Significance
The Tcra enhancer (Eα) is essential during thymocyte development for Tcra germline transcription, Vα-to-Jα rearrangement, and the generation of αβ T-lymphocytes. It has been assumed that Eα displays constitutive activity to drive the expression of rearranged Tcra genes in mature T cells. We show that Eα is dramatically inhibited in peripheral T-lymphocytes and is not reactivated in the context of antigen-mediated stimulation or T helper differentiation. We conclude that the major function of Eα is to coordinate the formation of a chromatin hub that drives germline transcription and primary Tcra rearrangements in DP thymocytes. Our results also imply the existence of an Eα-independent molecular mechanism that directs transcription of the rearranged Tcra locus in αβ T cells.
Keywords: enhancer, transcription, T-cell receptor, T-cell development
Abstract
The Tcra enhancer (Eα) is essential for Tcra locus germ-line transcription and primary Vα-to-Jα recombination during thymocyte development. We found that Eα is inhibited late during thymocyte differentiation and in αβ T lymphocytes, indicating that it is not required to drive transcription of rearranged Tcra genes. Eα inactivation resulted in the disruption of functional long-range enhancer-promoter interactions and was associated with loss of Eα-dependent histone modifications at promoter and enhancer regions, and reduced expression and recruitment of E2A to the Eα enhanceosome in T cells. Enhancer activity could not be recovered by T-cell activation, by forced expression of E2A or by the up-regulation of this and other transcription factors in the context of T helper differentiation. Our results argue that the major function of Eα is to coordinate the formation of a chromatin hub that drives Vα and Jα germ-line transcription and primary rearrangements in thymocytes and imply the existence of an Eα-independent mechanism to activate transcription of the rearranged Tcra locus in αβ T cells.
The generation of αβ T lymphocytes in the thymus depends on the assembly of V, D, and J gene segments at the Tcra and Tcrb loci by a developmentally regulated process known as V(D)J recombination (1). This process initiates at the Tcrb locus in immature CD4−CD8− double-negative (DN) thymocytes. DN thymocytes can be classified into four subpopulations (DN1-4) based on the expression of CD25 and CD44: DN1 (CD25−CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44−), and DN4 (CD25−CD44−) (2). Incomplete Tcrb DβJβ rearrangements are first detected in DN2 thymocytes, whereas completed VβDβJβ rearrangements are present in DN3 thymocytes. Successful Tcrb rearrangement permits TCRβ chain synthesis and assembly with the invariant pre-Tα chain to form the pre-TCR. Pre-TCR and Notch signaling then promote the differentiation of DN3 thymocytes to the DN4 and then the CD4+CD8+ double-positive (DP) stage, a process known as β-selection. Pre-TCR signaling also provokes the initiation of Tcra rearrangement in DN4 and DP thymocytes and inhibits the expression of Notch and pre-Tα (3, 4). As a consequence, in DP thymocytes pre-Tα is replaced by TCRα to form TCRαβ, allowing for positive and negative selection events that determine the pool of CD4+ and CD8+ single-positive (SP) thymocytes. SP thymocytes then migrate to the periphery as mature, naïve αβ T lymphocytes (2).
Tcra and Tcrd gene segments are organized into a single genetic locus, Tcra/Tcrd, which spans 1.7 megabases on mouse chromosome 14 (Fig. 1A). Tcra and Tcrd gene segments have different developmental programs, such that Tcrd is rearranged and expressed in DN2 and DN3 thymocytes, whereas Tcra rearrangement and expression begins in DN4 and DP thymocytes (1). It is well established that initiation of Vα-to-Jα rearrangements is regulated by the Tcra gene enhancer (Eα), which is itself activated by pre-TCR signaling in DN4 and early DP thymocytes (5–7). Eα is part of a locus control region (LCR) containing eight T-cell–specific DNaseI-hypersensitive sites located between Tcra and the ubiquitously expressed gene, Dad1 (8). Eα functions in cis to activate Tcra germ-line transcription, chromatin modifications and Vα-to-Jα rearrangement across 500 kb containing the most proximal Vα gene segments and the entire Jα gene segment cluster in early DP thymocytes. It also stimulates transcription of the rearranged Tcrd locus in γδ T lymphocytes (5, 6, 9). Other LCR elements act as insulator sequences that block Eα activity to maintain the distinct regulatory programs of the neighboring Tcra/Tcrd and Dad1 loci (10).
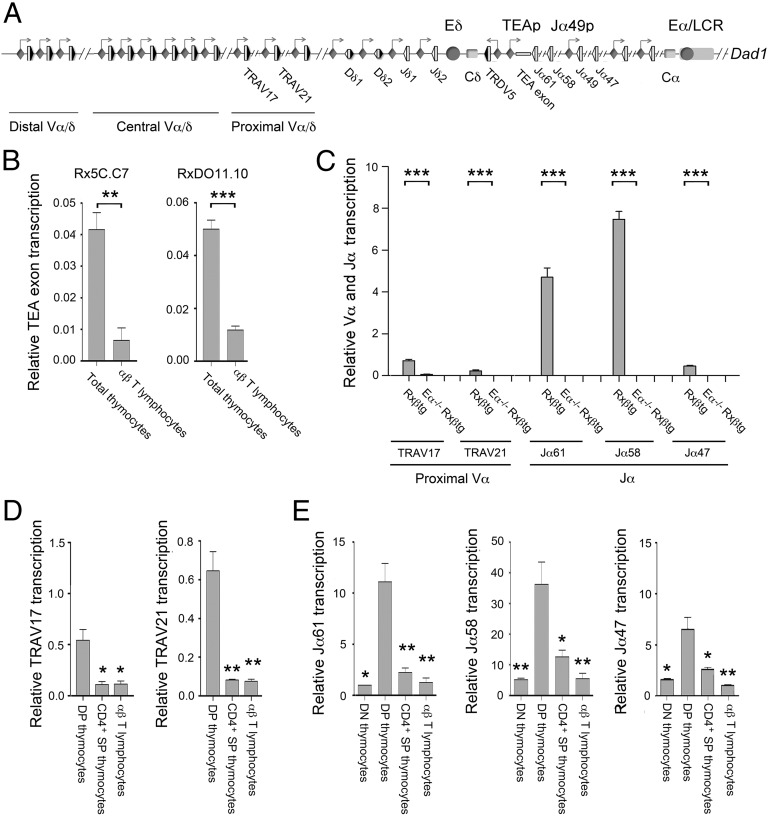
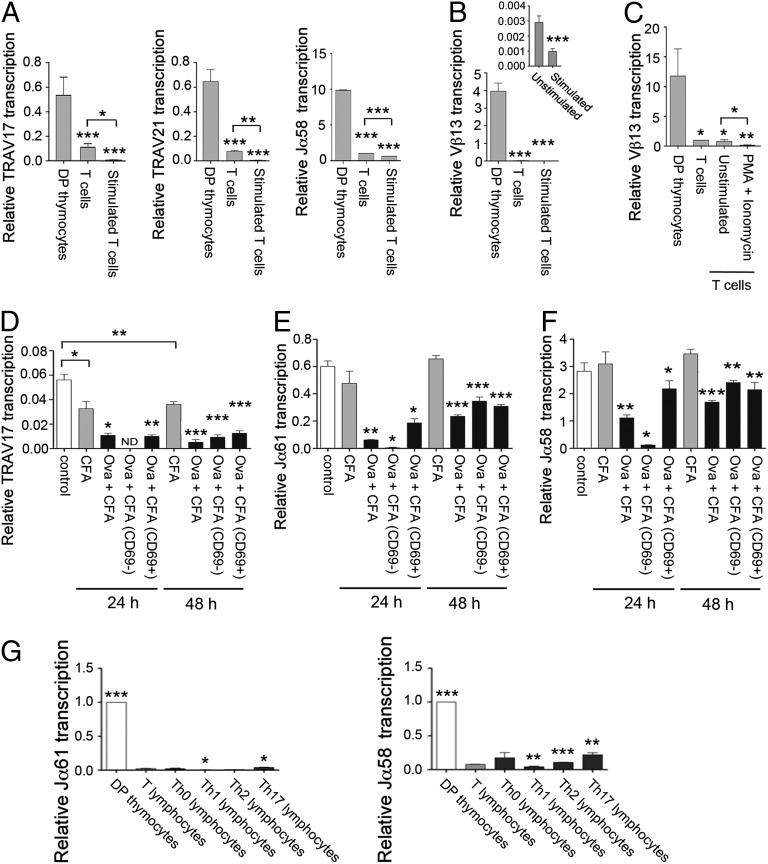
Fig. 1.
