Abstract
The connections existing between vessels and nerves go beyond the structural architecture of vascular and nervous systems to comprise cell fate determination. The analysis of functional/molecular links that interconnect endothelial and neural commitments requires a model in which the two differentiation programs take place at the same time in an artificial controllable environment. To this regard, this work presents an in vitro model to differentiate embryonic stem (ES) cells simultaneously into mature neurons and endothelial cells. Murine ES cells are differentiated within an artificial environment composed of PA6 stromal cells and a serum-free medium. Upon these basal culture conditions ES cells preferentially differentiate into neurons. The addition of basic fibroblast growth factor (FGF2) to the medium allows the simultaneous maturation of neurons and endothelial cells, whereas bone morphogenetic protein (BMP)4 drives endothelial differentiation to the disadvantage of neural commitment. The responsiveness of the system to exogenous cytokines was confirmed by genes expression analysis that revealed a significant up-regulation of endothelial genes in presence of FGF2 and a massive down-regulation of the neural markers in response to BMP4. Furthermore, the role played by single genes in determining endothelial and neural fate can be easily explored by knocking down the expression of the target gene with lentiviruses carrying the corresponding shRNA sequence. The possibility to address the neural and the endothelial fate separately or simultaneously by exogenous stimuli combined with an efficient gene silencing strategy make this model an optimal tool to identify environmental signals and genes pathways involved in both endothelial and neural specification.
Keywords: embryonic stem cells, endothelial differentiation, neural differentiation, stromal cells, shRNA, gene targeting
Introduction
In recent years, the similarities between vessels and nerves have become more apparent and extended to multiple biological aspects: from wiring, regulated by common guidance molecules, to parallelisms connecting neural and vascular cell fates up to endothelial cell-neuronal crosstalk in the vascular niche [1]. Thanks to guidance molecules able to affect both neural and vascular path-findings, nerves and vessels run aligned to each other and form neurovascular bundles in many locations of a developing organism [2]. The parallelism between vascular and nervous systems is also mirrored at the level of cell fate and commitment, where it is unveiled through the soluble and cell-associated signals present in the surrounding environment. In this respect, we previously identified an embryonic stem (ES) cell-derived population of uncommitted cells that retains the capacity to embrace endothelial or neural commitments depending on micro-environmental cues [3]. Neurovascular niches are specific microenvironments, present in the adult brain, in which the crosstalk occurring between neural stem cells and blood vessels guarantees stem cells self-renewal and survival [4]. These dynamic interplays, based on the mutual exchanges of molecules that affect both neural and vascular cell functions and known as angioneurins, seem to be involved in the progression of neurodegenerative diseases and of malignant gliomas [5, 6]. On these bases, considering the complexity of the neurovascular parallelism issue and the fragility of the in vivo models [7, 8], an in vitro model that resumes both vascular and nervous systems at the different stages of their maturation is auspicial, especially in view of therapeutic applications. To this purpose, ES cells represent the optimal strategy, since they are pluripotent cells able to spontaneously differentiate into the derivatives of the three germ layers [9]. Depending on the nature of the physical and biochemical elements present in the surrounding environment, ES cell commitment can be directed towards one specific cell type [10]. For instance, the ES-derived Flk1+ vascular precursors can give rise to endothelium both in chemically defined media [11, 12] and in co-cultures with OP9 stromal cells [13, 14]. Similarly, neurons can be differentiated from ES cells or in a chemically defined medium containing N2 and B27 supplements [15] or by using the PA6 stromal cells derived from skull bone marrow and known to efficiently sustain neural differentiation of ES cells [16]. Being serum components inhibitory of neural commitment, the omission of serum from the medium is an essential condition required to induce neural differentiation [16, 17]. On the contrary, serum is an important environmental element for endothelial commitment, always included in the differentiation culture medium in most of the used protocols. Thus, the neural and the endothelial commitments require apparently incompatible environments. Indeed, no protocols were so far reported that allow the simultaneous appearance of endothelium and neurons from differentiating stem cells. In this study we combine the stromal cells co-culture technique with the use of a chemically defined medium to obtain simultaneously endothelial and neural cells from differentiating ES cells. ES cells are induced to differentiate within an in vitro environment composed of a monolayer of PA6 cells in a serum-free medium. The addition of cytokines, such as fibroblast growth factor (FGF2) and epidermal growth factor (EGF), to the culture medium is enough to allow the maturation of endothelial cells without affecting neural commitment. The use of BMP4, a well-known neural differentiation inhibitor, causes the shift of the system towards the endothelial fate to the detriment of the neural one. The prevalence of the endothelial and/or neural commitment over other cell types potentiality is confirmed by gene expression analysis and immunofluorescence. Differently from the embryoid bodies model (EBs) [10], in which many cell types differentiate uncontrolled at the same time, this model represents a unique opportunity to study endothelial and neural differentiation separately or simultaneously, thus unveiling overlaps and parallelisms. Furthermore, the possibility to transduce the system with lentiviruses allows the execution of loss of function assays with the purpose to identify genes that can interfere with the endothelial and/or neural fate. The easy manipulation and modulation by the addition of exogenous recombinant molecules or gene silencing makes this system particularly suitable to study the molecular determinants of ES cells’ differentiation with particular regard to the endothelial and neural differentiation pathways and their possible points of contact.
Materials and methods
Cell lines and cultures
W4 mouse ES cells were kindly provided by Dr. A. Joyner [18]. R1 ES cells expressing the enhanced yellow fluorescent protein (YFP-ES) were obtained from Dr A. Nagy [19]. The knock in ES cells expressing the enhanced green fluorescent protein under the τ promoter (τ-EGFP) were gently provided by Dr. A.G. Smith [20] whereas stem cell leukemia (SCL) knockout and Flk1 null ES cells were obtained from Dr. C.G. Begley [21] and Dr. J. Rossant, respectively [22]. All ES cells lines were cultured as described [23]. Murine embryonic fibroblasts were purchased from American tissue culture collection (ATCC) (LGC Standards, Milano, Italy). PA6 stromal cells, kindly provided by Dr T. Schroeder, were cultured as described [24].
2D model of in vitro differentiation of ES cells
PA6 cells were grown in Lab-Tek II – CC2 chamber slides (Nunc, Naperville, IL, USA) or in tissue culture multiwell plates until confluence. ES cells were seeded onto PA6 monolayers at a density of 5000 cells/cm2 in N2B27 medium [20] (Invitrogen, Carlsbad, CA, USA), supplemented with 20 μg/ml insulin, 50 μg/ml bovine serum albumin and 200 μM ascorbic acid (Sigma, St. Louis, MO, USA). When required, 20 ng/ml of human recombinant FGF2, vascular endothelial growth factor (VEGF)-A165 and EGF or 5 ng/ml of murine BMP4 (R&D Systems, Minneapolis, MN, USA) were added at the onset of the culture and withdrawn after 3 days. Medium with fresh insulin was then replaced every 2 days.
