Abstract
AIM: To investigate the impact of minor abdominal surgery on the caecal microbial population and on markers of gut inflammation.
METHODS: Four week old piglets were randomly allocated to a no-surgery “control” group (n = 6) or a “transection surgery” group (n = 5). During the transection surgery procedure, a conventional midline incision of the lower abdominal wall was made and the small intestine was transected at a site 225 cm proximal to the ileocaecal valve, a 2 cm segment was removed and the intestine was re-anastomosed. Piglets received a polymeric infant formula diet throughout the study period and were sacrificed at two weeks post-surgery. Clinical outcomes including weight, stool consistency and presence of stool fat globules were monitored. High throughput DNA sequencing of colonic content was used to detect surgery-related disturbances in microbial composition at phylum, family and genus level. Diversity and richness estimates were calculated for the control and minor surgery groups. As disturbances in the gut microbial community are linked to inflammation we compared the gene expression of key inflammatory cytokines (TNF, IL1B, IL18, IL12, IL8, IL6 and IL10) in ileum, terminal ileum and colon mucosal extracts obtained from control and abdominal surgery groups at two weeks post-surgery.
RESULTS: Changes in the relative abundance of bacterial species at family and genus level were confined to bacterial members of the Proteobacteria and Bacteroidetes phyla. Family level compositional shifts included a reduction in the relative abundance of Enterobacteriaceae (22.95 ± 5.27 vs 2.07 ± 0.72, P < 0.01), Bacteroidaceae (2.54 ± 0.56 vs 0.86 ± 0.43, P < 0.05) and Rhodospirillaceae (0.40 ± 0.14 vs 0.00 ± 0.00, P < 0.05) following transection surgery. Similarly, at the genus level, changes associated with transection surgery were restricted to members of the Proteobacteria and Bacteroidetes phyla and included decreased relative abundance of Enterobacteriaceae (29.20 ± 6.74 vs 2.88 ± 1.08, P < 0.01), Alistipes (4.82 ± 1.73 vs 0.18 ± 0.13, P < 0.05) and Thalassospira (0.53 ± 0.19 vs 0.00 ± 0.00, P < 0.05). Surgery-associated microbial dysbiosis was accompanied by increased gene expression of markers of inflammation. Within the ileum IL6 expression was decreased (4.46 ± 1.60 vs 0.24 ± 0.06, P < 0.05) following transection surgery. In the terminal ileum, gene expression of TNF was decreased (1.51 ± 0.13 vs 0.80 ± 0.16, P < 0.01) and IL18 (1.21 ± 0.18 vs 2.13 ± 0.24, P < 0.01), IL12 (1.04 ± 0.16 vs 1.82 ± 0.32, P < 0.05) and IL10 (1.04 ± 0.06 vs 1.43 ± 0.09, P < 0.01) gene expression increased following transection surgery. Within the colon, IL12 (0.72 ± 0.13 vs 1.78 ± 0.28, P < 0.01) and IL10 (0.98 ± 0.02 vs 1.95 ± 0.14, P < 0.01) gene expression were increased following transection surgery.
CONCLUSION: This study suggests that minor abdominal surgery in infants, results in long-term alteration of the colonic microbial composition and persistent gastrointestinal inflammation.
Keywords: Surgery, Intestinal, Microbiota, Dysbiosis, Inflammation, Surgery, Bacteria, Infant, Pediatric, Transection
Core tip: Early colonization of the infant gut is increasingly recognized as impacting on health due to the influence of resident microbes on nutritional, immunological and physiological functions. However, medical and surgical interventions required during infancy, including abdominal surgery, have the potential to modify the microbiome. Using 454-pyrosequencing technology we have described microbial dysbiosis within the Proteobacteria and Bacteriodetes phylum and increased gut inflammatory cytokines following transection surgery in a juvenile pig model. This is the first study examining minor surgery-associated changes in the gut microbiome and inflammation and provides new insights into potential long-term consequences of abdominal surgery in infancy.
INTRODUCTION
The gut microbiota is essential to human health, yet the acquisition of this microbial community during infancy remains poorly understood. At birth, humans are essentially free of bacteria, with colonization of the gastrointestinal tract beginning during the birthing process as the newborn is exposed to maternal and environmental microbes[1]. The infant microbiota is marked by heterogeneity and instability until approximately 2-4 years of age[2,3], when it becomes more stable, resembling an adult microbiota[4]. The development of the microbiota is known to be strongly influenced by early extrinsic factors including mode of infant delivery[5], type of feeding[6,7] and antibiotic therapy[8-10]. Acute intestinal conditions including intestinal obstruction, perforation and intussusception may necessitate exploratory abdominal surgery and/or surgical intervention during the neonatal or infant period; however the impact of abdominal surgery on the microbiota has not previously been studied.