Tcra Eα-dependent transcription is inactivated in SP thymocytes and αβ T lymphocytes. (A) Schematic representation of Tcra/Tcrd genomic structure. The V, D, and J gene segments are represented by gray vertical rectangles, and the recombination signal sequences are shown as black (containing a 23-bp spacer) or white (containing a 12-bp spacer) triangles. Positions of the TEA exon, the Vα/δ, Dδ, Jδ, and Jα gene segments, and the Cδ and Cα regions are indicated. Arrows represent active sites for germ-line transcription. Promoters and enhancers are represented as diamonds and circles, respectively. The Tcra LCR is represented as a long horizontal rectangle. (B) Analysis of TEA exon transcription in Rx5C.C7 and RxDO11.10 thymocytes and T lymphocytes by RT-qPCR. (C) Analysis of proximal Vα and Jα transcription in DP thymocytes from Rxßtg and Eα−/− Rxßtg mice by RT-qPCR. (D) Analysis of TRAV17 and TRAV21 germ-line transcripts in RxDO11.10 sorted thymocyte populations and T lymphocytes by RT-qPCR. (E) Analysis of Jα transcription in RxDO11.10 sorted thymocyte populations and T lymphocytes by RT-qPCR. The results were normalized to those for Actb and represent the mean ± SEM of duplicate RT-qPCR from three independent experiments. Two-tailed Student’s t tests were used to determine the statistical significance between the values of the RT-qPCRs from total thymocytes vs. T lymphocytes (B), Rxßtg vs. Eα−/− Rxßtg thymocytes (C), and DP thymocytes vs. T lymphocytes (D and E) (*P < 0.05, **P < 0.005, and *** P < 0.0005).
Initial transcriptional activation of the Tcra locus occurs in the context of a chromatin hub through interactions between Eα and the Jα and proximal Vα promoters (Fig. 1A), with contacts mediated by enhancer- and promoter-bound transcription factors (TFs) and the CCCTC-binding factor (CTCF) and its associated cohesin (11, 12). Within this hub, Eα interacts with and activates the T early-α promoter (TEAp) associated with the T early α (TEA) exon and Jα61 gene segment, and the promoter associated with the Jα49 gene segment. Transcriptional elongation provides the recombinase proteins RAG-1 and RAG-2 accessibility to the Jα gene segments positioned downstream of these promoters (13–15). Eα similarly activates proximal Vα segments (9). Together, these activation events trigger primary Vα-to-Jα recombination, in which the proximal Vα gene segments rearrange to the most Eα-distal Jα gene segments (16, 17).
Pre-TCR signaling stimulates the assembly of an active Eα enhanceosome in DN4 and early DP thymocytes (6, 7). The minimal Eα segment with proper developmental regulation is 275-bp long (18). Within this segment, the Tα1-Tα2 region contains essential binding sites for cAMP response element binding-protein (CREB), T-cell factor 1 (TCF-1)/lymphocyte enhancer factor 1/(LEF-1), Runx-1, and Ets-1 (19), and cooperative binding among these factors is required to form the functional Tα1-Tα2 enhanceosome in vivo (20). Additional binding sites for nuclear factor of activated T cells (NFAT), CREB/ATF, Ets-1/Fli-1, early response factor 1 (Egr-1), Sp1, GATA-3, and E-proteins E2A and HEB flank Tα1-Tα2 both 5′ and 3′ (5′ Tα1 and Tα3-Tα4) (6, 7, 20, 21). Previous studies have shown that the constitutive TFs are already loaded onto the enhancer in DN3 thymocytes, and that they form a regulatory scaffold for the transient recruitment of pre-TCR induced TFs (NFAT, AP-1, and Egr-1) that activate the enhancer in DN4 and early DP thymocytes, as well as for the more stable recruitment of constitutive TFs in late DP thymocytes (7, 18, 21). Thus, Eα activity is regulated through the ordered assembly of different enhanceosomes during thymocyte development.
It has been assumed that Eα is important for transcription of rearranged Tcra genes in mature αβ T lymphocytes. Previous experiments using artificial transgenic reporter constructs in mice have shown that the activity of the 10.5-kb Tcra LCR appears to be decreased in peripheral αβ T lymphocytes compared with that in thymocytes (22). Because our previous work established Eα enhanceosome composition and function only as far as preselected resting DP thymocytes (7), we investigated Eα functionality at later developmental stages. Here, we describe the developmental regulation of Eα in its natural genomic context and when positioned at an ectopic location. We found that Eα function is strongly inhibited in SP thymocytes and peripheral αβ T lymphocytes indicating that this element is not necessary for the expression of the rearranged Tcra at these stages. Enhancer inactivation was accompanied by the disruption of long-range Eα-promoter interactions, and the loss of activating histone marks. Although enhancer inactivation was associated with the loss of E2A from the enhanceosome, our data argue that there must be additional limitations on Eα activity in αβ T cells. We conclude that Eα-independent mechanisms must exist to support the expression of rearranged Tcra genes in peripheral αβ T cells.
Results
Eα-Dependent Transcription Is Inhibited in SP Thymocytes and Mature αβ T Lymphocytes.
To assess Eα function in mature αβ T lymphocytes, we compared the abundance of Eα-dependent transcripts present in thymocytes and αβ T lymphocytes of Rag2−/− × TCRαβ transgenic mice (Fig. 1). Rag2−/− × 5C.C7 (Rx5C.C7) and Rag2−/− × DO11.10 (RxDO11.10) mice (23, 24) both express clonotypic TCRαβs that direct the differentiation of DP thymocytes toward the SP CD4+ lineage. Because the Tcra locus is maintained in unrearranged configuration, Tcra locus germ-line transcription can be readily assayed in mature T cells (25). Transcription of the TEA exon is known to be Eα-dependent in DP thymocytes (5, 6) (Fig. 1A). Although RT-mediated quantitative PCR (RT-qPCR) revealed similar expression of TEA transcripts in thymocytes of both mouse strains, expression was reduced by 80–85% in mature αβ T lymphocytes (Fig. 1B). Reduced transcription was also observed in mature αβ T lymphocytes of Rag2−/− × OT-I mice that express a clonotypic TCRαβ that directs differentiation of DP thymocytes toward the SP CD8+ lineage (Fig. S1A).
We similarly analyzed germ-line transcription of proximal Vα gene segments (TRAV17, TRAV21) and Jα gene segments (Jα61, Jα58, and Jα47). Primary Jα transcripts are typically spliced from a donor sequence immediately 3′ of the Jα segment to an acceptor at the 5′ end of the first Cα exon; Jα61 transcripts are atypically spliced using a more distant donor sequence (Fig. S1 B and C). We confirmed that all of these transcripts are Eα-dependent in DP thymocytes by evaluating transcript abundance in thymocytes of Rag2−/− and Eα−/− Rag2−/− mice in which DP differentiation was driven by a Tcrb transgene (25) (Rxßtg and Eα−/− Rxßtg, respectively) (Fig. 1C). Subsequent analysis of sorted thymocyte populations and peripheral T lymphocytes from RxDO11.10 mice revealed that TRAV17, TRAV21, Jα61, Jα58, and Jα47 transcripts were all strongly suppressed in SP thymocytes and αβ T lymphocytes (Fig. 1 D and E). Together, these data indicate that Eα-dependent Tcra locus germ-line transcription is down-regulated in SP thymocytes and mature αβ T lymphocytes.
Eα Is Inactivated in SP Thymocytes and Mature αβ T Lymphocytes.
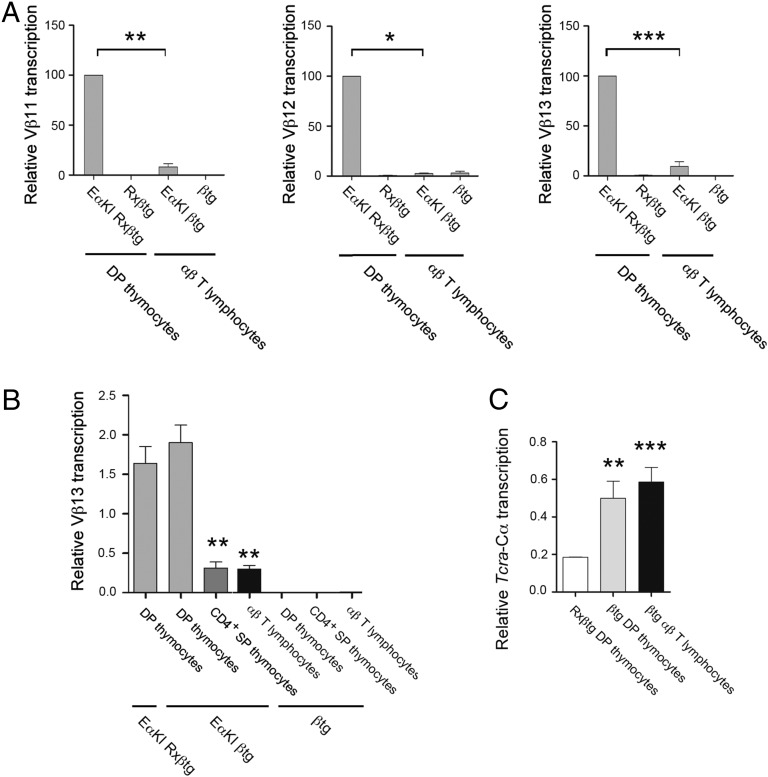
The inhibition of Eα-dependent germ-line Tcra transcription in SP thymocytes and αβ T lymphocytes could reflect inactivation of Eα or inactivation of other locus regulatory elements, including Vα and Jα promoters and the Tcra LCR. To distinguish between these possibilities, we analyzed EαKI alleles (26), in which Eα was introduced about 0.5 kb downstream of Vβ12 in the Tcrb locus. Previous experiments had shown that the introduced Eα (Eαi) is active in DP thymocytes, as evidenced by potent transcriptional activation of Vβ12 and the nearby Vβ13 and Vβ11 gene segments in DP thymocytes obtained from EαKI Rxßtg mice but not in DN3 thymocytes obtained from EαKI Rag2−/− mice (26). To assess Eαi activity in peripheral T lymphocytes, we compared the abundance of Vβ11, Vβ12, and Vβ13 transcripts in DP thymocytes of EαKI Rxßtg mice to that in peripheral T cells of EαKI βtg mice (Fig. 2A). The latter mice support complete αβ T lymphocyte development due to expression of the Tcrb transgene and an endogenous Tcra rearrangement. Expression of the Tcrb transgene also enforces Tcrb allelic exclusion, thereby maintaining the Vβ array in germ-line configuration and allowing analysis of Eαi activity. As expected, Eαi was a potent activator of Vβ11, Vβ12, and Vβ13 transcription in DP thymocytes (Fig. 2, compare EαKI Rxßtg to Rxßtg). However, this transcription was strongly inhibited in mature αβ T lymphocytes (Fig. 2A, compare EαKI Rxßtg to EαKI βtg) and in in CD4+ SP thymocytes (Fig. 2B, EαKI βtg). These data confirm that the activity of Eα is substantially down-regulated in SP thymocytes and in mature αβ T lymphocytes.
