Abstract
The antimicrotubular agent docetaxel is a widely used chemotherapeutic drug for the treatment of multiple solid tumors and is predominantly dependent on hepatic disposition. In this study, we evaluated drug uptake transporters capable of transporting radiolabeled docetaxel. By screening an array of drug uptake transporters in HeLa cells using a recombinant vaccinia-based method, five organic anion transporting polypeptides (OATPs) capable of docetaxel uptake were identified: OATP1A2, OATP1B1, OATP1B3, OATP1C1, and Oatp1b2. Kinetic analysis of docetaxel transport revealed similar kinetic parameters among hepatic OATP1B/1b transporters. An assessment of polymorphisms (SNPs) in SLCO1B1 and SLCO1B3 revealed that a number of OATP1B1 and OATP1B3 variants were associated with impaired docetaxel transport. A transwell-based vectorial transport assay using MDCKII stable cells showed that docetaxel was transported significantly into the apical compartment of double transfected-(MDCKIIOATP1B1/MDR1 and MDCKII-OATP1B3/MDR1) cells compared to single transfected-(MDCKII-OATP1B1 and MDCKII-OATP1B3) cells (p<0.05) or control (MDCKII-Co) cells (p<0.001). In vivo docetaxel transport studies in Slco1b2−/− mice showed ~>5.5-fold higher plasma concentrations (p<0.01) and ~3-fold decreased liver-to-plasma ratio (p<0.05) of docetaxel compared to wild-type mice. The plasma clearance of docetaxel in Slco1b2−/− mice was 83% lower than wild-type mice (p<0.05). In conclusion, this study demonstrates the important roles of OATP1B transporters to the hepatic disposition and clearance of docetaxel, and supporting roles of these transporters for docetaxel pharmacokinetics.
Keywords: OATP1B1, OATP1B3, Docetaxel, Disposition, Transporter
Introduction
Docetaxel is a widely used chemotherapeutic drug for the treatment of multiple solid tumors, including breast, lung, head and neck, stomach, and prostate cancer. There is wide variability in the pharmacokinetics of docetaxel with up to 10-fold difference in drug clearance amongst patients receiving the same treatment regimens (1). Furthermore, studies have indicated that a mere 50% decrease in docetaxel clearance increased more than 4-fold the odds of developing grade 4 neutropenia (2). Accordingly, this substantial interindividual variability in docetaxel exposure may have important ramifications for clinical efficacy and drug-mediated toxicity.
Recent studies suggest that drug disposition genes play a major role in the variability of docetaxel disposition (3-5). Furthermore, genetic variation, or polymorphisms, in drug disposition genes contributes to interindividual variability in chemotherapy response and toxicity (6-8). As the disposition of docetaxel is closely related to differences between an individual's ability to metabolize and eliminate this drug (3, 4, 6-8), a major cause of pharmacokinetic variability may be attribute to differential expression of polymorphic drug-metabolizing enzymes and/or drug-eliminating transporters, but it has still been unclear. Docetaxel is mainly metabolized by the hepatic phase I drug metabolizing enzyme CYP3A4 and, to a lesser extent, by CYP3A5 (9) and eliminated by Multidrug Resistance Protein 1 (MDR1, ABCB1, P-glycoprotein) (10). MDR1 is a membrane-localized and ATP-dependent drug efflux transporter with broad substrate specificity and primarily expressed in normal tissues such as the gastrointestinal tract, the liver, the kidney, and the brain (11, 12). Expression of MDR1 in such tissues results in reduced drug absorption (gastrointestinal tract), enhanced elimination into the bile (liver) and urine (kidney), and reduced accumulation of toxic agents (the brain) (11).
Docetaxel is predominantly dependent on hepatic disposition in humans; ~75% docetaxel is excreted in bile but only ~5% in urine (13). Until recently, little was known regarding the hepatic uptake of docetaxel. Organic anion-transporting polypeptides (OATPs) are primarily expressed in organs of importance to drug disposition such as the liver, intestine, kidney, and brain, and mediate the sodium-independent transport of a diverse range of amphipathic organic compounds including bile salts, steroid conjugates, thyroid hormones, anionic peptides, and numerous drugs (14, 15). Whereas OATP1A2, which is mainly expressed in brain, kidney and small intestine, is expressed in cholangiocytes of liver, but not in hepatocytes (16), OATP1B1 and OATP1B3, are expressed highly in the hepatocytes of the liver and their localization is restricted to the basolateral membrane of hepatocytes (17). Accordingly, these transporters facilitate the hepatocellular accumulation of compounds such as drugs before metabolism and biliary secretion, and thus may largely contribute to drug disposition. As hepatic uptake appears to be an important factor in docetaxel clearance, it would be of interest to evaluate hepatic uptake transporters as determinants of docetaxel disposition. Indeed, previous studies indicated that OATP1B3 is capable of transporting docetaxel in vitro (3, 18). Recently, a study also reported that differential expression of the OATP1B family in the human liver regulates the initial step in the elimination of docetaxel, before metabolism (5).
In this study, we identified multiple OATPs capable of transporting docetaxel by screening an array of drug uptake transporters in HeLa cells using a recombinant vaccinia-based method. Among them, we aimed to gain more insight into the importance of hepatic OATPs to the disposition of docetaxel. Thus, we studied the roles of OATP1B transporters in the hepatic uptake, clearance and plasma exposure of docetaxel in vivo and in vitro.
Materials and Methods
Chemicals and reagents
Radiolabeled [3H]docetaxel (5 Ci/mmol, 15 Ci/mmol; >98% purity) was obtained from Moravek Biochemicals and Radiochemicals (Brea, CA). Unlabeled docetaxel (>97% purity) was obtained from Sigma-Aldrich (St. Louis, MO). All other chemicals and reagents, unless stated otherwise, were obtained from Sigma-Aldrich Research and were of the highest grade available.
Cell culture and virus preparation
HeLa cells were purchased from American Type Culture Collection (Manassas, VA; July 2012) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (100 μg/ml) (Invitrogen, Grand Island, NY). The polarized MDCKII (Madin-Darby Canine Kidney II) cells were purchased from the Sigma-Aldrich (January 2013) and we generated several stable cell lines immediately after obtaining the cells. The original bank of MDCKII cells is from Public Health England (PHE) (London, UK) and PHE authenticates their cell line, including testing for mycoplasma by culture isolation, Hoechst DNA staining and PCR, culture testing for contaminant bacteria, yeast and fungi, species verification by DNA barcoding, and identity verification by DNA profiling. In addition, stable cell lines generated with the MDCKII cells for this study were confirmed by protein expression and transport activities using known specific substrates ([3H]estrone-3-sulfate for OATP1B1; [3H]cholecystokinin for OATP1B3). All MDCKII cells were cultured in DMEM containing high glucose and L-glutamine supplemented with 10 % heat-inactivated FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml) (Invitrogen) and all cells including HeLa cells were maintained in a 5% CO2 atmosphere at 37 °C in a humidified incubator. Preparation of viral stock of vtf-7 virus was prepared as described previously (19). Briefly, HeLa cells grown to near confluence in 25-cm tissue culture plates were infected with 1 plaque-forming unit (PFU)/10 cells. After an incubation period of 48 h at 37°C, the infected cells were pelleted, homogenized, and recovered through centrifugation, followed by tittering of viral stock as described by Blakely et al. (20).
Wild-type and variant transporter plasmid construction
Preparation of expression plasmids containing human and rat uptake transporters, including pEF6/V5-His/OATP1B1 and pEF6/V5-His/OATP1B3, has been described previously (21). Briefly, the full open reading frames (ORFs) of human and rat transporter cDNAs were obtained by PCR, using AmpliTaq® DNA polymerase (PerkinElmer, Waltham, MA), from cDNA libraries synthesized from a variety of human and rat tissue mRNA, respectively. A pEF6/V5-His/MDR1 expression plasmid was also obtained by the same method as above. Generation of OATP1B1 and OATP1B3 variant expression plasmids used for in vitro functional studies has been described previously (21, 22). For consistency of expression, all transporters were packaged into the pEF6/V5-His-TOPO® vector (Invitrogen). The expression plasmids for wild-type pcDNA3.1(+)/OATP1B1 and pcDNA3.1(+)/OATP1B3 were constructed by excising out the ORFs of the OATP1B1 and OATP1B3 cDNA sequences respectively from the plasmids pEF6/V5-His/OATP1B1 and pEF6/V5-His/OATP1B3 with Nhe1 and Xho1, and subcloning into pcDNA3.1(+) vector (Invitrogen). To generate an expression plasmid for pcDNA3.1-zeo(+)/MDR1 (wild-type), PCR was first performed with corresponding nucleotides containing start codon and internal EcoR1 site of MDR1 cDNA sequences using plasmid pEF6/V5-His/MDR1 as a template and PCR products (1-1182nts) were then subcloned into pcDNA3.1- zeo(+) vector (Invitrogen). Second, DNA fragments (1171-3843nts) were obtained by digesting MDR1 cDNA with EcoR1 and Not1 from plasmid pEF6/V5-His/MDR1. Finally, pcDNA3.1-zeo(+)/MDR1 (wt) plasmid containing the full ORFs (1-3843nts) of MDR1 was generated by inserting the second DNA fragments (1171-3843nts) into the first pcDNA3.1-zeo(+)/MDR1(1-1182nts) plasmid digested with the same enzymes described above. All plasmids including wild-types and variants were verified by sequencing in the DNA Sequencing Facility (VANTAGE) at Vanderbilt University Medical Center.