Colonization of the newborn intestine plays a key role in the development and fine-tuning of intestinal immune responses[11], and disruption of the gut microbiota has been linked to an increasing number of immune-related diseases, including inflammatory bowel disease, necrotizing enterocolitis (NEC), eczema, allergies and asthma[12]. Given the previously demonstrated impact of early life bacterial dysbiosis on future adult health, characterization of the impact of laparotomy in infancy on the development of the microbiome and inflammation is clinically relevant. The neonatal piglet surgical model is an excellent alternative model for the study of human gastrointestinal disease due to physiological[13,14], bacteriological[15] and immunological[16,17] similarities between pigs and humans. In addition, the accelerated ageing rate of pigs when compared with humans allows us to study the equivalent human timeframe of infancy to early childhood within a two week study period.
The primary aim of the present study was to use 454-pyrosequencing technology to assess the impact of minor abdominal surgery on the colonic microbiota in a juvenile pig model of intestinal transection surgery. Our secondary aim was to perform a multi-site assessment of molecular alterations in key gut inflammatory markers to assess if surgery-related microbial dysbiosis was associated with intestinal inflammation at two weeks following surgery.
MATERIALS AND METHODS
Animals
This study was approved by the Animal Ethics Committee of the Murdoch Childrens Research Institute. Weaned female three-week-old piglets (Landrace/Large White cross; Aussie Pride Pork) were transported to the University of Melbourne Centre for Animal Biotechnology and acclimatized prior to surgery. Piglets were fed a polymeric infant formula diet (Karicare De-Lact, Nutricia) supplemented to meet the daily requirements for piglets as described previously[18-23]. The diets were isocaloric and isonitrogenous among the groups and were administered on a per kilogram basis. Water was given twice daily. Piglets were housed separately throughout the study to allow accurate daily monitoring of food and water intake and collection of stool (analysed for consistency, presence of fat globules and presence of fatty acid crystals). Piglet weight was measured weekly before feeding.
Experimental design
Four week old piglets were randomly allocated to a no-surgery “control” group (n = 6) or a “transection” surgery group (n = 5). During the surgery procedure, a conventional midline incision of the lower abdominal wall was made and the small intestine was transected at a site 225 cm proximal to the ileocaecal valve, a 2 cm segment was removed and the intestine was re-anastomosed. The removal of 2 cm of intestine was equivalent to a mean of 0.11% of the initial intestinal length. Both groups received intramuscular amoxicillin (70 mg/kg; CSL Limited) 24 h pre-surgery, and for three days post-surgery in line with current clinical practice. In addition, both groups received oral rehydration salts (Sanofi-Aventis Australia) for three days post-surgery or equivalent date, with water and the polymeric infant formula diet re-introduced from the third day post-operation.
Sample collection
Animals in the transection group were sacrificed two weeks post-surgery and at an age-matched time in the control group. Ileum tissue was collected 8 cm distal to the anastomosis in the transection group and 217 cm proximal to the ileocaecal valve in the control group. Terminal ileum tissue was collected 7 cm proximal to the ileocaecal valve and colon tissue was collected 3 cm distal to the caecum in both groups. Samples from each site were snap frozen in liquid nitrogen. Colonic content was collected from the proximal colon.
High-throughput sequencing
The 16S rRNA amplicons from colonic content were generated using a previously outlined approach[24]. Amplicons were generated using one forward primer and a combination of four reverse primers as described previously[24]. Each primer contained a distinct multiple identifier (MID) allowing pooling of the amplicons and subsequent separation of the results for analysis. Duplicate PCR products were pooled and cleaned using Agencourt AMPure kit (Beckman Coulter, A63880). Quantification was completed using Quant-iT Picogreen quantification kit (Invitrogen, P7589) and the Nanodrop 3300 (Thermo Scientific). The V4 region of the 16S rRNA was sequenced at the Teagasc 454-Sequencing facility on a Genome Sequencer FLX platform (Roche Diagnostics Ltd.).
Bioinformatic analysis
Raw sequencing reads were quality trimmed using the RDP Pyrosequencing Pipeline applying the following criteria (1) exact matches to primer sequences and barcode tags; (2) no ambiguous bases (Ns); and (3) read-lengths no shorter than 150 base pairs. Trimmed FASTA sequences were then BLASTED[25] against the SILVA (v100) database for 16S reads[26]. Phylum, family and genus counts were extracted from MEGAN using a bit score cut-off of 86[26]. Clustering into operational taxonomical units (OTUs), alignments, chimera-checking and alpha diversities were implemented using the Qiime suite of tools. The relative abundance was determined for individual pigs as the number of reads for each species as a proportion of the total number of reads for that pig at the relevant level of phylum, family or genus.