Fig. 2.
Eαi is inactivated in SP thymocytes and αβ T lymphocytes. (A) Analysis of Vβ11, Vβ12, and Vβ13 transcription in EαKI Rxßtg and Rxßtg thymocytes and EαKI βtg and βtg T lymphocytes by RT-qPCR. (B) Analysis of Vβ13 transcription from EαKI Rxßtg DP thymocytes, EαKI βtg and βtg sorted thymocyte populations, and EαKI βtg and βtg T lymphocytes by RT-qPCR. (C) Analysis of Tcra-Cα transcription in Rxßtg DP thymocytes, sorted βtg DP thymocytes, and sorted βtg T lymphocytes by RT-qPCR. The results were normalized to those for Actb and represent the mean ± SEM of duplicate RT-qPCR from three independent experiments. Two-tailed Student’s t tests were used to determine the statistical significance between the values of the RT-qPCRs from EαKI Rxßtg or EαKI βtg DP thymocytes vs. EαKI βtg CD4+ SP thymocytes and T lymphocytes (A and B) or Rxßtg DP thymocytes vs. βtg DP thymocytes or αβ T lymphocytes (C) (*P < 0.05, **P < 0.005, and ***P < 0.0005).
Eα–TEAp Interactions Are Lost in Mature αβ T Lymphocytes.
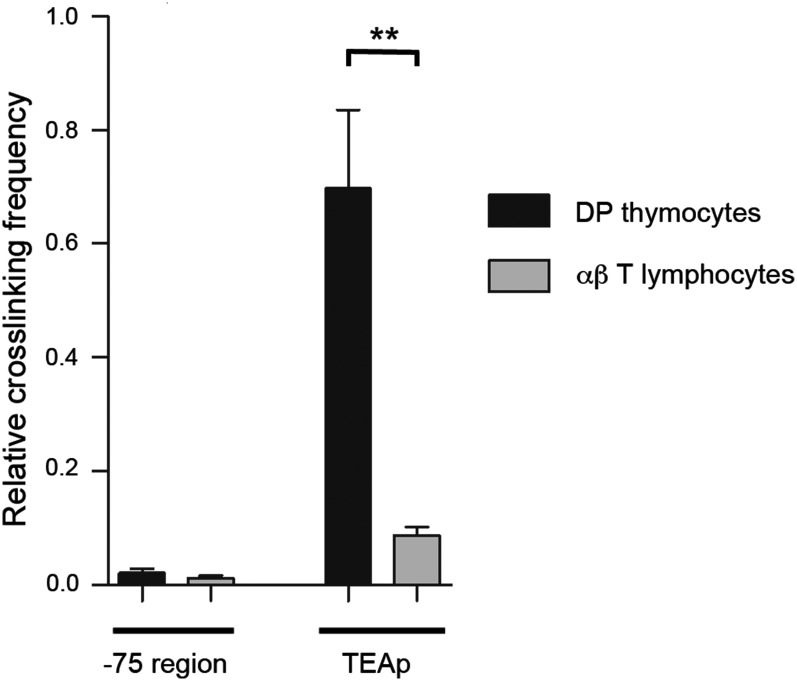
Eα activates transcription at Tcra locus promoters by direct physical interactions with DNA looping over long distances (11, 12). Therefore, we used chromosome conformation capture (3C) to analyze interactions between Eα and the TEAp across a distance of 73 kb in RxDO11.10 mice (Fig. 3 and Fig. S2). Eα interacted frequently with TEAp in DP thymocytes; this interaction was specific, because interaction was not detected with a DNA fragment only 2 kb further from Eα. Notably, the Eα–TEAp interaction was dramatically suppressed in αβ T lymphocytes (Fig. 3). These results indicate that Eα cannot establish contact with upstream promoters in αβ T lymphocytes.
Fig. 3.
The Eα-TEAp long-range interactions present in DP thymocytes are lost in αβ T lymphocytes. 3C analysis of physical contacts between Eα and TEAp or a genomic fragment located at −75 kb from the enhancer in DP thymocytes (black bars) and αβ T lymphocytes (gray bars) from RxDO11.10. Data were normalized to the ligation frequency between Eα and a neighboring HindIII fragment (Fig. S2 and Table S1). HindIII digestion efficiency and the relative interactions between Eα and the control fragment in DP thymocytes and T lymphocytes are shown in Fig. S2. The results represent the mean ± SEM of duplicate qPCR from three independent experiments. Two-tailed Student’s t tests were used to determine the statistical significance between the values of the qPCRs obtained after ligation of HindIII fragments from DP thymocytes vs. T lymphocytes (**P < 0.005).
Eα-Dependent Chromatin Modifications Are Lost in Mature αβ T Lymphocytes.
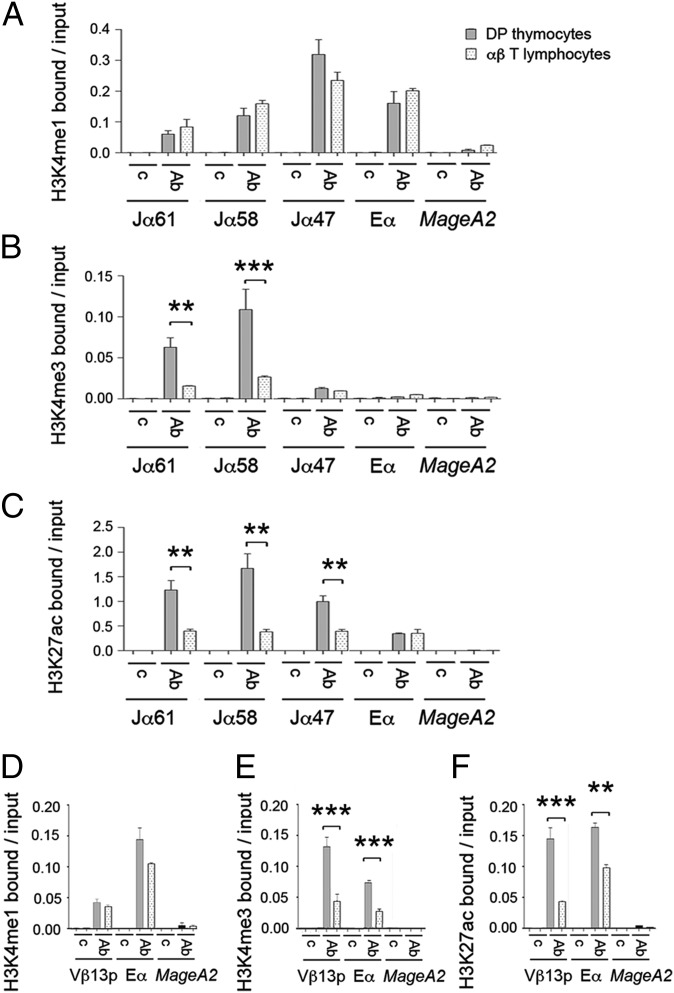
As an additional measure of Eα activity, we analyzed Eα-dependent histone H3 (H3) modifications in DP thymocytes and αβ T lymphocytes by quantitative chromatin immunoprecipitation (qChIP) (Fig. 4). We compared the pattern of H3K4me1 and H3K4me3 at Tcra in sorted DP thymocytes and T lymphocytes from RxDO11.10 mice (Fig. 4 A and B). Consistent with the known properties of enhancers and active promoters (27), in DP thymocytes we found that Eα is highly enriched for H3K4me1 and depleted of H3K4me3, whereas the opposite was true for the actively transcribed Jα61 and Jα58 gene segments. However, H3K4me3 was sharply reduced at Jα61 and Jα58 in αβ T lymphocytes (Fig. 4B). H3K27ac, another mark of active chromatin (28), was similarly suppressed at Jα61, Jα58, and Jα47 in αβ T lymphocytes compared with DP thymocytes (Fig. 4C). Gain of H3K4me1 and H3K4me3 at Eα has also been correlated with enhancer activity in the transition from DN3 to DP thymocytes (29). Because our qChIP experiments did not reveal any apparent change in H3K4me1 and H3K4me3 marks at Eα in DP thymocytes compared with T lymphocytes, we evaluated a public database that compares the presence of H3K4me1 and H3K4me3 by ChIP-sequencing in DP thymocytes and T lymphocytes from WT mice using the UCSC Genome Browser (30, 31). Evaluation of these data revealed that H3K4me1 and H3K4me3 marks are clearly inhibited in the Eα 3′ flanking region in T lymphocytes compared with DP thymocytes (Fig. S3). This difference may not have been detected in our qChIP analysis because we assayed a 500-bp region that includes the nucleosome depleted Tα1-Tα4 region and its 5′ flank (Fig. S3). We also used EαKI Rxßtg and EαKI βtg mice to analyze Eαi and Vβ gene segment chromatin structure on EαKI alleles (Fig. 4 D–F and Fig. S4). The H3K4me3 and H3K27ac marks were reduced at both the Vβ13p and Eαi in peripheral T lymphocytes compared with DP thymocytes (Fig. 4 E and F). Similar results were obtained for H3ac and H3K4me2 at Vβ13 and Eαi, and for H3K4me3 at Vβ12 (Fig. S4). All of these histone marks are Eαi-dependent, because they were not detected on WT Tcrb alleles (Rxßtg DP thymocytes and βtg αβ T lymphocytes, Fig. S4). Taken together, these chromatin analyses strongly support the notion that Eα function is inhibited in mature αβ T cells.