Measurement of docetaxel transport kinetics
Docetaxel transport kinetics were assessed in HeLa cells recombinantly expressing OATP1B/1b transporters. To measure transport kinetics, [3H]docetaxel uptake during the linear phase (first 3 min) was assessed in the presence of various concentrations of unlabeled compound. Transporter-dependent uptake was determined in parallel experiments as the difference in drug uptake between transporter and parental plasmid DNA-transfected HeLa cells. Michaelis-Menten-type nonlinear curve-fitting was used to estimate the maximal uptake rate (Vmax) and concentration at which half the maximal uptake occurs (Km) for OATP1B3 and Oatp1b2. For OATP1B1 uptake kinetics, weighted, non-linear curve fitting was performed using a two-component model comprising of a saturable process and a non-saturable process. All experiments were carried out in duplicate on at least two to three experimental days.
Generation of stably transfected MDCKII cells
MDCKII cell lines expressing hepatic uptake transporters or/and an efflux transporter as well as control cell (MDCKII-Co, MDCKII-MDR1, MDCKII-OATP1B1, MDCKII-OATP1B3, MDCKII-OATP1B1/MDR1 and MDCKII-OATP1B3/MDR1) were generated as follows: In order to generate the MDCKII-OATP1B1 and MDCKII-OATP1B3 single-transfected cell lines, MDCKII cells were transfected with the plasmids pcDNA3.1(+)/OATP1B1 or pcDNA3.1(+)/OATP1B3 using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. At 18-32h after transfection, the cells were split and divided into 10 cm culture dishes with fresh media containing G-418 sulfate (800 μg/mL; Mediatech, Manassas, VA). After additional selection with G-418 sulfate, single colonies of transfectants were screened for OATP1B1 and OATP1B3 protein expression by immunoblot analysis to detect cell clones with the desired protein expression. Cell clones with highest protein expression comparable with the expression of the control cell lines were chosen for further experiments. For generation of MDCKII-OATP1B1/MDR1 and MDCKII-OATP1B3/MDR1 double-transfected cell lines, single transfected MDCKII-OATP1B1 and MDCKII-OATP1B3 were re-transfected with the plasmid pcDNA3.1-zeo(+)/MDR1 using the same transfection reagent mentioned above. For generation of MDCKII-MDR1 cells, MDCKII cells were also transfected with the plasmid pcDNA3.1-zeo(+)/MDR1. After additional selection with zeocin (800 μg/mL; Invitrogen), single colonies of all transfectants were screened for MDR1 expression as well as OATP1B1 and OATP1B3 by immunoblot analysis to detect cell clones with the highest expression. All expression values were normalized to the protein β-actin. Consequently, cell clones with the highest uptake transporters and/or an efflux transporter expression were chosen and used for drug transport studies. Control cells (MDCKII-Co) lacking any insert were obtained by first transfecting the MDCKII cells with pcDNA3.1(+) vector only, followed by re-transfecting with pcDNA3.1-zeo (+)vector only.
Preparation of crude cell membrane fractions and cell surface proteins
Protein expression from MDCKII stable cells was assessed by immunoblot analysis of plasma membrane enriched preparations and biotinylated cell surface proteins. Crude cell membrane fractions were conducted as described previously with minor modification (23). Briefly, cells were scraped and collected in hypotonic lysis buffer (5 mM Hepes, pH 7.2, 300 mM mannitol, 1 mM DTT and 1x protease inhibitor cocktail (Roche, Indianapolis, IN)) and homogenized with pellet pestle (Thermo Scientific, Rockford, IL) and syringe (26G). The lysates were centrifuged at 800g for 10 min to remove cell debris and the supernatant was saved. The supernatant was then centrifuged at 30,000g at 4 °C for 30 min to obtain membrane pellets. The membrane pellets were dissolved in the same buffer without mannitol and protein concentration in pellets was determined using the bicinchoninic acid assay (BCA) protein assay kit (Thermo Scientific). Each 10 μg of extracted membrane proteins was used for immunoblot analysis. For the isolation of cell surface fractions, cell surface biotinylation was performed as described previously (21). Briefly, an equal number of each MDCKII stable cell was plated into 6 well culture plates. 24 hours later, each well was washed in ice-cold PBS- Ca2+/Mg2+ (138 mM NaCl2, 2.7 mM KCl, 1.5 mM KH2PO4, 1 mM MgCl2, 0.1 mM CaCl2, pH 7.4) and treated with sulfo-N-hydroxysuccinimide-SS-biotin (Thermo Scientific) for 1 h at 4 °C. The cells were washed with ice-cold PBS-Ca2+/Mg2+ containing 100 mM glycine and lysed with RIPA buffer (10 mM Tris base, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, pH 8.0) containing 1x protease inhibitor cocktail (Roche). Following centrifugation, 140 μL of streptavidin-agarose beads (Thermo Scientific) were added to 600 μl of cell lysate and incubated for 1 h at room temperature. Beads were washed four times with ice-cold lysis buffer, and biotinylated proteins were released from the beads by adding Laemmli sample buffer (Bio-Rad, Hercules, CA). Consequently, eluted biotinylated (cell surface-expressed) proteins were subjected to immunoblot analysis for detection of OATP1B1, OATP1B3 or MDR1 by specific antibodies.
Immunoblot analysis
To confirm protein expressions from each stable cell line generated, immunoblot analysis was performed as described previously (14). Briefly, crude cell membrane proteins (each 10 μg) and biotinylated fractions were loaded on 10% SDS polyacrylamide gels and transferred to nitrocellulose membranes (PerkinElmer). After preincubation in phosphate-buffered saline (PBS) containing 0.05% Tween 20 and 5% nonfat dry milk, the blots were first incubated with the polyclonal rabbit anti-OATP1B3 antibody (1:2,500 dilution) and re-probed with different antibodies later, such as the polyclonal rabbit anti-OATP1B1 antibody (1:2,500 dilution) and the monoclonal anti-MDR1 antibody (1:1000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) at room temperature for 2h. The blots were washed with PBS containing 0.05% Tween 20 for 15 min (3 X 5 min) and then incubated for 1h with the secondary antibody an anti-Rabbit IgG for OATP1B1 and OATP1B3 and an anti-mouse IgG antibody for MDR1 conjugated with horseradish peroxidase (HRP) (1:10,000 dilution; Promega, Madison, WI). The protein bands were detected using the western lightning plus-enhanced chemiluminescence ECL (PerkinElmer). As a loading control, the blots were stripped and re-probed with the monoclonal mouse anti-Na+/K+-ATPase α antibody (1:5,000 dilution; Santa Cruz Biotechnology), followed by incubation with an anti-mouse IgG antibody conjugated with HRP (1:10,000 dilution; Promega).
Transport assays
Recombinant vaccinia-based uptake transport assay
Transport assays using a recombinant vaccinia virus were conducted as described previously (19). Briefly, HeLa cells grown in 12-well plates (0.8 × 106 cells/well) were infected with vaccinia (vtf-7) at a multiplicity of infection of 10 PFU/cell in Opti-MEM I medium (Invitrogen) and allowed to adsorb for 30 min at 37°C. Cells in each well were then transfected with 1 μg transporter cDNA packaged in pEF6/V5-His-TOPO® vector (Invitrogen), along with Lipofectin® (Invitrogen) and incubated at 37°C for 16 h. The parental plasmid lacking any insert was used as control. Uptake (radioactivity) of docetaxel was measured after an incubation of 10 minutes. Total radioactivity was determined after the addition of cell lysates to vials containing 5 ml of Biodegradable Scintillation Cocktail (Amersham Biosciences, Buckinghamshire, England). Retained cellular radioactivity was quantified by liquid scintillation counter (PerkinElmer, Shelton, CT). Transport activities were expressed in percent compared to the vector control. All experiments were carried out in duplicate on at least 2-3 experimental days. In each set of experiments, taurocholate uptake into cells transfected to express sodium/taurocholate cotransporting polypeptide (NTCP) was included as a positive control for transfection and expression efficiency. Experience with this assay panel has demonstrated that inclusion of a single positive control is sufficiency, as relative activities of each transporter for reference substrates are consistent between experiments. In each set of experiments, docetaxel uptake into the cells transfected with the vector only was included as a negative control to ensure that results were not confounded by an effect of the transfection process.
