Abstract
How different pathways lead to the activation of a specific transcription factor (TF) with specific effects is not fully understood. We model context-specific transcriptional regulation as a modulatory network: triplets composed of a TF, target gene, and modulator. Modulators usually affect the activity of a specific TF at the posttranscriptional level in a target gene-specific action mode. This action may be classified as enhancement, attenuation, or inversion of either activation or inhibition. As a case study, we inferred, from a large collection of expression profiles, all potential modulations of NF-κB/RelA. The predicted modulators include many proteins previously not reported as physically binding to RelA but with relevant functions, such as RNA processing, cell cycle, mitochondrion, ubiquitin-dependent proteolysis, and chromatin modification. Modulators from different processes exert specific prevalent action modes on distinct pathways. Modulators from noncoding RNA, RNA-binding proteins, TFs, and kinases modulate the NF-κB/RelA activity with specific action modes consistent with their molecular functions and modulation level. The modulatory networks of NF-κB/RelA in the context epithelial–mesenchymal transition (EMT) and burn injury have different modulators, including those involved in extracellular matrix (FBN1), cytoskeletal regulation (ACTN1), and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long intergenic nonprotein coding RNA, and tumor suppression (FOXP1) for EMT, and TXNIP, GAPDH, PKM2, IFIT5, LDHA, NID1, and TPP1 for burn injury.
Key words: : integrative probabilistic model, modulator, modulatory network, NF-κB, transcription factor
1. Introduction
Transcription factors (TFs) have multiple functions and are involved in different processes. Proteins that modulate the activity of the specific TF, modulators, usually act at the posttranscriptional level with target gene-specific action modes: enhancement, attenuation, or inversion of either activation or inhibition. As opposed to binary regulatory networks, a modulatory network is composed of triplets: a specific TF, target gene, and modulator. Activation of TFs in different processes is mediated by different modulators, and thus leads to distinct target gene response patterns. As an extension of our previous study (Li et al., 2014) that uncovered the modulators of RelA among its binding proteins, we predicted functional modulators exerting their modulation on specific target genes at all possible posttranscriptional levels. Such modulators may include long noncoding RNAs, microRNAs, kinases, and RNA-binding proteins. The unconstrained modulatory network is a useful resource for studying the NF-κB/RelA-related transcriptional pathways.
NF-κB is a key human TF involved in diverse cellular processes, including immunity, inflammation, stress response, apoptosis (Jennewein et al., 2012), cell mobility and migration, cell cycle, and cell differentiation. Coupling of these cellular processes by NF-κB activity mediated by modulators plays critical roles in cell fate decision and is implicated in many diseases (Pahl and Szelenyi, 2002; Abraham, 2003; Chen, 2005; Ramdass et al., 2007; Csiszar et al., 2008).
Epithelial–mesenchymal transition (EMT) is an epithelial plasticity and metastasis process. During EMT, epithelial cells transform to mesenchymal cells by losing their cell polarity and adhesion, and gaining migratory and invasive properties. Three types of EMT are, respectively, involved in development, wound healing and fibrosis, and cancer progression. NF-κB has been reported to be essential for EMT and metastasis in a breast cancer model (Huber et al., 2004) and in a Hi-Myc transgenic mouse prostate cancer model (Jin et al., 2014). However, the mechanism of NF-κB involvement in epithelial plasticity and metastasis is not fully known. Similarly, burn injury activates the NF-κB/RelA pathways, but how the activity of NF-κB/RelA is modulated in burn injury is not known in detail. Here, we infer an unconstrained modulatory network of NF-κB/RelA (Li et al., 2014) and discuss its role in EMT and burn injury.
2. Methods
2.1. Probabilistic models for triplet prediction
The probabilistic model is modified from Babur et al. (2010). We further filtered the predicted modulators by the Shannon entropy of the distribution across the six action modes by each modulator (Li et al., 2014). Briefly, we approximated the probability that TG is highly expressed, p (high TG), as:
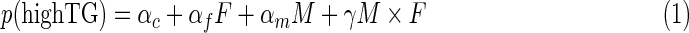 |
where F, M, and TG are the TF, its modulator, and the affected target gene, respectively. F, M, and TG are highly expressed if the ranked expression of the corresponding gene is in the upper tertile. Conversely, if it is in the lower tertile, F, M, and TG are set to low. The estimation of αc αf αm, and γ, action modes and entropy of each triplet can be found in Supplementary Material (available online at www.liebertpub.com/cmb).
2.2. Input data for inferring NF-κB/RelA modulatory network
We used gene expression profiles of 2158 tumor samples published by expO (expression project for oncology) to characterize each gene. As reported in our previous study (Li et al., 2014), we discretized the expression values by rank-ordering across genes and dividing the ranked 2158 expression values of each gene across experiments into 3 bins. We predicted the triplets on probeset level.
Modulators of RelA were predicted from the 15,373 annotated genes that have a p-call ratio at least 10% of all the expO microarrays. Among the annotated genes, there are 527 binding proteins of NF-κB/RelA (Li et al., 2014), which was used to validate the predicted modulators without constriction. The prediction is based on the list of 1182 target genes of RelA from Li et al. (2014), which had been derived from Pahl (1999) and Yang et al. (2013) and web resources by the Gilmore lab (Gilmore, 2006). We obtained 2283 probesets corresponding to 1069 candidate target genes and 27,867 probesets corresponding to 15,373 candidate modulator genes that were not target genes themselves with at least 10% p-call ratio (high-quality Affymetrix measurements). We considered the two probesets of RelA, 201783_s_at and 209878_s_individually, and ignored the third one, 230202_at, because of its low expression (Li et al., 2014).