Real-time reverse transcription PCR
The muscle layer was stripped from the ileum, terminal ileum and colon tissue leaving the mucosa, which comprised the epithelium and the lamina propria. Total RNA was extracted from 100 mg of intestinal mucosa using TRIzol (Invitrogen). Complementary DNA (cDNA) was synthesized with the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science). PCR primers were designed against pig gene sequences using Roche Universal ProbeLibrary Assay Design Centre (Roche Applied Science). Primer sequences and probe combinations are listed in Table 1. PCR reactions were performed in triplicate on the LightCycler 480. The 2-ΔΔCt method[27] was used to calculate relative changes in gene expression in the surgical group relative to the non-surgical control group, using RPL32 as a housekeeping gene.
Table 1.
List of primer sequences and Universal ProbeLibrary combinations used in this study
| Primer | Sequence 5’ to 3’ | UPL Probe |
| RPL32 Forward | AACTGGCCATCAGGGTCAC | #64 |
| RPL32 Reverse | CACAACTGGAACTCCTGTCTATTC | |
| TNF Forward | TTGTCGCTACATCGCTGAAC | #32 |
| TNF Reverse | CCAGTAGGGCGGTTACAGAC | |
| IL1β Forward | CCAATTCAGGGACCCTACC | #19 |
| IL1β Reverse | CATGGCTGCTTCAGAAACCT | |
| IL18 Forward | ACTTTACTTTGTAGCTGAAAACGATG | #85 |
| IL18 Reverse | TTTAGGTTCAAGCTTGCCAAA | |
| IL12 Forward | GAGGGTGAGTGAGTGCCTTG | #62 |
| IL12 Reverse | ACTCCGCCTAGGTTCGACTT | |
| IL8 Forward | TTCTTCTTTATCCCCAAACTGG | #41 |
| IL8 Reverse | CCACATGTCCTCAAGGTAGGA | |
| IL6 Forward | TGAACTCCCTCTCCACAAGC | #7 |
| IL6 Reverse | GGCAGTAGCCATCACCAGA | |
| IL10 Forward | TCCAGTTTTACCTGGAAGACG | #8 |
| IL10 Reverse | CCTTGATATCCTCCCCATCA |
Statistical analysis
Sequencing analysis was completed using Minitab Release 15.1.1.0 (Minitab Inc 2007). Parametric unpaired t-test was employed to identify significant differences in the percentage of assignable reads at phylum, family and genus levels between non-surgery control and transaction surgery groups. (GraphPad Prism Software 6.0). Parametric unpaired t-tests were also employed to test for statistical significance in the relative gene expression of inflammation markers between non-surgery control and transaction surgery groups. Statistical significance for all testing was accepted at P < 0.05.
RESULTS
Weight gain throughout the time course studied was comparable between the no-surgery control and transection surgery groups and both groups consumed equivalent energy per kilogram per day. There was no evidence of diarrhea or steatorrhea in any piglets in the transaction group and they were considered to be clinically well.
Microbial composition is significantly altered following transaction surgery
High throughput DNA sequencing was used to detect disturbances in microbial composition following transection surgery. Diversity and richness estimates were calculated for the control and transection surgery groups. The Chao 1 calculation is an estimator of phylotype richness in a dataset and the Shannon index of diversity reflects both the richness and the community evenness (i.e., proportional phylotype abundance)[28]. The overall alpha diversity of the colonic microbiota was unchanged following surgery as calculated by either the Chao 1 richness estimate (905 + 41 vs 892 ± 123) or the Shannon’s index for diversity (6.9 ± 0.2 vs 6.5 ± 0.6), and there was no difference in the number of observed species.
16S rRNA sequence data obtained from colonic content samples was analyzed to determine if transection surgery was associated with changes in the proportion of assignable reads at the phylum, family or genus level when compared with no surgery controls. There was no difference in composition at a phylum level between the control group and the transection group. Changes were observed however, at family and genus level (Tables 2 and 3). Changes in the relative abundance of bacterial species at family and genus level were confined to members of the Proteobacteria and Bacteroidetes phyla, with no difference in the relative abundance at family or genus level of species belonging to the Firmicute, Actinobacteria, Fusobacteria or Spirochaetes phylum.
Table 2.
