Abstract
AIM: To investigate the clinical significance of methyl-methanesulfonate sensitivity 19 (MMS19) expression in esophageal squamous cell carcinoma (ESCC).
METHODS: Between June 2008 and May 2013, specimens from 103 patients who underwent endoscopic biopsy for the diagnosis of ESCC at the endoscopy center of Sun Yat-Sen University Cancer Center were collected; 52 matched-normal esophageal squamous epithelium samples were biopsied as controls. MMS19 protein expression was measured by immunohistochemistry. Of the 103 cases of ESCC, 49 received radical surgery following neoadjuvant chemoradiotherapy consisting of concurrent radiation in a total dose of 40 Gy and two cycles of chemotherapy with vinorelbine and cisplatin. Relationships between MMS19 expression, clinicopathologic characteristics and chemoradiotherapy response were analyzed.
RESULTS: The MMS19 protein could be detected in both the cytoplasm and nucleus of most specimens. High cytoplasmic expression of MMS19 was detected in 63.1% of ESCC samples, whereas high nuclear expression of MMS19 was found in 35.0%. High cytoplasmic MMS19 expression was associated with regional lymph node metastases (OR = 11.3, 95%CI: 2.3-54.7; P < 0.001) and distant metastases (OR = 13.1, 95%CI: 1.7-103.0; P = 0.002). Furthermore, high cytoplasmic MMS19 expression was associated with a response of ESCC to chemoradiotherapy (OR = 11.5, 95%CI: 3.0-44.5; P < 0.001), with a high cytoplasmic MMS19 expression rates in 79.3% and 25.0% of patients from the good chemoradiotherapy response group and poor response group, respectively. Nuclear MMS19 expression did not show any significant association with clinicopathologic characteristics or chemoradiotherapy response in ESCC.
CONCLUSION: The results of our preliminary study suggest that MMS19 may be a potential new predictor of metastasis and chemoradiotherapy response in ESCC.
Keywords: Chemoradiotherapy, Esophageal squamous cell carcinoma, Metastases, Methyl-methanesulfonate sensitivity 19, Surgery
Core tip: Methyl-methanesulfonate sensitivity 19 (MMS19) was first identified as a DNA repair protein, and recently as a part of cytoplasmic Fe-S assembly machinery that produce proteins involved in maintenance of genomic stability, such as DNA polymerase, DNA repair proteins, and DNA nuclease/helicase. However, the clinical significance of MMS19 protein expression in esophageal cancer has not been reported. This study shows that MMS19 is abnormally expressed in esophageal cancer, and the elevated cytoplasmic MMS19 expression is associated with lymph node and distant metastases, and response to chemoradiotherapy in esophageal squamous cell carcinoma.
INTRODUCTION
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive tumors, ranking fourth among the top ten cancer-related deaths in China[1,2]. In China, the histology of esophageal cancer is mainly ESCC, whereas esophageal adenocarcinoma is rare[3]. Because of the high recurrence and metastasis rates, the five-year survival rate of ESCC treated with surgery alone is poor, only approximately 25%, and in such circumstances, surgery plus radiotherapy and/or chemotherapy is increasingly adopted for locally advanced esophageal cancer[4]. The results from phase III randomized trials of chemoradiotherapy (CRT) prior to surgery are encouraging; however, these studies reveal that only patients who are sensitive to CRT will ultimately benefit from the multimodality treatment[5-7]. Thus, the identification of patients who can benefit from CRT is crucial for the success of the combined treatment of CRT followed by surgery. However, there is currently no biomarker that can predict response of ESCC to CRT. Therefore, the discovery of biomarkers that can predict sensitivity of ESCC to CRT is an urgent need in clinical practice.
The methyl-methanesulfonate sensitivity 19 (MMS19) gene, also named as MMS19L or hMMS19, encodes a multifunctional protein involved in DNA metabolism and the maintenance of genomic stability[8]. Nucleotide excision repair (NER) plays a vital role in the development of carcinogen-induced cancers[9,10] and in tumor resistance to chemo- and radiotherapy[11,12]. By interacting with the core transcription factors of NER, MMS19 can affect NER functions[13,14]. In addition, Fe-S proteins are crucial for genomic instability[15], which is a hallmark of cancer[16]. As a part of the cytoplasmic Fe-S assembly machinery, MMS19 facilitates the transfer of the Fe-S cluster to target Fe-S proteins, which include DNA polymerase δ, xeroderma pigmentosum group D, Fanconi anemia pathway component J (also known as BACH1 or BRIP1)[17], DNA2 nuclease/helicase, RNase L inhibitor (also known as ABCE1), and endonuclease three-like glycosylase 2[18]. Thus, MMS19 is suggested to be involved in maintaining genomic stability[17,18].