2.3. Enrichment analysis of the action modes of the triplets composed of specific groups of predicted modulators
Among predicted modulators, typical modulators with defined biochemical properties, including RNA-binding proteins, cytoskeleton proteins, kinase, microRNAs, and TFs, were extracted from Gene Ontology (GO) term annotation and literature mining. For biological processes and pathway action mode enrichment analysis, we grouped the predicted modulators into their respective enriched GO terms and removed the smaller set of modulators with 50% or greater overlap with genes in other GO term and kept the sets with defined gene set size range.
Overrepresentation of the predicted modulators in six action modes was assessed by hypergeometric cumulative distribution function by comparing the action modes of the triplets comprising the modulators and target genes from different processes to the background action mode distribution of all triplets.
2.4. Modulatory network in EMT and burn injury
Differentially expressed genes of EMT were obtained based on time-course GSE17708 (Sartor et al., 2010) of IGFB1-treated A549 cells. We used genes with anti log2 ratio significantly greater than 1 with p<0.01 between control and 72 hours after IGFB1 treatment. Differentially expressed genes were mapped to the above predicted general modulatory network. The EMT modulatory network composed of the modulators with at least six TGs was then visualized with Cytoscape. The modulatory network of burn injury was constructed in the same way based on GSE 19743 (Zhou et al., 2010). Genes were considered to be differentially expressed using a fold ratio of 2 and p<0.01 with Kolmogorov–Smirnov test between control and burn injury.
3. Results
3.1. Inferring the unconstrained NF-κB/RelA modulatory network
As an extension of our previous study (Li et al., 2014), we predicted all possible modulators of NF-κB/RelA not limited to its identified binding proteins with all genes as candidate modulators. We used the 1182 target genes of NF-κB/RelA (Li et al., 2014) as candidate TGs. Each (M, TF, TG) triplet was considered independently based on the probabilistic model, where the conditional probabilities of high expression of a TG were calculated dependent on the expression of the modulator and TF. Action modes of modulators were categorized into enhancement, attenuation, or inversion of activation or inhibition of the TF. (For more detail, please refer to Supplementary Material and Li et al., 2014.) Benjamini-Hochberg p-value correction was used to reduce the false discovery rate. At a false discovery rate under 1%, 17,687 triplets and 5759 modulators with one of the six defined action modes were predicted (see Supplementary Tables S1 and S2). Of the predicted modulators, 296 overlapped with the 527 binding proteins (see Methods) among all the considered candidate modulator genes with p=4.2e-11 from hypergeometric test, indicating the relevance of our predicted modulators.
3.2. Characterization of the predicted modulators of RelA
GO term analysis of the predicted NF-κB/RelA modulators confirmed their functional involvement in biological processes characteristic of NF-κB/RelA. Enriched GO categories include cell cycle, RNA splicing, cytoskeleton organization, ubiquitin-specific protein catabolic process, chromatin modification, and JNK cascade shared with GO analysis of existing binding proteins in Supplementary Table S3. Other enriched processes include DNA replication, Wnt receptor signaling pathways, and collagen fibril organization.
A connected network with 3189 nodes and 30,435 edges was reconstructed by mapping the newly identified 5759 NF-κB/RelA-associated proteins onto the global protein–protein interaction network by using the Reactome FI Cytoscape Plugin (Wu et al., 2010). Pathway enrichment of the connected protein network shows that the most enriched pathways are mRNA processing, cell cycle, integrin pathway, pathways in cancer, cytoskeleton organization, and ubiquitin mediated proteolysis. In addition to uncovered pathways enriched by the binding modulators of RelA, new enriched pathways were also discovered (See Table 1), such as beta1 integrin cell surface interaction (Scatena et al., 1998), NGF (Foehr et al., 2000), PDGFR-beta (Romashkova and Makarov, 1999), FGFR (Drafahl et al., 2010), and EGFR pathways (Yang et al., 2012). Most of these can activate the NF-κB/RelA activity.
Table 1.