Relative abundance of bacterial species at the family level in colonic content samples obtained from no-surgery control animals (n = 6) or animals who have undergone a transaction surgery (n = 5)
| Phylum level | Family level | Control (Rel. Abundance) | Transection (Rel. Abundance) | P value |
| Proteobacteria | Enterobacteriaceae | 22.95 + 5.27 | 2.07 + 0.72a | 0.0104 |
| Desulfovibrionaceae | 2.89 + 1.26 | 2.95 + 1.16 | 0.9716 | |
| Moraxellaceae | 0.69 + 0.55 | 8.81 + 8.72 | 0.4501 | |
| Pseudoalteromonadaceae | 0.55 + 0.43 | 1.42 + 1.42 | 0.5844 | |
| Vibrionaceae | 0.52 + 0.37 | 6.03 + 5.91 | 0.4038 | |
| Bacteriodetes | Rikenallaceae | 6.00 + 1.87 | 3.70 + 1.31 | 0.3416 |
| Porphyromonadaceae | 2.33 + 0.76 | 4.50 + 1.78 | 0.3080 | |
| Prevotellaceae | 2.76 + 0.39 | 8.05 + 2.60 | 0.1105 | |
| Bacteroidaceae | 2.54 + 0.56 | 0.86 + 0.43a | 0.0432 | |
| Firmicutes | Ruminococcaceae | 19.97 + 2.01 | 15.26 + 4.71 | 0.3962 |
| Veillonellaceae | 14.19 + 4.12 | 22.40 + 7.73 | 0.3834 | |
| Lachnospiraceae | 7.63 + 2.31 | 6.19 + 2.00 | 0.6475 | |
| Peptostreptococcaceae | 3.19 + 1.06 | 1.52 + 1.08 | 0.2991 | |
| Erysipeolotrichales Incertae Sedis | 1.78 + 0.61 | 1.03 + 0.53 | 0.3809 | |
| Clostridiaceae | 1.45 + 0.35 | 0.73 + 0.36 | 0.1843 | |
| Lactobacillaceae | 1.09 + 0.45 | 1.58 + 0.83 | 0.6197 | |
| Streptococcaceae | 0.13 + 0.06 | 1.56 + 1.54 | 0.4056 | |
| Actinobacteria | Micrococcinea | 0.07 + 0.07 | 1.54 + 1.54 | 0.3926 |
| Fusobacteria | Fusobacteriaceae | 0.98 + 0.44 | 3.85 + 3.09 | 0.4079 |
| Spirochaetes | Spirochaetaceae | 1.83 + 1.15 | 0.09 + 0.08 | 0.1898 |
| Other | 6.49 + 1.09 | 5.84 + 1.41 | 0.7245 |
Data are expressed as mean + SEM (%).
P < 0.05 vs control. Bacterial species whose relative abundance was < 1% in both the control and transaction surgery group are compiled within the “other” category. Significant changes in bacteria whose relative abundance was < 1% are detailed within the text.
Table 3.
Relative abundance of bacterial species at the genus level in colonic content samples obtained from no-surgery control animals (n = 6) or animals who have undergone a transaction surgery (n = 5)
| Phylum | Genus level | Control (Rel. Abundance) | Transection (Rel. Abundance) | P value |
| Proteobacteria | Members of the Enterobacteriaceae | 29.20 + 6.74 | 2.88 + 1.08a | 0.0108 |
| Desulfovibrio | 2.08 + 0.68 | 2.74 + 0.78 | 0.5389 | |
| Bilophila | 1.48 + 0.81 | 1.13 + 0.95 | 0.7847 | |
| Psychrobacter | 0.90 + 0.72 | 9.13 + 9.01 | 0.4136 | |
| Vibrio | 0.62 + 0.46 | 5.89 + 5.70 | 0.4105 | |
| Pseudoalteromonas | 0.15 + 0.12 | 1.47 + 1.47 | 0.4192 | |
| Bacteriodetes | Alistipes | 4.82 + 1.73 | 0.18 + 0.13a | 0.0441 |
| Prevotella | 3.61 + 0.59 | 11.11 + 3.68 | 0.1110 | |
| Bacteroides | 3.25 + 0.71 | 1.24 + 0.63 | 0.0625 | |
| Parabacteroides | 2.88 + 0.65 | 5.59 + 2.41 | 0.3313 | |
| Lachnospitaceae Incertae sedis | 2.50 + 1.02 | 1.10 + 0.47 | 0.2548 | |
| Firmicutes | Megasphaera | 9.71 + 3.87 | 14.18 + 4.01 | 0.4434 |
| Phascolartobacterium | 4.07 + 1.00 | 6.90 + 1.89 | 0.2330 | |
| Rumminococcaceae Incertae sedis | 2.94 + 0.71 | 1.78 + 0.47 | 0.2106 | |
| peptostreptococcaceae incertae sedis | 2.79 + 1.23 | 1.80 + 1.49 | 0.6208 | |
| Subdoligranulum | 2.49 + 0.99 | 1.28 + 0.57 | 0.3264 | |
| Acidaminococcus | 2.40 + 0.71 | 3.53 + 1.82 | 0.5866 | |
| Erysipelotrichales incertae sedis | 2.28 + 0.77 | 1.48 + 0.78 | 0.4859 | |
| Lactobacillus | 1.96 + 0.74 | 2.15 + 1.16 | 0.8958 | |
| Clostridium | 1.86 + 0.45 | 1.05 + 0.52 | 0.2732 | |
| Anaerotruncus | 2.23 + 0.71 | 0.97 + 0.31 | 0.1501 | |
| Oribacterium | 0.63 + 0.25 | 1.88 + 0.88 | 0.2341 | |
| Lactococcus | 0.06 + 0.06 | 1.60 + 1.58 | 0.3860 | |
| Mitsuokella | 0.05 + 0.05 | 1.90 + 1.82 | 0.3664 | |
| Actinobacteria | Microccoccaceae | 0.07 + 0.07 | 1.38 + 1.38 | 0.3959 |
| Fusobacteria | Fusobacterium | 1.27 + 0.60 | 5.43 + 4.39 | 0.3986 |
| Spirochaetes | Treponema | 2.29 + 1.44 | 0.13 + 0.11 | 0.1951 |
| Uncult. Clone | EU774397 | 1.43 + 0.73 | 0.05 + 0.05 | 0.1178 |
| AF371921 | 1.43 + 0.47 | 2.18 + 0.99 | 0.5217 | |
| AF371920 | 0.24 + 0.14 | 1.11 + 0.79 | 0.3315 | |
| Other | 8.05 + 0.69 | 6.66 + 1.21 | 0.3549 |
Data are expressed as mean + SEM (%).