Fig. 4.
Comparative histone H3 modification analyses in DP thymocytes and αβ T lymphocytes. Analysis of Tcra Jα and Eα histone H3K4me1 (A), H3K4me3 (B), and H3K27ac (C) modifications in DP thymocytes and T lymphocytes from RxDO11.10 mice by qChIP. Analysis of Vβ13p and Eαi histone H3K4me1 (D), H3K4me3 (E), and H3K27ac (F) modifications in DP thymocytes from EαKI Rxßtg mice and T lymphocytes from EαKI βtg by qChIp. MageA2, negative control (49). Data represent the mean ± SEM of duplicate qPCR analysis of three independent experiments. Two-tailed Student’s t test were used to determine the statistical significance between the values of the qChIPs from DP thymocytes vs. T lymphocytes (**P < 0.005 and ***P < 0.0005).
Transcription of Rearranged Tcra Genes Is Not Sensitive to Eα Inactivation.
Because Eα appears to be inactivated by the SP stage, our data imply that transcription of rearranged Tcra genes must become Eα independent. To evaluate whether inactivation of Eα impacts the expression of the rearranged Tcra locus we compared the abundance of Tcra-Cα transcripts from the unrearranged Tcra locus in sorted preselected DP thymocytes of Rxßtg mice with that of the rearranged Tcra locus in sorted postselected DP thymocytes and peripheral T lymphocytes of βtg mice (Fig. 2C). Transcription of heterogeneously rearranged Tcra alleles in postselection DP thymocytes was substantially higher than from unrearranged alleles in preselection DP thymocytes and high-level transcription was maintained in peripheral T cells. Therefore, transcription of the rearranged Tcra locus is not down-regulated in αβ T cells, despite Eα inactivation, suggesting that transcription of the rearranged alleles becomes Eα-independent.
Eα Cannot Be Reactivated by Stimulation Through the TCR or T Helper Differentiation.
Because Eα can be activated in vivo by pre-TCR signaling in thymocytes and in vitro by stimulation of thymocyte-derived cell lines (7), we asked whether Eα could similarly be activated by TCR signaling in mature T lymphocytes (Fig. 5 A–F). We incubated T lymphocytes from RxDO11.10 mice for 24 h in the presence of plate-bound anti-CD3ε and anti-CD28. This activation was effective based on strong down-regulation of TCRβ and up-regulation of CD25 and CD69 (Fig. S5A). However, TRAV17, TRAV21, and Jα58 transcripts were suppressed rather than induced by this activation (Fig. 5A). Analysis of Vβ13 transcription in stimulated EαKI βtg T lymphocytes also demonstrated suppression rather than induction of transcription (Fig. 5B). Furthermore, in vitro treatment of T lymphocytes with PMA plus ionomycin, which mimics pre-TCR/TCR signaling and can potently induce Eα-dependent Jα transcription in Scid-adh and Jurkat cells (7), also failed to activate (but rather inhibited) Vβ13 transcription in T lymphocytes (Fig. 5C).
Fig. 5.
Eα and Eαi are not activated by TCR-mediated cell stimulation and Th differentiation in T lymphocytes. (A) Analysis of TRAV17, TRAV21, and Jα58 germ-line transcripts in sorted DP thymocytes and unstimulated and stimulated T lymphocytes from RxDO11.10 mice by RT-qPCR. (B) Analysis of Vβ13 transcription in EαKI Rxßtg thymocytes and unstimulated and stimulated EαKI βtg T lymphocytes by RT-qPCR. (C) Analysis of Vβ13 transcription in EαKI Rxßtg DP thymocytes, EαKI βtg T lymphocytes, and EαKI tgβ T lymphocytes left in complete medium (unstimulated) or incubated with PMA and ionomycin (PMA + Ionomycin) by RT-qPCR. Analysis of TRAV17 (D), Jα61 (E), and Jα58 (F) transcripts present in unstimulated and in vivo stimulated RxDO11.10 T lymphocytes by RT-qPCR. Indicated transcripts were compared in T lymphocytes from noninjected mice (control), CFA-injected mice (CFA), or Ova 323–339 + CFA-injected mice (Ova + CFA). Indicated transcripts were compared in sorted CD69− and CD69+ T populations from Ova 323–339 + CFA-immunized mice. (G) Analysis of Gata3, Tcfe2a, Jα61, and Jα58 transcripts in sorted DP thymocytes, T lymphocytes, and Th0, Th1, Th2, and Th17 populations from RxDO11.10 mice by RT-qPCR. The results were normalized to those for Actb and represent the mean ± SEM of duplicate RT-qPCR from three independent experiments. Two-tailed Student’s t tests were used to determine the statistical significance between the values of the RT-qPCRs from DP thymocytes vs. T lymphocytes (A-C), from unstimulated vs. stimulated T lymphocytes (A–F), and from T lymphocytes vs. DP thymocytes or Th populations (G) (*P < 0.05, **P < 0.005, and ***P < 0.0005).
We also activated T lymphocytes in vivo by immunizing RxDO11.10 mice with 200 μg of OVA peptide 323–339 (Ova 323–339) in CFA. Such activation was effective as judged by CD69 induction 24 or 48 h later (Fig. S5B). However, as was observed in vitro, TRAV17, Jα61, and Jα58 transcription was further suppressed rather than induced in vivo (Fig. 5 D–F). Moreover, this suppression was even apparent in sorted CD69+ T lymphocytes (Fig. 5 D–F).
To evaluate whether the combinations of TFs present naturally during T helper (Th) differentiation could rescue Eα activity, we differentiated RxDO11.10 T cells in vitro to Th populations. As expected, differentiated Th1 cells expressed high levels of Tbet, Th2 expressed elevated Gata3 transcripts and protein, and Th17 cells specifically expressed Rorgt (Fig. S6 A and B) (32). Nevertheless, Jα61 and Jα58 transcription remained low in all Th populations (Fig. 5G), and Eα-flanking H3K4me1 and H3K4me3 marks were low in Th2 cells (Fig. S3). These data indicate that Eα remains inactive in the face of a broad range of peripheral T-cell activation and differentiation cues.
E2A Binding to the Eα Enhanceosome Is Reduced in αβ T Lymphocytes.