Enriched Pathways of the Protein Network Composed of the Predicted Modulators Without Added Linkers as Analyzed by the Reactome FI Cytoscape Plugin
| Gene seta | Protein from network | FDR | Overlapb |
|---|---|---|---|
| Processing of capped intron-containing pre-mRNA(R) | 79 | <1.00e-03 | N |
| Mitotic M-M/G1 phases(R) | 110 | <5.00e-04 | N |
| Translation(R) | 77 | <3.33e-04 | Y |
| Axon guidance(R) | 119 | 2.50e-04 | Y |
| Focal adhesion(K) | 95 | 2.00e-04 | Y |
| Cell cycle(K) | 66 | 3.33e-04 | Y |
| Influenza infection(R) | 72 | 5.71e-04 | N |
| Class I MHC-mediated antigen processing & presentation(R) | 111 | 5.00e-04 | N |
| Mitotic G1-G1/S phases(R) | 68 | 6.67e-04 | Y |
| Extracellular matrix organization(R) | 80 | 6.00e-04 | N |
| Integrin signaling pathway(P) | 76 | 5.45e-04 | N |
| Spliceosome(K) | 64 | 5.00e-04 | Y |
| Protein processing in endoplasmic reticulum(K) | 78 | 5.38e-04 | N |
| PDGFR-beta signaling pathway(N) | 61 | 5.71e-04 | N |
| Cell cycle checkpoints(R) | 60 | 5.33e-04 | Y |
| Nonsense-mediated decay(R) | 55 | 6.88e-04 | N |
| Regulation of mitotic cell cycle(R) | 46 | 6.47e-04 | Y |
| Host interactions of HIV factors(R) | 62 | 7.22e-04 | N |
| NGF signaling via TRKA from the plasma membrane(R) | 94 | 1.16e-03 | Y |
| Mitotic G2-G2/M phases(R) | 46 | 1.45e-03 | N |
| S phase(R) | 55 | 1.76e-03 | N |
| Ubiquitin-mediated proteolysis(K) | 64 | 1.73e-03 | Y |
| Regulation of DNA replication(R) | 41 | 1.74e-03 | Y |
| Pathways in cancer(K) | 127 | 1.67e-03 | N |
| HTLV-I infection(K) | 106 | 2.56e-03 | N |
| Beta1 integrin cell surface interactions(N) | 37 | 2.46e-03 | N |
| PLK1 signaling events(N) | 28 | 2.56e-03 | N |
| Signaling by PDGF(R) | 85 | 3.25e-03 | N |
| CDC42 signaling events(N) | 38 | 3.34e-03 | N |
| ErbB1 downstream signaling(N) | 49 | 3.70e-03 | N |
| ECM-receptor interaction(K) | 43 | 5.19e-03 | N |
| mTOR signaling pathway(N) | 35 | 5.06e-03 | N |
| Synthesis of DNA(R) | 47 | 4.94e-03 | Y |
| Ribosome(K) | 45 | 5.09e-03 | Y |
| Regulation of mRNA stability by proteins that bind AU-rich elements(R) | 43 | 5.57e-03 | Y |
| Ras pathway(P) | 34 | 6.42e-03 | N |
| Regulation of actin cytoskeleton(K) | 86 | 6.35e-03 | Y |
| Signaling events mediated by hepatocyte growth factor receptor (c-Met)(N) | 40 | 7.84e-03 | N |
| Renal cell carcinoma(K) | 36 | 9.05e-03 | N |
| Stabilization and expansion of the E-cadherin adherens junction(N) | 25 | 9.58e-03 | N |
| Insulin signaling pathway(K) | 60 | 9.59e-03 | Y |
| Signaling by EGFR(R) | 78 | 1.00e-02 | Y |
| Factors involved in megakaryocyte development and platelet production(R) | 55 | 9.66e-03 | Y |
| Aurora B signaling(N) | 24 | 9.58e-03 | N |
The letter in parentheses following each pathway gene set name corresponds to the source of the pathway annotations: B, BioCarta; C, CellMap; K, KEGG; N, NCI PID; and R, Reactome.
Overlap: N, new or nonoverlapping pathway; Y, overlapping pathway between binding and nonrestrained modulators.
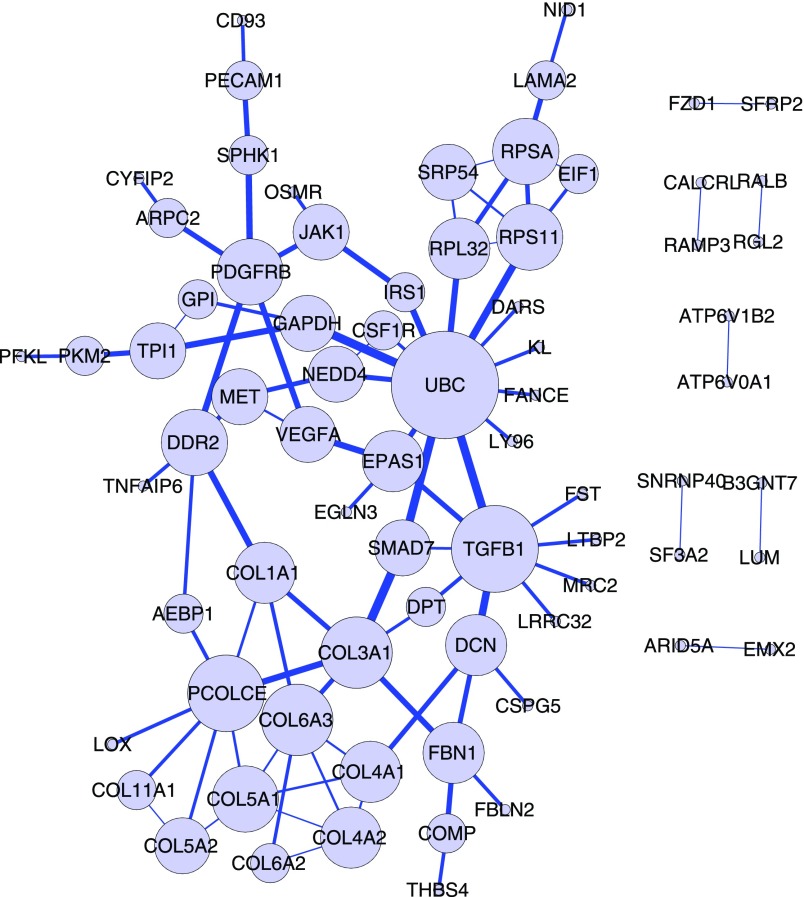
These results demonstrate the relevance of predicted modulators to the biological context of RelA association. The predicted modulators and their involved triplets provide insight into how these pathways cross-talk with NF-κB/RelA pathway and served as important general hypotheses for further validation. Figure 1 shows the protein network of predicted NF-κB/RelA modulators mapped onto Reactome, where each modulator has no less than five targets. The complete list of the modulators with action mode distribution and entropy can be found in Supplementary Table S2.