Indirect immunofluorescence and image analysis
Differentiated ES cells were fixed with phosphate-buffered saline 4% paraformaldehyde (PFA) and incubated with primary antibodies for 1 hr at 37°C in a moistchamber. The following primary antibodies were used: anti-βIII-tubulin, MAP2, γ-aminobutyric acid (GABA), smooth muscle actin (SMA) and anti-Actinin (Sigma), anti-S100, GFAP and anti-Insulin (Dako, Glostrup, Denmark), anti-VE-cadherin (R&D Systems), anti-stage specific embryonic antigen (SSEA)-1,Tyrosine Hydroxylase and anti-Olig2 (Millipore, Billerica, MA, USA), anti-Flk-1, Pecam-1, intercellular adhesion molecule (ICAM)-2, CxcR4 and monoclonal anti-nestin (BD Biosciences, San Jose, CA, USA), anti-synaptic vesicle glycoprotein (SV)2 (Developmental Studies Hybridoma Bank, Iowa City, IA, USA), anti-Prominin-1 (eBiosciences, San Diego, CA, USA), anti-BLBP (brain lipidic binding protein), Nanog, Oct4 and polyclonal anti-nestin (Abcam, Cambridge, UK), anti-GFP (Invitrogen). Images were captured by using a Leica TCS SP2 AOBS confocal microscope, analysed with Leica Confocal Software (LCS; Leica Microsystems, Wetzlar, Germany) and elaborated with Imaris software (Bitplane AG, Zurich, Switzerland) to make a 3D reconstruction. To quantify VE-cadherin+ endothelial cells and βIII-tubulin+ neurons, sequential images, covering the entire surface of a chamber slide well (0.7 cm2), were acquired with a Leica AF6000LX workstation. The area occupied by endothelial cells or neurons was measured with ImageJ software. To identify proliferating cells, 10 μM of the thymidine analogue EdU was added to the culture medium for 1 hr and detected on fixed cells by using Click-iT EdU imaging kit (Invitrogen) by following the manufacturer’s instructions.
Cytofluorimetric absolute cell count and cell cycle analysis
Single cells suspension was obtained by incubating the cell culture with Accutase (PAA, Pashing, Austria) for 10 sec. at 37°C. For the absolute cell count, a sample of cells was mixed with an equal amount of Flow-count fluorospheres (Beckman Coulter, Brea, CA, USA) and analysed with a FACSCalibur flow cytometer (Beckman Coulter). Proliferation rate and DNA content were evaluated by using Click-iT EdU flow cytometry assay kit (Invitrogen) and propidium iodide (PI) staining. Briefly, after 1 hr incubation with 10 μM EdU, cell suspensions were prepared from ES-PA6 co-cultures and processed for EdU detection following the manufacturer’s instructions. Cell DNA was labelled by a 3 hrs treatment with 50 μg/ml PI and 100 μg/ml Ribonuclease A. Data were acquired with a CyAn ADP flow cytometer (Dako) and analysed with FlowJo software (Tree Star, Ashland, OR, USA).
Low-density arrays, real-time PCR and gene expression analysis
Total RNA was extracted with Trizol reagent (Invitrogen) and purified with DNA-free DNase I (Ambion, Austin, TX, USA). RNAs were quantified with RNA 6000 Nano Assay kit in an Agilent 2100 bioanalyser (Agilent Technologies, Santa Clara, CA, USA) and reverse transcribed using High-Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). cDNAs were then loaded on a TaqMan Custom Array (Applied Biosystems) by following the manufactory instructions and PCR amplification was carried out in a 7900 real-time PCR system (Applied Biosystems). The expression of Flk1, Ace, Tal1, T and VE-cadherin genes, along with the house-keeping gene Tbp, were analysed by real-time PCR using TaqMan probes (Applied Biosystems). Expression data were analysed with SDS 2.3 software.
Lentiviral-mediated shRNA knockdown
Five different shRNA MISSION RNA interference vectors were obtained for each target gene from Sigma (listed in Table S1). Lentiviral particles were produced by transfecting 293T cells with a second generation packaging system as previously described [25, 26]. Viral titre was calculated by infecting HeLa cells with increasing volumes of the virus preparations and by counting viable cells after puromycin selection. The optimal multiplicity of infection (MOI) for ES cells was established at 20 MOI/cell as result of a dose-curve experiment performed by infecting cells with increasing amounts of GFP-pLKO lentivirus that revealed 90% of GFP+ cells. ES cells were infected with lentiviral shRNAs at the onset of culture and afterward positively selected with 2.5 μg/ml puromycin. To measure silencing efficiency, the expression of the target genes was analysed by real-time PCR 7 days after infection and compared to the expression levels measured in cells transduced with lentiviruses carrying MISSION non-target shRNA control vector.
Results
FGF2 and EGF drive endothelial differentiation of ES cells in a neural suited culture system
To better analyse the plastic correlation existing between vascular and neural commitments [3, 27], an in vitro model of endothelial and neural cells type simultaneous specifications was obtained by using ES cells. To compromise between the two opposite tissue culture requirements necessary for obtaining endothelium and neurons from ES cells, i.e. presence and absence of serum, respectively [28, 29], we set up an in vitro serum-free culture system of ES cells’ differentiation in which ectoderm induction is sustained by the neural fate-promoting activity of the stromal PA6 cells [16], whereas the lack of mesoderm determination, due to the absence of serum, is overcome by the addition of soluble cytokines to the medium. When ES cells were seeded at low density onto a PA6 monolayer in N2B27 medium [15] in the absence of serum and of any other growth factor (–GFs), they mainly differentiated into neurons which were detectable by an anti-βIII-tubulin antibody in immunofluorescence assay (–GFs, Fig. 1A). Conversely, when FGF2 and EGF were added to the medium at the onset of the culture (+GFs), many VE-cadherin+ endothelial cells developed along with neurons (+GFs, Fig. 1A). As demonstrated by the 3D reconstruction (Fig. 1B), the endothelial branches intermingled with neurons and made points of contact with crossing axons, thus making this system useful for studying vascular/nervous relationships (Fig. 1C, arrowheads). The efficiency of endothelial and neural differentiation was quantified by measuring the areas covered by VE-cadherin+ and βIII-tubulin+ cells, respectively, in both experimental conditions. The measurements, normalized over the +GFs condition, clearly confirmed that endothelial cells differentiated only upon GFs stimulation, whereas neurons developed in both conditions (Fig. 2A). The fact that, in the presence of GFs, many neurons developed shorter axons than upon basal conditions (Fig. 1A, +GFs versus–GFs) might account for the difference between the +GFs and –GFs measurements of areas. Indeed, the quantification by FACS of the neurons developed in the presence or in the absence of the cytokines confirmed that neural differentiation was not diminished but rather supported by GFs (Fig. S1). Nevertheless, the area measurement method was preferred to flow cytometry because of the poor performance of the anti-VE-cadherin antibodies tested for FACS analysis. Since FGF2 and EGF are potent mitogens, we wondered if the increase of endothelial cells number was merely due to a proliferative effect and not to a differentiative stimulus. Therefore, we analysed the cell growth rate in response to GFs, at different time-points, by flow cytometry analysis. ES-derived cells can be distinguished from PA6 cells because their different physical parameters and therefore they can be counted separately (Fig. S2A). As shown in Fig. 2B, ES-derived cells increased in number during time, both in the absence and in the presence of GFs. However, the doubling of cells registered at day 7 in the presence of growth factors cannot account for the 10-fold increase of VE-cadherin+ endothelial cells quantified after GFs stimulation (Fig. 2A), thus suggesting that FGF2 and EGF played an active role in the endothelial differentiation process. After an initial doubling consequent to GFs addition, the number of PA6 cells did not change during time, both in the absence and in the presence of GFs (Fig. S2B). Since the persistence of a small percentage of undifferentiated ES cells occurs in 2D neural differentiation models [15], we decided to test the presence of undifferentiated cells in our culture system by looking at the expression of the stemness markers Nanog and Otc4. An ES cell line that constitutively expresses YFP-ES was used to distinguish ES-derived cells from PA6 cells. After 6 days of differentiation, clusters of Nanog+ and Oct4+ cells were detected mainly in the presence of GFs (Fig. 2C), thus suggesting that FGF2 and EGF preserved the expression of the transcription factors that are required to sustain the stemness status in undifferentiated ES cells (Fig. S3). In line with the proliferative effect elicited by GFs on the total cell population (Fig. 2B), many cells, including the Oct4+ undifferentiated ES cells and the nestin+ neural precursors, incorporated into the DNA the thymidine analogue EdU (Fig. 2D, see enlargements). As expected, the mature endothelium and neurons did not incorporate EdU, having already accomplished their differentiation program. These data suggest that, in the ES-PA6 co-culture system, besides supporting the differentiation of both neural and endothelial cells, GFs also contribute to maintain a pluripotent stem cells compartment by preserving the expression of Oct4 and Nanog and by supporting ES cells self-renewal.