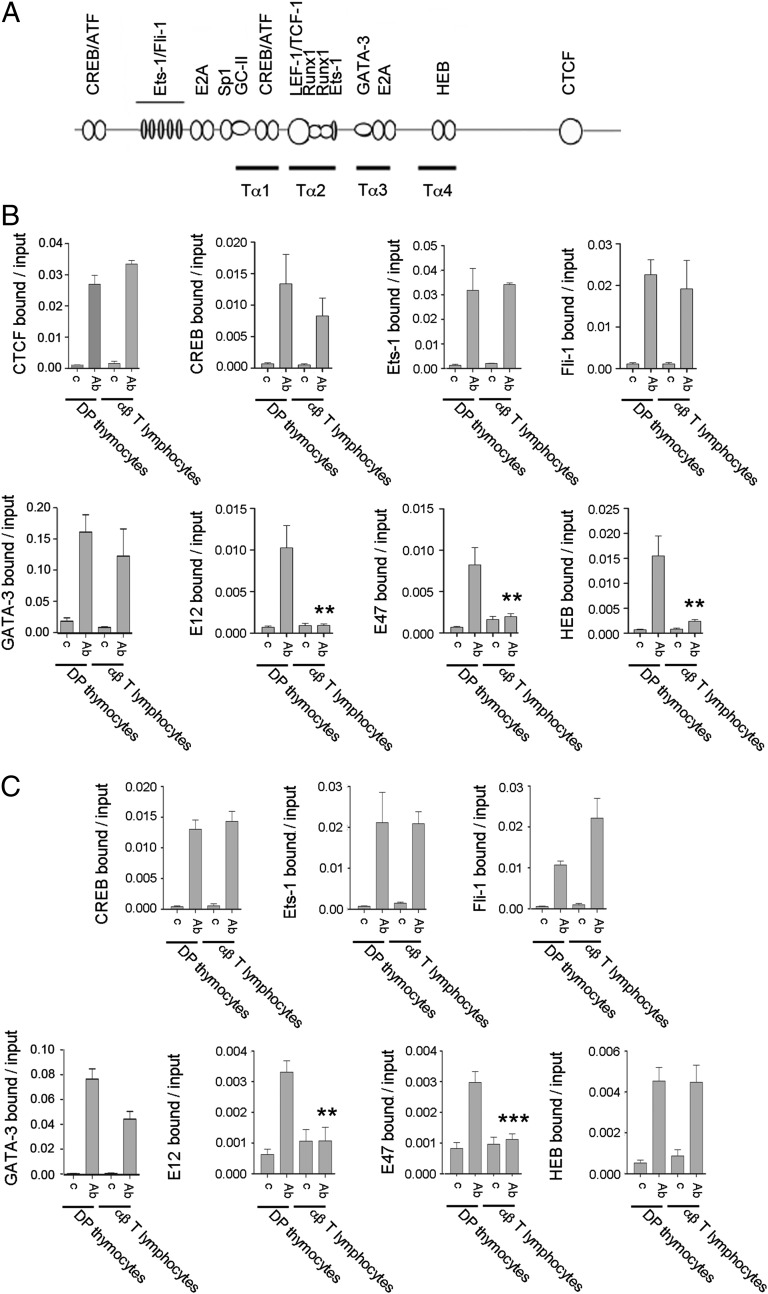
To evaluate the molecular mechanism for Eα inactivation in αβ T lymphocytes, we analyzed the binding of CTCF and constitutive TFs to Eα and Eαi in DP thymocytes and αβ T lymphocytes by qChIP (Fig. 6A). Analysis of RxDO11.10 mice revealed no loss of CTCF binding to Eα and TEAp in T lymphocytes (Fig. 6B and Fig. S7A). DP thymocytes from EαKI Rxßtg and αβ T cells from EαKI βtg mice were then used to analyze constitutive TF binding to Eα (Fig. 6B) and to Eαi (Fig. 6C). We found no differences in CREB, Ets-1, Fli-1, and GATA-3 binding in DP thymocytes and αβ T lymphocytes in any of the genomic contexts analyzed for both Eα and Eαi. However, we found dramatic reductions in the binding of E2A TFs E12 and E47 at sites positioned 5′ Tα1 and in Tα3-Tα4 (21). Reduced binding of E2A in T lymphocytes is consistent with reduced expression of Tcfe2a (encoding E2A TFs) and increased expression of Id2 in T lymphocytes compared with that in DP thymocytes (Fig. S8) (33) (www.immgen.org). Notably, HEB bound equivalently in both cell types to Tα4 in the context of Eαi (Fig. 6C) but bound better in DP thymocytes than in T lymphocytes to Tα4 in the context of Eα (Fig. 6B). Selective loss of HEB from Eα but not from Eαi must reflect the distinct genomic contexts of the two enhancers.
Fig. 6.
E2A is not present at the Eα enhanceosome assembled in αβ T lymphocytes. (A) Diagram depicts the location of the Tα1-Tα4 regions as well as the described TF binding sites. (B and C) For CTCF q ChIPs, chromatin was prepared from sorted DP and T lymphocytes from RxDO11.10 mice. For all other TF ChIPs, DP thymocyte chromatin was prepared from EαKI Rxßtg thymocytes and T lymphocyte chromatin was prepared from EαKI βtg T lymphocytes. Immunoprecipitation was performed with the indicated control (c) or specific Abs (Ab). DNA purified from the input and Ab-bound fractions was used as a template for qPCR to evaluate the presence of Eα (B) and Eαi (C). Data represent the mean ± SEM of duplicate qPCR from three independent experiments. Two-tailed Student’s t tests were used to determine the statistical significance between the values of the qChIPs from DP thymocytes vs. T lymphocytes (*P < 0.05, **P < 0.005, and ***P < 0.0005).
One potential caveat to our conclusion that E2A and HEB are lost from Eα in mature T lymphocytes is that we compared Eα occupancy in DP thymocytes from mice with an unrearranged Tcra locus to T lymphocytes from mice with a rearranged Tcra locus (Fig. 6A). To ensure that Eα occupancy is not influence by locus rearrangement, we compared Eα TF binding in DP thymocytes from Rxßtg mice (unrearranged Tcra locus) to DP thymocytes from βtg mice (rearranged Tcra locus) (Fig. S7B). The binding of E2A, HEB, and other TFs was indistinguishable in the two DP populations, indicating that the loss of E2A and HEB binding detected in T lymphocytes was not dependent on rearrangement status. Our results suggest that reduced binding of E2A and HEB correlates with enhancer inactivation in mature αβ T lymphocytes.
E2A Activates Specific Eα-Dependent Promoters.
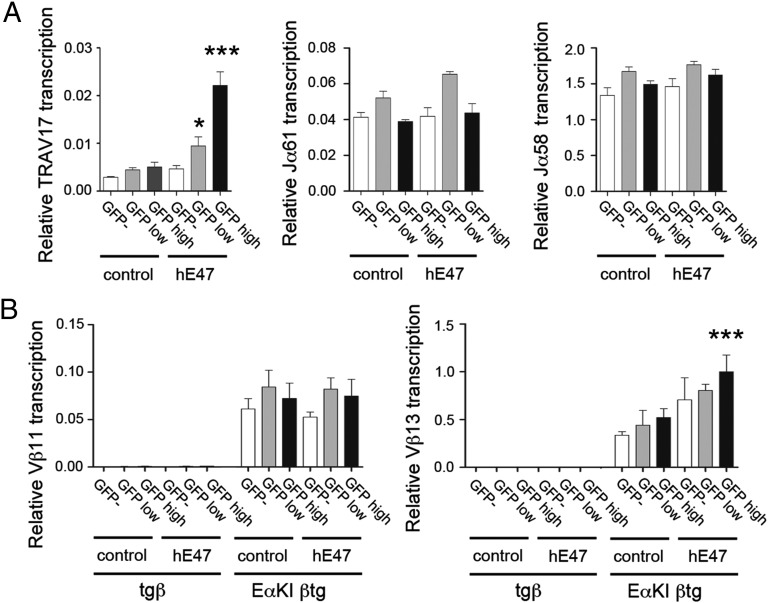
To assess whether restoration of E2A would be sufficient to recover Eα function, we forced its expression in αβ T lymphocytes by retroviral transduction (Fig. 7). We transduced T lymphocytes from RxDO11.10 mice with MigR vectors directing the expression of human rE47 (hE47), sorted GFP-negative, GFP-low, and GFP-high cells, and confirmed that GFP-high cells expressed hE47 transcripts and displayed elevated binding of E47 to Eα (Fig. S9 A and B). Forced expression of hE47 induced TRAV17 transcripts in a dose-dependent manner, but did not induce Jα61 or Jα58 transcripts (Fig. 7A) and did not induce Tcra-Cα transcripts in either RxDO11.10 or βtg mice (Fig. S9 C and D). Failure to induce Jα61 or Jα58 transcripts could not be overcome by stimulation of MigR-hE47-transduced T lymphocytes with PMA + ionomycin (Fig. S9E). Thus, neither hE47 alone or in combination with inducible TFs can activate Eα-dependent Jα transcription in T lymphocytes. Consistently with these results, induction of Tcfe2a transcription in Th2 and Th17 populations that have similar levels of Tcfe2a to those present in preselected DP thymocytes did not activate Jα61 and Jα58 transcripts (Fig. S6C). Forced expression of hE47 also modestly enhanced Vβ13 expression but had no effect on Vβ11 gene expression in EαKI βtg T lymphocytes (Fig. 7B). Because the effects of hE47 overexpression were generally modest and promoter-specific and were not observed on total Tcra-Cα transcripts, they are more likely to reflect the activities of this TF at the individual promoters rather than at Eα. We conclude that E2A down-regulation may contribute to Eα inactivation after the DP stage, but that Eα inactivation must be enforced by additional mechanisms as well.
Fig. 7.
E47 can partially rescue specific Eα-dependent transcription in αβ T lymphocytes. (A) Forced expression of hE47 by retroviral transduction of T lymphocytes from RxDO11.10 mice induces TRAV17 transcription but not Jα61 and Jα58 transcription. (B) Forced expression of hE47 by retroviral transduction of T lymphocytes from EαKI βtg mice induces Vβ13 but not Vβ11 gene segment transcription. GFP−, GFP-low, and GFP-high–transduced populations with MigR (control) or MigR-hE47 (hE47) were sorted, and the indicated transcripts were quantified by RT-qPCR. The results were normalized to those for Actb and represent the mean ± SEM of duplicate RT-qPCR from three independent experiments. Two-tailed Student’s t tests were used to determine the statistical significance between the RT-qPCR values from MigR- and MigR-hE47-transduced T lymphocytes by comparing MigR-control GFP− cells with MigR-hE47 GFP− cells, MigR-control GFP-low cells with MigR-hE47 GFP-low cells, and MigR-control GFP-high cells with MigR-hE47 GFP-high cells (*P < 0.05 and ***P < 0.0005).