FIG. 1.
Protein network of the top RelA modulators. The network was generated by Reactome FI Cytoscape Plugin with all modulators with at least five target genes as input. Edge width is proportional to the betweenness coefficient of the edge. Node size is proportional to the linker degree of each modulator.
The 10 modulators with the highest number of target genes are SLC37A4, VEGFA, COL16A1, GLT8D2, CCDC72, PDPN, FOXP1, FBN1, SRPX, and COL5A2 (see Table 2). Since our model cannot fully discriminate modulator and target genes without binding constraint, some modulators, such as VEGFA, SLC37A4, and collagen proteins, may also be target genes of NF-κB. Moreover, the inferred modulators also contain nonbinding modulators that modulate the activity of NF-κB through pathways. However, the biological relevance of the top modulators, including VEGFA, GLT8D2, PDPN, FOXP1, and FBN1, was validated by literature mining (see Table 2).
Table 2.
Top-Ten Modulators of RelA
| Number of TGs per action mode | ||||||||
|---|---|---|---|---|---|---|---|---|
| Top-10 modulators | IA | IE | II | AI | AE | AA | Validation reference | Comments |
| SLC37A4 | 6 | 15 | 0 | 0 | 26 | 11 | Jun et al., 2012 | Mutation in macrophages can lead to deregulated energy metabolism in congenital neutropenia syndrome and decreased trafficking of macrophages. |
| VEGFA | 3 | 3 | 0 | 0 | 25 | 2 | Marumo et al., 1999; Grosjean et al., 2006 | Activates NF-κB/RelA in bovine retinal microvascular endothelial cells and in human umbilical vein endothelial cells |
| COL16A1 | 8 | 1 | 0 | 0 | 4 | 17 | Han et al., 1998 | As collagen family members may regulate the activity of NF-κB in collagen-induced rheumatoid arthritis |
| COL5A2 | 9 | 0 | 4 | 0 | 10 | 1 | ||
| GLT8D2 | 12 | 0 | 5 | 0 | 7 | 6 | Wei et al., 2013 | GLT8D2 increases apoB100 protein expression in hepatocytes; apoB100 is a target of NF-κB/RelA. |
| CCDC72 | 10 | 1 | 0 | 5 | 0 | 12 | Charpin et al., 2013 | Is an immunity-related protein |
| PDPN | 12 | 0 | 0 | 0 | 11 | 4 | Maruyama et al., 2014 | Suppresses TNF-alpha secretion, and NF-κB pathways in the thioglycollate-induced macrophages in flamed conditions |
| FOXP1 | 15 | 1 | 0 | 0 | 0 | 11 | Green et al., 2009 | Associated with differential NF-κB activity in follicular lymphoma |
| FBN1 | 8 | 1 | 7 | 0 | 8 | 1 | Merk et al., 2011 | Decreased activation of NF-κB was observed with the FBNI (C1039G/+) mutation in the aorta of the Marfan mouse |
| SRPX | 14 | 0 | 5 | 0 | 6 | 0 | Miljkovic-Licina et al., 2009; Lu et al., 2011 | Are involved in angiogenesis |
AA, activation attenuation; AE, activation enhancement; AI, activation inversion; IA, inhibition activation; IE, inhibition enhancement.
Among the predicted modulators, typical biomedical properties are identified, including RNA-binding proteins, cytoskeleton proteins, kinase, noncoding RNAs, and TFs. For example, the predicted modulators are enriched in kinase with p=2.66e-7. Nineteen of the 30 kinase modulators (Choudhary et al., 2011) of canonical NF-κB/RelA pathways were predicted (p=0.014). The predicted modulators are also enriched in TFs with p=3.8e-5, and in binding TFs with p=0.0046. GO terms analysis also shows enrichment in RNA binding. There is moderate enrichment in the small number of annotated noncoding RNA genes (p=0.02).
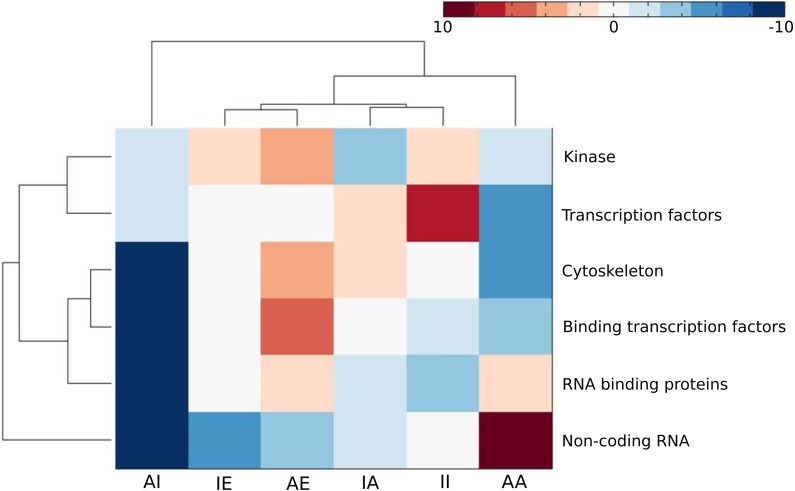
Certain classes of modulators have a prevalent action mode (see Fig. 2, Table 3, and Supplementary Table S4). Noncoding RNA modulators of RelA are mostly enriched in activation attenuation (AA). Kinase modulators are mostly enriched in activation enhancement (AE). TFs show the most enrichment in inhibition inversion (II). Binding TFs are enriched in AE. As modulators, the cytoskeleton proteins are enriched in AE. For depletion, TFs show depletion in AA.
