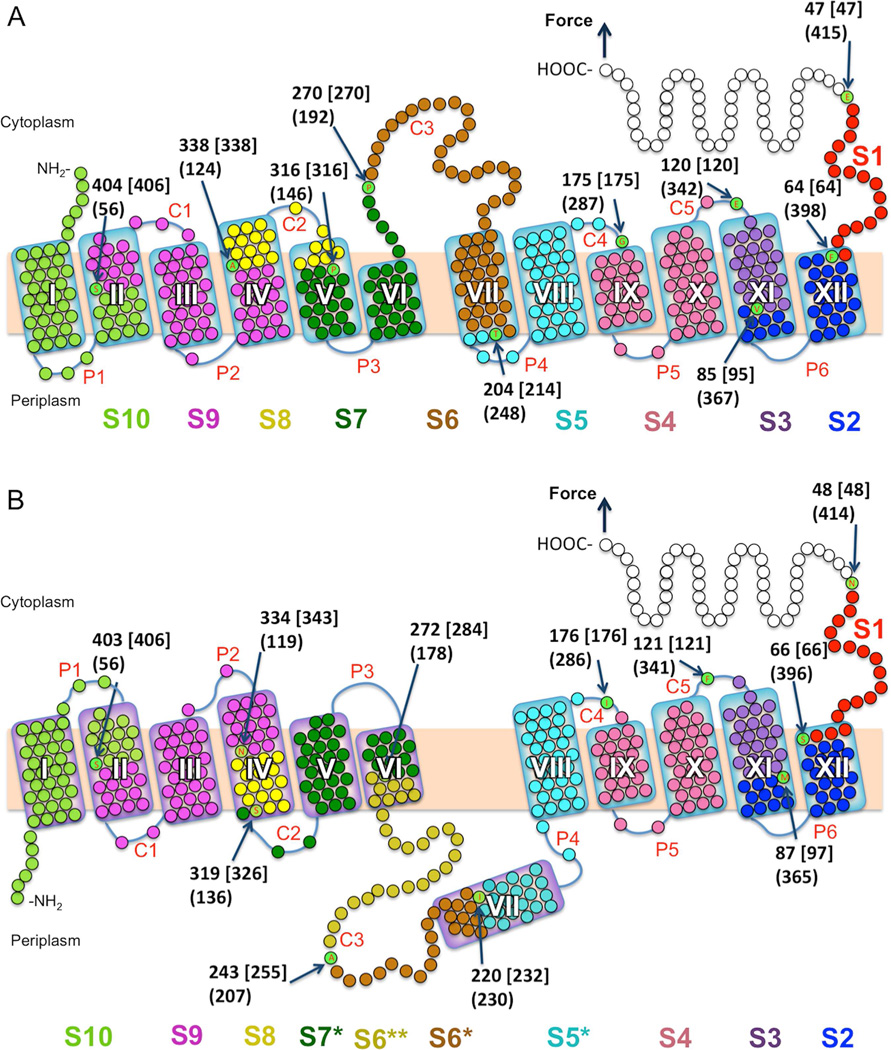
Figure 4. Effect of PE and PG on the stable structural segments in LacY.
(A and B) Structural segments were mapped to the secondary structures of WT LacY in native (A) and inverted (B) topologies. Stable structural segments mapped in (A) and (B) were detected in the class 1 and class 2 FD curves, respectively. (A) Secondary structure model of LacY (PDB ID code 1PV7) mapped with 10 (class 1 FD curves) structural segments S1–S10 (native topology) and (B) with 11 (class 2 FD curves) structural segments S1–S4, S5*, S6*, S6**, S7*, S8, S9, S10 (inverted topology). Each mean contour length of a force peak class (see histograms of Figure 2, 3) assigns the beginning of a structural segment (arrows pointing to aa). The numbers at arrows show the mean contour lengths of a force peak class (in aa), numbers in square brackets indicate the aa position counted from the C-terminal end, and numbers in parentheses give the aa position from the N-terminal end. Each of these numbers distinguishes the end of the previous and the beginning of a next stable structural segment. The length of polyGly tag and His-tag was considered for mapping. If the beginning/end of a stabilizing structural segment locates on the mica-facing side of the membrane or within the membrane the thickness of the membrane is taken into account. Transmembrane helices are labeled I-XII, cytoplasmic loops are labeled C1–C5 and periplasmic loops are labeled P1–P6.