Discussion
Although Eα is accepted to be important for transcription of rearranged Tcra genes in mature αβ T lymphocytes, previous experiments using artificial transgenic reporter constructs suggested that the activity of a 10.5-kb Tcra LCR fragment containing Eα was partially inhibited in peripheral αβ T lymphocytes compared with thymocytes (22). This observation, together with our previous data establishing the changing composition and functionality of Eα as thymocytes transit from the DN to the DP stage across the β-selection checkpoint (7), prompted us to address enhancer function at late stages of αβ T-cell maturation. Here we found that Eα function is dramatically impaired in SP thymocytes and αβ T lymphocytes, and we precisely map this effect to the 551-bp fragment of Eα present on EαKI alleles. Although Eα can be activated by the binding of inducible TFs in response to pre-TCR signals associated with β-selection during thymocyte development, the different combinations of TFs that are present during TCR-mediated activation or Th differentiation could not rescue its activity. Enhancer inactivation was associated with reduced binding of E2A; however, enforced expression of this TF, although able to activate transcription from specific Eα-dependent promoters, could not induce enhancer function. We propose that the major Eα function is to coordinate the formation of a chromatin hub to drive primary rearrangements in DP thymocytes.
Our demonstration of Eα inactivation in peripheral T lymphocytes is in general agreement with a previous study using a human CD2 (hCD2) reporter gene driven by the Tcra LCR (22). However, that study found the LCR to be active in SP thymocytes and down-regulated thereafter; in contrast, our data revealed that Eα is down-regulated in SP thymocytes, suggesting that down-regulation is linked to positive selection. Moreover, the extent of enhancer down-regulation in T lymphocytes detected in our study was much more substantial than that documented using the Tcra LCR-hCD2 reporter. We believe that the differences between the two studies are a consequence of the artificial nature of the hCD2 transgenic reporter system. In support of this notion, the Tcra LCR-hCD2 reporter was highly expressed in B cells, whereas a Tcra LCR-human β-globin reporter was not (22). More recent studies revealed variable expression of the same Tcra LCR-hCD2 construct in B cells differentiated from transfected ES cells (34). Hence, features of the hCD2 reporter dictate the developmental profile of Tcra LCR-hCD2 activity.
Our previous studies of Eα occupancy indicated that three different enhanceosomes are assembled in DN3, DN4/early DP thymocytes, and small DP thymocytes (7). In DN3 thymocytes, the constitutive TFs CREB, Ets-1, GATA-3, Fli-1, and E2A are bound to Eα as demonstrated by ChIP assays (7, 31, 33). In addition, because an intact TCF/LEF binding site is essential for the assembly of the Tα1-Τα2 enhanceosome (6, 20), TCF-1/LEF-1 and Runx-1 must be bound as well. Thus, we assume that the entire set of TFs that constitute the Tα1-Tα4 enhanceosome are bound in DN3 thymocytes. In fact, Eα is as sensitive to DNase I digestion in DN3 thymocytes as it is in DP thymocytes, even though it is inactive in the former and active in the latter (6, 18, 35). In DN4/early DP thymocytes, previous ChIP experiments demonstrated that pre-TCR-inducible TFs as well as increased Ets-1 and p300 are recruited to Eα (7). These events are thought to be critical for enhancer activation and the induction of germ-line transcription and primary Vα-to-Jα rearrangements. Small DP thymocytes display stable binding of CREB, Fli-1, HEB, Runx1, and TCF-1, and strongly increased binding of Ets-1, E2A, and GATA-3 (7, 31, 36–38). Finally, the newly described inactive Eα enhanceosome assembled in αβ T lymphocytes is defined by continued binding of CREB, Fli-1, HEB, Ets-1, GATA-3, and TCF-1 as demonstrated by ChIP experiments (39). We presume Runx-1 remains bound as well, because the recruitment of Ets-1 to Tα1-Tα2 depends on its cooperative binding with Runx-1 (20, 40). However, there is diminished binding of E2A. Based on our studies, we can now define four different enhanceosomes during αβ T-cell development, two active and two inactive. Both inactive enhanceosomes display little binding of E2A.
Notably, in contrast to its role in peripheral αβ T cells, Eα is active and critical for Tcrd gene expression in peripheral γδ T lymphocytes (5). Hence, a different Eα enhanceosome must be assembled in peripheral γδ compared with those in αβ T lymphocytes. Although the structure of this enhanceosome is unknown, it should be noted that there are distinct subsets of γδ T lymphocytes that differentially express Id proteins (41), suggesting that there may be multiple γδ-specific Eα enhanceosomes.
Prior work had established that some Tcra locus promoters, notably TEAp and proximal V gene segment promoters like TRAV17 and TRAV21, are Eα-dependent in DP thymocytes, although the promoters of more distal V gene segments display activities that are Eα-independent (42). However, all such conclusions were drawn by analysis of the Tcra locus in the germ-line configuration. Our current data indicate that the activities of the Eα-dependent germ-line promoters are substantially diminished no later than the SP stage, but that the transcription of heterogeneously rearranged Tcra alleles remains strong in peripheral T cells. Presumably, the same V promoters that are Eα-dependent in unrearranged configuration must become Eα-independent following rearrangement. The molecular basis for this transition is not known. One possibility is that the rearranged locus adopts a different conformation or chromatin configuration, perhaps due to the deletion of intergenic sequences, that might facilitate the intrinsic activities of rearranged Vα promoters or that might unmask a novel enhancer activity in the locus. Any novel enhancer activity would have to be located upstream of the first V gene segment, TRAV1, or downstream of the last functional Jα segment, Jα2, to ensure its retention in the rearranged locus. A second possibility is that expression of the TCR–CD3 complex in resting cells might transduce tonic signals that can promote Eα-independent activation of the rearranged Vα promoters, activation of a novel enhancer, or both. With this in mind, it is notable that basal LAT-dyacylglycerol-RasGRP1 signals are necessary to maintain normal transcription of the rearranged Tcra locus and surface TCR–CD3 expression in Jurkat cells and naïve primary T cells (43). Moreover, rearranged Tcra transcription is responsive to treatments with phorbol esters and calcium ionophores (44–46). Locus reconfiguration and TCR-dependent signals are not necessarily mutually exclusive possibilities and could even operate synergistically to promote the transcription of rearranged Tcra genes. Additional work will clearly be required to decipher the molecular mechanisms responsible for rearranged Tcra gene expression in T cells.
Materials and Methods
Mouse Strains.
Rag2−/−, Rxßtg, DO11.10 and 5C.C7 were described (23–25, 47) and obtained from Taconic Farms. EαKI Rxßtg, βtg, and EαKI βtg mice were described (26). Animals were maintained in pathogen-free conditions within the Animal Houses at the Instituto de Parasitologia y Biomedicina “López-Neyra” - Consejo Superior de Investigaciones Científicas (CSIC) and at Duke University. Five- to 10-wk-old mice were used in all experiments. Animal use adhered to CSIC and Duke University Bioethical Guidelines.
Cell Isolation and Sorting.
For thymocyte cell sorting, total thymocytes were stained with fluorochrome-conjugated CD4 and CD8 Abs (BD Pharmingen). Samples were sorted using a FACSaria III (BD). Sorted thymocyte populations were 99% pure based on flow cytometry reanalysis. For T lymphocyte preparations, cell suspensions from LN and spleens were passed consecutively through two sterilized nylon wool columns or sorted using a FITC-conjugated TCRβ Ab (clone H57-597 from BD Pharmingen) using a FACSaria III (BD). Nylon wool-enriched T lymphocyte suspensions were around 85–90% pure and sorted T cells were 99% pure based on flow cytometry reanalysis. No γδ T lymphocytes were detected in the T lymphocyte preparations from RxDO11.10, βtg, and EαKI βtg mice based on flow cytometry analysis with a PE-conjugated γδ Ab (Miltenyi Biotec).
TCR-Mediated T Lymphocyte Stimulation.
For in vitro TCR-mediated cell stimulation, 1–1.5 × 106 T lymphocytes were suspended in RPMI medium 1640 supplemented with 10% (vol/vol) FCS, 50 μM 2-mercaptoethanol, and 20 ng/mL murine IL-2 (Peprotech) and stimulated with 1 μg/mL and 2 μg/mL of plate-bound CD3ε (145-2C11) and CD28 (37.51) Abs, respectively, in 24-well plates for 24 or 48 h at 37 °C with 5% CO2. Alternatively, 1–1.5 × 106 T lymphocytes were stimulated with 20 ng/mL of PMA and 0.5 μg/mL of ionomycin 24-well plates for 48 h at 37 °C with 5% CO2. For in vivo TCR-mediated cell stimulation, RxDO11.10 mice were s.c. injected with 200 μg of OVA peptide 323–339 (Ova 323–339) emulsified in CFA in a volume of 100 μL distributed between three sites on the back at the tail base. Mice were killed 24 or 48 h after immunization, and T lymphocytes were obtained as described above.
In Vitro Differentiation of Th0, Th1, Th2, and Th17 T Cells.