Transwell-based vectorial transport assay
Vectorial transport assays were conducted as described previously with minor modifications (24). MDCKII cells were seeded onto 12-Transwell (diameter 12 mm; pore size 0.4 μm; Corning Incorporated, Corning, NY) at an initial density of 0.4 X 106 cells per well and grown for 3 days. Twenty-four hours before transport experiments, the cells were treated with 10 mM sodium butyrate (Sigma-Aldrich) to increase protein expression. Transepithelial resistance was measured in each well using a millicell voltammeter (model ERS; Millipore, Bedford, MA); wells registering a resistance of 200 Ω or greater, after correcting for the resistance obtained in control blank wells, were used in the transport experiments (23). [3H]Docetaxel was dissolved in the uptake buffer Opti-MEM 1 medium (Invitrogen) to a final concentration of 0.1 μM without addition of unlabeled docetaxel. After washing with pre-warmed uptake buffer, 0.8 mL of uptake buffer without [3H]docetaxel was added to the apical compartments of the cell monolayers. The same amount (0.8 mL) of uptake buffer containing [3H]docetaxel was then added to the basolateral compartments of the cell monolayers. Cells were then incubated at 37°C for given time points. After 0.5, 1 and 2h, aliquots (50 μL) were removed from both the apical and basolateral compartments and plates were placed back to the incubator. After 3h incubation, additional 50 μL were taken from both compartments and docetaxel radioactivity in media obtained at each time point was then measured by liquid scintillation counter (PerkinElmer). Finally, the cells were washed three times with ice-cold PBS buffer and were lysed with 0.2% SDS solution containing 1x protease inhibitor cocktail (Roche). Cell lysates were then used to determine the protein concentrations by BCA assay kit (Thermo Scientific). To measure the activity of intracellular docetaxel, cells in transwells were incubated for 0.5, 1, 2, and 3h after adding radiolabeled docetaxel (final conc. 0.1 μM). The transwell plates were taken out of the incubator at each time point. Cells were then washed, lysed, followed by measuring the radioactivity of docetaxel and determining the protein concentration as described above. All experiments were performed more than three times in > triplicates.
Docetaxel distribution in Slco1b2 −/− mice
Male Slco1b2 −/− mice, 21-24 weeks of aged and male WT littermate controls were used to determine docetaxel distribution. The radiolabeled [3H]docetaxel (1 mg/kg, specific activity 30 Ci/mmol) dissolved in ethanol/0.9% saline solution was injected intravenously into the tail vein of groups of four mice. Blood samples from each mouse were drawn from the saphenous vein at 5 and 15 min post-injection. After 30 min, mice were anesthetized with isofluorane, blood was removed by cardiac puncture and livers were harvested, weighed and homogenized with PBS (1% wt/vol bovine serum albumin). Total radioactivity was determined after the addition of plasma (25 μl) or liver homogenates (500 μl) to vials containing 5 ml of Ultima Gold Scintillation Cocktail (PerkinElmer LAS Canada Inc., Woodbridge, ON, Canada). Docetaxel clearance after IV injection was calculated as dose/AUC, where AUC is the area under the plasma concentration-time profile from t = 0 to ∞. The protocols for the animal experiments were approved by The University of Western Ontario Animal Care Committee.
Data fitting and statistical analysis
Parameters for saturation kinetics (Vmax and Km) were estimated by nonlinear curve fitting using Prism (GraphPad Software, Inc., San Diego, CA) or Scientist (Micromath Scientific Software, St. Louis, MO). Determination of statistical differences between group parameters was determined using Student's t test, Mann-Whitney U test, one way ANOVA, analysis of variance (using Tukey-Kramer multiple comparison test), or Fisher's exact test, as appropriate. A p value of <0.05 was taken to be the minimum level of statistical significance.
Results
Docetaxel uptake is mediated by multiple OATP transporters in vitro
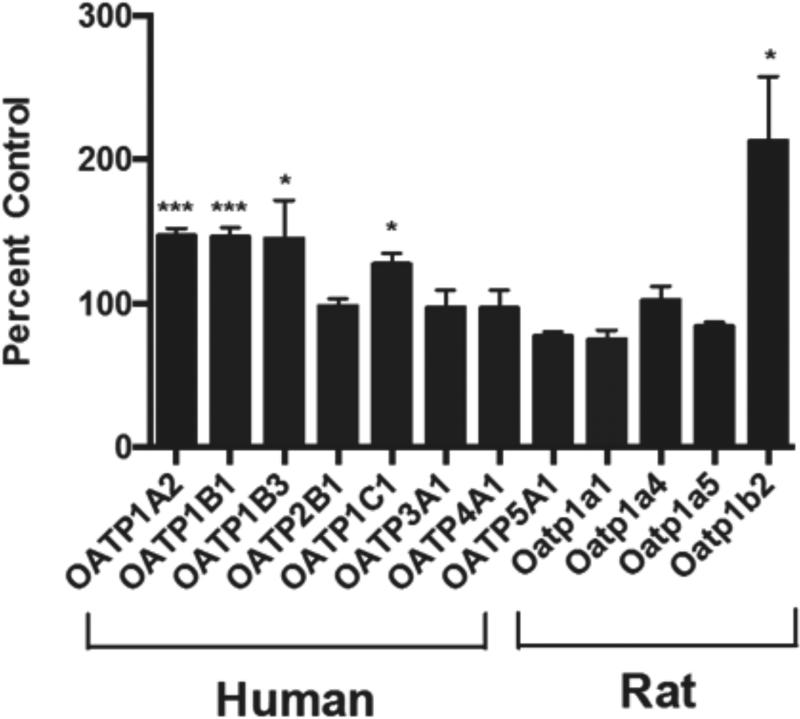
A panel of uptake transporter expression plasmids was expressed individually in HeLa cells using recombinant vaccinia and evaluated for docetaxel transport. After 10 min incubation, we identified several members of the human OATP family capable of docetaxel uptake, including OATP1A2, OATP1B1, OATP1B3, and OATP1C1 (Fig. 1). Rat Oatp1b2, orthologous to human OATP1B1 and OATP1B3, also showed efficient docetaxel uptake. However, not all OATPs were capable of transporting docetaxel as OATP2B1, OATP3A1, OATP4A1 and OATP5A1 demonstrated no significant docetaxel uptake. In addition, other known drug uptake transporters such as the organic anion transporters (OATs), organic cation transporters (OCTs), as well as bile acid uptake transporters such as sodium/taurocholate cotransporting polypeptide (NTCP) and apical sodium-dependent bile acid transporter (ASBT), exhibited no docetaxel transport.
Figure 1.
Multiple OATPs transport docetaxel in vitro. A panel of OATP drug uptake transporters was assessed for docetaxel (0.1 μM) transport activity at 10 min. Vector control (pEF6) was used as a reference. Multiple human OATPs, including OATP1A2, OATP1B1, OATP1B3, and OATP1C1 are capable of transporting docetaxel in vitro. Rat Oatp1b2 also transports docetaxel in vitro. Data are expressed as percentage of cellular uptake compared to vector control (mean ± S.E., n = 6). *, p < 0.05; ***, p < 0.001.