FIG. 2.
Heatmap of the clustered enrichment (red) or depletion (blue) of the action modes of different molecular functional groups of modulators.
Table 3.
−log10(p-Value) of the Enriched Action Modes and log10(p-Value) of the Depleted Action Modes of Modulators with Different Molecular Functions
| Modulators | ||||||
|---|---|---|---|---|---|---|
| Action modes | IA | IE | II | AI | AE | AA |
| Noncoding RNA | −2.35 | −5.21 | −0.417 | −17 | −3.77 | 13.1 |
| RNA binding | −1.45 | −0.874 | −3.93 | −17 | 1.15 | 1.99 |
| Kinase | −2.81 | 2.63 | 1.26 | −0.917 | 2.88 | −2.27 |
| Transcription factors | 1.45 | −0.39 | 6.55 | −1.71 | 0.581 | −5.41 |
| Binding transcription factors | −0.604 | −0.354 | −1.09 | −17 | 4.87 | −3.44 |
| Cytoskeleton | 1.1 | 0.398 | 0.727 | −17 | 2.79 | −5.47 |
Minus sign represents depletion, and plus sign (omitted) represents enrichment.
3.3. Different pathways modulate distinct NF-κB/RelA-dependent processes
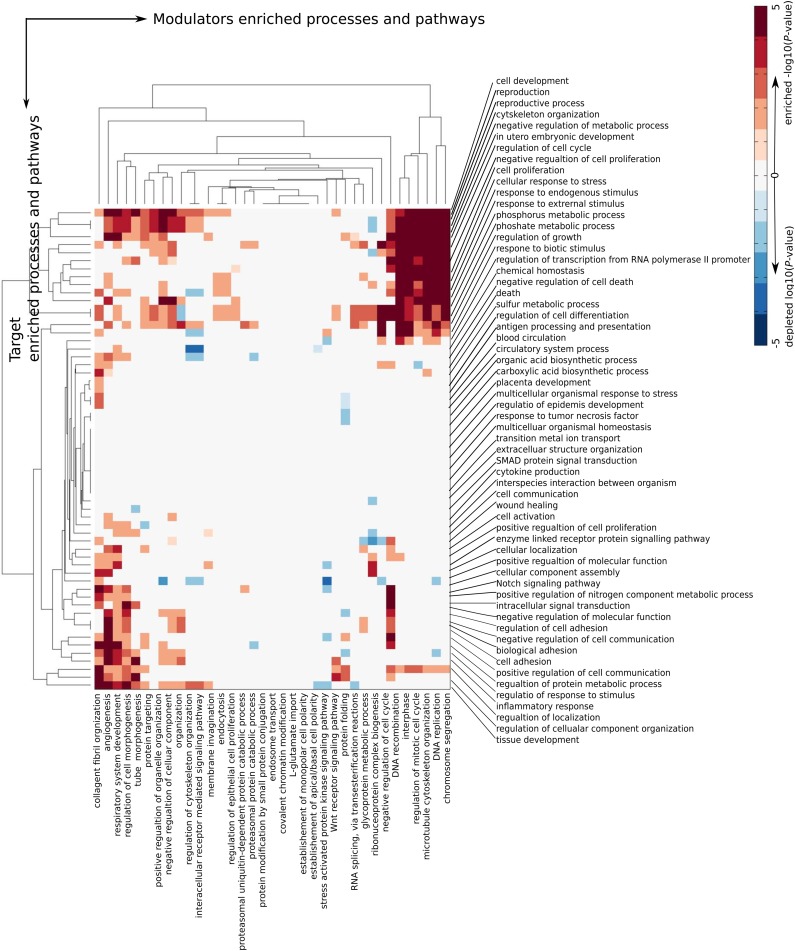
We characterized how modulators from different processes affect different groups of RelA TGs by directly using the enriched −log (p-value) or depleted (log p-value) of each specific action mode category. We grouped the predicted modulators by GO terms, removing overlaps of 50% or more. We considered 39 groups of 50–600 modulators and 60 groups of 10–100 target genes. Of all triplets, we predicted 4541 IA, 1163 IE, 790 II, 62 AI, 4647 AE, and 6484 AA. For the AE enrichment map (see Fig. 3), there is a prevalent enrichment of AE action mode between different pathways and processes, which may reflect their high cooperation. Consistently, more than half of the between-processes action modes are depleted in AA (see Figs. 3 and 4). The AE heatmap classified the processes involved by modulators into two major clusters. The bigger cluster includes processes in cell morphogenesis, cell signaling, cellular component organization, Wnt receptor signaling pathway, RNA splicing, and protein folding and targeting. The smaller cluster includes modulators enriched in cell cycle and DNA recombination and replication. We noticed three major submodules of modulator- and target gene-enriched processes. The largest submodule is built from modulators involved in negative regulation of cell cycle, DNA recombination and replication, interphase, and chromosome segregation, and target genes involved in cellular development, reproductive process, metabolic process regulation, embryonic development, cell proliferation, and response to stress and phosphorylation process. This is consistent with the strict cell cycle regulation in development, reproduction, cell proliferation, and cellular response to stress.
FIG. 3.