At present, some studies have reported that single-nucleotide polymorphisms of the MMS19 gene are associated with the risk of pancreatic cancer[19], chemotherapy toxicity of non-small-cell lung cancer[20], and chemotherapy response of osteosarcoma[21]. These polymorphisms could increase cancer susceptibility by altering conserved amino acids[22] and could affect cancer prognosis by modulating gene expression[23]. However, the cellular expression level of MMS19 protein in cancer and its clinical significance have not been reported. In this study, we investigated the cellular expression level and distribution of MMS19 in ESCC and the relationships of MMS19 expression with the clinicopathologic factors and CRT response of ESCC.
MATERIALS AND METHODS
The study was performed in accordance with the Declaration of Helsinki of the World Medical Association and was approved by the Medical Ethics Committee of Sun Yat-Sen University Cancer Center. All patients signed an informed consent form for this investigation.
Patients
Between June 2008 and May 2013, specimens from 103 ESCC patients who underwent endoscopic biopsy for diagnosis at the endoscopy center of Sun Yat-Sen University Cancer Center were collected. As controls, 52 samples of normal esophageal squamous epithelia (NESE) were biopsied from ≥ 5 cm from the primary lesion in the same patients. Patients who received any anticancer treatments before diagnosis were excluded. The biopsied specimens were diagnosed by two pathologists. Tumor staging was performed based on the combined results of physical examination, endoscopic ultrasonography, CT scan of the chest and abdomen, and color ultrasonography scan of the abdomen and neck. The tumors were staged according to AJCC (2002). The patients were aged from 42 to 83 years (median 59 years), including 84 men and 19 women. Two patients were classified as stage I, 25 patients as stage II, 58 patients as stage III and 18 patients as stage IV. Among the 103 ESCC patients, a cohort of 49 patients with thoracic esophageal carcinoma staged IIb and III received neoadjuvant CRT followed by surgery.
Neoadjuvant chemoradiotherapy and surgery
Radiation treatment planning was designed according to CT simulation or three-dimensional conformal radiation therapy. The patients were treated with 6 or 8 MV photons delivered in a total dose of 40 Gy (20 fractions of 2 Gy per fraction in 4 wk) in anteroposterior fields including esophageal tumors and enlarged lymph nodes, with 3-cm proximal and distal margins, and an 0.8-cm radial margin. The patients received two cycles of vinorelbine and cisplatin.Vinorelbine (25 mg/m2) was administered intravenously on days 1, 8, 22 and 29, and cisplatin (75 mg/m2) was infused on day 1 and day 22 (or 25 mg/m2 on days 1-4 and 22-25). Total thoracic esophagectomy through a right thoracotomy with radical mediastinal and abdominal lymph node dissection was performed ≥ 4-6 wk after the completion of CRT.
Evaluation of histopathologic response to preoperative CRT
For evaluation of response to CRT, surgical cancer samples from 49 patients who underwent CRT and surgery were obtained. The histopathologic response to CRT was evaluated by two experienced pathologists according to previously published criteria[24,25]. The percentage of residual viable tumor cells was estimated, and each patient was subsequently allocated to one of the following four groups: complete response group, no residual tumor cells; major response group, < 10% residual tumor cells; partial response group, 10%-50% of residual tumor cells; minor response group, > 50% of residual tumor cells. For the statistical analysis, the patients were divided into two groups according to CRT response: a good response group, consisting of patients with a complete or major response; and a poor response group, including patients with a partial or minor response.
Immunohistochemical staining
Immunohistochemical staining was performed on 4-μm-thick paraffin sections. The sections were deparaffinized in xylene and rehydrated through graded alcohol. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 10 min. For epitope retrieval, the tissue slides were immersed in EDTA buffer (pH 8.0) and heated for 2.5 min on high power in a microwave oven. After washing, the tissue slides were incubated with an anti-MMS19 antibody (16015-1-AP; Proteintech, Chicago, IL, United States) at a dilution of 1:50 for 50 min at 37 °C in a moist chamber. Subsequently, the secondary antibody (K5007, Real Envision/HRP; Dako of Agilent Technologies, Santa Clara, CA, United States) was applied to the tissue section for 30 min at 37 °C. Finally, 3.3'-diaminobenzidine was used for color development and hematoxylin for counterstaining. Negative control slides in the absence of primary antibody were included for each batch of staining.
Cytoplasmic and nuclear MMS19 staining was evaluated separately. The immunochemistry staining for the MMS19 protein was evaluated under 400× high-power magnification. The positively stained cells in five separate areas of epithelial or intratumoral regions were counted. The quantification of MMS19 expression was performed according to a previous study[26]. The percentage of cells positively stained was scored as follows: 0 ≤ 5%, 1 = 6%-25%; 2 = 26%-50%; 3 = 51%-75%; 4 > 75%. The staining intensity was scored as follows: 0 = no staining, 1 = weak, 2 = moderate, 3 = strong. For each case, the final score for MMS19 immunostaining was calculated by multiplying the percentage score of positive cells with the staining intensity score. Immunostaining was independently evaluated by two experienced pathologists who had no knowledge of the patients’ clinicopathologic information. If different scores for the same sample were made by the two pathologists, the sample was revaluated and, if needed, discussed to decide a final score. Then, a composite score scaled as 0, 1, 2, 3, 4, 6, 8, 9, and 12 was obtained. Based on the final score, each case was divided into a high expression group (≥ 6) or a low expression group (< 6).