Stable structural segments of LacY in class 1 are the following (Figure 4A): The first force peak at 47 aa is assigned as segment S1 (red) established by the C-terminus; the second force peak at 64 aa segment S2 (dark blue) established by the short periplasmic loop P6 and helix XII; force peak at 85 aa as segment S3 (purple) established by helix XI; force peak at 120 aa as segment S4 (magenta) established by loop C5, helix X, loop P5, and helix IX; force peak at 175 aa as S5 (light blue) established by loop C4, helix VIII and loop P4; force peak at 204 aa as S6 (brown) established by the C-terminal half of loop C3 and helix VII; force peak at 270 aa as S7 (dark green) established by the N-terminal half of loop C3, helix VI, loop P3, and periplasmic half of helix V; force peak at 316 aa as S8 (yellow) established by the cytoplasmic half of helix IV, loop C2, and the cytoplasmic end of helix V; force peak at 338 aa as S9 (violet) of the cytoplasmic end of helix II, loop C1, helix III, loop P2, and the periplasmic half of helix IV; and force peak at 404 aa as S10 (pale green) of the N-terminus, helix I, loop P1, and the periplasmic half of helix II.
The class 2 FD curves recorded for about half the molecules in PG and as a minor component in PE:PG represent an altered conformational state of LacY. Thus, the force peaks of class 2 FD curves were mapped on to the secondary structure of perturbed LacY conformers based upon the inverted model proposed by Dowhan and co-workers (Bogdanov et al., 2002; Bogdanov et al., 2008; Vitrac et al., 2013; Wang et al., 2002) (Figure 4B). The first force peaks of the class 2 FD curves have the same contour lengths of 48, 66, 87, and 121 aa as detected for class 1 FD curves. Mapped to the inverted topology of LacY (Figure 4B) these force peaks are assigned the same C-terminal segments S1, S2, S3, and S4 as observed with native LacY. However, the next force peaks are assigned to very different segments. The force peak at 176 aa is assigned a new segment S5* (light blue) formed by half of helix VII, loop P4, helix VIII and loop C4; the force peak at 220 aa to a new segment S6* (brown) containing the C-terminal half of loop C3 and only half of helix VII; the force peak at 243 aa forms a new stable segment S6** (ochre) containing half of loop C3 and half of helix VI, and the force peak at 272 aa forms a new structural segment S7* established by loop C2, helix V, loop P3 and half of helix VI. The next force peaks of the FD curves detected at 319, 334, and 403 aa occur at the same position as detected for class 1 FD curves. Thus, the last three structural segments S8, S9, and S10 of the N-terminal helix bundle, although inverted, are the same length as those of native LacY, while the C-terminal bundle remains essentially unchanged. Most remarkably, stable structural segments S5*, S6*, S6**, and S7* are drastically different from those detected predominantly with WT LacY in PE:PG. Since the perturbed segments correspond to transmembrane helices VI, VII, VIII, periplasmic loop P4 and the middle cytoplasmic loop (Figures 4A and 4B) and in view of the biochemical observations of Dowhan and coworkers (Bogdanov and Dowhan, 2012; Bogdanov et al., 2002; Bogdanov et al., 2008; Vitrac et al., 2013; Wang et al., 2002), the SMFS findings are clearly consistent with the model developed previously of the lipid-dependent inversion of the LacY topology (Figure 4B).