Fig 1.
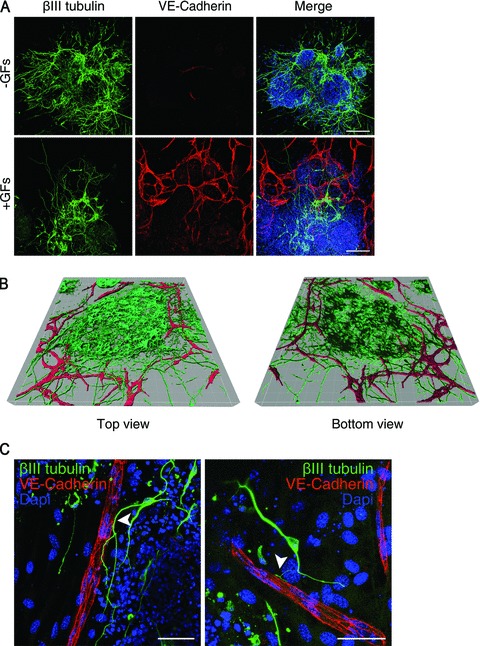
Differentiation of mouse ES cells into neurons and endothelial cells. (A) Immunofluorescent stainings of ES-PA6 cells co-cultures performed with anti-βIII-tubulin and anti-VE-cadherin antibodies at day 7. Upon basic culture conditions (–GFs), ES cells mainly differentiate into neurons, whereas, in presence of FGF2 and EGF (+GFs), mature endothelial cells arise and form vessel-like structures. Scale bars: 150 μm. (B) Sequential images of ES-derived βIII-tubulin+ neurons surrounded by VE-cadherin+ ramified vessels-like structures were captured by confocal microscope and elaborated into a 3D reconstruction by Imaris software. (C) High magnification images of βIII-tubulin+ neurons that run along and make contacts with VE-cadherin+ endothelium (arrowheads). Scale bars: 50 μm.
Fig 2.
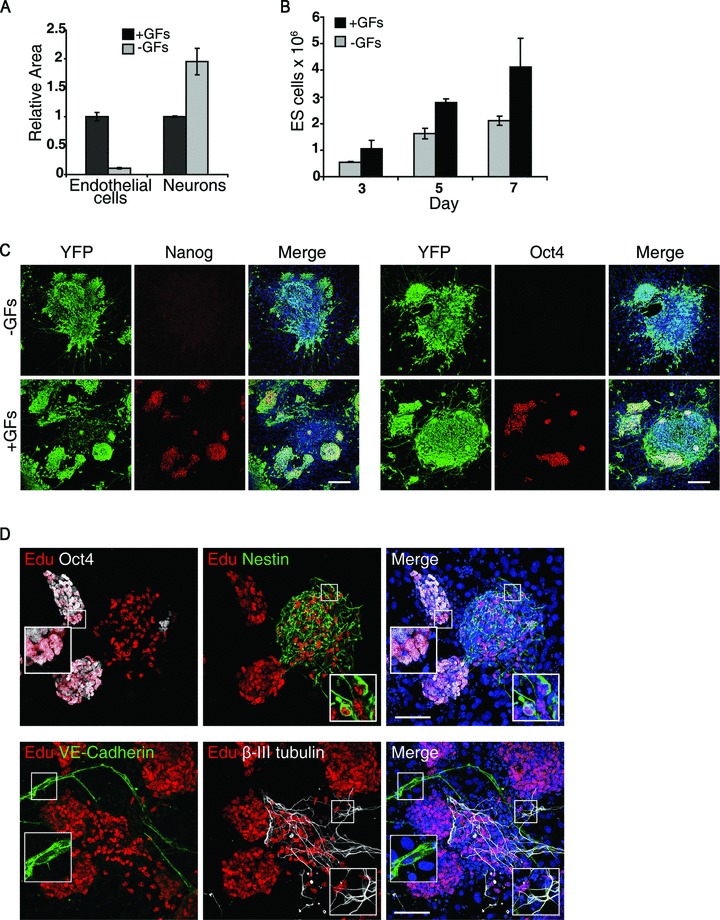
Characterization of ES/PA6 co-culture system: analysis of neural, endothelial, stemness components and of GFs-induced cell proliferation. (A) Neural and endothelial components were quantified by measuring the area occupied by βIII-tubulin+ neurons and VE-cadherin+ cells in the presence (+GFs) or in the absence (–GFs) of growth factors. The resulting data were normalized to the condition +GFs. (B) Analysis of cell growth rate. Absolute counting of the ES cells present in the co-cultures was performed by flow cytofluorometry at different time-points both in the presence and in the absence of growth factors. (C) ES cells that constitutively express YFP (YFP-ES) were co-cultured with PA6 cells in the presence or in the absence of GFs. The expression of the stemness markers Nanog and Oct4 was evaluated by immunofluorescence at day 7 of differentiation. ES-derived cells are distinguishable from feeders because their YFP expression. Scale bars: 150 μm. (D) Characterization of ES-derived proliferating cells after 7 days of culture in the presence of GFs. The dividing cells that have incorporated the thymidine analogue EdU into the DNA are visualized in red. Enlargements show that undifferentiated ES cells (Oct4+) and neural precursors (nestin+) are dividing whereas VE-cadherin+ and βIII-tubulin+ cells are not. Scale bars: 75 μm.
Characterization of the ES-derived immature and mature endothelial cells and neurons
To better characterize the phenotype of the differentiated endothelial and neural cells, immunofluorescent stainings were performed to detect endothelial or neural markers 7 days after having co-cultured YFP-ES cells with PA6 cells in the presence of GFs. Other than VE-cadherin, ES-derived endothelial cells also expressed Flk-1, ICAM-2 and CD31, though the latter is also expressed by undifferentiated ES cells [30] (Fig. 3A).
Fig 3.