P < 0.05 vs control. Bacterial species whose relative abundance was < 1% in both the control and transaction surgery group are compiled within the “other” category. Significant changes in bacteria whose relative abundance was < 1% are detailed within the text.
As can be seen in Table 2, family level compositional shifts included a 10-fold reduction in the relative abundance of Enterobacteriacceae and a 3-fold reduction in the relative abundance of Bacteriodaceae following transection surgery. The proportion of Bifidobacteriaceae was reduced 10-fold in surgical samples (0.40 ± 0.16 vs 0.04 ± 0.03, P = 0.0712) and although Rhodospirillaceae was present in 5/6 control samples, Rhodospirillaceae was not detected amongst any of the surgical samples (0.40 ± 0.14 vs 0.00 ± 0.00, P = 0.0370). Similarly at the genus level, changes associated with transection surgery were restricted to members of the Proteobacteria and Bacteroidetes phyla (Table 3). Specifically transection surgery resulted in a 10-fold decrease in the relative abundance of members of the Enterobacteriaceae, a 27-fold decrease in the relative abundance of Alistipes and a two-fold decrease in the relative abundance of Bacteroides. The relative abundance of Thalassospira (0.53 ± 0.19 vs 0.00 ± 0.00, P = 0.0379) and Bifidobacterium (0.51 ± 0.21 vs 0.06 ± 0.04, P = 0.0838) was also decreased (though not significantly) following surgery. Thalassospira was detected in all control samples but was undetectable in the surgical samples.
A tissue-specific pattern of inflammation occurs following transaction surgery
Disturbances of the gut microbial community are closely linked to inflammation[29], therefore given the detected bacterial dysbiosis in the transection surgery model we next examined changes in the mucosal gene expression of key inflammatory cytokines within the ileum, terminal ileum and colon (Figure 1). Within the ileum, a decrease in interleukin 6 (IL6) gene expression followed surgery (P = 0.0383; Figure 1A). In the terminal ileum, gene expression of the pro-inflammatory cytokine tumor necrosis factor (TNF) was decreased (P = 0.0072), and interleukin 18 (IL18) and interleukin 12 (IL12) gene expression was increased following surgery when compared with controls (P = 0.0123 and P = 0.0430 respectively; Figure 1B). Gene expression of the anti-inflammatory cytokine interleukin 10 (IL10) was increased in terminal ileum samples from the surgery group when compared with controls (P = 0.0054). Similar to the terminal ileum IL12 and IL10 gene expression was increased within the colon following surgery (P = 0.0055 and P = 0.0001 respectively; Figure 1C).
Figure 1.
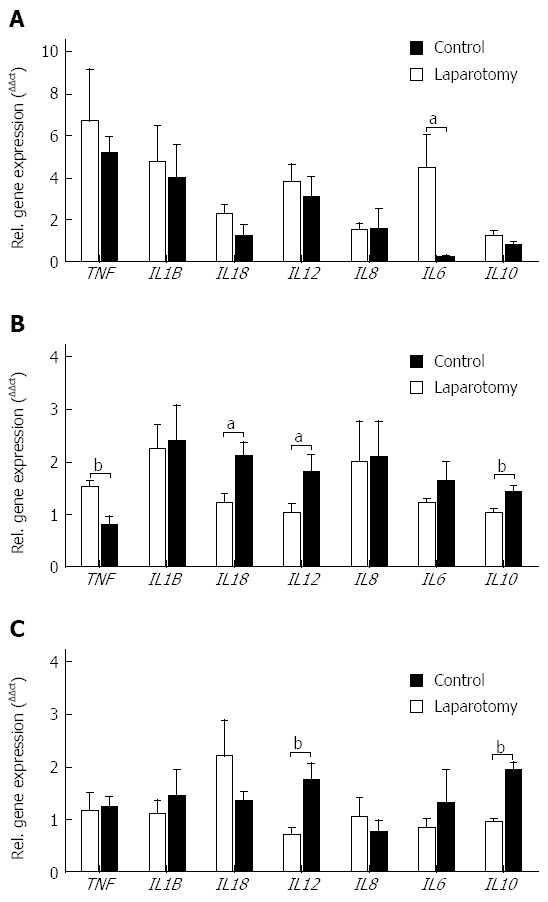
Relative gene expression of pro- and anti-inflammatory cytokines in (A) ileum, (B) terminal ileum and (C) colon from no-surgery control and transection surgery groups. Mean ± SEM. aP < 0.05, bP < 0.01 vs control.