DISCUSSION
In order to transport sugar, LacY goes through a series of conformational changes that enable alternating access of the sugar-binding site to either side of the membrane by opening and closing of aqueous periplasmic and cytoplasmic cavities in reciprocal fashion (Kaback et al., 2011; Smirnova et al., 2011). Thus, LacY is structurally highly flexible and populates multiple conformations (Madej et al., 2012; Smirnova et al., 2011). In the native E. coli membrane (70–80% PE and 20–30% PG plus cardiolipin) or synthetic PE:PG (3:1) membranes, LacY is fully functional (Viitanen et al., 1986); however, upon replacement of PE with PG, the ability of LacY to catalyze active transport is abrogated (Bogdanov and Dowhan, 1995). Studies with a conformationally specific monoclonal antibody and site-directed alkylation studies with single-Cys mutants indicate that LacY either synthesized and assembled in vivo or reconstituted into proteoliposomes undergoes drastic but reversible structural changes driven by either an increase or decrease in PE levels (Bogdanov and Dowhan, 1998, 2012; Bogdanov et al., 1996; Bogdanov et al., 2008; Vitrac et al., 2013). These structural changes likely cause transmembrane helix VII to become an extramembraneous domain on the periplasmic side of the membrane. Moreover, the N-terminal helix bundle becomes inverted (Figure 4B). Here we used SMFS to study this phenomenon, and the findings reinforce the previous findings of Dowhan and colleagues (Bogdanov and Dowhan, 1998, 2012; Bogdanov et al., 2008; Dowhan and Bogdanov, 2012; Vitrac et al., 2013).
SMFS reveals that only a small percentage of LacY in PE:PG proteoliposomes is in an altered conformational state while about half the molecules in PG membranes are in a perturbed state. The main difference between the two states is that 11 stable segments are observed with perturbed LacY in PG while only ten stable segments are present with native LacY in PE:PG (Figure 4). In addition, most force peaks detected for the class 2 FD curves show a much wider contour length distribution compared to the force peaks detected for the class 1 FD curves (Figure 2 and 3). This increase in width suggests greater conformational variability and flexibility of the structural segments stabilizing the inverted LacY topology and possibly indicates the coexistence of multiple intermediate states (Bippes et al., 2013). The suggestion is consistent with the finding of Vitrac et al (Vitrac et al., 2013) that different structural elements of the N-terminal bundle in LacY behave asynchronously in response to varying the PE concentration of the proteoliposomes.
The anionic nature of PG makes it an important player in determining the LacY topogenesis (Bogdanov and Dowhan, 2012; Vitrac et al., 2013). Transmembrane helix VII of LacY contains two residues (Asp 237 and Asp 240) in the middle of membrane that may be negatively charged. This possibility together with low hydrophobicity makes helix VII an “Achilles' heel” of LacY with respect to folding and topogenesis (Bogdanov et al., 2014; Bogdanov et al., 2002; Bogdanov et al., 2008). Accordingly, when reconstituting LacY in PE:PG (3:1), electrostatic repulsion between the negatively charged Asp residues in helix VII and anionic PG is suppressed by zwitterionic PE (Bogdanov and Dowhan, 2012; Vitrac et al., 2013) so that LacY almost doesn’t “feel” the electrostatic field imposed by PG. However, in the absence of PE, electrostatic repulsion between PG and the Asp residues in helix VII may dominate LacY folding. In terms of the free energy landscape of the LacY topology, PG may tilt the landscape and favor a thermodynamic equilibrium between native LacY and the inverted conformer (Figure 5) as determined by the balance between anionic and net neutral lipids. Therefore the maintenance of either correct or inverted conformation or co-existence dual topologies can be simply determined by lipid-protein interactions that thermodynamically balance the relative strength of hydrophobic forces and charge effects (Bogdanov and Dowhan, 2012).