Statistical analysis
The statistical analyses were performed using SPSS 20.0 software (IBM Corp., Armonk, NY, United States). Data are expressed as mean ± SE. The differences in MMS19 expression levels between the different groups and the correlations between MMS19 expression and clinicopathologic characteristics as well as response to CRT were analyzed by the χ2 test. Spearman’s rank correlation (r) was used to determine whether there was a positive or negative correlation. Two-tailed P < 0.05 was considered statistically significant. The statistical methods of this study were reviewed by Qing Liu from Sun Yat-Sen University Cancer Center.
RESULTS
Different expression levels of MMS19 in biopsied NESE and ESCC tissues
Using immunohistochemistry, the MMS19 protein was detected in both the cytoplasm and nucleus of most endoscopic biopsied specimens (Figure 1), which is consistent with its cellular functions[13,18]. Cytoplasmic MMS19 staining in NESE was mainly found in the basal and suprabasal layers, with a gradually decreased staining from the basal layer to the superficial layer. In contrast, nuclear MMS19 staining in NESE was scattered throughout the entire layer (Figure 1A and B). The intensity of MMS19 staining was typically homogeneous within an ESCC specimen, but varied considerably among different tumors (Figure 1D and E). Figure 1C and F show weak staining in both the cytoplasm and nucleus of NESE and ESCC, respectively.
Figure 1.
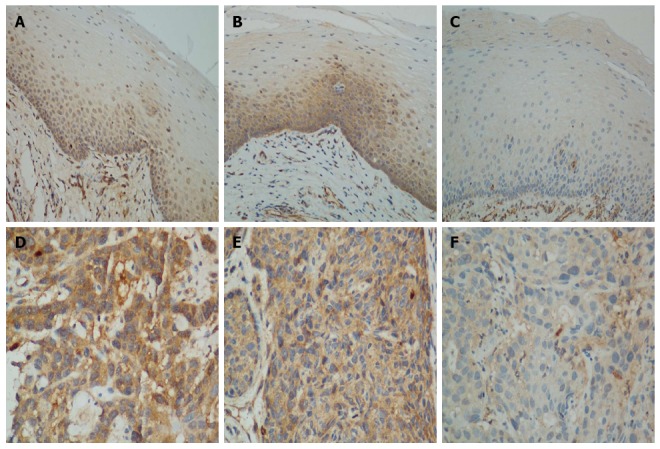
Methyl-methanesulfonate sensitivity 19 immunohistochemistry. Normal esophageal squamous epithelium with A: Strong nuclear but weak cytoplasmic staining; B: Strong cytoplasmic staining in the basal and suprabasal layers, with scattered strong nuclear staining in the normal epithelium area; C: Weak staining in both the cytoplasm and nucleus (Magnification × 200); Esophageal squamous cell carcinoma with D: Strong staining in the cytoplasm and nucleus; E: Strong staining in the cytoplasm and weak staining in the nucleus; F: Weak staining in both the cytoplasm and nucleus (Magnification × 400).
The mean scores for cytoplasmic MMS19 expression in the high expression group and low expression group were 7.78 ± 0.27 and 2.79 ± 0.21, respectively. Whereas, the mean scores of nuclear MMS19 expression in the high expression group and low expression group were 6.86 ± 0.32 and 2.68 ± 0.14, respectively. High cytoplasmic expression of MMS19 was detected in 63.1% of the ESCC samples, which was significantly higher than the 15.4% in NESE (P < 0.001, Table 1). High nuclear expression of MMS19 was found in 35.0% of the ESCC specimens, which was significantly lower than the 69.2% found in NESE (P < 0.001, Table 1).
Table 1.
Methyl-methanesulfonate sensitivity 19 expression n (%)
| Tissue | Cases (n) |
Cytoplasmic expression |
P value |
Nuclear expression |
P value | ||
| High | Low | High | Low | ||||
| NESE | 52 | 8 (15.4) | 44 (84.6) | < 0.001 | 36 (69.2) | 16 (30.8) | < 0.001 |
| ESCC | 103 | 65 (63.1) | 38 (36.9) | 36 (35.0) | 67 (65.0) | ||
ESCC: Esophageal squamous cell carcinoma; High: Including composite score of 6, 8, 9 and 12; NESE: Normal esophageal squamous epithelium; Low: Including composite score of 0, 1, 2, 3 and 4.