DISCUSSION
Immediately after birth, the newborn gut environment is colonized by facultative anaerobic bacteria such as Enterobacteriaceae and Streptococcaceae[11]. These bacteria gradually consume the oxygen in the intestine and produce new metabolites, preparing the intestinal environment for the establishment of a strict anaerobic population dominated by Bifidobacterium, Clostridium and Bacteroides sp., genera that may contribute to neonatal gut maturation[11]. However, medical and surgical interventions required during infancy, including abdominal surgery, have the potential to modify the microbiome. As, microbial colonization of the infant gut plays a key role in the development and fine-tuning of the intestinal immune responses[11], it is essential to detail the effect of minor abdominal surgery performed during the infant period, on the major microbial community and associated inflammation.
Our current study details alterations in the colonic microbial community in response to abdominal surgery. We chose to focus on the colonic microbiota as (1) the colonic microbial community represents the largest, and most widely studied microbial community in both health and disease states; and (2) any alterations to the colonic microbial community are likely to influence the immune response via interactions with the gut-associated lymphoid tissue. In the current study, whilst there was no observed effect on the overall richness or diversity of the colonic microbiota, there were alterations in specific bacterial genera that are likely to be of clinical relevance. Similarly, studies of irritable bowel syndrome also describe significant associations between quantitative differences in specific bacterial components of the gut microbiome and disease symptoms[30,31] in the face of unchanged overall microbial diversity. In particular, we observed alterations only within the Proteobacteria and Bacteriodetes phylum and report reductions in the relative abundance of family level members Enterobacteriacceae, Rhodaspirillaceae and Bacteroidaceae and genus level reductions in the relative abundance of Enterobacteriaceae, Thalassospira, Alistipes and Bacteroides following transection surgery.
The reduction in the relative abundance of Bacteroides sp. is of particular clinical relevance as, in early life, the intestinal microbiota of healthy infants displays a large abundance of Bacteriodes sp.[32]. Bacteroides sp. play a fundamental role in host metabolic processes[33] and prevents the colonisation of the gut by potential pathogens[34]. Depletions in Bacteroides species are observed in children with inflammatory conditions including IgE mediated food allergy[35] and inflammatory bowel disease[36]. Furthermore, Bacteroides is postulated to play a crucial role in the development of gastrointestinal-associated lymphoid tissues and in the modulation of T-helper Th1/Th2/T-regulatory balance[37]. Therefore reduction in relative abundance of Bacteroides in the gut following intestinal transaction surgery may, at least in-part, explain the observed increase in IL12 and IL18 gene expression within the terminal ileum. IL12 and IL18 form a link between innate resistance and adaptive immunity and may be produced by a wide range of immune cells in response to bacteria and bacterial products[38].
Interestingly, we observed markers of an active gut inflammatory response at two weeks post-surgery that was not limited to the area within the immediate proximity of the intestinal transection and re-anastomosis. Specifically, we observed increased gene expression of IL12 and IL10 in both terminal ileum and colon, and increased IL18 and decreased TNF gene expression in terminal ileum. Whilst it is not possible to ascertain the underlying mechanism responsible for the wide-spread increase in inflammatory mediators observed in our juvenile pig model, we postulate surgery-related alterations in the small intestine microbiota may influence inflammatory marker expression within the ileum and terminal ileum. Alternatively, surgery-related factors including bowel handling could have contributed to the inflammatory response observed. However, bowel handling has been reported to induce TNF, IL6 and IL1B production[39,40], cytokines that remained unchanged or were decreased in this study (Figure 1).
Early colonization of the infant gut is increasingly recognized as impacting on health due to the influence of resident microbes on nutritional, immunological and physiological functions. However, medical and surgical interventions required during infancy, including abdominal surgery, have the potential to modify the microbiome, sometimes permanently. We have described microbial dysbiosis within the Proteobacteria and Bacteriodetes phylum and increased gut inflammatory cytokines following surgery in a juvenile pig model. This is the first study examining changes in the gut microbiome and in gut inflammation that occur following transaction surgery in infancy and provides new insights into potential long-term consequences of abdominal surgery in infancy.
ACKNOWLEDGMENTS
The authors acknowledge Professors Gerald Fitzgerald and Paul Ross for their continued support and thank Magdy Sourial, Shane Osterfield and Dr Andrew French for expert technical assistance with the animals, Dr Jean-Pierre Scheerlinck and The University of Melbourne Centre for Animal Biotechnology for the use of their facilities, and Drs Orla O’Sullivan and Eva Rosberg-Cody for assistance with high-throughput DNA sequencing.