Enrichment (red) and depletion (blue) of activation enhancement (AE) action mode between cross-processes modulation.
FIG. 4.
Enrichment (red) and depletion (blue) of activation attenuation (AA) action mode between cross-process modulation.
For the AA heatmap, four clusters of modulator-enriched processes were noticed. The enriched modulator processes represent two opposite dominant action mode patterns: three AA-depleted clusters and one AA-enriched cluster. The only AA-enriched cluster of modulator processes includes RNA processing and translational elongation.
3.4. RelA-centered modulatory network in the EMT and burn injury
In total, 3238 differentially expressed probesets were obtained directly from Sartor et al. (2010) by comparing control and 72 hours after TGFB1 treatment. After mapping the differentially expressed probesets to both the modulators and TGs composing above 17,687 triplets of NF-κB/RelA, we extracted 749 triplets, which were called the modulatory network of NF-κB/RelA of the EMT. We then extracted the modulators each with at least 5 target genes, and 173 triplets composed of 21 modulators and 45 target genes were obtained (Supplementary Table S5) and visualized using Cytoscape (Fig. 5). We note that EMT-associated NF-κB/RelA modulators include FOXP1, encoded by a tumor suppressor gene involved in cardiac development; FBN1 and CCDC80, encoded by interacting genes involved in extracellular matrix formation; ACTN1, important in formation of actin polymers; DOCK10, a Rho GTPase associated with cytoskeletal motility; and MALAT1, a long intergenic non-protein-coding RNA. These functions suggest how the EMT state modulates the activity of the ubiquitous NF-κB/RelA in a cell-state-dependent manner.
FIG. 5.
Modulatory network involved in epithelial–mesenchymal transition. For visualization, the modulatory network was restricted to modulators with at least six target genes at probeset level. As a predicted modulator, VEGFA and its target genes were removed as VEGFA is a target itself. Diamonds are target genes and circles are modulators of RelA. For edge color, dark green represents inhibition attenuation (IA); light green, inhibition enhancement (IE); yellow, inhibition inversion (II); orange, activation inversion (AI); dark orange, activation enhancement (AE); and red, activation attenuation (AA). Edge widths are proportional to the −log10(p-value of γ). For node color, green indicates downregulated genes and red indicates upregulated genes in EMT.
Burn injury response also involves activity of NF-κB/RelA as indicated by the enrichment of the 4228 DEGs in the 1182 TGs of RelA (Li et al., 2014) with p=3.1e-6. Similarly, after mapping the differentially expressed genes to the 17,687 triplets of NF-κB/RelA, we extracted 81 triplets. The top interesting differentially expressed modulators with at least three TGs include TXNIP, GAPDH, PKM2, IFIT5, LDHA, NID1, and TPP1. Among them, TXNIP has been reported to be transcriptional repressor of mTOR (Jin et al., 2011). GAPDH is one of the top modulators constrained to the binding proteins of RelA (Li et al., 2014). Literature mining implicates their association with NF-κB/RelA (see Table 4). The function of the modulators may suggest how burn injury modulates the activity of NF-κB/RelA in leukocytes. We noticed the significant difference of rewiring of NF-κB/RelA-centered modulatory networks in the EMT and burn injury.
Table 4.
Top Modulators of NF-κB/RelA in Modulatory Network of Burn Injury
| Number of TGs per action mode | ||||||||
|---|---|---|---|---|---|---|---|---|
| Top modulators | IA | IE | II | AI | AE | AA | Validation reference | Comments |
| TXNIP | 6 | 15 | 0 | 0 | 26 | 11 | Jin et al., 2011 | Is a transcriptional repressor of mTOR and also activate NF-κB/RelA |
| GAPDH | 3 | 3 | 0 | 0 | 25 | 2 | Li et al., 2014 | Is one of the top predicted modulators constrained to the binding proteins of RelA |
| PKM2 | 8 | 1 | 0 | 0 | 4 | 17 | Yang et al., 2012 | Is upregulated through NF-κB after EGFR stimulation |
| IFIT5 | 12 | 0 | 5 | 0 | 7 | 6 | Zhang et al., 2013 | Enhances immune signaling pathway |
| LDHA | 10 | 1 | 0 | 5 | 0 | 12 | Nair et al., 2013 | Increased levels combined with RelA expression were synergistically associated with recurrence and death of non-small-cell lung cancer patients. |
| NID1 | 12 | 0 | 0 | 0 | 11 | 4 | Koziol et al., 2012 | Is a substrate of MT1-MMP protease in TNF-alpha-activated endothelial cells |
| TPP1 | 15 | 1 | 0 | 0 | 0 | 11 | Vuillemenot et al., 2014 | TPP1 treatment on CLN2 disease can reduce the inflammation of brain |
4. Discussion
We used the approach developed in our previous study, where the candidate modulators of RelA were constrained to the binding proteins to infer the modulators of NF-κB/RelA without restrictions on the candidate modulator genes. We inferred a network of 17,687 M-TF-TG triplets, approximately twice the size of the constrained network (Li et al., 2014), and predicted 5759 potential modulators of NF-κB/RelA from genome-wide genes with at least one TG. The results have been validated by enrichment of predicted modulators, in the physically binding modulators in nucleus (Li et al., 2014). Additional validation of the method comes from the literature-based analysis of the functions of the top predicted modulators. The 17,687 M-TF-TG triplets provide more diverse testable hypotheses for experimental validation and further analysis.