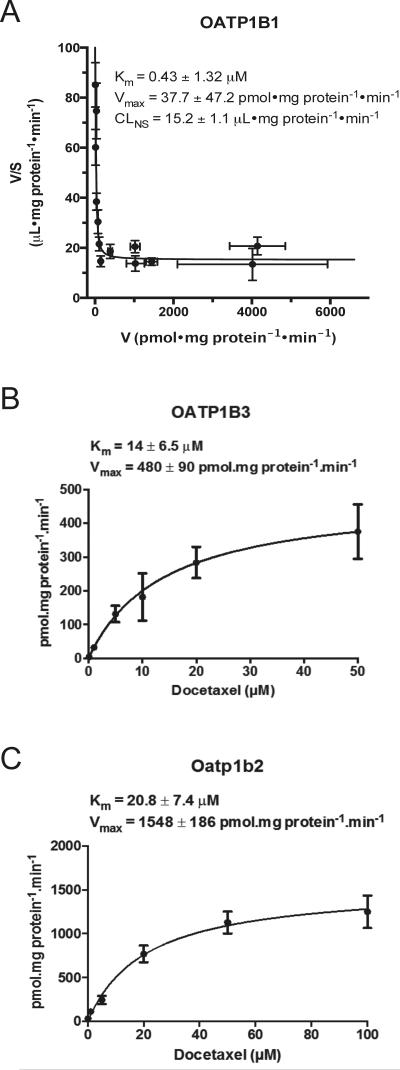
Time course experiments indicated significant accumulation of docetaxel over control in OATP1B1-, OATP1B3-, and rat Oatp1b2-expressing HeLa cells up to 30 minutes after incubation. To gain a better understanding of the pharmacokinetic parameters of docetaxel transport mediated by hepatic OATP transporters, kinetic analysis was performed. Radiolabeled docetaxel uptake was assessed in the presence of varying concentrations of unlabeled docetaxel (0.1-50 μM for OATP1B3; 0.1-300 uM for OATP1B1; 0.1-100 uM for Oatp1b2) for estimation of Km and Vmax. Interestingly, kinetic analysis for OATP1B1 suggested 2 kinetic components for transport. An Eadie-Hofstee plot together with curve fitting indicated that, for OATP1B1, transport kinetics at concentrations up to 300 μM are described by a high affinity, low capacity component (Km 0.43 ± 1.32 μM, Vmax 37.7 ± 47.2 pmol/mg/min) together with a non-saturable component that is low affinity, high capacity (Fig. 2A). Transport of docetaxel into HeLa cells transfected with OATP1B3 or Oatp1b2 was time dependent and saturable with a Km of 14 ± 6.5 μM and 20.8 ± 7.4 μM, respectively and a maximum velocity Vmax of 480 ± 90, and 1548 ± 186 pmol/mg/min, respectively (Figs. 2B-C).
Figure 2.
Docetaxel transport kinetics. An Eadie-Hofstee plot at a broad range of concentrations (0.1-300 μM) (A) for OATP1B1 showed a high affinity, low capacity component (Km 0.43 ± 1.32 μM, Vmax 37.7 ± 47.2 pmol/mg/min) together with a non-saturable component that is low affinity, high capacity (V—reaction rate; S—substrate concentration). Michaelis-Menten-type nonlinear curve-fitting are demonstrated for OATP1B3 (0.1-50 μM) (B) and Oatp1b2 (0.1-100 μM) (C). Data are expressed as mean ± S.E. (n=8 for OATP1B3 and Oatp1b2; n=12 for OATP1B1).
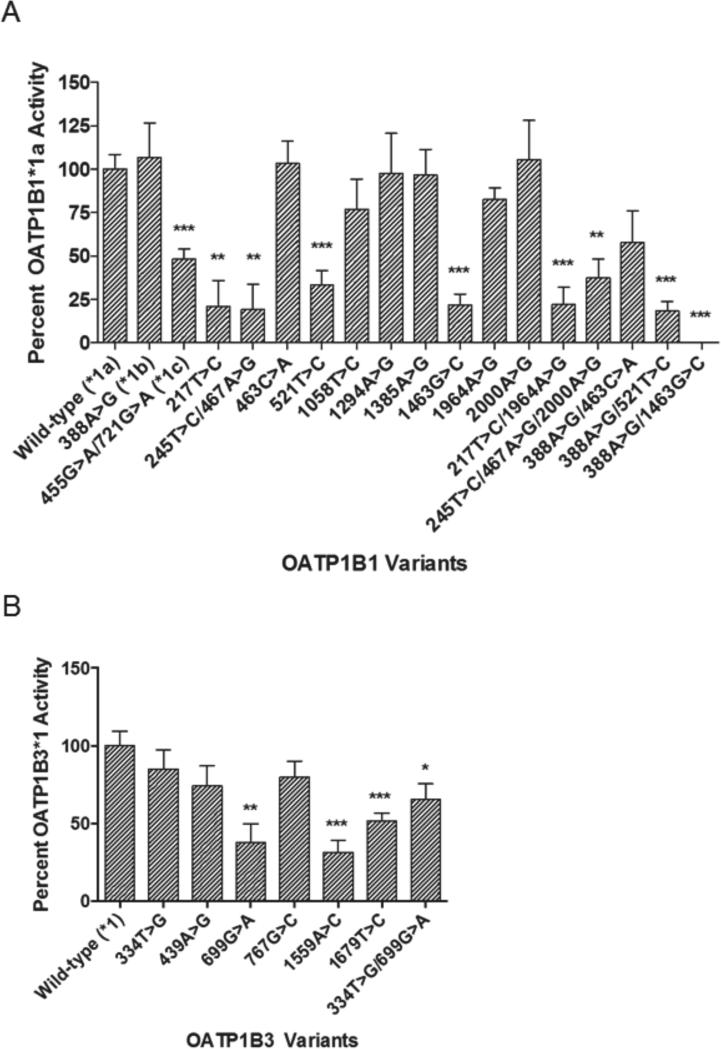
OATP1B1 and OATP1B3 variants differentially transport docetaxel in vitro
Each panel of OATP1B1 (Supplementary Table 1) and OATP1B3 (Supplementary Table 2) nonsynonymous variants was expressed in HeLa cells and evaluated for differential transport of docetaxel in vitro. A number of variants were associated with significantly impaired docetaxel uptake when compared to the wild-type reference alleles, SLCO1B1*1a or SLCO1B3*1. Pertaining to SLCO1B1 encoding OATP1B1, most notably, the common alleles, SLCO1B1 521T>C (*5) and 388 A>G/521T>C (*15), demonstrated significantly reduced docetaxel uptake (Fig. 3A). The two reference alleles, SLCO1B1 388G>A (*1a) and SLCO1B1 388A>G (*1b), exhibited equivalent docetaxel transport activity. Regarding SLCO1B3 encoding OATP1B3, several nonsynonymous SNPs were associated with impaired docetaxel transport in vitro, including SLCO1B3 699G>A, 1559A>C, 1679T>C, and 334T>G/699G>A (Fig. 3B).
Figure 3.
OATP1B1 and OATP1B3 variants differentially transport docetaxel in vitro. Uptake of radiolabeled docetaxel (0.1 μM) at 20 min by HeLa cells transfected with OATP1B1 or OATP1B3 variants was assessed relative to wild-type OATP1B1*1a and OATP1B3*1, respectively. The commonly occurring SLCO1B1 variants 521T>C (*5) and 388A>G/521T>C (*15) were associated with significantly impaired docetaxel transport. There was no significant differences in transport between the two reference alleles SLCO1B1 388G>A (*1a) and SLCO1B1 388A>G (*1b) (A). Several SLCO1B3 variants, including 699G>A, 1559A>C, 1679T>C, and 334T>G/699G>A, were associated with impaired docetaxel transport (B). Values are expressed as percentage of cellular uptake by OATP1B1*1a or OATP1B3*1 (mean ± SE, n = 6). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
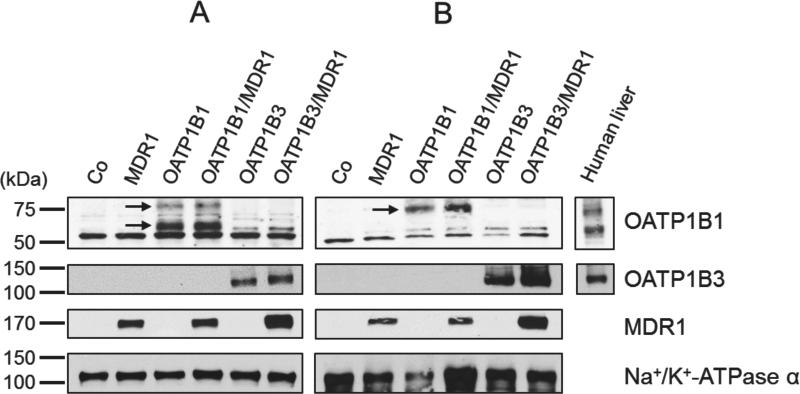
Protein expression of all stable MDCKII cell lines
To investigate the directional transport activity of docetaxel, we generated MDCKII cell lines stably expressing hepatic uptake transporters OATP1B1 or OATP1B3 and/or an efflux transporter MDR1. OATP1B1, OATP1B3, and MDR1 protein expression from all stable cells are shown in Figure 4. All cell lines expressed the proteins of predicted molecular weights, whereas no significant amounts of any protein of interest were detectable in the MDCKII-Co cells lacking any insert. OATP1B1 in crude cell membrane fractions (Fig. 4A) showed one glycosylated form with an apparent molecular weight of 84 kDa and one unglycosylated form (58 kDa), but only glycosylated form 85 kDa was detected in cell surface proteins (Fig. 4B). These results support the previous reports that 58 kDa of OATP1B1 is an ER-attached immature form (14, 25). In this study, we did not detect constitutive OATP1B1 or OATP1B3 protein expression (Figs. 4A and 4B) as well as OATP1A2 in control cells (MDCKII-Co) by immunoblot analysis, consistent with previous reports (26-28). Furthermore, transcriptional expression of constitutive OATP1A2 and OATP1B1 had been previously noted in MDCK cells, but not in MDCKII cells (29).