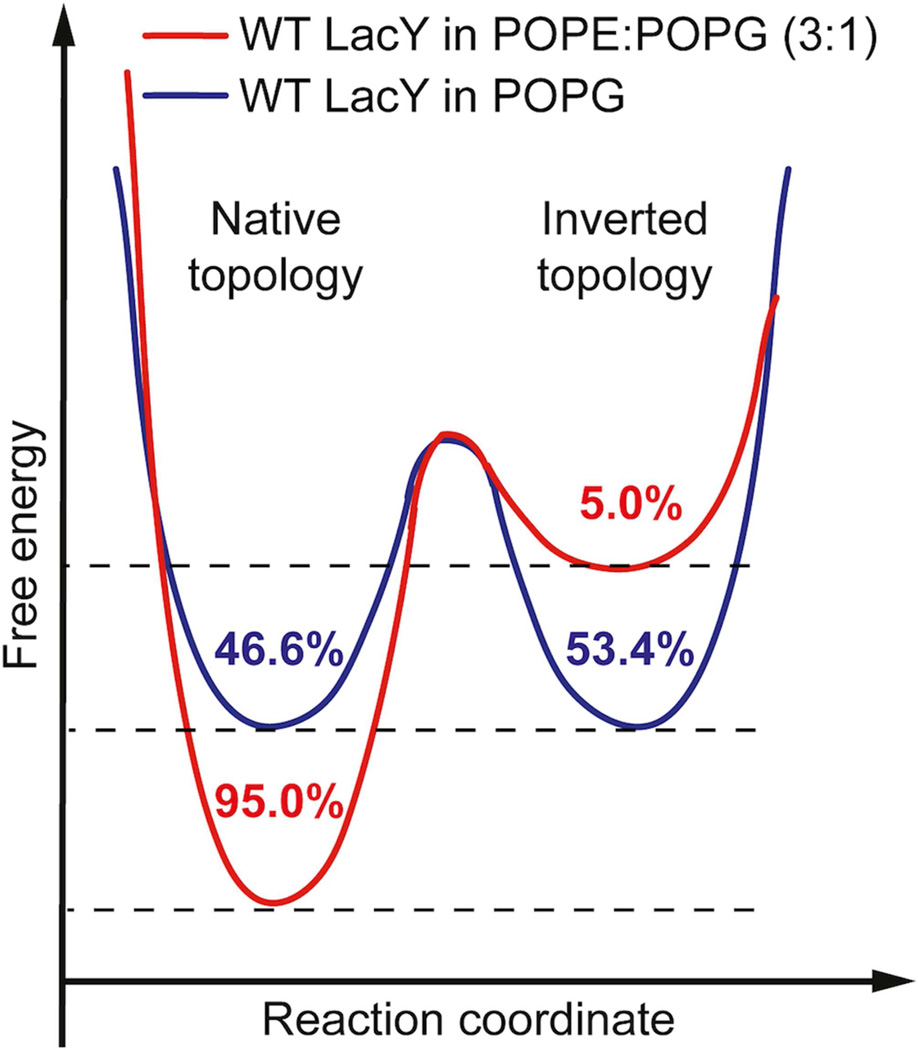
Figure 5. Schematic model of the free energy landscape of the native and inverted LacY topology in the presence and absence of PE.
In the mixture of PE and PG (ratio 3:1) LacY preferably (95%) shows the native fold required for lactose transport. The free energy landscape (red landscape), which has been adapted according to (Bogdanov and Dowhan, 2012), is shaped so that the inverted topology is rarely adopted by LacY (5%). In the absence of PE (blue landscape), the anionic PG alters the energy landscape of LacY so that native and inverted topologies become energetically favorable. Thus, embedded in PG the probabilities of LacY to adopt the native or the inverted conformation are almost equal. Values give the probabilities of detecting one of both LacY topologies.
Qualitatively, the SMFS findings are in general agreement with the data obtained biochemically (Bogdanov and Dowhan, 2012; Bogdanov et al., 2002; Bogdanov et al., 2008; Vitrac et al., 2013; Wang et al., 2002). However, a quantitative difference between SMFS and site-directed alkylation is apparent. Specifically, in PG membranes SMFS reveals ≈50:50 ratio between LacY molecules in the native versus the perturbed state, while site-directed alkylation exhibits 100% inversion. Notably, SMFS does not involve the mutations introduced for site-directed alkylation, which might be responsible for the quantitative differences, and SMFS, as a single-molecule technique, avoids problems involving signal averaging and dilution.
Alternatively, the low lipid to protein ratio of 5:1 (w:w) used in our studies in contrast to the much higher ratios used for site-directed alkylation could influence the thermodynamic balance (or equilibrium) between the two conformational states of LacY. An increase in dual topological conformers for LacY in PG only proteoliposomes was observed as the lipid to protein ratio was decreased below 50:1, and at a 5:1 ratio (w:w), a 35:65 ratio of native to inverted conformer was observed with site-directed alkylation (H. Vitrac and W. Dowhan, personal communication). In any event, it is most important that SMFS further confirms that the topogenesis of proteins can be probabilistic by nature and the balance of forces near equilibrium is an important decision-making step in thermodynamically driven membrane protein topogenesis (Bogdanov and Dowhan, 2012).