The identified modulators are functionally enriched in previously identified pathways as related to NF-κB/RelA activity, such as mRNA processing, cell cycle, translation, spliceosome, ubiquitin-mediated proteolysis, ribosome, regulation of mRNA stability by proteins that bind AU-rich elements, and regulation of actin organization. Newly predicted interesting pathways modulating NF-κB/RelA were also identified, which include focal adhesion, MHC-mediated antigen processing and presentation, extracellular matrix organization, integrin signaling pathways, PDGFR-beta signaling pathway, nonsense-mediated decay, host interactions of HIV factors, NGF signaling via TRKA from the plasma membrane, mTOR signaling pathway, CDC42 pathway, Ras pathway, and factors involved in megakaryocyte development and platelet production. Most of these pathways are related to NF-κB/RelA as supported by the literature.
We developed a new statistical approach to study how these different pathways may cooperate through NF-κB/RelA based on the 17,687 M-TF-TG triplets. We discovered modules enriched in action modes that point to how different processes modulating the activity of NF-κB/RelA. We generated an EMT context-specific modulatory subnetwork comprising extracellular matrix formation, actin polymerization pathways, and long intergenic non-protein-coding RNA regulated by EMT (Ijaz et al., 2014). This information may provide potential drug targets for EMT-related cancer progression. By comparing the top modulators of the EMT to the burn injury modulatory networks, we found that top modulators in burn injury tend to be more metabolic related.
In conclusion, predicting all modulators of RelA (the main component of NF-κB complex), including those in both cytoplasm and nucleus, and the M-TF-TG triplets resulted in an integrated genome-wide functional interaction network between different processes. This network contains potential new mechanistic insight into the cooperation of the different processes modifying NF-κB activity and specificity. Analysis of the module organization of the enriched action modes of the cross-group modulatory subnetwork built from enriched groups of modulators and target genes may reveal mechanisms involved in the cooperation between different biological processes, such as immunity, inflammation, cancer, development, and others. The new mechanisms of RelA-dependent gene expression and related signaling pathways that cross talk with NF-κB may provide guidance for the design of targeted therapeutics. NF-κB/RelA modulators involved in burn injury and EMT may shed light on how distinct processes differentially regulate NF-κB/RelA activity. Since NF-κB/RelA can trigger EMT and is involved in cancer progression, the novel NF-κB/RelA modulators we discovered, involved in EMT, may be potential drug targets for cancer progression. In future work, we will apply our approach to inferring the modulatory network composed of all genome-wide TFs.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/NHLBI Grants N01-HV-00245 and NIAID AI062885 (to A.R.B.), by National Natural Science Foundation of China Grants Nos. 31271412 and 31371340 (to X.L.) and No. 61273324 (to M.Z.), and by a training fellowship from the Keck Center Computational Cancer Biology Training Program of the Gulf Coast Consortia (CPRIT Grant No. RP140113, PI-Rathindra Bose; to X.L., A.S.K., and A.R.B.). The funders had no role in the preparation of the article.
Author Disclosure Statement
The authors declare that they have no conflicts of interest.
References
- Abraham E.2003. Nuclear factor-kappaB and its role in sepsis-associated organ failure. J. Infect. Dis. 187Suppl 2, S364–S369 [DOI] [PubMed] [Google Scholar]
- Babur O., Demir E., Gonen M., et al. 2010. Discovering modulators of gene expression. Nucleic Acids Res. 38, 5648–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpin C., Arnoux F., Martin M., et al. 2013. New autoantibodies in early rheumatoid arthritis. Arthritis Res. Ther. 15, R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.2005. Is NF-kappaB a culprit in type 2 diabetes? Biochem. Biophys. Res. Commun. 332, 1–3 [DOI] [PubMed] [Google Scholar]
- Choudhary S., Rosenblatt K.P., Fang L., et al. 2011. High throughput short interfering RNA (siRNA) screening of the human kinome identifies novel kinases controlling the canonical nuclear factor-kappaB (NF-kappaB) activation pathway. J. Biol. Chem. 286, 37187–37195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csiszar A., Wang M., Lakatta E.G., and Ungvari Z.2008. Inflammation and endothelial dysfunction during aging: role of NF-kappaB. J. Appl. Physiol. 105, 1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drafahl K.A., McAndrew C.W., Meyer A.N., et al. 2010. The receptor tyrosine kinase FGFR4 negatively regulates NF-kappaB signaling. PLoS One 5, e14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foehr E.D., Lin X., O'Mahony A., et al. 2000. NF-kappa B signaling promotes both cell survival and neurite process formation in nerve growth factor-stimulated PC12 cells. J. Neurosci. 20, 7556–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore T.D.2006. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene 25, 6680–6684 [DOI] [PubMed] [Google Scholar]
- Green M.R., Gandhi M.K., Courtney M.J., et al. 2009. Relative abundance of full-length and truncated FOXP1 isoforms is associated with differential NFkappaB activity in follicular lymphoma. Leuk. Res. 33, 1699–1702 [DOI] [PubMed] [Google Scholar]