Figure 4.
Immunoblot analyses of OATP1B1, OATP1B3 and MDR1 expression in MDCKII-control cells (Co), single-(OATP1B1, OATP1B3, MDR1) and double-(OATP1B1/MDR1, OATP1B3/MDR1) cell lines. Crude cell membrane fractions (A) and biotinylated cell surface proteins (B) were subjected to 10% SDS polyacrylamide gels and then transferred onto nitrocellulose membranes. The blots were probed or re-probed with different antibodies, including OATP1B1, OATP1B3, MDR1, and Na+/K+-ATPase α antibodies. Whereas OATP1B1 in crude cell membrane fractions showed one glycosylated form 84 kDa and one unglycosylated form 58 kDa, only a glycosylated form 85 kDa of OATP1B1 was detected in cell surface proteins (arrows). OATP1B3 showed one band with approximately 120 kDa and MDR1 one with 170 kDa. Human liver extracts were used as a positive control.
Vectorial docetaxel transport is dependent on OATP1B uptake and MDR1 efflux
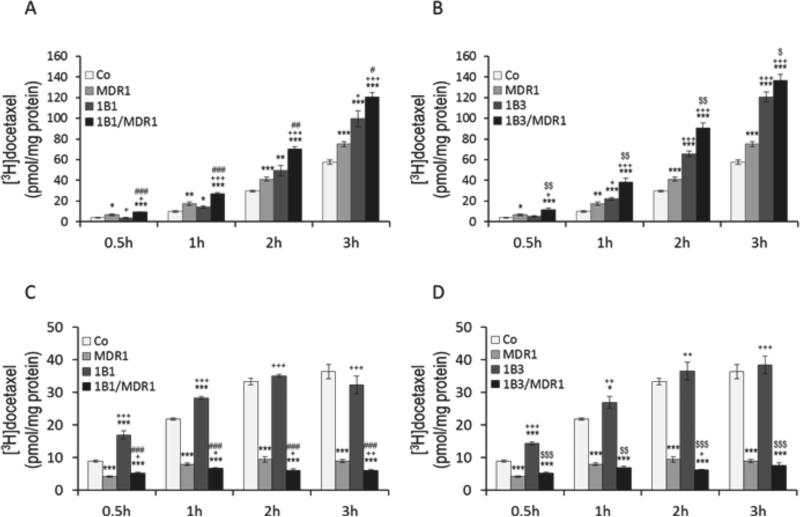
For vectorial transport assay, [3H]docetaxel was administered to the basal compartment of cell monolayers of control (MDCKII-Co), single-(MDCKII-OATP1B1, MDCKII-OATP1B3, MDCKII-MDR1), and double-(MDCKII-OATP1B1/MDR1, MDCKII-OATP1B3/MDR1) transfected cells. There was no significant difference between MDCKII-Co and MDCKIIOATP1B1 or MDCKII-OATP1B3 cells at incubation of 0.5h, but after 0.5h the apical docetaxel accumulation in MDCKII-OATP1B1 and MDCKII-OATP1B3 cells was significantly higher compared to MDCKII-Co cells (Figs. 5A and 5B). In contrast, at 0.5h docetaxel transported into the apical compartment of double-transfected MDCKII-OATP1B1/MDR1 and MDCKIIOATP1B3/MDR1 cells was significantly higher than in single-transfected MDCKII-OATP1B1 and MDCKII-OATP1B3 cells, respectively, (p < 0.001 for OATP1B1; p < 0.01 for OATP1B3). This pattern continued up to 3h. Interestingly, the apical docetaxel was significantly higher in MDCKII-MDR1 cells expressing MDR1only than in MDCKII-Co cells over time (p < 0.05).
Figure 5.
Vectorial transport and intracellular accumulation of radiolabeled docetaxel. [3H]Docetaxel (0.1 μM) was administered to the basal compartment of monolayers of MDCKII-control (Co), single-(MDR1, OATP1B1, OATP1B3) and double-(OATP1B1/MDR1, OATP1B3/MDR1) cell lines. After 0.5, 1, 2, and 3h incubation, translocation of docetaxel into the apical compartment (A, B) and intracellular accumulation of docetaxel (C, D) are shown. Whereas apical docetaxel was significantly higher in MDCKII-OATP1B1/MDR1 and -OATP1B3/MDR1 cells compared to MDCKII-OATP1B1 and -OATP1B3 cells respectively at all time points, intracellular docetaxel was significantly lower in MDCKII-OATP1B1/MDR1 and –OATP1B3/MDR1 cells. Data are expressed as mean ± S.E. (n = 6 for vectorial transport study; n = 4 for intracellular accumulation study). *p < 0.05, **p < 0.01, and ***p < 0.001 versus MDCKII-Co cells; +p < 0.05, ++p < 0.01, and +++p < 0.001 versus MDCKII-MDR1 cells; #p < 0.05, ##p < 0.01, and ###p < 0.001 versus MDCKII-OATP1B1cells; $p < 0.05, $$p < 0.01, and $$$p < 0.001 versus MDCKII-OATP1B3 cells.
To examine intracellular docetaxel, [3H]docetaxel was administrated to the basal compartment of cell monolayers in transwell and cells were incubated for 0.5, 1, 2, and 3h. The transwell plates were taken out of the incubator at each time point and cells were then analyzed for intracellular accumulation of docetaxel. As shown in Figures 5C and 5D, intracellular accumulation of docetaxel was significantly increased in MDCKII-OATP1B1 and MDCKIIOATP1B3 cells over MDCKII-Co cells at 0.5 h and 1 h time points, but uptake appeared to be saturable at later time points , reflecting efficient active uptake at early time points. Intracellular docetaxel was significantly lower in the double-transfected MDCKII-OATP1B1/MDR1 (p < 0.001) and MDCKII-OATP1B3/MDR1 cells (p < 0.01) compared to the single-transfected MDCKII-OATP1B1 and MDCKII-OATP1B3 cells at all time points, respectively. Accordingly, considerably higher amounts of docetaxel were found in the apical compartment of double-transfected MDCKII-OATP1B1/MDR1 and MDCKII-OATP1B3/MDR1 cells compared to single-transfected MDCKII-OATP1B1 and MDCKII-OATP1B3 cells (Figs. 5A and 5B). Vectorial transport studies were also conducted with a 4h incubation time. In this experiment, cellular translocation of docetaxel was markedly greater when [3H]docetaxel was administrated to the basal side of all cultured cells including single and double transfected cells and its presence measured on the apical side (basal-to-apical) compared to addition of docetaxel to the opposite compartment (apical-to-basal). Moreover, the amount of docetaxel transported into the basal side (apical-to-basal) was maintained at baseline over time.