Relationships of MMS19 expression in biopsied ESCC tissues with clinicopathologic features
First, associations of cytoplasmic MMS19 expression with clinicopathologic features were investigated. The results showed that high cytoplasmic MMS19 expression was significantly associated with regional lymph node metastases (LNM) (OR = 11.25, 95%CI: 2.31-54.73; P < 0.001) and distant metastases (DM) (OR = 13.10, 95%CI: 1.67-103.00; P = 0.002), suggesting that cytoplasmic MMS19 expression might be involved in cancer metastasis. The Spearman correlation coefficients of high cytoplasmic MMS19 expression with LNM and DM were 0.35 and 0.299, respectively, indicating that higher levels of MMS19 expression are positively correlated with ESCC metastasis. There was no significant association of cytoplasmic MMS19 expression with other clinicopathologic features, including histologic grade, invasion depth, patient age, tumor stage, or sex (Table 2). Nuclear MMS19 expression did not show any significant association with clinicopathologic parameters (Table 2).
Table 2.
Associations of MMS19 expression with clinicopathologic features of esophageal squamous cell carcinoma
| Characteristic | Cases (n) |
Cytoplasmic MMS19 |
Nuclear MMS19 |
||||
| High | Low | P value | High | Low | P value | ||
| Total | 103 | 65 | 38 | 36 | 67 | ||
| Sex | |||||||
| Male | 84 | 55 | 29 | 0.295 | 32 | 52 | 0.159 |
| Female | 19 | 10 | 9 | 4 | 15 | ||
| Age (yr) | |||||||
| < 55 | 33 | 18 | 15 | 0.216 | 14 | 19 | 0.275 |
| ≥ 55 | 70 | 47 | 23 | 22 | 48 | ||
| Site | |||||||
| Upper thoracic | 10 | 7 | 3 | 0.686 | 2 | 8 | 0.508 |
| Middle thoracic | 47 | 31 | 16 | 16 | 31 | ||
| Lower thoracic | 46 | 27 | 19 | 18 | 28 | ||
| Differentiation | |||||||
| Well | 20 | 13 | 7 | 0.785 | 6 | 14 | 0.871 |
| Moderate | 52 | 34 | 18 | 19 | 33 | ||
| Poor | 31 | 18 | 13 | 11 | 20 | ||
| TNM stage | |||||||
| I + II | 27 | 14 | 13 | 0.158 | 8 | 19 | 0.500 |
| III + IV | 76 | 51 | 25 | 28 | 48 | ||
| Invasion depth | |||||||
| T1 + T2 | 25 | 17 | 8 | 0.560 | 7 | 18 | 0.402 |
| T3 + T4 | 78 | 48 | 30 | 29 | 49 | ||
| LNM | |||||||
| No | 12 | 2 | 10 | < 0.001 | 4 | 8 | 0.900 |
| Yes | 91 | 63 | 28 | 32 | 59 | ||
| DM | |||||||
| No | 85 | 48 | 37 | 0.002 | 32 | 53 | 0.212 |
| Yes | 18 | 17 | 1 | 4 | 14 | ||
DM: Distant metastases; ESCC: Esophageal squamous cell carcinoma; High: Including composite score of 6, 8, 9 and 12; LNM: Lymph node metastases; Low: Including composite score of 0, 1, 2, 3 and 4; MMS19: methyl-methanesulfonate sensitivity 19.
Relationship of MMS19 expression in biopsied specimens with CRT response of resected ESCC
According to the histopathologic response to preoperative CRT, 24 cases showed complete response, 5 cases showed a major response, 9 cases showed a partial response, and 11 cases showed a minor response. Thus the good and poor response groups included 29 and 20 cases, respectively. Then, relationships of MMS19 expression with CRT response of ESCC were investigated. In the good response group, high cytoplasmic expression of MMS19 was observed in 23/29 (79.3%) patients. In contrast, high MMS19 expression was found in only 5/20 (25.0%) patients in the poor response group, and the difference in MMS19 expression between the two groups was statistically significant (OR = 11.5, 95%CI: 2.97-44.51; P < 0.001, Table 3). The Spearman correlation coefficient of high cytoplasmic MMS19 expression with a good response was 0.539, suggesting that high cytoplasmic expression of MMS19 is positively correlated with a good response to preoperative CRT. This result suggested that MMS19 might be a potential new biomarker to predict tumor response to preoperative CRT. However, nuclear MMS19 expression was not associated with CRT response, with a rate of high nuclear expression of 31.0% (9/29) in the good response group and 45.0% (9/20) in the poor response group (Table 3).
Table 3.