COMMENTS
Background
Early colonization of the infant gut is increasingly recognized as impacting on health due to the influence of resident microbes on nutritional, immunological and physiological functions. However, medical and surgical interventions required during infancy, including abdominal surgery, have the potential to modify the microbiome. This is the first study examining minor surgery-associated changes in the gut microbiome and inflammation and provides new insights into potential long-term consequences of abdominal surgery in infancy.
Research frontiers
Colonization of the newborn intestine plays a key role in the development and fine-tuning of intestinal immune responses, and disruption of the gut microbiota has been linked to an increasing number of immune-related diseases, including inflammatory bowel disease, necrotizing enterocolitis, eczema, allergies and asthma. The current research hotspot is to characterize intervention-related alterations in microbial communities and assess the impact of these changes on future clinical outcome.
Innovations and breakthroughs
This is the first study to apply 454-pyrosequencing technology to assess the impact of minor abdominal surgery on the colonic microbial population using a juvenile pig model of intestinal transaction surgery. The authors observed reduced abundance of family and genus level members of the Proteobacteria and Bacteriodetes phylum, concurrent with unresolved gastrointestinal inflammation within the ileum, terminal ileum and colon. Importantly, this study provides new insights into potential long-term consequences of abdominal surgery in infancy.
Applications
Whilst for the majority of intestinal surgeries, the benefits of the operative procedure far outweigh the potential risks of altering the gut microbiota and increasing the level of inflammatory cytokines, this study suggests that a straight-forward and uncomplicated laparotomy with minimal bowel resection and re-anastomosis may have secondary unforeseen circumstances that may have a long-term impact on the health of pediatric patients.
Terminology
The term colonic microbiota is the name given to the microbe population living within the caecum.
Peer-review
This is the first study examining minor surgery-associated changes in the gut microbiome and inflammation and provides new insights into potential long-term consequences of abdominal surgery in infancy. This is a well-conducted and well written study. The experiments are described in detail, the results are shown nicely and the figures are impressive.
Footnotes
Supported by Victorian Government’s Operational Infrastructure Support program; The PC lab is supported in part by grants from Science Foundation Ireland in the form of a center grant (Alimentary Pharmabiotic Centre; No. SFI/12/RC/2273 and No. 12/RC/2273 and a PI grand to PCR No.11/PI/1137; FF is in receipt of an Irish Research Council EMBARK scholarship and is a Teagasc Walsk Fellow.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: May 28, 2014
First decision: July 21, 2014
Article in press: October 21, 2014
P- Reviewer: Triantafyllou K, Zhu X S- Editor: Qi Y L- Editor: A E- Editor: Liu XM
References
- 1.Tapiainen T, Ylitalo S, Eerola E, Uhari M. Dynamics of gut colonization and source of intestinal flora in healthy newborn infants. APMIS. 2006;114:812–817. doi: 10.1111/j.1600-0463.2006.apm_488.x. [DOI] [PubMed] [Google Scholar]
- 2.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 5.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 6.Jacquot A, Neveu D, Aujoulat F, Mercier G, Marchandin H, Jumas-Bilak E, Picaud JC. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J Pediatr. 2011;158:390–396. doi: 10.1016/j.jpeds.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005;243:141–147. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 8.Kalenić S, Francetić I, Polak J, Zele-Starcević L, Bencić Z. Impact of ampicillin and cefuroxime on bacterial colonization and infection in patients on a neonatal intensive care unit. J Hosp Infect. 1993;23:35–41. doi: 10.1016/0195-6701(93)90128-m. [DOI] [PubMed] [Google Scholar]
- 9.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, Shirakawa T, Sonomoto K, Nakayama J. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. 2009;56:80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 11.Marques TM, Wall R, Ross RP, Fitzgerald GF, Ryan CA, Stanton C. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol. 2010;21:149–156. doi: 10.1016/j.copbio.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol. 2012;28:63–69. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moughan PJ, Birtles MJ, Cranwell PD, Smith WC, Pedraza M. The piglet as a model animal for studying aspects of digestion and absorption in milk-fed human infants. World Rev Nutr Diet. 1992;67:40–113. doi: 10.1159/000419461. [DOI] [PubMed] [Google Scholar]
- 14.Miller ER, Ullrey DE. The pig as a model for human nutrition. Annu Rev Nutr. 1987;7:361–382. doi: 10.1146/annurev.nu.07.070187.002045. [DOI] [PubMed] [Google Scholar]