Oatp1b2−/− mice significantly alter docetaxel disposition
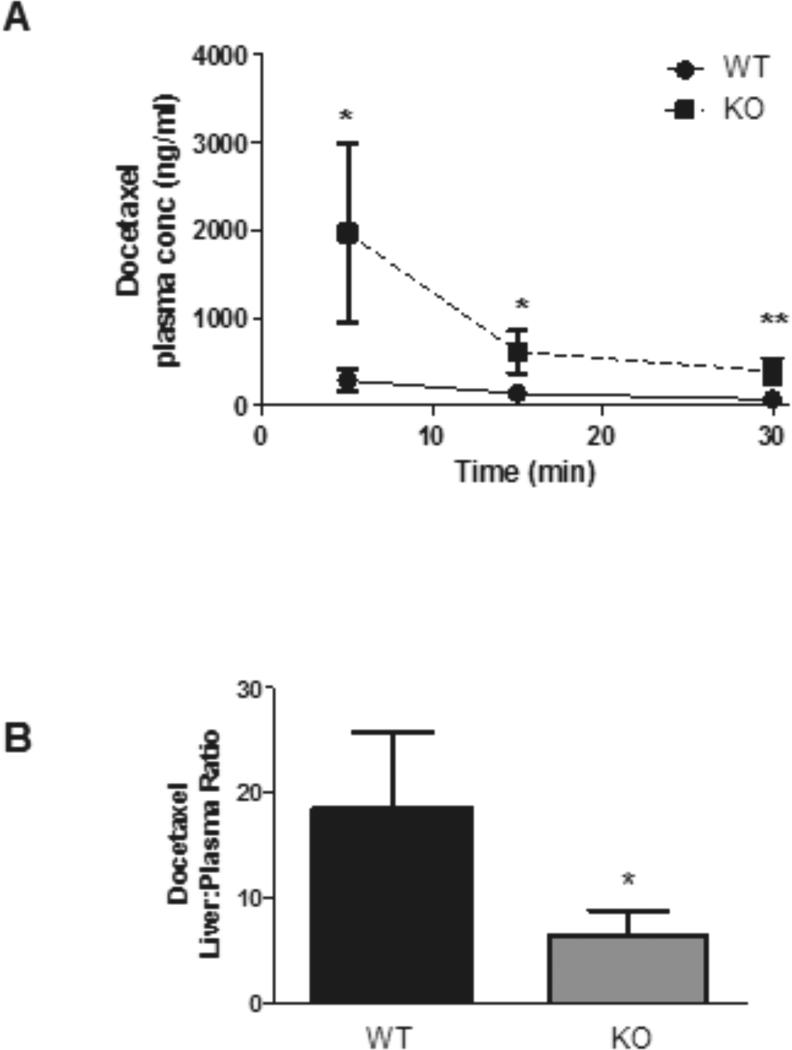
We performed experiments in Slco1b2−/− mice to assess the role of hepatic OATP1B to the in vivo disposition of docetaxel. As the initial paper describing Slco1b2−/− mice demonstrated modest gender-dependent differences in Slco1a4 and Slco2b1 expression in female Slco1b2−/− mice only (30), we utilized male Slco1b2−/− mice for docetaxel transport experiments to eliminate any confounding by compensatory effects in other Oatp transporters. Male Slco1b2−/− mice (age 21-24 weeks) and male wild-type littermate controls were used for docetaxel distribution experiments. As shown in Fig. 6A, there were significantly higher plasma docetaxel levels in mice lacking Oatp1b2 expression at given-time points assessed. The plasma AUC in Slco1b2−/− mice was ~5.5-fold greater than in wild-type mice (mean docetaxel plasma AUC ± S.D.: wild-type, 340 ± 149 ng·hr/ml; Slco1b2−/−, 62 ± 8 ng·hr/ml, p < 0.05). While there was no significant difference in docetaxel liver concentrations in Slco1b2−/− mice compared to wild-type mice (data not shown), the liver-to-plasma ratio was ~3-fold decreased (Fig. 6B) in Slco1b2−/− mice (p < 0.05). The plasma clearance of docetaxel in Slco1b2−/− mice was 83% lower than wild-type mice (mean plasma clearance ± S.D.: wild-type mice, 473 ± 124 ml/h; Slco1b2−/− mice, 73 ± 35 ml/h/kg; p < 0.05).
Figure 6.
Docetaxel disposition studies in Slco1b2−/− mice. Slco1b2−/− mice (KO) have significantly higher docetaxel plasma AUC than wild-type mice (WT) (A). Measured after a single intravenous dose 1 mg/kg 3H-docetaxel (30 Ci/mmol). Blood was sampled from the saphenous vein of each mouse at 5, 15 and 30 minutes post-injection. Data are expressed as mean ± SD. *, p < 0.05; **, p < 0.01. (n=4 per group). Slco1b2−/− mice have a significantly lower docetaxel liver:plasma ratio compared to wild-type mice (B). Liver-to-plasma ratio comparing wild-type and Slco1b2−/− mice was shown. Data are expressed as mean ± SD. *, p < 0.05; **, p < 0.01. (n= 4/group; 21-24 weeks).
Discussion
In this study, we conducted a systematic evaluation to define relevant OATP transporters responsible for docetaxel uptake and clearance. We identified multiple OATPs, including human OATP1A2, OATP1B1, OATP1B3, and OATP1C1, capable of transporting docetaxel in vitro. Rat Oatp1b2 was also capable of docetaxel transport in vitro and in vivo. As OATP1B1 and OATP1B3 are expressed primarily at the basolateral membrane of hepatocytes (17), this would suggest potential important roles for these transporters in the hepatic uptake of docetaxel. Kinetic analysis of docetaxel transport revealed similar kinetic parameters among OATP1B/1b transporters. In addition, a MDCKII stable cell system revealed an effective transcellular transport activity of docetaxel mediated by recombinant human OATP1B and MDR1 transporters in double transfected cells.
Contrary to our findings, previous studies demonstrated that docetaxel is a substrate for OATP1B3, not OATP1B1, in studies of Xenopus laevis oocyte transporter expression system (3, 18). The reason underlying the discrepancies are unclear but may be related to intrinsic differences in expression systems or time points at which uptake were determined. In addition, we previously indicated differential hepatic expression patterns for OATP1B1 and OATP1B3: in immunohistochemical analysis using normal human liver sections, OATP1B1 exhibited a diffuse staining pattern throughout liver sections while OATP1B3 showed a more restricted staining pattern with the greatest degree of expression being confined to perivenous distribution (14). As OATP1B1 and OATP1B3 are important determinants of hepatic uptake of drug substrates, it is unclear whether the differences in hepatic expression have significant clinical implications for drug disposition. Congruent with our results, a recent study reported by de Graan et al. (5) indicated that docetaxel uptake was facilitated by OATP1B1 and OATP1B3 in HEK293 or CHO cells expressing these transporters while only OATP1B3 constructs in Xenopus laevis oocytes, but not OATP1B1, exhibited docetaxel transport, illustrating that differences in expression systems may account for discrepancies in docetaxel substrate specificity.
Mice deficient in Oatp transporters have been generated and functionally characterized for in vivo transport of endogenous or xenobiotic OATP substrates (30, 31). Mouse Oatp1b2 is orthologous to human OATP1B1 and OATP1B3 (32). Slco1b2−/− mice are fertile, develop normally, and have no overt phenotypic abnormalities (30). However, these mice significantly altered the hepatic disposition of the prototypical OATP drug substrates pravastatin and rifampinin vivo (30). Another Oatp mouse model, Slco1a/1b−/− mice, significantly decreased the hepatic uptake of drug substrates such as methotrexate and fexofenadine (31). These studies illustrate the important roles of these animal models for understanding the in vivo roles of hepatic OATP transporters in drug disposition.
Accordingly, we further extended the findings from our in vitro experiments in Slco1b2−/− mice to assess the role of hepatic OATPs to the in vivo disposition of docetaxel. The plasma AUC in Slco1b2−/− mice was ~5.5-fold greater than in wild-type mice while the liver-to-plasma ratio was ~3-fold decreased in Slco1b2−/− mice. Importantly, the plasma clearance of docetaxel in Slco1b2−/− mice was 83% lower than wild-type mice. While a previous study demonstrated a dramatically increased docetaxel AUC and 18-fold decreased clearance in Slco1b2−/− mice (5), follow up studies from this group have confirmed that the contribution of Oatp1b transporters to docetaxel disposition is more in line with results from our study (33). Moreover, it is known that docetaxel is a substrate of the efflux transporter MDR1 (P-gp) (34). However, studies conducted in P-gp knockout mice (Mdr1a/1b−/−) indicated that the AUC of docetaxel following intravenous administration was similar in knockout and wild-type mice, suggesting mechanisms other than efflux contribute significantly to hepatic clearance of docetaxel (35). Finally, our vectorial transport assay in vitro showed a higher docetaxel transport in the apical compartment of the cell monolayers of the double-transfected OATP1B and MDR1 cells compared to the single-transfected MDR1 cells. Collectively, our studies suggest that hepatic OATP1B transporters are critical to the hepatic clearance of docetaxel, strongly supporting the liver as a major organ for docetaxel distribution and hepatic OATPs plays significant roles in docetaxel disposition.
The various processes mediating drug elimination, either through metabolic breakdown or excretion, can substantially impact interindividual variability in drug handling. Polymorphisms in CYP3A4 and MDR1 have been identified and characterized (36, 37). Several studies found no significant differences between ABCB1 genotype and docetaxel clearance in Asian or Caucasian cancer patients (3, 38). Another study assessed variants in CYP3A4/5 for associations with docetaxel pharmacokinetics (3). The combination of the CYP3A4*1B and CYP3A5*1A alleles (CYP3A4/5*2 haplotype) was associated with a 64% increase in docetaxel clearance independent of sex and CYP3A activity but this haplotype was only noted in 6/92 patients and likely only explains a very minor proportion of interindividual variability in docetaxel pharmacokinetics.