EXPERIMENTAL PROCEDURES
Materials
Talon superflow resin was purchased from BD Clontech (Palo Alto, CA). Dodecyl-β-D-maltopyranoside (DDM) and n-octyl-β-D-glucoside were from Anatrace (Maumee, OH), and synthetic phospholipids (POPE and POPG) were from Avanti Polar Lipids, Inc. (Alabaster, AL). All other materials were of reagent grade obtained from commercial sources.
LacY Engineering, Expression and Purification
Engineering, expression and purification of LacY was carried out as described previously (Serdiuk et al., 2014). Briefly, a 36 aa “polyGly” followed by a 8 His-tag extension [GSM(G11)EAVEEAVEEA(Gly11)S(His8)] was engineered to the LacY C-terminus using QuikChange® II PCR and plasmid pT7-5/LacY as template. WT LacY was purified from E. coli XL1-Blue (Stratagene, La Jolla, CA) transformed with pT7-5 plasmids harboring given mutant genes by using Co(II) affinity chromatography. LacY eluted from the Co(II)-Talon column was concentrated and washed with 50 mM sodium phosphate (NaPi) (pH 7.5)/0.01% w/w DDM on an Amicon Ultra-15 concentrator with a 30K cutoff (Millipore, Billerica, MA). All protein preparations were >95% pure as judged by silver staining after sodium dodecylsulfate polyacrylamide (SDS) gel electrophoresis. Samples with protein concentrations of 5–10 mg/ml in 50 mM NaPi buffer (pH 7.5)/2–4% w/w DDM, were frozen in liquid nitrogen and stored at –80°C.
Reconstitution of Purified Proteins into Proteoliposomes
Reconstitution was performed described previously (Serdiuk et al., 2014). Briefly, synthetic phospholipids (POPE:POPG at ratio 3:1 or POPG) were used for protein reconstitution by the dilution method (Viitanen et al., 1986). Purified LacY in 0.02% w/w DDM was mixed with phospholipids dissolved in 1.2% w/w octyl-β-D-glucopyranoside at a lipid to protein ratio of 5:1 (w/w). The mixture was kept on ice for 20 min and then quickly diluted 50 times in 50 mM NaPi (pH 7.5). Proteoliposomes were collected by centrifugation at 100000g for 1 h and subjected to 2 cycles of freeze-thaw/sonication before use.
SMFS
SMFS was performed on WT LacY reconstituted in POPE:POPG (3:1) or POPG in buffer solution (50 mM KPi, pH=7.2) at ≈25°C as described (Serdiuk et al., 2014). Briefly, SMFS measurements were done using an 850 nm laser-equipped AFM (NanoWizard II, JPK Instruments) and Si3N4 cantilevers (OMCL RC800PCA, Olympus) having a nominal spring constant of 0.05 N/m. Cantilever calibration was performed in buffer solution using the equipartition theorem at the beginning and at the end of each experiment (te Riet et al., 2011). SMFS was performed retracting the cantilever at different speeds (500, 700, 1’000, 3’000, 4’500, 6’000 nm/s), with negligible hydrodynamic effects (Janovjak et al., 2005). To record high-frequency data at pulling speeds >1’000 nm/s, an external 16-bit data acquisition card (NI PCI-6221, National Instruments) was used (Bippes et al., 2009).
SMFS Data Analysis
Mechanically fully unfolded and stretched LacY [417 aa elongated by a 36 aa long polyGly tail and an His8-tag] exhibit the force peak patterns extended to the distance ≈110–120 nm. To ensure that we only analyzed FD curves recording the unfolding of LacY from the terminal end, we selected the FD curves corresponding to the fully unfolded and stretched LacY polypeptide. Therefore, FD curves showing force peak patterns extending to distances >110 nm were accounted. Every selected FD curve was fitted with the wormlike- chain (WLC) model with a persistence length of 0.4 nm and a contour length of 0.36 nm per aa (Bustamante et al., 1994; Rief et al., 1997). The WLC fit of each unfolding force peak shows the number of aa unfolded and stretched (contour length) and a force that is required to mechanically unfold a stable structural segment (rupture force). Contour lengths and rupture forces of all peaks were statistically analyzed. Histograms showing how frequently a force peak was detected at a given contour length were generated. To calculate the frequency, all counts of force peaks were divided through the total number of FD curves analyzed. Since every FD curve has many force peaks, the total frequency of all peaks of a histogram summed up to >>100%, but the probability of each individual force peak class being ≤100%. Contour length histograms were fitted with the Gaussian mixture model as described (Kawamura et al., 2013). Accordingly to this model, each i-th observed contour length li stems from one force peak class s = 1,…., M with probability πs or from background noise with probability π0. Ten force peaks classes were found for the class 1 and eleven force peak classes were found for the class 2 FD curves recorded from WT LacY. The contour length for a given force peak class s was described by a Gaussian distribution with mean contour length µs and variance σs2. Because it is unknown to which force peak class a contour length belongs, the probability density f of li can be presented as a mixture of Gaussians and background noise with weights πs and π0 correspondingly.