Clinical features of patients receiving neoadjuvant chemoradiotherapy followed by surgery, n
| Characteristic | Good response | Poor response | P value |
| (n = 29) | (n = 20 ) | ||
| Age (yr) | |||
| < 55 | 11 | 11 | 0.238 |
| ≥ 55 | 18 | 9 | |
| Sex | |||
| Male | 24 | 15 | 0.763 |
| Female | 5 | 5 | |
| Tumor size (cm) | |||
| < 5 | 12 | 7 | 0.652 |
| ≥ 5 | 17 | 13 | |
| Site | |||
| Upper thoracic | 3 | 2 | 0.405 |
| Middle thoracic | 14 | 6 | |
| Lower thoracic | 12 | 12 | |
| Differentiation | |||
| Well | 5 | 4 | 0.936 |
| Moderate | 16 | 10 | |
| Poor | 8 | 6 | |
| TNM stage | |||
| IIb | 8 | 6 | 0.857 |
| III | 21 | 14 | |
| Cytoplasm | |||
| High | 23 | 5 | < 0.001 |
| Low | 6 | 15 | |
| Nucleus | |||
| High | 9 | 9 | 0.319 |
| Low | 20 | 11 |
High: Including composite score of 6, 8, 9 and 12; Low: Including composite score of 0, 1, 2, 3 and 4; TNM: Tumor-node-metastasis.
The relationships of CRT response with clinicopathologic features were also analyzed. However, there was no relationship between preoperative CRT response and clinicopathologic features, including tumor size, tumor site, differentiation, or Tumor-node-metastasis stage (Table 3). This result indicates that no clinicopathologic features should be used for predicting preoperative CRT response.
DISCUSSION
The results of the present study show, for the first time, that MMS19 expression in ESCC is upregulated in the cytoplasm and downregulated in the nucleus. The abnormal cellular distribution of MMS19 protein suggests that MMS19 is involved in the development and progression of ESCC. Furthermore, we found that MMS19 protein expression is associated with LNM and DM. More importantly, we found that cytoplasmic MMS19 protein expression is associated with the CRT response of ESCC. In clinical management, the therapeutic strategy for ESCC is primarily based on whether metastases exist, which is the most important determinant of patient outcome[27-30]. Furthermore, multimodality treatment only benefits patients who are sensitive to CRT[5-7]. Thus results of this study suggest that MMS19 has the potential to be a new biomarker for predicting metastasis and CRT response in ESCC.
In the present study, we found that the subcellular distribution of high MMS19 expression is changed from the nucleus in NESE to the cytoplasm in ESCC. Although the mechanism for this change in MMS19 expression in ESCC is not yet clear, a similar phenomenon has been reported for the DNA repair genes Ape1/ref-1 and JWA[31,32]. The aberrant subcellular distribution of the MMS19 protein may implicate that the DNA repair function of MMS19 in the nucleus is attenuated. Conversely, as a cytoplasmic Fe-S assembly machinery component in the cytoplasm, MMS19 would promote the synthesis of many Fe-S proteins participating in DNA metabolism in the nucleus. Thus, we hypothesize that, as a consequence, DNA mutations will accumulate in cancer cells due to the impaired DNA repair function, with cell division and proliferation accelerating as a result of the increased DNA metabolism, exerting adaptive pressure on these cells[33-35]. Previous studies have reported that rapidly proliferating esophageal cancer cells are more sensitive to CRT[36,37] and that DNA damage in cancer cells is associated with the sensitivity of cancer to CRT[38,39]. Our finding that ESCC with higher cytoplasmic MMS19 expression is much more sensitive to preoperative CRT is in accord with these studies. Furthermore, ESCC with higher MMS19 expression will accumulate an array of mutations, facilitating cancer metastasis.
Previous studies reported that DNA repair genes are associated with chemo- or radiotherapy response. The low nuclear expressions of ERCC1 and XRCC1 are associated with a good chemotherapy response in non-small-cell lung cancer[40-42] and gastric cancer[32], respectively, whereas high nuclear expression is associated with the radio-resistance of laryngeal cancer[43]. Furthermore, high nuclear expression of RRM1 is significantly associated with a lower disease control rate in non-small-cell lung cancer[44]. However, in the present study, we found that cytoplasmic MMS19 expression, but not nuclear MMS19 expression, is associated with CRT response. In addition to a role in DNA repair, MMS19 in the nucleus is also involved in mitotic segregation[45], histone modification[46], and interaction with regulator of telomere elongation helicase 1[17]. One reason that our study did not reveal an association between nuclear MMS19 and CRT response and metastasis may be that in the situation of abnormally expressed MMS19 in ESCC, the cytoplasmic function, but not the nuclear function of MMS19 plays the dominant role, underlying the development and progression of cancer cells.
In conclusion, the results demonstrate that MMS19 is abnormally expressed in esophageal squamous cell cancer. Elevated cytoplasmic MMS19 expression was associated with regional LNM, DM and a good preoperative chemoradiotherapy response of ESCC. These results provide novel preliminary evidence that MMS19 is involved in a mechanism of cancer development and progression, and has the potential to serve as a tumor biomarker that predicts metastasis and chemoradiotherapy response in ESCC.
ACKNOWLEDGMENTS
We acknowledge pathologists Jia Fu and Jia-Bin Lu in the department of pathology of Sun Yat-Sen University Cancer Center for their evaluation of the immunohistochemical expression of MMS19.