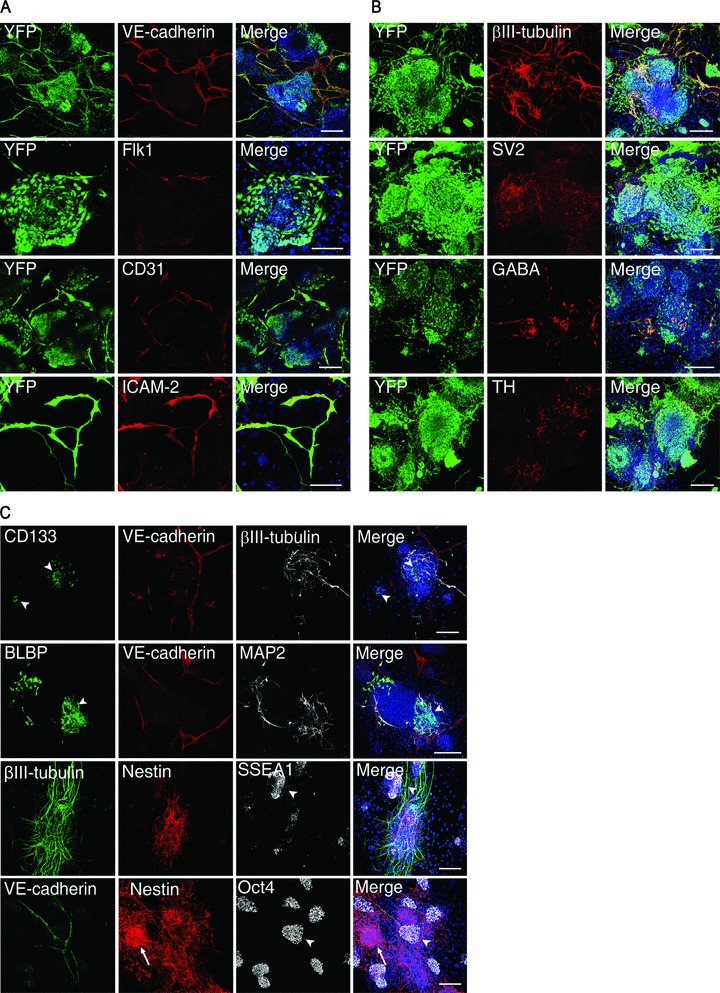
Characterization of ES-derived endothelial cells and neurons. After 7 days of co-culture with PA6 cells in the presence of GFs, YFP-ES cells were analysed by immunofluorescence and confocal microscopy to detect the expression of (A) the endothelial markers VE-cadherin, Flk1, CD31 and ICAM-2, (B) the neural markers βIII-tubulin, SV2 (synaptic vesicles antigen), GABA and tyrosine hydroxylase. (C) Co-immunostainings performed with anti-VE-cadherin and anti-βIII-tubulin or anti-MAP2 antibodies in association with anti-CD133, anti-BLBP, anti-nestin, anti-SSEA-1 and anti-Oct4 antibodies unveil the presence of uncommitted precursors and undifferentiated ES cells within the system. All the scale bars are 150 μm except for Flk1 and ICAM-2 images that are 100 μm.
The degree of neural maturation achieved by the differentiating neural cells was revealed by the expression of the functional synaptic protein SV2, GABA and tyrosine hydroxylase (Fig. 3B), thus showing the capacity of this system to give rise to both gabaergic and dopaminergic neurons, respectively.
The persistence of undifferentiated ES cells within the system questioned about the presence of precursors and their spatial relationship with the correspondent mature progenies. Indeed, clusters of endothelial-committed cells expressing CD133 [31] and of neural precursors identified by the radial glia markers nestin and BLBP [32–35] were detected within the colonies along with undifferentiated SSEA-1+ and Oct4+ ES cells, thus accounting for the cells that were not expressing endothelial or neuronal markers (Fig. 3C). These observations indicate that in our system mature neurons and endothelium coexist with their correspondent precursors, thus creating a unique opportunity to study the different phases of the corresponding differentiation programs at the same time.
Effect of distinct growth factors and cytokines on endothelial or neural differentiation
The addition of FGF2 and EGF to the N2B27 serum-free medium is crucial for the induction of endothelial differentiation of ES cells co-cultured with PA6 cells (Fig. 1A). To better analyse the role played by each of these two cytokines and to explore the effect of other possible inducers, FGF2, EGF, VEGF and BMP4 were separately added to the medium at the onset of the culture. After 7 days, ES-derived endothelial cells and neurons were visualized by immunofluorescence with an anti-VE-cadherin and anti-βIII-tubulin antibodies (Fig. 4A) and quantified by measuring the corresponding immunostained areas (Fig. 4B). Among the cytokines tested, FGF2 and BMP4 efficiently induced endothelial differentiation while EGF and VEGF exerted only a mild action. With the exception of the neural inhibitor BMP4 [15], none of the cytokines compromised neural differentiation, being the slight decrease of the relative area measured in presence of FGF2 imputable to axons extensions and not to the number of differentiated neurons, as discussed above. To measure the proliferation activity of the cells in response to single cytokines stimulation, the incorporation of the thymidine analogue EdU, combined with the cell-cycle analysis, was evaluated by FACS. As shown in the dot plots of Figure 5, FGF2 and EGF significantly increased the number of proliferating cells compared to the basal conditions (–GFs). Specifically, FGF2 and EGF, either administered singularly or combined, increased the number of cells that, having entered the S and G2 cell cycle phases, were replicating their genome. Conversely, VEGF and BMP4 did not significantly affect cell proliferation rate.
Fig 4.
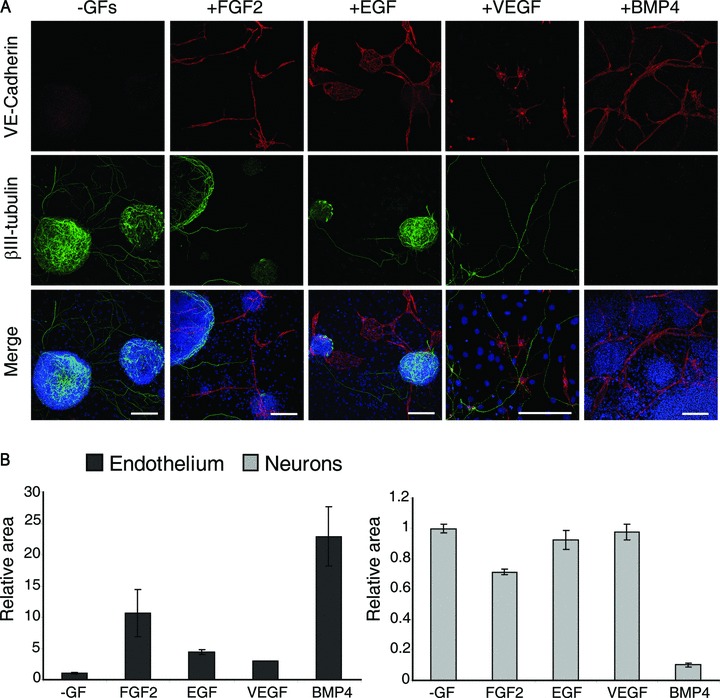
Endothelial and neural differentiation upon stimulation with single cytokines. (A) Endothelial cells and neurons derived from ES cells in basal conditions (–GFs) or in the presence of FGF2, EGF, VEGF or BMP4 were detected by immunofluorescence with anti-VE-cadherin and anti-βIII-tubulin antibodies and quantified by measuring the immunostained areas (B). Both FGF2 and BMP4 induce endothelial commitment but only BMP4 inhibits neurons maturation. Scale bars: 150 μm except for EGF and VEGF that are 200 and 100 μm, respectively.
Fig 5.
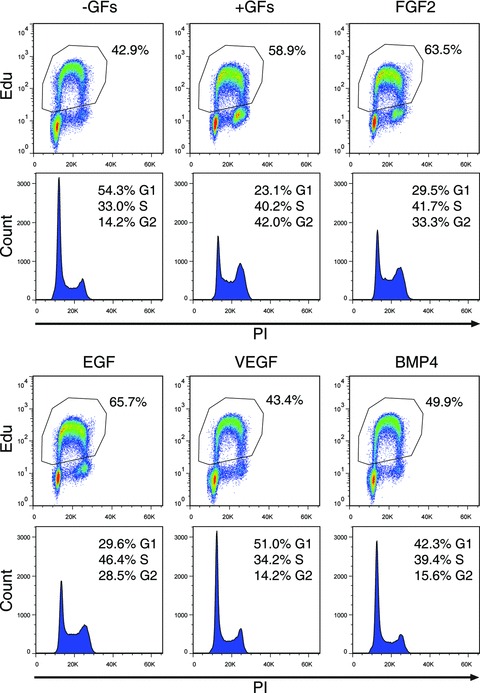
Analysis of cell proliferation rate in response to single cytokines treatment. Cell proliferation was studied by incubating cells with the thymidine analogue EdU in the presence of single cytokines. The incorporation of the tracer into the DNA was detected thank to a fluorescent ligand and measured by flow cytometry. DNA content was evaluated by PI staining. For each treatment, the percentage of the EdU+ proliferating cells is shown in the dot plot (upper panel), whereas the histogram underneath shows the different phases of cell cycle. Percentages of the cells that enter G1, S and G2 stages are indicated.