- Grosjean J., Kiriakidis S., Reilly K., et al. 2006. Vascular endothelial growth factor signalling in endothelial cell survival: a role for NFkappaB. Biochem. Biophys. Res. Commun. 340, 984–994 [DOI] [PubMed] [Google Scholar]
- Han Z., Boyle D.L., Manning A.M., and Firestein G.S.1998. AP-1 and NF-kappaB regulation in rheumatoid arthritis and murine collagen-induced arthritis. Autoimmunity 28, 197–208 [DOI] [PubMed] [Google Scholar]
- Huber M.A., Azoitei N., Baumann B., et al. 2004. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Invest. 114, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijaz T., Pazdrak K., Kalita M., et al. 2014. Systems biology approaches to understanding Epithelial Mesenchymal Transition (EMT) in mucosal remodeling and signaling in asthma. World Allergy Organ. J. 7, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein C., Karl S., Baumann B., et al. 2012. Identification of a novel pro-apoptotic role of NF-kappaB in the regulation of TRAIL- and CD95-mediated apoptosis of glioblastoma cells. Oncogene 31, 1468–1474 [DOI] [PubMed] [Google Scholar]
- Jin H.O., Seo S.K., Kim Y.S., et al. 2011. TXNIP potentiates Redd1-induced mTOR suppression through stabilization of Redd1. Oncogene 30, 3792–3801 [DOI] [PubMed] [Google Scholar]
- Jin R., Yi Y., Yull F.E., et al. 2014. NF-kappaB gene signature predicts prostate cancer progression. Cancer Res. 74, 2763–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun H.S., Cheung Y.Y., Lee Y.M., et al. 2012. Glucose-6-phosphatase-beta, implicated in a congenital neutropenia syndrome, is essential for macrophage energy homeostasis and functionality. Blood 119, 4047–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol A., Gonzalo P., Mota A., et al. 2012. The protease MT1-MMP drives a combinatorial proteolytic program in activated endothelial cells. FASEB J. 26, 4481–4494 [DOI] [PubMed] [Google Scholar]
- Li X., Zhao Y., Tian B., et al. 2014. Modulation of gene expression regulated by the transcription factor NF-kappaB/RelA. J. Biol. Chem. 289, 11927–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Proto J.D., Guo L., et al. 2011. NF-kappaB negatively impacts the myogenic potential of muscle-derived stem cells. Mol. Ther. 20, 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumo T., Schini-Kerth V.B., and Busse R.1999. Vascular endothelial growth factor activates nuclear factor-kappaB and induces monocyte chemoattractant protein-1 in bovine retinal endothelial cells. Diabetes 48, 1131–1137 [DOI] [PubMed] [Google Scholar]
- Maruyama Y., Maruyama K., Kato Y., et al. 2014. The effect of podoplanin inhibition on lymphangiogenesis under pathological conditions. Invest. Ophthalmol. Vis. Sci. 55, 4813–4822 [DOI] [PubMed] [Google Scholar]
- Merk D.R., Chin J.T., Dake B.A., et al. 2011. miR-29b participates in early aneurysm development in Marfan syndrome. Circ. Res. 110, 312–324 [DOI] [PubMed] [Google Scholar]
- Miljkovic-Liciana M., Hammel P., Garrido-Urbani S., et al. 2009. Sushi repeat protein X-linked 2, a novel mediator of angiogensis. FASEB J. 23, 4105–4116 [DOI] [PubMed] [Google Scholar]
- Nair V.S., Gevaert O., Davidzon G., et al. 2013. NF-kappaB protein expression associates with (18)F-FDG PET tumor uptake in non-small cell lung cancer: a radiogenomics validation study to understand tumor metabolism. Lung Cancer 83, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl A., and Szelenyi I.2002. Asthma therapy in the new millennium. Inflamm. Res. 51, 273–282 [DOI] [PubMed] [Google Scholar]
- Pahl H.L.1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- Ramdass B., Maliekal T.T., Lakshmi S., et al. 2007. Coexpression of Notch1 and NF-kappaB signaling pathway components in human cervical cancer progression. Gynecol. Oncol. 104, 352–361 [DOI] [PubMed] [Google Scholar]
- Romashkova J.A., and Makarov S.S.1999. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature 401, 86–90 [DOI] [PubMed] [Google Scholar]
- Sartor M.A., Mahavisno V., Keshamouni V.G., et al. 2010. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics 26, 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scatena M., Almeida M., Chaisson M.L., et al. 1998. NF-kappaB mediates alphavbeta3 integrin-induced endothelial cell survival. J. Cell Biol. 141, 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuillemenot B.R., Kennedy D., Cooper J.D., et al. 2014. Nonclinical evaluation of CNS-administered TPP1 enzyme replacement in canine CLN2 neuronal ceroid lipofuscinosis. Mol. Genet. Metab. 114, 281–293 [DOI] [PubMed] [Google Scholar]
- Wei H.S., Wei H.L., Zhao F., et al. 2013. Glycosyltransferase GLT8D2 positively regulates ApoB100 protein expression in hepatocytes. Int. J. Mol. Sci. 14, 21435–21446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Feng X., and Stein L.2010. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 11, R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Mitra A., Dojer N., et al. 2013. A probabilistic approach to learn chromatin architecture and accurate inference of the NF-κB/RelA regulatory network using ChIP-Seq. Nucleic Acids Res. 41, 7240–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Xia Y., Cao Y., et al. 2012. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell. 48, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Liu X., Chen W., and Chen L.2013. IFIT5 potentiates anti-viral response through enhancing innate immune signaling pathways. Acta Biochim. Biophys. Sin. 45, 867–874 [DOI] [PubMed] [Google Scholar]
- Zhou B., Xu W., Herndon D., et al. 2010. Analysis of factorial time-course microarrays with application to a clinical study of burn injury. Proc. Natl. Acad. Sci. USA 107, 9923–9928 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.