RxDO11.10 T lymphocytes were differentiated in vitro to Th0, Th1, Th2, and Th17 as described with minor modifications (48). In brief, 1 × 106 RxDO11.10 T cells were cultured in 1 mL of RPMI medium 1640 supplemented with 10% (vol/vol) FCS, 50 μM 2-mercaptoethanol in 24-well plates with plate-coated anti-CD3ε at 2 μg/mL in the presence of soluble anti-CD28 antibody at 1 μg/mL. For Th0 differentiation, cells were incubated in the presence of 20 ng/mL of IL-2; for Th1 differentiation, cells were incubated in the presence of 20 ng/mL of IL-2, 20 ng/mL of IL-12 (Peprotech), and 10 μg/mL of anti-IL-4 antibody (Biolegend); for Th2 differentiation, cells were incubated in the presence of 20 ng/mL IL-2, 100 ng/mL IL-4, and 10 μg/mL anti-IFNγ and 10 μg/mL anti-IL-12 (Biolegend); for Th17 differentiation, cells were incubated in the presence of 1 ng/mL of TGFβ and 100 ng/mL of IL-6 (Peprotech), and 10 μg/mL of anti-IFNγ and 10 μg/mL of anti-IL-12 (Biolegend). After 3 d, cells were transferred to new wells in the same medium and cultured for three more days.
RT-qPCR.
Total RNA was isolated from cells using peqGOLD TriFast (Peqlab) using the manufacturer’s protocol. The RNA was digested with 10 U of DNase I using the DNase I Treatment and Removal kit (Ambion) as recommended by the manufacturer. After DNase I inactivation, 200 U of SuperScript III Reverse Transcriptase (Invitrogen) and 300 ng of hexaprimers, in the presence of 60 U of the ribonuclease inhibitor RNAseOUT (Invitrogen), were used to synthesize cDNA from 2 to 5 μg of total RNA for 1 h at 50 °C, and resuspended in 100 μL of water. For qPCR, templates equivalent to 100–400 ng of RNA were assessed using iTaq Universal Sybr Green Supermix (Bio-Rad) in a Bio-Rad CFX96 thermocycler (Bio-Rad) or Kapa Sybr Fast (Kapa) in a Roche LightCycler 1.5 thermocycler (Roche). The following PCR programs were used: 95 °C for 5 min, followed by 46 cycles of 95 °C for 30 s, 59.5 °C (for proximal Vα, TEA exon, Jα, Tcfe2a, and Gata3 transcripts) or 59 °C (for Vβ transcripts) for 30 s, and 72 °C for 45 s. All PCR reactions were run in duplicate. The sequence primers used are listed in Table S1. The efficiency of the used primers was tested on genomic DNA (for Vα and Vβ transcripts) or cDNA templates (for Jα and Actb transcripts). Expression levels were normalized to levels of Actb in each sample.
3C.
3C experiments were performed as described with minor modifications (12). Briefly, 3 × 106 cells per mL were fixed in RPMI 1640 with 10% (vol/vol) FCS, 1.5% (vol/vol) formaldehyde for 10 min at 26 °C in a water bath, and fixation was stopped with glycine (0.125 M) in ice. Cells were washed twice in cold Hank’s buffered salt solution. In total, 107 sorted DP thymocytes or T lymphocytes from RxDO11.10 mice were lysed in 5 mL of 10 mM Tris pH 8.0, 10 mM NaCl, 5 mM MgCl2, 0.2% (vol/vol) Nonidet P-40, 1× complete protease inhibitor (Roche), and 1 mM PMSF for 30 min on ice to obtain the cellular nuclei. The nuclei were pelleted and resuspended in 0.5 mL 1.2× digestion buffer (NEB2, New England Biolabs) and permeabilized with 0.5% (wt/vol) SDS for 1 h at 37 °C, shaking at 900 rpm. After adding 3.3% (vol/vol) Triton X-100, nuclei were incubated for an additional 1 h at 37 °C, shaking at 900 rpm. Five microliters of each sample were reserved for further analysis of the digestion efficiency (undigested sample). Then, 500 U of HindIII (New England Biolabs) were added to each sample and incubated at 37 °C overnight, shaking at 900 rpm. Five hundred units of HindIII were added again to each sample, and the samples were incubated at 37 °C for 6 h, shaking at 900 rpm. Five microliters of each sample was reserved for further analysis of the digestion efficiency (digested sample). The restriction enzyme was inactivated by the addition of 1.5% (wt/vol) SDS and incubation at 65 °C for 30 min, shaking at 900 rpm. The reactions were diluted by adding 7 mL of 1.15x T4 ligase buffer (New England Biolabs) and 1% (vol/vol) Triton X-100, and were incubated for 1 h at 37 °C, mixing by vortexing every 10 min. Five hundred units T4 ligase (New England Biolabs) was added and incubated overnight at 16 °C. Three hundred micrograms of DNase-free proteinase K was added per sample and incubated overnight at 65 °C. Three hundred micrograms of RNase A were added per sample and incubated for 1 h at 37 °C. DNA was isolated by phenol/chloroform extraction, two chloroform extractions, and ethanol precipitation. The DNA pellet was resuspended in 150 μL of water, and the digestion efficiency was determined by qPCR on both undigested and digested purified DNA samples. The restriction efficiency was calculated as described in Fig. S2. The DNA sample concentration was compared with that of a reference sample of genomic DNA of known concentration using the Eα primers listed in Table S1. The efficiency and linearity of 3C primers (Table S1) were tested on templates obtained by HindIII digestion and ligation of genomic PCR products. To quantify each interaction, 3C samples were compared with serial dilutions of the control templates to obtain the crosslinking frequency between Eα and each fragment. Data were normalized to the crosslinking frequency between Eα and the control fragment to calculate the relative crosslinking frequencies. The following PCR program was used: 95 °C for 5 min, followed by 46 cycles of 95 °C for 30 s, 61 °C for 30 s, and 72 °C 45 s using iTaq Universal Sybr Green Supermix (Bio-Rad) in a Bio-Rad CFX96 thermocycler (Bio-Rad). All PCR reactions were run in duplicate.
qChIP.
To perform qChIP experiments to analyze histone modifications, either mononucleosomes were prepared and immunoprecipitated as described (26), or genomic DNA from 5 × 106 cells was fragmented by sonication and immunoprecipitated as described (7). Mononucleosomes were immunoprecipitated with 5 µg each of H3K4me3 (04-745), H3K4me2 (07-030), or H3ac (06-599) Abs (Millipore), or control rabbit IgG (ab-105-c) Abs (R&D Systems) (Fig. S4), and sonicated chromatin fragments were immunoprecipitated with 5 μg each of H3K27ac (ab4729), H3K4me3 (ab8580), or H3K4me1 (ab8895) Abs (Abcam), or control rabbit IgG (ab46540) Abs (Abcam) (Figs. 4 and 6). qChIP experiments to analyze TF binding, except for those for GATA-3, were performed as reported (7). For these experiments, sonicated chromatin fragments from 5 × 106 cells were immunoprecipitated with 5 μL of CTCF rabbit antiserum (07-729, Millipore), 10 µg each of HEB (sc-357), E12 (sc-762), E47 (sc-763), Fli-1 (sc-356), CREB-1 (sc-186), Est-1 (sc-350) Abs (Santa Cruz Biotechnology), or rabbit IgG control Ab (ab46540) (Abcam) and protein A agarose beads (16-156, Millipore). For GATA-3 immunoprecipitations from sonicated chromatin fragments from 5 × 106 cells, 2.5 μg of specific (sc-268; Santa Cruz Biotechnology) or control mouse IgG1 (M9035 clone MOPC31) Abs (Sigma-Aldrich) and 25 μL of anti-mouse secondary antibody-coupled magnetic beads (Dynabeads) were used as reported (36). Immunoprecipitated DNA was resuspended in 50 μL of water. For quantification of immunoprecipitated DNA by qPCR, 5 μL of Ab-bound DNA or 5 μL of input DNA resuspended in 250 μL of water were amplified in a LightCycler 2.0 thermocycler or Bio-Rad CFX96 thermocycler using the primers listed in Table S1. The efficiency of the primers was tested on genomic DNA. Analysis of MageA2 and Oct2 were used as negative controls (7, 21, 49). H3Ac, H3K4me2, and H3K4me3 levels were normalized to those of the Actb promoter in each sample. The following PCR programs were used: 95 °C for 5 min, followed by 46 cycles of 95 °C for 30 s, 59.5 °C for 30 s, and 72 °C for 45 s (Biorad CFX96 thermocycler) or 95 °C for 3 min, followed by 55 cycles of 95 °C for 10 s, 59.5 °C for 20 s, and 72 °C for 1 s (LightCycler 2.0).
Retroviral Transduction of T Lymphocytes.
The mouse stem cell virus-based bicistronic retroviral MigR-IRES-GFP (MigR) and the MigR-hE47 vectors were generous gifts from J. Aster, Harvard University, Cambridge, MA, and B. Kee, Chicago University, Chicago, respectively. Retroviral transduction of T lymphocytes was performed as reported (50) with minor modifications. For retrovirus production, 0.4–0.8 × 105 293T cells per well in six-well plates were transiently cotransfected with 4 µg of pCAG-ECO MLV envelope plasmid (35817 from Addgene), 4 µg of MigR1-hE47, and 1.6 µg of gag-pol plasmid using the LipoD293 DNA In Vitro Transfection Reagent (Signagen Laboratories). Two to 3 d after transfection, virus supernatants were collected. To transduce T lymphocytes, T cells were stimulated in vitro for 48 h as described above in the presence of 20 ng/mL of murine IL-2 to induce proliferation. In total, 106 proliferating T lymphocytes were resuspended with 1 mL of virus supernatant containing 20 ng/mL of IL-2 and 8 μg/mL of polybrene and were then plated in a well of a 24-well plate. The plate was centrifuged for 90 min at 2,000 × g at 32 °C with no brake. The infection procedure was repeated the next day. Transduced T lymphocytes were cultured in RPMI medium 1640 containing 10% (vol/vol) FCS and 20 ng/mL of IL-2 for 6–7 d at 37 °C with 5% CO2. Cells were then collected and sorted based on GFP expression in GFP−–, GFP-low–, and GFP-high–expressing cells using a FACSaria III (BD).