| (Equation 1) |
where ϕ(li, µs, σs2) is the probability density of the Gaussian distribution and g(li) the background noise. To find parameters of the model (π, µ, σ2) the expectation optimization algorithm was applied (Dempster et al., 1977). The most probable force peak class si was assigned to any given contour length li with the Bayes classifier by setting (Schwarz, 1978):
| (Equation 2) |
All detected contour lengths were assigned to the force peak classes (Serdiuk et al., 2014).
Supplementary Material
ACKNOWLEDGEMENTS
We are greatly indebted to William Dowhan and Mikhail Bogdanov for their very helpful editorial input. We are also indebted to Irina Smirnova for purifying and providing LacY, and we thank Johannes Thoma and Stefan Weiser for assistance and critical discussion. This work was supported the NCCR Molecular Systems Engineering of the Swiss National Science Foundation (SNF; Grant 200021_134521 to D.J.M.), the European Union Marie Curie Actions program through the ACRITAS Initial Training Network (FP7-PEOPLE-2012-ITN, Project 317348), National Institutes of Health Grants DK51131, DK069463, and GM073210, as well as National Science Foundation Grant MCB-1129551 (to H.R.K.).
Abbreviations
- aa
amino acids
- AFM
atomic force microscopy
- FD curve
force-distance curve
- LacY
lactose permease
- KPi
potassium phosphate
- MFS
major facilitator superfamily
- polyGly
polyglycine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3[phospho-rac-(1-glycerol)
- S1–S10
structural segments 1–10
- SD
standard deviation
- SMFS
single-molecule force spectroscopy
- WLC
worm-like-chain.
Variables and constants
- D
distance
- F
force
- ϕ
probability density of the Gaussian distribution
- g
background noise of force peak histograms
- li
i-th observed contour length
- M
number of force peak classes
- Δµ̃H+
electrochemical H+ gradient
- µs
mean contour length
- n
number of FD curves
- rsv
most probable loading rate
- π0
weight of background noise
- πs
weights of Gaussians
- s
force peak class
- σs2
variance
- v
pulling velocity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information including figures and Supplemental Experimental Procedures and can be found with this article online at xxx will be completed later xxx.
AUTHOR CONTRIBUTIONS
T.S., H.R.K., and D.J.M. designed research; T.S. and S.A.M. performed research; J.S. contributed new reagents/analytic tools; T.S., H.R.K. and D.J.M. analyzed data; and T.S., H.R.K., and D.J.M. wrote the paper.