ES cells co-cultured with PA6 cells: analysis of gene expression in absence or presence of FGF2 and BMP4
To study the progression from ES cells to neural and endothelial cells at the mRNA level, a gene expression analysis was performed by using TaqMan low-density arrays. Besides genes representative of endothelial and neuronal phenotypes, markers representative of different other cell types were included such as glia, stem cells, muscle, epithelial, pancreatic cells and markers for germ layers specification as well (see Table S2 for a complete list). Among 83 genes analysed in two independent experiments, those found differentially expressed with a fold change ≥2 were considered significantly modulated. At first, the progression of the ES cells differentiation was analysed during time in the absence of growth factors by comparing the gene expression levels obtained after 6 days of culture versus those measured at day 3 of differentiation. This analysis revealed that most of the neural genes were up-regulated, whereas the endothelial genes were down-regulated (Fig. 6A, B). Genes representative of fibroblasts (Col1a2 and Fap), haematopoietic (Ly6a/Sca1) and cardiac cells (Myl2), were down-regulated, thus confirming the predominance of the neural commitment over the endothelial and any other potential of differentiation (Fig. 6C). As expected, the expression of the stem cells markers Oct4 (Pouf1) and Rex1 (Zfp42), along with the epiblast marker Fgf5, significantly decreased as the differentiation progressed towards mature neurons. On the contrary, genes representative of mesoderm and endoderm (Gsc and T, Foxa2 and Cxcr4, respectively) were up-regulated, suggesting that the growth factor-serum free medium N2B27 is not selective to non-neuronal cell types but, instead, it preserves other cell types potentiality without allowing their final accomplishment, as long as no signals are provided by the environment [15].
Fig 6.
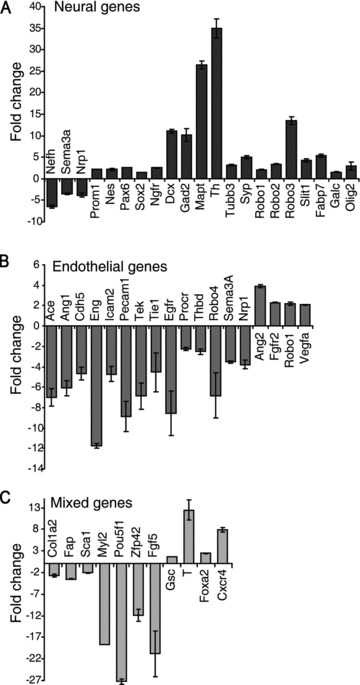
Analysis of genes expression upon basal conditions. Co-cultures of ES and PA6 cells were grown for 3 and 6 days in the absence of growth factors. Total RNAs were purified and used to analyse genes expression by TaqMan low-density arrays. Data are normalized against the gene expression levels measured at day 3 of differentiation. (A) With the exception of NefH, Nrp1 and Sema3A, all neural genes analysed are activated. (B) All endothelial markers are down-regulated, except Ang2, FGF-R2, Robo1 and VegfA. (C) Markers of fibroblasts (Col1a2 and Fap), haematopoietic cells (Sca1), cardiac muscle cells (Myl2), undifferentiated stem cells (Oct4 and Rex1) and epiblast cells (Fgf5) are down-regulated whereas the mesoderm (Gsc and T) and endoderm (Foxa2 and Cxcr4) genes are up regulated.
We next induced endothelial differentiation with FGF2 and, after 6 days of culture, we analysed gene expression levels in stimulated versus not stimulated cells. Upon FGF2 stimulation, most of the endothelial genes examined were up-regulated, indicating the progression of the differentiation program towards the endothelial fate. Importantly, none of the neural genes was down-regulated, but rather the expression of a significant number of neural markers increased (Fig. 7A), thus confirming that FGF2 did not compromise neural commitment. The observed contribution of GFs to the maintenance of a stemness component (Fig. 2C) was confirmed at gene expression level by the up-regulation of Pou5f1 and Zfp42 mRNAs (Fig. 7B). Despite the up-regulation of all three germ layers transcripts, no genes representative of mature cell types, other than endothelial or neural cells, were modulated by FGF2, except Myog, Acta1 and Neurog3 (Fig. S4). Nevertheless, no mature corresponding actinin+ skeletal muscle and insulin+ pancreatic cells were ever detected by immunofluorescence analysis (Fig. S5). On the contrary, few GFAP+ and SMA+ cells were instead visualized, thus revealing the possibility to differentiate also the accessory cells of the neural and vascular systems, i.e. glia and smooth muscle cells. Altogether these data confirm that, in the ES/PA6 co-culture system, the addition of FGF2 allows the simultaneous maturation of ES-derived endothelium and neurons by excluding any other cell type that does not belong to the vascular or nervous system.
Fig 7.
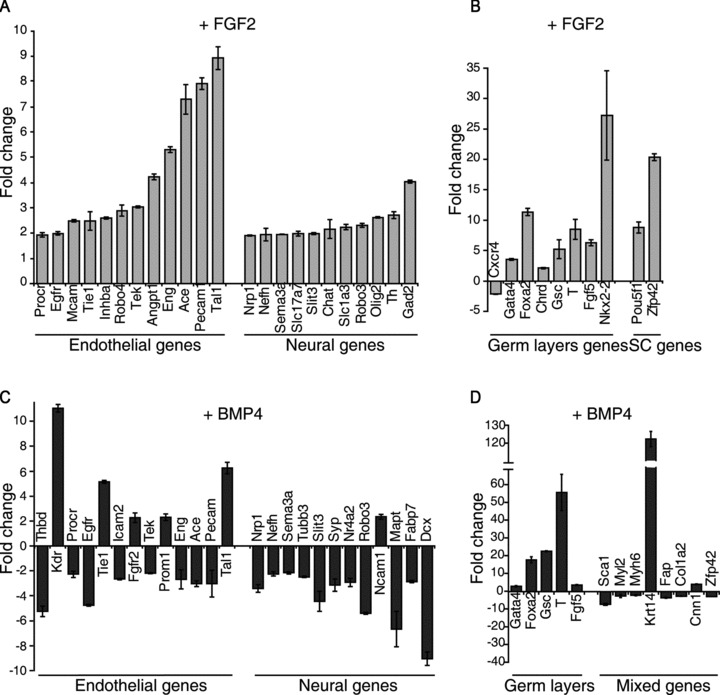
Analysis of gene expression upon FGF2 or BMP4 stimulation. Total RNAs were purified from ES-PA6 cells co-cultures grown in presence of FGF2 (A and B) or BMP4 (C and D) and used to analyse genes expression by TaqMan low-density arrays. Data are normalized against –GFs culture condition. (A) FGF2 activates the expression of most of the endothelial genes and even reinforced the expression of some neural genes. (B) The three germ layers and stem cells markers are up-regulated. (C) In the presence of BMP4, early endothelial genes (Flk1, Tal1, Prom1, Tie1 and Fgfr2) are up-regulated, the mature endothelial markers are un-affected or down-regulated, whereas all the neural genes are down-regulated. (D) Among the germ layers genes, the mesoderm markers T and Gsc are the most affected by BMP4 treatment. Genes representing haematopoietic (Sca1), cardiac muscles (Myl2 and Myh6), fibroblasts (Fap and Col1a2) and stem cells (Zfp42) are suppressed whereas the pericytes marker Cnn1 is slightly up-regulated. According to what previously observed [16], BMP4 induces the activation of K14 transcript.