COMMENTS
Background
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive, malignant neoplasms. Surgery plus radiotherapy and/or chemotherapy is an effective treatment method for locally advanced esophageal cancer. However, there is no biomarker to predict the response of ESCC to chemoradiotherapy in clinical practice. Methyl-methanesulfonate sensitivity 19 (MMS19) is a multifunctional protein involved in DNA metabolism and genomic stability maintenance. Studies have demonstrated that single-nucleotide polymorphisms of the MMS19 gene are associated with the risk of pancreatic cancer, chemotherapy toxicity of non-small-cell lung cancer, and chemotherapy response of osteosarcoma. So far, the clinical significance of the expression of MMS19 in ESCC is not clear.
Research frontiers
Fe-S proteins such as DNA glycosylases, primases, DNA helicases, and nuclease are crucial for genomic instability, which is a hallmark of cancer. As a part of the cytoplasmic Fe-S assembly machinery, MMS19 facilitates the transfer of the Fe-S cluster to target Fe-S proteins, such as DNA polymerase, Dna2 nuclease/helicase, RNase L inhibitor, and endonuclease three-like glycosylase 2. However, the roles of the components of this machinery in cancer have rarely been explored.
Innovations and breakthroughs
Previous studies investigated the relationships of single-nucleotide polymorphisms of the MMS19 gene with cancer. We discovered, for the first time, that MMS19 is abnormally expressed in esophageal squamous cell cancer. Abnormally elevated cytoplasmic MMS19 expression is associated with regional lymph node metastases, distant metastases, and a good preoperative chemoradiotherapy response of ESCC. These results suggest that cytoplasmic Fe-S assembly machinery may play an important role in cancer development and progression, revealing new mechanisms of carcinogenesis and therapeutic targets.
Applications
The study results provide preliminary evidence that MMS19 has the potential to serve as a novel tumor biomarker for predicting metastasis and chemoradiotherapy response in ESCC.
Terminology
Methyl-methanesulfonate is an alkylating agent that can lead to DNA damage.
Peer-review
This is basically an interesting paper assessing a novel prognostic biomarker in ESCC with some potential to open up new lines of research.
Footnotes
Ethics approval: The study was reviewed and approved by the Medical Ethics Committee of Sun Yat-Sen University Cancer Center.
Informed consent: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest: We declare no competing commercial, personal, political, intellectual, or religious interests in relation to the submitted work.
Data sharing: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: November 5, 2014
First decision: November 26, 2014
Article in press: January 30, 2015
P- Reviewer: Kim BW, Nilsson M, Shim CS, Yi YX S- Editor: Qi Y L- Editor: AmEditor E- Editor: Ma S
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Liu WL, Guo XZ, Zhang LJ, Wang JY, Zhang G, Guan S, Chen YM, Kong QL, Xu LH, Li MZ, et al. Prognostic relevance of Bmi-1 expression and autoantibodies in esophageal squamous cell carcinoma. BMC Cancer. 2010;10:467. doi: 10.1186/1471-2407-10-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, Wei W, Inoue M, Tanaka H. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233–242. doi: 10.2188/jea.JE20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariette C, Piessen G, Triboulet JP. Therapeutic strategies in oesophageal carcinoma: role of surgery and other modalities. Lancet Oncol. 2007;8:545–553. doi: 10.1016/S1470-2045(07)70172-9. [DOI] [PubMed] [Google Scholar]
- 5.Medical Research Council Oesophageal Cancer Working Group. Surgical resection with or without preoperative chemotherapy in oesophageal cancer: a randomised controlled trial. Lancet. 2002;359:1727–1733. doi: 10.1016/S0140-6736(02)08651-8. [DOI] [PubMed] [Google Scholar]
- 6.Urba SG, Orringer MB, Turrisi A, Iannettoni M, Forastiere A, Strawderman M. Randomized trial of preoperative chemoradiation versus surgery alone in patients with locoregional esophageal carcinoma. J Clin Oncol. 2001;19:305–313. doi: 10.1200/JCO.2001.19.2.305. [DOI] [PubMed] [Google Scholar]
- 7.Walsh TN, Noonan N, Hollywood D, Kelly A, Keeling N, Hennessy TP. A comparison of multimodal therapy and surgery for esophageal adenocarcinoma. N Engl J Med. 1996;335:462–467. doi: 10.1056/NEJM199608153350702. [DOI] [PubMed] [Google Scholar]
- 8.Papatriantafyllou M. DNA Metabolism: MMS19: CIA agent for DNA-linked affairs. Nat Rev Mol Cell Biol. 2012;13:538. doi: 10.1038/nrm3411. [DOI] [PubMed] [Google Scholar]