Serdiuk et al. structurally localize the interactions that stabilize single lactose permeases (LacY) in phospholipid membranes. In the absence of phosphatidylethanolamine LacY adopts perturbed conformations, thus suggesting an alternating topology. Drastic changes are located at helices VI and VII and the intervening loop.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- Bippes C, Zeltina A, Casagrande F, Ratera M, Palacin M, Muller DJ, Fotiadis D. Substrate binding tunes conformational flexibility and kinetic stability of an amino acid antiporter. J. Biol. Chem. 2009;28:18651–18663. doi: 10.1074/jbc.M109.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bippes CA, Ge L, Meury M, Harder D, Ucurum Z, Daniel H, Fotiadis D, Muller DJ. Peptide transporter DtpA has two alternate conformations, one of which is promoted by inhibitor binding. Proc. Natl. Acad. Sci. U S A. 2013;110:E3978–E3986. doi: 10.1073/pnas.1312959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bippes CA, Muller DJ. High-resolution atomic force microscopy and spectroscopy of native membrane proteins. Rep. Progr. Phys. 2011;74:086601. [Google Scholar]
- Bogdanov M, Dowhan W. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J. Biol. Chem. 1995;270:732–739. doi: 10.1074/jbc.270.2.732. [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J. 1998;17:5255–5264. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W. Lipid-dependent generation of dual topology for a membrane protein. J. Biol. Chem. 2012;287:37939–37948. doi: 10.1074/jbc.M112.404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W, Vitrac H. Lipids and topological rules governing membrane protein assembly. Biochim. Biophys. Acta. 2014;1843:1475–1488. doi: 10.1016/j.bbamcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Heacock PN, Dowhan W. Study of polytopic membrane protein topological organization as a function of membrane lipid composition. Methods Mol. Biol. 2010;619:79–101. doi: 10.1007/978-1-60327-412-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J. Biol. Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J. Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Calamia J, Manoil C. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc. Natl. Acad. Sci. U S A. 1990;87:4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaptal V, Kwon S, Sawaya MR, Guan L, Kaback HR, Abramson J. Crystal structure of lactose permease in complex with an affinity inactivator yields unique insight into sugar recognition. Proc. Natl. Acad. Sci. U S A. 2011;108:9361–9366. doi: 10.1073/pnas.1105687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wilson TH. The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J. Biol. Chem. 1984;259:10150–10158. [PubMed] [Google Scholar]
- Dempster A, Laird N, D R. Maximum likelihood from incomplete data via the EM algorithm (with discussions) J. R. Statist. Soc. B. 1977;39:1–38. [Google Scholar]
- Dowhan W. Lipids and extracellular materials. Annu Rev Biochem. 2014;83:45–49. doi: 10.1146/annurev-biochem-010314-112017. [DOI] [PubMed] [Google Scholar]
- Dowhan W, Bogdanov M. Molecular genetic and biochemical approaches for defining lipid-dependent membrane protein folding. Biochim. Biophys. Acta. 2012;1818:1097–1107. doi: 10.1016/j.bbamem.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu. Rev. Biochem. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- Foster DL, Boublik M, Kaback HR. Structure of the lac carrier protein of Escherichia coli. J. Biol. Chem. 1983;258:31–34. [PubMed] [Google Scholar]
- Ge L, Perez C, Waclawska I, Ziegler C, Muller DJ. Locating an extracellular K+-dependent interaction site that modulates betaine-binding of the Na+-coupled betaine symporter BetP. Proc Natl Acad Sci U S A. 2011;108:E890–E898. doi: 10.1073/pnas.1109597108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc. Natl. Acad. Sci. U S A. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janovjak H, Kessler M, Oesterhelt D, Gaub H, Muller DJ. Unfolding pathways of native bacteriorhodopsin depend on temperature. Embo J. 2003;22:5220–5229. doi: 10.1093/emboj/cdg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janovjak H, Struckmeier J, Muller DJ. Hydrodynamic effects in fast AFM single-molecule force measurements. Eur. Biophys. J. 2005;34:91–96. doi: 10.1007/s00249-004-0430-3. [DOI] [PubMed] [Google Scholar]