- 15.Buzoianu SG, Walsh MC, Rea MC, O’Sullivan O, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt maize) for 31 days. Appl Environ Microbiol. 2012;78:4217–4224. doi: 10.1128/AEM.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler JE, Weber P, Sinkora M, Baker D, Schoenherr A, Mayer B, Francis D. Antibody repertoire development in fetal and neonatal piglets. VIII. Colonization is required for newborn piglets to make serum antibodies to T-dependent and type 2 T-independent antigens. J Immunol. 2002;169:6822–6830. doi: 10.4049/jimmunol.169.12.6822. [DOI] [PubMed] [Google Scholar]
- 17.Scharek L, Guth J, Reiter K, Weyrauch KD, Taras D, Schwerk P, Schierack P, Schmidt MF, Wieler LH, Tedin K. Influence of a probiotic Enterococcus faecium strain on development of the immune system of sows and piglets. Vet Immunol Immunopathol. 2005;105:151–161. doi: 10.1016/j.vetimm.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 18.Lapthorne S, Pereira-Fantini PM, Fouhy F, Wilson G, Thomas SL, Dellios NL, Scurr M, O’Sullivan O, Ross RP, Stanton C, et al. Gut microbial diversity is reduced and is associated with colonic inflammation in a piglet model of short bowel syndrome. Gut Microbes. 2013;4:212–221. doi: 10.4161/gmic.24372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira-Fantini PM, Thomas SL, Wilson G, Taylor RG, Sourial M, Bines JE. Short- and long-term effects of small bowel resection: a unique histological study in a piglet model of short bowel syndrome. Histochem Cell Biol. 2011;135:195–202. doi: 10.1007/s00418-011-0778-2. [DOI] [PubMed] [Google Scholar]
- 20.Healey KL, Bines JE, Thomas SL, Wilson G, Taylor RG, Sourial M, Pereira-Fantini PM. Morphological and functional changes in the colon after massive small bowel resection. J Pediatr Surg. 2010;45:1581–1590. doi: 10.1016/j.jpedsurg.2010.02.040. [DOI] [PubMed] [Google Scholar]
- 21.Stephens AN, Pereira-Fantini PM, Wilson G, Taylor RG, Rainczuk A, Meehan KL, Sourial M, Fuller PJ, Stanton PG, Robertson DM, et al. Proteomic analysis of the intestinal adaptation response reveals altered expression of fatty acid binding proteins following massive small bowel resection. J Proteome Res. 2010;9:1437–1449. doi: 10.1021/pr900976f. [DOI] [PubMed] [Google Scholar]
- 22.Pereira-Fantini PM, Thomas SL, Taylor RG, Nagy E, Sourial M, Fuller PJ, Bines JE. Colostrum supplementation restores insulin-like growth factor -1 levels and alters muscle morphology following massive small bowel resection. JPEN J Parenter Enteral Nutr. 2008;32:266–275. doi: 10.1177/0148607108316197. [DOI] [PubMed] [Google Scholar]
- 23.Pereira-Fantini PM, Nagy ES, Thomas SL, Taylor RG, Sourial M, Paris MC, Holst JJ, Fuller PJ, Bines JE. GLP-2 administration results in increased proliferation but paradoxically an adverse outcome in a juvenile piglet model of short bowel syndrome. J Pediatr Gastroenterol Nutr. 2008;46:20–28. doi: 10.1097/01.mpg.0000304449.46434.06. [DOI] [PubMed] [Google Scholar]
- 24.Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, Brigidi P. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. 2014;20:908–922. doi: 10.3748/wjg.v20.i4.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeffery IB, O’Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EM, Simrén M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 32.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 33.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto-Osaki T, Kamiya S, Sawamura S, Kai M, Ozawa A. Growth inhibition of Clostridium difficile by intestinal flora of infant faeces in continuous flow culture. J Med Microbiol. 1994;40:179–187. doi: 10.1099/00222615-40-3-179. [DOI] [PubMed] [Google Scholar]
- 35.Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X, Yuan L, Wang Y, Sun J, Li L, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aomatsu T, Imaeda H, Fujimoto T, Takahashi K, Yoden A, Tamai H, Fujiyama Y, Andoh A. Terminal restriction fragment length polymorphism analysis of the gut microbiota profiles of pediatric patients with inflammatory bowel disease. Digestion. 2012;86:129–135. doi: 10.1159/000339777. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez B, Prioult G, Hacini-Rachinel F, Moine D, Bruttin A, Ngom-Bru C, Labellie C, Nicolis I, Berger B, Mercenier A, et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol Ecol. 2012;79:192–202. doi: 10.1111/j.1574-6941.2011.01207.x. [DOI] [PubMed] [Google Scholar]
- 38.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 39.Wehner S, Schwarz NT, Hundsdoerfer R, Hierholzer C, Tweardy DJ, Billiar TR, Bauer AJ, Kalff JC. Induction of IL-6 within the rodent intestinal muscularis after intestinal surgical stress. Surgery. 2005;137:436–446. doi: 10.1016/j.surg.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Kalff JC, Schraut WH, Simmons RL, Bauer AJ. Surgical manipulation of the gut elicits an intestinal muscularis inflammatory response resulting in postsurgical ileus. Ann Surg. 1998;228:652–663. doi: 10.1097/00000658-199811000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]