Thank to its properties, BMP4 was used as molecular signal to deprive the system of neurons without precluding endothelial commitment (Fig. 4). Indeed, upon BMP4 treatment, most of the neural markers were down-regulated, the early endothelial markers Flk1, Tie1, Prominin-1 and Tal1 showed a significant up-regulation, while the late endothelial markers did not (Fig. 7C). These data, along with the increased expression of the mesoderm markers brachyury (T) and goosecoid (Gsc) (Fig. 7D), suggest that, compared to FGF2, BMP4 plays an earlier role on endothelial fate determination, by acting at the level of mesoderm and haemangioblasts induction. In agreement with the observed role of BMP4 in preserving the epidermal potentiality of ES-derived ectoderm precursors [16], the early epidermal marker keratin 14 was strongly up-regulated. Since ES cells were differentiated in a co-culture system with feeder cells, we wondered if the PA6-mRNA component could bias the output of the gene expression analysis. Therefore, we analysed the levels of gene expression of pure PA6 cells cultures stimulated or not with FGF2 (Fig. S6). Out of 83 genes analysed, 45 were not expressed at all, while only 9 were slightly modulated by FGF2. Given the fact that, within the ES/PA6 co-cultures the feeder component is only one tenth of the total cells as quantified by FACS analysis, the possible interference by PA6-mRNAs was hence considered not relevant.
Effect on endothelial or neural differentiation after lentiviral mediated gene knockdown
The massive gene up-regulation observed upon FGF2 stimulation leads to the question about the possible active role played by some of these genes in the differentiation process. Therefore, the role played by Ace (angiotensin converting enzyme), brachyury (T), Flk1 and Tal1 genes in the ES cells differentiation program was analysed by knocking-down their expression with lentiviral vectors carrying the correspondent shRNAs. Being aware of possible off-target effects, five different shRNA sequences were assayed for each gene. Gene knockdown efficiency was evaluated by real-time PCR after 7 days of culturing ES cells in the presence of GFs. Down-regulation of the target gene was expressed as fold change of its mRNA level over the one obtained with control cells transduced with lentiviral particles carrying a non-targeting shRNA (NTshRNA) (Fig. 8A). For each gene, the effects exerted on ES-derived endothelial cells and neurons by the two most efficient shRNAs were evaluated by immunofluorescence staining with anti-VE-cadherin and anti-βIII-tubulin antibodies. Compared to the well-ramified endothelial network developed by ES cells transduced with NTshRNA, silencing of Flk1, Ace and Tal1 generated only few aberrant VE-cadherin+ branches or scattered endothelial cells, whereas an almost absolute absence of endothelium was caused by suppression of brachyury expression (Fig. 8B). In such conditions, the quantification of VE-cadherin mRNA by real-time PCR confirmed that VE-cadherin expression was indeed strongly down-regulated (Fig. 8C). The quantification of the areas covered by neurons and endothelial cells revealed that silencing the expression of Flk1 and Tal1 impaired only endothelial differentiation whereas knocking down Ace and brachyury also affected the maturation of βIII-tubulin+ neurons. As proof of concept of the shRNA approach stands the fact that the endothelial defects observed in Flk1 and Tal1-shRNA-transduced cells perfectly reproduce the phenotype obtained with Flk1−/– and Tal1−/– ES cells differentiated upon the same culture conditions (Fig. S7).
Fig 8.
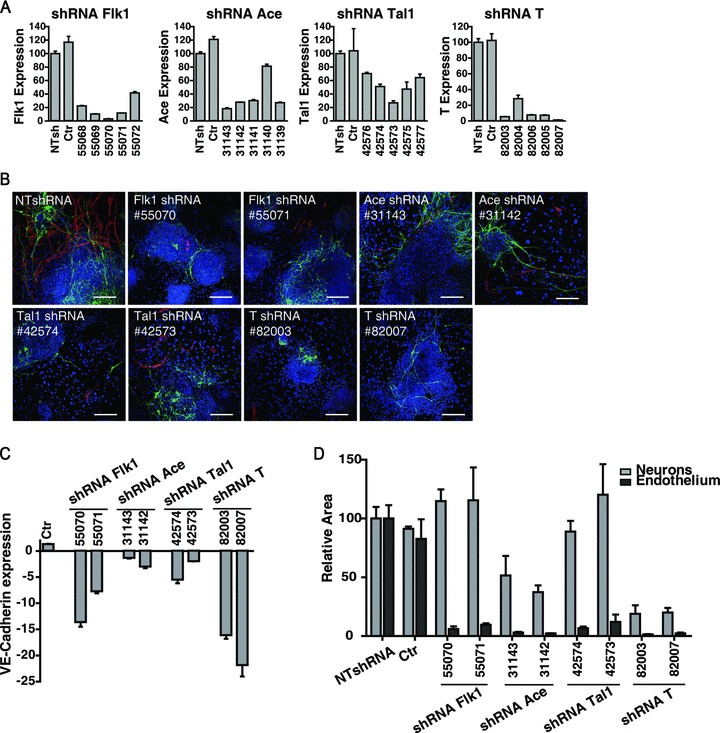
Silencing gene expression in ES/PA6 co-cultures: effects on endothelial and neural differentiation. ES cells were induced to differentiate into neurons and endothelium in the presence of GFs. The expression of Flk1, Ace, Tal1 and T genes was silenced by using lentiviral vectors carrying the corresponding shRNAs. (A) For each target gene, five different shRNA sequences were assayed and the efficiency of gene silencing was measured by real-time PCR. Down regulation of each target gene was expressed as relative expression of its mRNA level over cells transduced with lentiviral particles carrying a NTshRNA. Not infected cells were also considered (Ctr). Each shRNA sequence is indicated by a number code (Table S1). (B) Endothelial cells and neurons obtained from ES cells silenced with the two most effective shRNAs of each target gene, are detected by immunofluorescence with anti-VE-cadherin and anti-βIII-tubulin antibodies. (C) The shRNA-mediated inhibition of endothelial differentiation was quantified by measuring the levels of VE-cadherin mRNA by real-time PCR. Down-regulation of VE-cadherin gene is expressed as fold change of its mRNA level over cells transduced with NTshRNA. (D) The effect exerted on endothelial and neural commitment by the two most effective shRNAs of each target gene was quantified by measuring the area occupied by VE-cadherin+ cells and βIII-tubulin+ neurons, respectively. The data are normalized to the measurements made on cells silenced with NTshRNA.