Statistical Analysis.
Statistical analysis was performed using Prism 5.0 software (GraphPad). Two-tailed Student’s t tests were used to compare the means between the samples and their respective controls. The P values are represented in the figures by asterisks (*P < 0.05, **P < 0.005, and ***P < 0.0005). The absence of an asterisk indicates that the change relative to control was not statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Q.-J. Li (Duke University) for providing Rx5C.C7 mice, Dr. J. C. Aster for providing the gag-pol and MigR1-GFP plasmids, Dr. B. Kee (University of Chicago) for providing the MigR1-hE47 plasmids, and Dr. C. Suñé for critical reading of the manuscript and helpful discussions. We also thank J. López Ros for technical support, S. J. Guerrero for help with the FACS, C. Sánchez and B. Ruiz for their help with our mouse colony, and F. Matesanz for her help with bioinformatic analysis of published data. This work was supported by the Spanish government (Grants BFU2009-08796 and BFU2013-44660R to C.H.-M. and a predoctoral FPI fellowship to B.d.B.), the Andalusian government (Grants CTS-6587 and CVI-4526 to C.H.-M. and a postdoctoral contract to B.d.B.), the European Regional Development Fund (ERDF/FEDER to C.H.-M.), and the National Institutes of Health (Grant R37 GM41052 to M.S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406551112/-/DCSupplemental.
References
- 1.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 3.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24(1):53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Ciofani M, et al. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol. 2004;172(9):5230–5239. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- 5.Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR α enhancer in αβ and γδ T cells. Immunity. 1997;7(4):505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 6.Hernández-Munain C, Sleckman BP, Krangel MS. A developmental switch from TCR δ enhancer to TCR α enhancer function during thymocyte maturation. Immunity. 1999;10(6):723–733. doi: 10.1016/s1074-7613(00)80071-0. [DOI] [PubMed] [Google Scholar]
- 7.del Blanco B, García-Mariscal A, Wiest DL, Hernández-Munain C. Tcra enhancer activation by inducible transcription factors downstream of pre-TCR signaling. J Immunol. 2012;188(7):3278–3293. doi: 10.4049/jimmunol.1100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz P, Cado D, Winoto A. A locus control region in the T cell receptor α/δ locus. Immunity. 1994;1(3):207–217. doi: 10.1016/1074-7613(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 9.Hawwari A, Krangel MS. Regulation of TCR δ and α repertoires by local and long-distance control of variable gene segment chromatin structure. J Exp Med. 2005;202(4):467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortiz BD, Cado D, Winoto A. A new element within the T-cell receptor α locus required for tissue-specific locus control region activity. Mol Cell Biol. 1999;19(3):1901–1909. doi: 10.1128/mcb.19.3.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih HY, et al. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci USA. 2012;109(50):E3493–E3502. doi: 10.1073/pnas.1214131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seitan VC, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476(7361):467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abarrategui I, Krangel MS. Regulation of T cell receptor-α gene recombination by transcription. Nat Immunol. 2006;7(10):1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 14.Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor α recombination. EMBO J. 2007;26(20):4380–4390. doi: 10.1038/sj.emboj.7601866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Y, et al. Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination. J Exp Med. 2010;207(13):2809–2816. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawwari A, Bock C, Krangel MS. Regulation of T cell receptor α gene assembly by a complex hierarchy of germline Jα promoters. Nat Immunol. 2005;6(5):481–489. doi: 10.1038/ni1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP. Defect in rearrangement of the most 5′ TCR-J α following targeted deletion of T early α (TEA): Implications for TCR α locus accessibility. Immunity. 1996;5(4):331–342. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- 18.Balmelle N, Zamarreño N, Krangel MS, Hernández-Munain C. Developmental activation of the TCR α enhancer requires functional collaboration among proteins bound inside and outside the core enhancer. J Immunol. 2004;173(8):5054–5063. doi: 10.4049/jimmunol.173.8.5054. [DOI] [PubMed] [Google Scholar]
- 19.Roberts JL, Lauzurica P, Krangel MS. Developmental regulation of VDJ recombination by the core fragment of the T cell receptor α enhancer. J Exp Med. 1997;185(1):131–140. doi: 10.1084/jem.185.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernández-Munain C, Roberts JL, Krangel MS. Cooperation among multiple transcription factors is required for access to minimal T-cell receptor α-enhancer chromatin in vivo. Mol Cell Biol. 1998;18(6):3223–3233. doi: 10.1128/mcb.18.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Blanco B, Roberts JL, Zamarreño N, Balmelle-Devaux N, Hernández-Munain C. Flexible stereospecific interactions and composition within nucleoprotein complexes assembled on the TCR α gene enhancer. J Immunol. 2009;183(3):1871–1883. doi: 10.4049/jimmunol.0803351. [DOI] [PubMed] [Google Scholar]
- 22.Harrow F, Ortiz BD. The TCRα locus control region specifies thymic, but not peripheral, patterns of TCRα gene expression. J Immunol. 2005;175(10):6659–6667. doi: 10.4049/jimmunol.175.10.6659. [DOI] [PubMed] [Google Scholar]
- 23.Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250(4988):1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 24.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176(4):1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinkai Y, et al. Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259(5096):822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 26.Jackson A, Kondilis HD, Khor B, Sleckman BP, Krangel MS. Regulation of T cell receptor β allelic exclusion at a level beyond accessibility. Nat Immunol. 2005;6(2):189–197. doi: 10.1038/ni1157. [DOI] [PubMed] [Google Scholar]
- 27.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pekowska A, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30(20):4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei G, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35(2):299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamane H, Paul WE. Early signaling events that underlie fate decisions of naive CD4+ T cells toward distinct T-helper cell subsets. Immunol Rev. 2013;252(1):12–23. doi: 10.1111/imr.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyazaki M, et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12(10):992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahiji A, et al. Complete TCR-α gene locus control region activity in T cells derived in vitro from embryonic stem cells. J Immunol. 2013;191(1):472–479. doi: 10.4049/jimmunol.1300521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spicuglia S, et al. TCRα enhancer activation occurs via a conformational change of a pre-assembled nucleo-protein complex. EMBO J. 2000;19(9):2034–2045. doi: 10.1093/emboj/19.9.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformation of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:476–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dose M, et al. β-Catenin induces T-cell transformation by promoting genomic instability. Proc Natl Acad Sci USA. 2014;111(1):391–396. doi: 10.1073/pnas.1315752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lepoivre C, et al. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics. 2013;14:914–934. doi: 10.1186/1471-2164-14-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinke FC, et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4+ T cell fate and interact with Runx3 to silence Cd4 in CD8+ T cells. Nat Immunol. 2014;15(7):646–656. doi: 10.1038/ni.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wotton D, Ghysdael J, Wang S, Speck NA, Owen MJ. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994;14(1):840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narayan K, et al. Immunological Genome Project Consortium Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat Immunol. 2012;13(5):511–518. doi: 10.1038/ni.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawwari A, Krangel MS. Role for rearranged variable gene segments in directing secondary T cell receptor α recombination. Proc Natl Acad Sci USA. 2007;104(3):903–907. doi: 10.1073/pnas.0608248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markegard E, et al. Basal LAT-diacylglycerol-RasGRP1 signals in T cells maintain TCRα gene expression. PLoS ONE. 2011;6(9):e25540. doi: 10.1371/journal.pone.0025540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindsten T, June CH, Thompson CB. Transcription of T cell antigen receptor genes is induced by protein kinase C activation. J Immunol. 1988;141(5):1769–1774. [PubMed] [Google Scholar]
- 45.Wilkinson MF, MacLeod CL. Induction of T-cell receptor-α and -β mRNA in SL12 cells can occur by transcriptional and post-transcriptional mechanisms. EMBO J. 1988;7(1):101–109. doi: 10.1002/j.1460-2075.1988.tb02788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallory MJ, et al. Signal- and development-dependent alternative splicing of LEF1 in T cells is controlled by CELF2. Mol Cell Biol. 2011;31(11):2184–2195. doi: 10.1128/MCB.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68(5):855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 48.Li J. 2011 In vitro differentiation of mouse Th0, Th1, Th2, and Th17 from naïve CD4 T cells. Bio-protocol 1(22):e157 ( www.bio-protocol.org/e157)
- 49.Hao B, Krangel MS. Long-distance regulation of fetal Vδ gene segment TRDV4 by the Tcrd enhancer. J Immunol. 2011;187(5):2484–2491. doi: 10.4049/jimmunol.1100468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong S, Malecek K, Pérez-Garcia A, Krogsgaard M. Retroviral transduction of T-cell receptors in mouse T-cells. J Vis Exp. 2010;44(44) doi: 10.3791/2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.