- 9.Cheng L, Sturgis EM, Eicher SA, Spitz MR, Wei Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer. 2002;94:393–397. doi: 10.1002/cncr.10231. [DOI] [PubMed] [Google Scholar]
- 10.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42:65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 11.Sarries C, Haura EB, Roig B, Taron M, Abad A, Scagliotti G, Rosell R. Pharmacogenomic strategies for developing customized chemotherapy in non-small cell lung cancer. Pharmacogenomics. 2002;3:763–780. doi: 10.1517/14622416.3.6.763. [DOI] [PubMed] [Google Scholar]
- 12.Xu Z, Chen ZP, Malapetsa A, Alaoui-Jamali M, Bergeron J, Monks A, Myers TG, Mohr G, Sausville EA, Scudiero DA, et al. DNA repair protein levels vis-à-vis anticancer drug resistance in the human tumor cell lines of the National Cancer Institute drug screening program. Anticancer Drugs. 2002;13:511–519. doi: 10.1097/00001813-200206000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Seroz T, Winkler GS, Auriol J, Verhage RA, Vermeulen W, Smit B, Brouwer J, Eker AP, Weeda G, Egly JM, et al. Cloning of a human homolog of the yeast nucleotide excision repair gene MMS19 and interaction with transcription repair factor TFIIH via the XPB and XPD helicases. Nucleic Acids Res. 2000;28:4506–4513. doi: 10.1093/nar/28.22.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatfield MD, Reis AM, Obeso D, Cook JR, Thompson DM, Rao M, Friedberg EC, Queimado L. Identification of MMS19 domains with distinct functions in NER and transcription. DNA Repair (Amst) 2006;5:914–924. doi: 10.1016/j.dnarep.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Brosh RM. DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 2012;40:4247–4260. doi: 10.1093/nar/gks039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 17.Gari K, León Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science. 2012;337:243–245. doi: 10.1126/science.1219664. [DOI] [PubMed] [Google Scholar]
- 18.Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337:195–199. doi: 10.1126/science.1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McWilliams RR, Bamlet WR, de Andrade M, Rider DN, Cunningham JM, Petersen GM. Nucleotide excision repair pathway polymorphisms and pancreatic cancer risk: evidence for role of MMS19L. Cancer Epidemiol Biomarkers Prev. 2009;18:1295–1302. doi: 10.1158/1055-9965.EPI-08-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Gao G, Li X, Ren S, Li A, Xu J, Zhang J, Zhou C. Association between single nucleotide polymorphisms (SNPs) and toxicity of advanced non-small-cell lung cancer patients treated with chemotherapy. PLoS One. 2012;7:e48350. doi: 10.1371/journal.pone.0048350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai SB, Chen HX, Bao YX, Luo X, Zhong JJ. Predictive impact of common variations in DNA repair genes on clinical outcome of osteosarcoma. Asian Pac J Cancer Prev. 2013;14:3677–3680. doi: 10.7314/apjcp.2013.14.6.3677. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Spitz MR, Amos CI, Lin J, Schabath MB, Wu X. An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res. 2004;64:2251–2257. doi: 10.1158/0008-5472.can-03-2800. [DOI] [PubMed] [Google Scholar]
- 23.Vaclavikova R, Nordgard SH, Alnaes GI, Hubackova M, Kubala E, Kodet R, Mrhalova M, Novotny J, Gut I, Kristensen VN, et al. Single nucleotide polymorphisms in the multidrug resistance gene 1 (ABCB1): effects on its expression and clinicopathological characteristics in breast cancer patients. Pharmacogenet Genomics. 2008;18:263–273. doi: 10.1097/FPC.0b013e3282f60a91. [DOI] [PubMed] [Google Scholar]
- 24.Becker K, Mueller JD, Schulmacher C, Ott K, Fink U, Busch R, Böttcher K, Siewert JR, Höfler H. Histomorphology and grading of regression in gastric carcinoma treated with neoadjuvant chemotherapy. Cancer. 2003;98:1521–1530. doi: 10.1002/cncr.11660. [DOI] [PubMed] [Google Scholar]
- 25.Brücher BL, Becker K, Lordick F, Fink U, Sarbia M, Stein H, Busch R, Zimmermann F, Molls M, Höfler H, et al. The clinical impact of histopathologic response assessment by residual tumor cell quantification in esophageal squamous cell carcinomas. Cancer. 2006;106:2119–2127. doi: 10.1002/cncr.21850. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, He Y, Gao J, Fan L, Li Z, Yang G, Chen H. Caveolin-1 expression level in cancer associated fibroblasts predicts outcome in gastric cancer. PLoS One. 2013;8:e59102. doi: 10.1371/journal.pone.0059102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ando N, Ozawa S, Kitagawa Y, Shinozawa Y, Kitajima M. Improvement in the results of surgical treatment of advanced squamous esophageal carcinoma during 15 consecutive years. Ann Surg. 2000;232:225–232. doi: 10.1097/00000658-200008000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komukai S, Nishimaki T, Watanabe H, Ajioka Y, Suzuki T, Hatakeyama K. Significance of immunohistochemically demonstrated micrometastases to lymph nodes in esophageal cancer with histologically negative nodes. Surgery. 2000;127:40–46. doi: 10.1067/msy.2000.102754. [DOI] [PubMed] [Google Scholar]