There has been significant effort in studying polymorphisms in SLCO genes as potential determinants of interindividual variability in drug disposition. To date, the functional consequences of polymorphism in the hepatic uptake transporter OATP1B1 have received the most attention. A common SNP, 521T>C (Val174Ala; SLCO1B1*5), has frequencies of approximately 15%, 2% and 15% in Caucasians, African-Americans and Asians, respectively (21, 39). The apparent mechanisms for impaired function for many functionally relevant OATP1B1 variants, including the 521T>C variant, are due to either decreased protein expression or impaired trafficking of the protein to the membrane (21). The pharmacokinetics of a growing number of drugs appear to be dependent on SLCO1B1 521T>C genotype including pitavastatin, rosuvastatin, fexofenadine, ezetimibe, and nateglinide (40-44). Notably, this allele was associated with a significant increased risk for simvastatin-induced myopathy in a genomewide association study (45). More recently, functionally relevant nonsynonymous polymorphisms have also been identified in SLCO1B3 and associated with impaired in vitro transport of substrates such as cholecystokinin (CCK8) and rosuvastatin (22). Two variants, 1559A>C and 1679T>C, were associated with significantly reduced total and cell surface protein expression. However, the clinical relevance of genetic heterogeneity in SLCO1B3 to drug disposition is unclear at this time.
We explored the potential roles of SLCO1B1 and SLCO1B3 SNPs to docetaxel disposition. Either of two reference alleles, A388 (OATP1B1*1a) or G388 (OATP1B1*1b), demonstrated equivalent transport activity. A number of SLCO1B1 SNPs were associated with impaired docetaxel transport in vitro. In particular, the commonly occurring 521T>C (*5) variant, as well as the *15 variant (*1b+*5), were both associated with significantly impaired docetaxel transport activity. Furthermore, several SCLO1B3 variants, including 1559A>C and 1679T>C, were also associated with significantly impaired docetaxel transport. Interestingly, as docetaxel has been demonstrated to be an OATP1B3 substrate, SLCO1B3 SNPs have also been evaluated in relation to docetaxel disposition and response. Baker et al. (3) found no signification association between six nonsynonymous SNPs in SLCO1B3 and docetaxel clearance in a population of Caucasian adult oncology patients. In contrast, Kiyotani et al. (46) identified a noncoding intronic SNP in SLCO1B3 (rs11045585) that was associated with a significantly higher risk for docetaxel-induced leucopenia/neutropenia in a population of Japanese adult oncology patients. Finally, de Graan et al. (5) found no significant differences in docetaxel clearance amongst a predominantly white adult oncology population (n=141) when assessed for commonly occurring polymorphisms in SLCO1B1 or 1B3. As OATP1B1 and OATP1B3 are both expressed at the basolateral membrane of hepatocytes and can facilitate docetaxel uptake, it is possible that a potential loss of docetaxel uptake associated with polymorphic variant in either OATP1B1 or 1B3 may be compensated for by the other OATP1B transporter, thereby mitigating the influence of the polymorphism on hepatic clearance. However, additional clinical PK:PG correlative studies in larger populations of patients may clarify the relevance of OATP1B polymorphisms to the wide variability in docetaxel pharmacokinetics.
In conclusion, we describe important roles for hepatic OATPs, including OATP1B1 and OATP1B3, to the disposition of docetaxel. Through a series of in vitro and in vivo experiments, we demonstrate that hepatic OATP1B transporters play significant roles in the hepatic uptake, clearance and plasma exposure of docetaxel. Moreover, we indicate that commonly occurring SLCO1B1 and SLCO1B3 variants differentially transport docetaxel in vitro, which may contribute to the oft witnessed wide interindividual variability in docetaxel disposition and response. Accordingly, our findings reveal important insights into the relevance of hepatic OATPs to the clinical pharmacology of docetaxel. In addition, we suggest that docetaxel is transported by multiple OATPs, which may also play important roles on docetaxel disposition, pharmacokinetics and toxicities.
Supplementary Material
Acknowledgments
Financial support: This project was supported by grants from Hyundai Hope on Wheels (to R.H. Ho), NIH R01 GM099924 (to R.H. Ho) and Canadian Institutes of Health Research (CIHR) grants (MOP-8975 and DSEN-PREVENT FRN-117588) to R.B. Kim.
Footnotes
Disclosure Statement: The authors have no conflicts of interest to disclose.
References
- 1.Baker SD, Sparreboom A, Verweij J. Clinical pharmacokinetics of docetaxel : recent developments. Clinical pharmacokinetics. 2006;45:235–52. doi: 10.2165/00003088-200645030-00002. [DOI] [PubMed] [Google Scholar]
- 2.Bruno R, Hille D, Riva A, Vivier N, ten Bokkel Huinnink WW, van Oosterom AT, et al. Population pharmacokinetics/pharmacodynamics of docetaxel in phase II studies in patients with cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1998;16:187–96. doi: 10.1200/JCO.1998.16.1.187. [DOI] [PubMed] [Google Scholar]
- 3.Baker SD, Verweij J, Cusatis GA, van Schaik RH, Marsh S, Orwick SJ, et al. Pharmacogenetic pathway analysis of docetaxel elimination. Clinical pharmacology and therapeutics. 2009;85:155–63. doi: 10.1038/clpt.2008.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jabir RS, Naidu R, Annuar MA, Ho GF, Munisamy M, Stanslas J. Pharmacogenetics of taxanes: impact of gene polymorphisms of drug transporters on pharmacokinetics and toxicity. Pharmacogenomics. 2012;13:1979–88. doi: 10.2217/pgs.12.165. [DOI] [PubMed] [Google Scholar]
- 5.de Graan AJ, Lancaster CS, Obaidat A, Hagenbuch B, Elens L, Friberg LE, et al. Influence of polymorphic OATP1B-type carriers on the disposition of docetaxel. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:4433–40. doi: 10.1158/1078-0432.CCR-12-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim KP, Ahn JH, Kim SB, Jung KH, Yoon DH, Lee JS, et al. Prospective evaluation of the drug-metabolizing enzyme polymorphisms and toxicity profile of docetaxel in Korean patients with operable lymph node-positive breast cancer receiving adjuvant chemotherapy. Cancer chemotherapy and pharmacology. 2012;69:1221–7. doi: 10.1007/s00280-011-1816-4. [DOI] [PubMed] [Google Scholar]
- 7.Tsai SM, Lin CY, Wu SH, Hou LA, Ma H, Tsai LY, et al. Side effects after docetaxel treatment in Taiwanese breast cancer patients with CYP3A4, CYP3A5, and ABCB1 gene polymorphisms. Clinica chimica acta; international journal of clinical chemistry. 2009;404:160–5. doi: 10.1016/j.cca.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 8.Tran A, Jullien V, Alexandre J, Rey E, Rabillon F, Girre V, et al. Pharmacokinetics and toxicity of docetaxel: role of CYP3A, MDR1, and GST polymorphisms. Clinical pharmacology and therapeutics. 2006;79:570–80. doi: 10.1016/j.clpt.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 9.van Herwaarden AE, Wagenaar E, van der Kruijssen CM, van Waterschoot RA, Smit JW, Song JY, et al. Knockout of cytochrome P450 3A yields new mouse models for understanding xenobiotic metabolism. The Journal of clinical investigation. 2007;117:3583–92. doi: 10.1172/JCI33435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wils P, Phung-Ba V, Warnery A, Lechardeur D, Raeissi S, Hidalgo IJ, et al. Polarized transport of docetaxel and vinblastine mediated by P-glycoprotein in human intestinal epithelial cell monolayers. Biochemical pharmacology. 1994;48:1528–30. doi: 10.1016/0006-2952(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annual review of biochemistry. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 12.Leveque D, Jehl F. P-glycoprotein and pharmacokinetics. Anticancer research. 1995;15:331–6. [PubMed] [Google Scholar]
- 13.Cortes JE, Pazdur R. Docetaxel. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1995;13:2643–55. doi: 10.1200/JCO.1995.13.10.2643. [DOI] [PubMed] [Google Scholar]
- 14.Ho RH, Tirona RG, Leake BF, Glaeser H, Lee W, Lemke CJ, et al. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology. 2006;130:1793–806. doi: 10.1053/j.gastro.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 15.Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochimica et biophysica acta. 2003;1609:1–18. doi: 10.1016/s0005-2736(02)00633-8. [DOI] [PubMed] [Google Scholar]