Discussion
In this study we set up an in vitro model of ES cells differentiation that allows the simultaneous maturation of endothelial cells and neurons. ES cells are induced to differentiate within an artificial system reconstructed by combining two elements, both known for their neural promoting activity: the PA6 stromal cells and the chemically defined serum-free N2B27 medium [16, 20]. As expected, upon these culture conditions, neural fate overcomes any other potential. To induce endothelial commitment without disturbing neural predetermination, a strategy alternative to the use of serum is mandatory, being serum a well-known neural differentiation inhibitor [17]. The addition of recombinant FGF2 and EGF to the medium best accomplishes the purpose of triggering endothelial fate without precluding neural maturation, thus leading to the differentiation of both endothelium and neurons at the same time. Importantly, the lack of other mature cell types (Figs S4 and S5) suggests the confinement of the ES cells commitment to neural and endothelial fates. Besides being always included in the growth factors cocktail used to sustain endothelial commitment in EBs [36–38], FGF2 is also required to efficiently induce haemangioblasts from mesoderm precursors [39]. In accordance with these data, we observed that, other than a well-documented proliferative effect (Figs 2B and 5), the addition of FGF2 to the ES/PA6 co-culture medium has its major impact on the ES cells differentiative potential, as indeed confirmed by the massive activation of those endothelial markers that, in basal condition, are instead suppressed (Figs 7A and 6B). The challenging task to identify the mechanism by which FGF2 induces endothelial differentiation in our system leads to two possible paths, one PA6-dependent and the second that would directly target ES cells bypassing the stromal feeder layer. A possible active involvement of PA6 cells, suggested by the change of morphology that these stromal cells undergo when exposed to FGF2 (personal observation), may work through an FGF2-dependent production and secretion of inductive signals, likely a balanced cocktail of molecules both soluble and matrix bound, considering that recombinant VEGF and EGF singularly added to the culture medium are not effective. Alternatively, FGF2 might directly intervene on the induction of endothelial precursors from ES cells, being FGF2 a well-known mesoderm inducer [40]. Upon growth factors stimulation, the persistence of small clusters of undifferentiated and intermediately differentiated cells, identified by SSEA-1 and Oct4 or CD133, nestin and BLBP, respectively (Fig. 3C), suggests that GFs contribute to maintain a reservoir of uncommitted precursors, thus making asynchronous the onset of mature neurons and endothelium. This intriguing characteristic, also observed in the N2B27 monoculture system [15], allows the study of the different phases of neural and endothelial differentiation altogether, being all represented within the same temporal window. The possibility to develop simultaneously endothelium and neurons comprehensive of their precursors within the same in vitro culture system represents a precious tool to approach neurovascular parallelism. Interactions occurring between endothelial cells and neurons can be visualized and analysed (Fig. 1C) and the possible relationships connecting neural precursors, supporting stroma and endothelium can be unveiled thank to the malleability of this artificial niche. The investigative potentiality of this model is indeed enriched by the possibility to efficiently transduce ES cells with lentiviral vectors at the culture onset. Thus, the lentiviral approach represents an efficient strategy to identify genes and molecular pathways involved in endothelial and neural fate determination.
One of the major features of the ES/PA6 co-culture system consists in the possibility to differentiate only neurons or neurons plus endothelium by simply choosing to include or not FGF2 to the serum-free N2B27 medium. Taking advantage of the double role played by BMP4 as mesoderm inducer [41, 42] and neural differentiation inhibitor [15, 16], the addition of recombinant BMP4 to the medium represents the successful strategy to give rise to endothelial cells only. The observation that BMP4 up-regulates the mesoderm markers brachyury, goosecoid and Flk1without affecting the expression of late endothelial markers suggests that BMP4 action is mainly focused onto mesoderm induction (Fig. 7C and D). This is in agreement with what observed in the EBs model, where the efficient maturation of haemangioblasts is arrested to the formation of mesoderm precursors unless FGF2 is sequentially added to the medium after BMP4 exposure [39]. Nevertheless, unlikely the EBs model, our co-culture system can count on the possible paracrine support from PA6 cells, a contribution that could be significant enough to allow the formation of vessels-like structures upon BMP4 treatment, without inducing the massive endothelial gene activation obtained in response to recombinant FGF2 (Fig. 7A).
Conclusion
In the ES/PA6 co-culture system neural differentiation spontaneously occurs in the absence of growth factors, exogenous BMP4 triggers the maturation of endothelium, whereas FGF2 drives both commitments, neural and endothelial, at the same time. The simultaneous progression of the endothelial and neural differentiation programs represents a unique opportunity to address the vascular/nervous relationships at the level of cell fate and commitment. Being an artificial system composed by a limited number of components, the role played by the environment on cell fate determination is easily addressable by the controlled manipulation of each of its elements. To this regard, the contribution given by PA6 cells to ES cells potentiality, either in response to exogenous stimuli or to gene expression modulation, is an important topic worth to be addressed.
Acknowledgments
We thank Dr. A. Joyner, A. Nagy, A. G. Smith, C. G. Begley and Dr. J. Rossant for providing us with 129/SvEv W4, R1-YFP, τ-EGFP knock in, SCL and Flk1 knockout ES cell lines, respectively, and Dr. T. Schroeder for supplying us with PA6 feeder cells. We also thank Dr. G. Serini and L. Primo for helpful suggestions and comments. This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC); Regione Piemonte Ricerca Sanitaria Finalizzata 2006, 2007, 2008; Ricerca Industriale e Sviluppo precompetitivo 2006; grants PRESTO, Piattaforme Tecnologiche per le Biotecnologie – Grant Druidi (to F.B.); Ministero della Salute Programma di Ricerca Finalizzata 2006 and Programma Straordinario di Ricerca Oncologica 2006.
Conflicts of interest
The authors confirm that there are no conflicts of interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1 Quantification by FACS analysis of the neurons differentiated within ES-PA6 co-cultures.
Fig. S2 Cytofluorimetric absolute cells count of ES-derived cells and PA6 cells.
Fig. S3 biochemical detection and quantification of Oct4 protein in ES-PA6 co-cultures.
Fig. S4 analysis of gene expression upon FGF2 or BMP4 stimulation.
Fig. S5 characterization by immunofluorescence analysis of ES-PA6 cocultures for the presence of glia, endoderm, pancreatic and smooth muscle cells.
Fig. S6 Gene expression analysis in pure PA6 cell cultures stimulated with FGF2.
Fig. S7 2D in vitro differentiation ofFlk1-/- and Tal1-/- ES cell lines.
Table S1 List of shRNA MISSION clones used to stably knock-down the expression of Flk1, Ace, Tal1 and T.
Table S2 list of the genes used to customize the Taqman Low Density Array.