- 29.Kato H, Tachimori Y, Watanabe H, Igaki H, Nakanishi Y, Ochiai A. Recurrent esophageal carcinoma after esophagectomy with three-field lymph node dissection. J Surg Oncol. 1996;61:267–272. doi: 10.1002/(SICI)1096-9098(199604)61:4<267::AID-JSO6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Matsubara T, Ueda M, Takahashi T, Nakajima T, Nishi M. Localization of recurrent disease after extended lymph node dissection for carcinoma of the thoracic esophagus. J Am Coll Surg. 1996;182:340–346. [PubMed] [Google Scholar]
- 31.Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, Broshears J, Koch M. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7:824–830. [PubMed] [Google Scholar]
- 32.Wang S, Wu X, Chen Y, Zhang J, Ding J, Zhou Y, He S, Tan Y, Qiang F, Bai J, et al. Prognostic and predictive role of JWA and XRCC1 expressions in gastric cancer. Clin Cancer Res. 2012;18:2987–2996. doi: 10.1158/1078-0432.CCR-11-2863. [DOI] [PubMed] [Google Scholar]
- 33.Bayani J, Selvarajah S, Maire G, Vukovic B, Al-Romaih K, Zielenska M, Squire JA. Genomic mechanisms and measurement of structural and numerical instability in cancer cells. Semin Cancer Biol. 2007;17:5–18. doi: 10.1016/j.semcancer.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Raptis S, Bapat B. Genetic instability in human tumors. EXS. 2006:303–320. doi: 10.1007/3-7643-7378-4_13. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Imdahl A, Jenkner J, Ihling C, Rückauer K, Farthmann EH. Is MIB-1 proliferation index a predictor for response to neoadjuvant therapy in patients with esophageal cancer? Am J Surg. 2000;179:514–520. doi: 10.1016/s0002-9610(00)00386-x. [DOI] [PubMed] [Google Scholar]
- 37.Beardsmore DM, Verbeke CS, Davies CL, Guillou PJ, Clark GW. Apoptotic and proliferative indexes in esophageal cancer: predictors of response to neoadjuvant therapy [corrected] J Gastrointest Surg. 2003;7:77–86; discussion 86-87. [PubMed] [Google Scholar]
- 38.Turesson I, Carlsson J, Brahme A, Glimelius B, Zackrisson B, Stenerlöw B. Biological response to radiation therapy. Acta Oncol. 2003;42:92–106. doi: 10.1080/02841860310004959. [DOI] [PubMed] [Google Scholar]
- 39.Alexander BM, Wang XZ, Niemierko A, Weaver DT, Mak RH, Roof KS, Fidias P, Wain J, Choi NC. DNA repair biomarkers predict response to neoadjuvant chemoradiotherapy in esophageal cancer. Int J Radiat Oncol Biol Phys. 2012;83:164–171. doi: 10.1016/j.ijrobp.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 40.Hwang IG, Ahn MJ, Park BB, Ahn YC, Han J, Lee S, Kim J, Shim YM, Ahn JS, Park K. ERCC1 expression as a prognostic marker in N2(+) nonsmall-cell lung cancer patients treated with platinum-based neoadjuvant concurrent chemoradiotherapy. Cancer. 2008;113:1379–1386. doi: 10.1002/cncr.23693. [DOI] [PubMed] [Google Scholar]
- 41.Olaussen KA, Dunant A, Fouret P, Brambilla E, André F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Zhang J, Wang R, Luo X, Chen H. The platinum-based treatments for advanced non-small cell lung cancer, is low/negative ERCC1 expression better than high/positive ERCC1 expression? A meta-analysis. Lung Cancer. 2010;70:63–70. doi: 10.1016/j.lungcan.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 43.Nix P, Greenman J, Stafford N, Cawkwell L. Expression of XRCC1 and ERCC1 proteins in radioresistant and radiosensitive laryngeal cancer. Cancer Ther. 2004;2:47–53. [Google Scholar]
- 44.Lee JJ, Maeng CH, Baek SK, Kim GY, Yoo JH, Choi CW, Kim YH, Kwak YT, Kim DH, Lee YK, et al. The immunohistochemical overexpression of ribonucleotide reductase regulatory subunit M1 (RRM1) protein is a predictor of shorter survival to gemcitabine-based chemotherapy in advanced non-small cell lung cancer (NSCLC) Lung Cancer. 2010;70:205–210. doi: 10.1016/j.lungcan.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Ito S, Tan LJ, Andoh D, Narita T, Seki M, Hirano Y, Narita K, Kuraoka I, Hiraoka Y, Tanaka K. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell. 2010;39:632–640. doi: 10.1016/j.molcel.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 46.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]