- Kaback HR, Smirnova I, Kasho V, Nie Y, Zhou Y. The alternating access transport mechanism in LacY. J. Membr. Biol. 2011;239:85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura S, Gerstung M, Colozo AT, Helenius J, Maeda A, Beerenwinkel N, Park PS, Muller DJ. Kinetic, energetic, and mechanical differences between dark-state rhodopsin and opsin. Structure. 2013;21:426–437. doi: 10.1016/j.str.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrov A, Janovjak H, Sapra KT, Muller DJ. Deciphering molecular interactions of native membrane proteins by single-molecule force spectroscopy. Annu. Rev. Biophys. Biomol. Struct. 2007;36:233–260. doi: 10.1146/annurev.biophys.36.040306.132640. [DOI] [PubMed] [Google Scholar]
- Kedrov A, Krieg M, Ziegler C, Kuhlbrandt W, Muller DJ. Locating ligand binding and activation of a single antiporter. EMBO Rep. 2005;6:668–674. doi: 10.1038/sj.embor.7400455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kasho V, Smirnova I, Finer-Moore JS, Kaback HR, Stroud RM. Structure of sugar-bound LacY. Proc. Natl. Acad. Sci. U S A. 2014;111:1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej MG, Soro SN, Kaback HR. Apo-intermediate in the transport cycle of lactose permease (LacY) Proc. Natl. Acad. Sci. U S A. 2012;109:E2970–E2978. doi: 10.1073/pnas.1211183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger MD, Saier MH., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller DJ, Engel A. Atomic force microscopy and spectroscopy of native membrane proteins. Nat. Protoc. 2007;2:2191–2197. doi: 10.1038/nprot.2007.309. [DOI] [PubMed] [Google Scholar]
- Muller DJ, Kessler M, Oesterhelt F, Moller C, Oesterhelt D, Gaub H. Stability of bacteriorhodopsin alpha-helices and loops analyzed by single-molecule force spectroscopy. Biophys. J. 2002;83:3578–3588. doi: 10.1016/S0006-3495(02)75358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PS, Sapra KT, Kolinski M, Filipek S, Palczewski K, Muller DJ. Stabilizing effect of Zn2+ in native bovine rhodopsin. J. Biol. Chem. 2007;282:11377–11385. doi: 10.1074/jbc.M610341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Sapra KT, Balasubramanian GP, Labudde D, Bowie JU, Muller DJ. Point mutations in membrane proteins reshape energy landscape and populate different unfolding pathways. J. Mol. Biol. 2008;376:1076–1090. doi: 10.1016/j.jmb.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra KT, Besir H, Oesterhelt D, Muller DJ. Characterizing molecular interactions in different bacteriorhodopsin assemblies by single-molecule force spectroscopy. J. Mol. Biol. 2006;355:640–650. doi: 10.1016/j.jmb.2005.10.080. [DOI] [PubMed] [Google Scholar]
- Schwarz G. Estimating the dimension of a model. Ann. Statist. 1978;6:461–464. [Google Scholar]
- Serdiuk T, Madej MG, Sugihara J, Kawamura S, Mari SA, Kaback HR, Muller DJ. Substrate-induced changes in the structural properties of LacY. Proc. Natl. Acad. Sci. U S A. 2014;111:E1571–E1580. doi: 10.1073/pnas.1404446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto-Young D, Chen CC, Wilson TH. Effect of different phospholipids on the reconstitution of two functions of the lactose carrier of Escherichia coli. J. Membr. Biol. 1985;84:259–267. doi: 10.1007/BF01871389. [DOI] [PubMed] [Google Scholar]
- Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50:9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riet J, Katan AJ, Rankl C, Stahl SW, van Buul AM, Phang IY, Gomez-Casado A, Schon P, Gerritsen JW, Cambi A, et al. Interlaboratory round robin on cantilever calibration for AFM force spectroscopy. Ultramicroscopy. 2011;111:1659–1669. doi: 10.1016/j.ultramic.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Viitanen P, Newman MJ, Foster DL, Wilson TH, Kaback HR. Purification, reconstitution, and characterization of the lac permease of Escherichia coli. Methods Enzymol. 1986;125:429–452. doi: 10.1016/s0076-6879(86)25034-x. [DOI] [PubMed] [Google Scholar]
- Vitrac H, Bogdanov M, Dowhan W. In vitro reconstitution of lipid-dependent dual topology and postassembly topological switching of a membrane protein. Proc. Natl. Acad. Sci. U S A. 2013;110:9338–9343. doi: 10.1073/pnas.1304375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocher M, Fung JJ, Kobilka BK, Muller DJ. Ligand-specific interactions modulate kinetic, energetic, and mechanical properties of the human beta(2) adrenergic receptor. Structure. 2012a;20:1391–1402. doi: 10.1016/j.str.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocher M, Zhang C, Rasmussen SG, Kobilka BK, Muller DJ. Cholesterol increases kinetic, energetic, and mechanical stability of the human beta2-adrenergic receptor. Proc. Natl. Acad. Sci. U S A. 2012b;109:E3463–E3472. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.