References
- 1.Segura I, De Smet F, Hohensinner PJ, et al. The neurovascular link in health and disease: an update. Trends Mol Med. 2009;15:439–51. doi: 10.1016/j.molmed.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Larrivee B, Freitas C, Suchting S, et al. Guidance of vascular development: lessons from the nervous system. Circ Res. 2009;104:428–41. doi: 10.1161/CIRCRESAHA.108.188144. [DOI] [PubMed] [Google Scholar]
- 3.Gualandris A, Noghero A, Geuna M, et al. Microenvironment drives the endothelial or neural fate of differentiating embryonic stem cells coexpressing neuropilin-1 and Flk-1. FASEB J. 2009;23:68–78. doi: 10.1096/fj.08-112847. [DOI] [PubMed] [Google Scholar]
- 4.Riquelme PA, Drapeau E, Doetsch F. Brain micro-ecologies: neural stem cell niches in the adult mammalian brain. Philos Trans R Soc Lond B Biol Sci. 2008;363:123–37. doi: 10.1098/rstb.2006.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barami K. Relationship of neural stem cells with their vascular niche: implications in the malignant progression of gliomas. J Clin Neurosci. 2008;15:1193–7. doi: 10.1016/j.jocn.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Zacchigna S, Lambrechts D, Carmeliet P. Neurovascular signalling defects in neurodegeneration. Nat Rev Neurosci. 2008;9:169–81. doi: 10.1038/nrn2336. [DOI] [PubMed] [Google Scholar]
- 7.Mukouyama YS, Shin D, Britsch S, et al. Sensory nerves determine the pattern of arterial differentiation and blood vessel branching in the skin. Cell. 2002;109:693–705. doi: 10.1016/s0092-8674(02)00757-2. [DOI] [PubMed] [Google Scholar]
- 8.Nagy N, Mwizerwa O, Yaniv K, et al. Endothelial cells promote migration and proliferation of enteric neural crest cells via beta1 integrin signaling. Dev Biol. 2009;330:263–72. doi: 10.1016/j.ydbio.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang NS, Varghese S, Elisseeff J. Controlled differentiation of stem cells. Adv Drug Deliv Rev. 2008;60:199–214. doi: 10.1016/j.addr.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noghero A, Bussolino F, Gualandris A. Role of the microenvironment in the specification of endothelial progenitors derived from embryonic stem cells. Microvasc Res. 79:178–83. doi: 10.1016/j.mvr.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita J, Itoh H, Hirashima M, et al. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–6. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- 12.Yurugi-Kobayashi T, Itoh H, Schroeder T, et al. Adrenomedullin/cyclic AMP pathway induces Notch activation and differentiation of arterial endothelial cells from vascular progenitors. Arterioscler Thromb Vasc Biol. 2006;26:1977–84. doi: 10.1161/01.ATV.0000234978.10658.41. [DOI] [PubMed] [Google Scholar]
- 13.Hirashima M, Kataoka H, Nishikawa S, et al. Maturation of embryonic stem cells into endothelial cells in an in vitro model of vasculogenesis. Blood. 1999;93:1253–63. [PubMed] [Google Scholar]
- 14.Yamashita JK, Takano M, Hiraoka-Kanie M, et al. Prospective identification of cardiac progenitors by a novel single cell-based cardiomyocyte induction. FASEB J. 2005;19:1534–6. doi: 10.1096/fj.04-3540fje. [DOI] [PubMed] [Google Scholar]
- 15.Ying QL, Stavridis M, Griffiths D, et al. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–6. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H, Mizuseki K, Nishikawa S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 17.Darmon M, Stallcup WB, Pittman QJ. Induction of neural differentiation by serum deprivation in cultures of the embryonal carcinoma cell line 1003. Exp Cell Res. 1982;138:73–8. doi: 10.1016/0014-4827(82)90092-1. [DOI] [PubMed] [Google Scholar]
- 18.Auerbach W, Dunmore JH, Fairchild-Huntress V, et al. Establishment and chimera analysis of 129/SvEv- and C57BL/6-derived mouse embryonic stem cell lines. Biotechniques. 2000;29:1024–8. doi: 10.2144/00295st04. , 30, 32. [DOI] [PubMed] [Google Scholar]
- 19.Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–41. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 21.Robb L, Elwood NJ, Elefanty AG, et al. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 1996;15:4123–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Shalaby F, Ho J, Stanford WL, et al. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell. 1997;89:981–90. doi: 10.1016/s0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 23.Gualandris A, Annes JP, Arese M, et al. The latent transforming growth factor-beta-binding protein-1 promotes in vitro differentiation of embryonic stem cells into endothelium. Mol Biol Cell. 2000;11:4295–308. doi: 10.1091/mbc.11.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeder T, Meier-Stiegen F, Schwanbeck R, et al. Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech Dev. 2006;123:570–9. doi: 10.1016/j.mod.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Moffat J, Grueneberg DA, Yang X, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 26.Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 27.Ii M, Nishimura H, Sekiguchi H, et al. Concurrent vasculogenesis and neurogenesis from adult neural stem cells. Circ Res. 2009;105:860–8. doi: 10.1161/CIRCRESAHA.109.199299. [DOI] [PubMed] [Google Scholar]
- 28.Suter DM, Krause KH. Neural commitment of embryonic stem cells: molecules, pathways and potential for cell therapy. J Pathol. 2008;215:355–68. doi: 10.1002/path.2380. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita JK. Differentiation of arterial, venous, and lymphatic endothelial cells from vascular progenitors. Trends Cardiovasc Med. 2007;17:59–63. doi: 10.1016/j.tcm.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Li ZJ, Wang ZZ, Zheng YZ, et al. Kinetic expression of platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31) during embryonic stem cell differentiation. J Cell Biochem. 2005;95:559–70. doi: 10.1002/jcb.20436. [DOI] [PubMed] [Google Scholar]
- 31.Fons P, Herault JP, Delesque N, et al. VEGF-R2 and neuropilin-1 are involved in VEGF-A-induced differentiation of human bone marrow progenitor cells. J Cell Physiol. 2004;200:351–9. doi: 10.1002/jcp.20076. [DOI] [PubMed] [Google Scholar]
- 32.Anthony TE, Klein C, Fishell G, et al. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–90. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 33.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–84. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park D, Xiang AP, Zhang L, et al. The radial glia antibody RC2 recognizes a protein encoded by Nestin. Biochem Biophys Res Commun. 2009;382:588–92. doi: 10.1016/j.bbrc.2009.03.074. [DOI] [PubMed] [Google Scholar]
- 35.Wiese C, Rolletschek A, Kania G, et al. Nestin expression–a property of multi-lineage progenitor cells. Cell Mol Life Sci. 2004;61:2510–22. doi: 10.1007/s00018-004-4144-6. [DOI] [PubMed] [Google Scholar]
- 36.Feraud O, Cao Y, Vittet D. Embryonic stem cell-derived embryoid bodies development in collagen gels recapitulates sprouting angiogenesis. Lab Invest. 2001;81:1669–81. doi: 10.1038/labinvest.3780380. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy M, D’Souza SL, Lynch-Kattman M, et al. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–87. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vittet D, Prandini MH, Berthier R, et al. Embryonic stem cells differentiate in vitro to endothelial cells through successive maturation steps. Blood. 1996;88:3424–31. [PubMed] [Google Scholar]
- 39.Pearson S, Sroczynska P, Lacaud G, et al. The stepwise specification of embryonic stem cells to hematopoietic fate is driven by sequential exposure to Bmp4, activin A, bFGF and VEGF. Development. 2008;135:1525–35. doi: 10.1242/dev.011767. [DOI] [PubMed] [Google Scholar]
- 40.Villegas SN, Canham M, Brickman JM. FGF signalling as a mediator of lineage transitions–evidence from embryonic stem cell differentiation. J Cell Biochem. 110:10–20. doi: 10.1002/jcb.22536. [DOI] [PubMed] [Google Scholar]
- 41.Era T, Izumi N, Hayashi M, et al. Multiple mesoderm subsets give rise to endothelial cells, whereas hematopoietic cells are differentiated only from a restricted subset in embryonic stem cell differentiation culture. Stem Cells. 2008;26:401–11. doi: 10.1634/stemcells.2006-0809. [DOI] [PubMed] [Google Scholar]
- 42.Winnier G, Blessing M, Labosky PA, et al. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–16. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Quantification by FACS analysis of the neurons differentiated within ES-PA6 co-cultures.
Fig. S2 Cytofluorimetric absolute cells count of ES-derived cells and PA6 cells.
Fig. S3 biochemical detection and quantification of Oct4 protein in ES-PA6 co-cultures.
Fig. S4 analysis of gene expression upon FGF2 or BMP4 stimulation.
Fig. S5 characterization by immunofluorescence analysis of ES-PA6 cocultures for the presence of glia, endoderm, pancreatic and smooth muscle cells.
Fig. S6 Gene expression analysis in pure PA6 cell cultures stimulated with FGF2.
Fig. S7 2D in vitro differentiation ofFlk1-/- and Tal1-/- ES cell lines.
Table S1 List of shRNA MISSION clones used to stably knock-down the expression of Flk1, Ace, Tal1 and T.
Table S2 list of the genes used to customize the Taqman Low Density Array.