- 16.Iusuf D, van de Steeg E, Schinkel AH. Functions of OATP1A and 1B transporters in vivo: insights from mouse models. Trends in pharmacological sciences. 2012;33:100–8. doi: 10.1016/j.tips.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica; the fate of foreign compounds in biological systems. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- 18.Smith NF, Acharya MR, Desai N, Figg WD, Sparreboom A. Identification of OATP1B3 as a high-affinity hepatocellular transporter of paclitaxel. Cancer biology & therapy. 2005;4:815–8. doi: 10.4161/cbt.4.8.1867. [DOI] [PubMed] [Google Scholar]
- 19.Ho RH, Leake BF, Roberts RL, Lee W, Kim RB. Ethnicity-dependent polymorphism in Na+-taurocholate cotransporting polypeptide (SLC10A1) reveals a domain critical for bile acid substrate recognition. The Journal of biological chemistry. 2004;279:7213–22. doi: 10.1074/jbc.M305782200. [DOI] [PubMed] [Google Scholar]
- 20.Blakely RD, Clark JA, Rudnick G, Amara SG. Vaccinia-T7 RNA polymerase expression system: evaluation for the expression cloning of plasma membrane transporters. Analytical biochemistry. 1991;194:302–8. doi: 10.1016/0003-2697(91)90233-j. [DOI] [PubMed] [Google Scholar]
- 21.Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. The Journal of biological chemistry. 2001;276:35669–75. doi: 10.1074/jbc.M103792200. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz UI, Meyer zu Schwabedissen HE, Tirona RG, Suzuki A, Leake BF, Mokrab Y, et al. Identification of novel functional organic anion-transporting polypeptide 1B3 polymorphisms and assessment of substrate specificity. Pharmacogenetics and genomics. 2011;21:103–14. doi: 10.1097/FPC.0b013e328342f5b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim RB, Fromm MF, Wandel C, Leake B, Wood AJ, Roden DM, et al. The drug transporter P-glycoprotein limits oral absorption and brain entry of HIV-1 protease inhibitors. The Journal of clinical investigation. 1998;101:289–94. doi: 10.1172/JCI1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahrmayr C, Konig J, Auge D, Mieth M, Munch K, Segrestaa J, et al. Phase I and II metabolism and MRP2-mediated export of bosentan in a MDCKII-OATP1B1-CYP3A4-UGT1A1-MRP2 quadruple-transfected cell line. British journal of pharmacology. 2013;169:21–33. doi: 10.1111/bph.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konig J, Cui Y, Nies AT, Keppler D. Localization and genomic organization of a new hepatocellular organic anion transporting polypeptide. The Journal of biological chemistry. 2000;275:23161–8. doi: 10.1074/jbc.M001448200. [DOI] [PubMed] [Google Scholar]
- 26.Kopplow K, Letschert K, Konig J, Walter B, Keppler D. Human hepatobiliary transport of organic anions analyzed by quadruple-transfected cells. Molecular pharmacology. 2005;68:1031–8. doi: 10.1124/mol.105.014605. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki M, Suzuki H, Ito K, Abe T, Sugiyama Y. Transcellular transport of organic anions across a double-transfected Madin-Darby canine kidney II cell monolayer expressing both human organic anion-transporting polypeptide (OATP2/SLC21A6) and Multidrug resistance-associated protein 2 (MRP2/ABCC2). The Journal of biological chemistry. 2002;277:6497–503. doi: 10.1074/jbc.M109081200. [DOI] [PubMed] [Google Scholar]
- 28.Konig J, Cui Y, Nies AT, Keppler D. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. American journal of physiology Gastrointestinal and liver physiology. 2000;278:G156–64. doi: 10.1152/ajpgi.2000.278.1.G156. [DOI] [PubMed] [Google Scholar]
- 29.Goh LB, Spears KJ, Yao D, Ayrton A, Morgan P, Roland Wolf C, et al. Endogenous drug transporters in in vitro and in vivo models for the prediction of drug disposition in man. Biochemical pharmacology. 2002;64:1569–78. doi: 10.1016/s0006-2952(02)01355-2. [DOI] [PubMed] [Google Scholar]
- 30.Zaher H, Meyer zu Schwabedissen HE, Tirona RG, Cox ML, Obert LA, Agrawal N, et al. Targeted disruption of murine organic anion-transporting polypeptide 1b2 (Oatp1b2/Slco1b2) significantly alters disposition of prototypical drug substrates pravastatin and rifampin. Molecular pharmacology. 2008;74:320–9. doi: 10.1124/mol.108.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Steeg E, Wagenaar E, van der Kruijssen CM, Burggraaff JE, de Waart DR, Elferink RP, et al. Organic anion transporting polypeptide 1a/1b-knockout mice provide insights into hepatic handling of bilirubin, bile acids, and drugs. The Journal of clinical investigation. 2010;120:2942–52. doi: 10.1172/JCI42168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer WJ, Altheimer S, Cattori V, Meier PJ, Dietrich DR, Hagenbuch B. Organic anion transporting polypeptides expressed in liver and brain mediate uptake of microcystin. Toxicology and applied pharmacology. 2005;203:257–63. doi: 10.1016/j.taap.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Sparreboom A, Mathijssen RH. Hepatic uptake transporters and docetaxel disposition in mice-letter. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:4167. doi: 10.1158/1078-0432.CCR-14-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardelmeijer HA, Ouwehand M, Buckle T, Huisman MT, Schellens JH, Beijnen JH, et al. Low systemic exposure of oral docetaxel in mice resulting from extensive first-pass metabolism is boosted by ritonavir. Cancer research. 2002;62:6158–64. [PubMed] [Google Scholar]
- 35.van Waterschoot RA, Lagas JS, Wagenaar E, Rosing H, Beijnen JH, Schinkel AH. Individual and combined roles of CYP3A, P-glycoprotein (MDR1/ABCB1) and MRP2 (ABCC2) in the pharmacokinetics of docetaxel. International journal of cancer Journal international du cancer. 2010;127:2959–64. doi: 10.1002/ijc.25279. [DOI] [PubMed] [Google Scholar]
- 36.Zhou SF, Di YM, Chan E, Du YM, Chow VD, Xue CC, et al. Clinical pharmacogenetics and potential application in personalized medicine. Current drug metabolism. 2008;9:738–84. doi: 10.2174/138920008786049302. [DOI] [PubMed] [Google Scholar]
- 37.Chinn LW, Kroetz DL. ABCB1 pharmacogenetics: progress, pitfalls, and promise. Clinical pharmacology and therapeutics. 2007;81:265–9. doi: 10.1038/sj.clpt.6100052. [DOI] [PubMed] [Google Scholar]
- 38.Cox MC, Low J, Lee J, Walshe J, Denduluri N, Berman A, et al. Influence of garlic (Allium sativum) on the pharmacokinetics of docetaxel. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:4636–40. doi: 10.1158/1078-0432.CCR-06-0388. [DOI] [PubMed] [Google Scholar]
- 39.Ho RH, Choi L, Lee W, Mayo G, Schwarz UI, Tirona RG, et al. Effect of drug transporter genotypes on pravastatin disposition in European- and African-American participants. Pharmacogenetics and genomics. 2007;17:647–56. doi: 10.1097/FPC.0b013e3280ef698f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung JY, Cho JY, Yu KS, Kim JR, Oh DS, Jung HR, et al. Effect of OATP1B1 (SLCO1B1) variant alleles on the pharmacokinetics of pitavastatin in healthy volunteers. Clinical pharmacology and therapeutics. 2005;78:342–50. doi: 10.1016/j.clpt.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Lee E, Ryan S, Birmingham B, Zalikowski J, March R, Ambrose H, et al. Rosuvastatin pharmacokinetics and pharmacogenetics in white and Asian subjects residing in the same environment. Clinical pharmacology and therapeutics. 2005;78:330–41. doi: 10.1016/j.clpt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, et al. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clinical pharmacology and therapeutics. 2005;77:468–78. doi: 10.1016/j.clpt.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 43.Oswald S, Giessmann T, Luetjohann D, Wegner D, Rosskopf D, Weitschies W, et al. Disposition and sterol-lowering effect of ezetimibe are influenced by single-dose coadministration of rifampin, an inhibitor of multidrug transport proteins. Clinical pharmacology and therapeutics. 2006;80:477–85. doi: 10.1016/j.clpt.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, He YJ, Han CT, Liu ZQ, Li Q, Fan L, et al. Effect of SLCO1B1 genetic polymorphism on the pharmacokinetics of nateglinide. British journal of clinical pharmacology. 2006;62:567–72. doi: 10.1111/j.1365-2125.2006.02686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. The New England journal of medicine. 2008;359:789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 46.Kiyotani K, Mushiroda T, Kubo M, Zembutsu H, Sugiyama Y, Nakamura Y. Association of genetic polymorphisms in SLCO1B3 and ABCC2 with docetaxel-induced leukopenia. Cancer science. 2008;99:967–72. doi: 10.1111/j.1349-7006.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.