Significance
Coral reefs are highly productive ecosystems that raise a conundrum called “Darwin’s paradox”: How can high production flourish in low-nutrient conditions? We show here that in three abundant Caribbean sponges, the granules that have been commonly observed in sponge tissue for decades are polyphosphate granules. These granules can account for up to 40% of the total phosphorus (P) in sponge tissue. This finding has important implications for understanding P sequestration and recycling in the reef environment. We provide evidence that these granules are of bacterial origin and propose a P sequestration pathway by microbial symbionts and the sponge hosts. Considering the ancient origin of both partners, this process may have had an impact on the P cycle in Earth’s early history.
Keywords: phosphorus sequestration, sponge, microbial symbionts, polyphosphate
Abstract
Marine sponges are major habitat-forming organisms in coastal benthic communities and have an ancient origin in evolution history. Here, we report significant accumulation of polyphosphate (polyP) granules in three common sponge species of the Caribbean coral reef. The identity of the polyP granules was confirmed by energy-dispersive spectroscopy (EDS) and by the fluorescence properties of the granules. Microscopy images revealed that a large proportion of microbial cells associated with sponge hosts contained intracellular polyP granules. Cyanobacterial symbionts cultured from sponges were shown to accumulate polyP. We also amplified polyphosphate kinase (ppk) genes from sponge DNA and confirmed that the gene was expressed. Based on these findings, we propose here a potentially important phosphorus (P) sequestration pathway through symbiotic microorganisms of marine sponges. Considering the widespread sponge population and abundant microbial cells associated with them, this pathway is likely to have a significant impact on the P cycle in benthic ecosystems.
Marine sponges are sessile, benthic dwellers in the reef environment and harbor a high abundance of symbiotic microorganisms that can account for up to 40% of their body volume (1). High-throughput sequencing of bacterial 16S ribosomal RNA genes has revealed the enormous diversity and stability of the sponge bacterial community, which is distinct from that of the surrounding environment (1–3). Metagenomic and transcriptomic studies of the sponge microbiome have provided molecular evidence of the functional roles for these microbial cells (4, 5), including nutrient cycling between hosts and symbionts. Symbiotic cyanobacteria can transfer photosynthetic carbohydrates to sponges, presumably for the benefit of their hosts (6). Other studies have confirmed the presence of nitrogen and sulfur nutrient cycles mediated by the symbiotic microbial community (1, 4). Although phosphorous (P) is one of the potential limiting elements in oligotrophic environments, the P cycle inside marine sponges has been barely studied (1).
It is argued that P is the ultimate limiting nutrient in marine oligotrophic ecosystems (7, 8), and its biogeochemical cycle is strongly tied to the natural microbial community (9). Here, we report the observation of significant amounts of polyphosphate (polyP) granules with microbial origins in sponge mesohyl tissue and discuss the implications for the P biogeochemical cycle in the local environment.
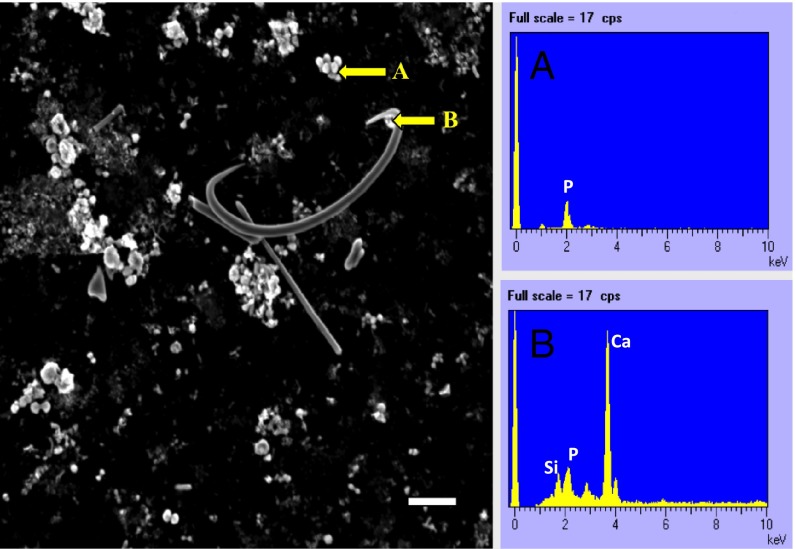
We collected samples of three abundant marine sponges Xestospongia muta, Ircinia strobilina, and Mycale laxissima from Conch Reef in Key Largo, Florida, during June 2011. Sponge tissues sections of ∼8 μm in thickness were stained with 10 μM of the fluorescent dye DAPI. Fluorescence microscopy images showed an abundance of yellow fluorescent granules (Fig. 1A), corresponding to the shifting fluorescent emission of polyP after DAPI staining (10). To confirm the identity of the granules further, SEM-energy dispersive spectroscopy (EDS) was applied to the granules separated from sponge tissue by Percoll density gradient. [It is reported that polyP granules are not well penetrated by embedding reagents and tend to be removed by microtome blades under traditional transmission EM thin-section protocols (11)]. Most of the granules were oval in shape, with various sizes ranging from 0.5 to 3 μm. They were visible as white, reflective bodies under SEM. A typical element point analysis on the granules yielded strong P signals, with a signature X-ray wavelength emitted from innermost shell of the atom (Kα) peak at 2.01 keV (Fig. 2B). The spectra for some granules showed association of the P signal with cation ions, such as calcium (Ca), with the signature peak at 3.69 keV (Fig. 2C), suggesting possible formation of apatite or phosphite precursors in the sponge mesohyl.
Fig. 1.
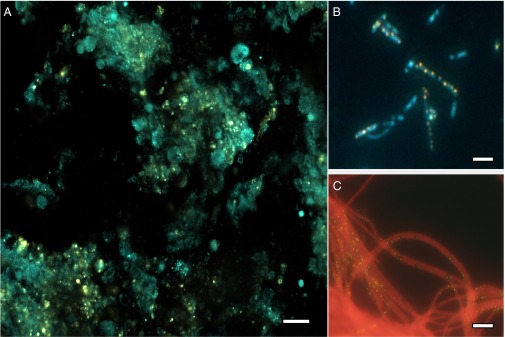
Yellow fluorescent granules observed in DAPI-stained samples. (A) X. muta tissue section under a confocal fluorescent microscope. (Scale bar: 20 μm.) (B) Bacterial cell fraction of the sponge X. muta under an epifluorescent microscope. (Scale bar: 1 μm.) (C) Enrichment culture of the filamentous cyanobacterial Leptolyngbya isolated from sponge I. stroblina tissue. (Scale bar: 1 μm.)
Fig. 2.
SEM image of a sponge Percoll fraction obtained from M. laxissima and the energy dispersive X-ray spectra from the corresponding spots indicated by the yellow arrows. (Scale bar: 10 μm.) (A) PolyP granules (spectra peak at 2.01 keV indicates P signal). (B) PolyP granules close to a spicule [three major spectra peaks were identified: a 1.74-keV peak for silicon (Si), a 2.01-keV peak for P, and a 3.69-keV peak for Ca].
The amount of polyP in the tissues of three sponge species was determined by extraction and measurement of fluorescent emission with DAPI staining (12). To make direct comparisons, we calculated the proportion of P in the form of polyp to the total P (Table 1). Results showed the polyP pool could account for up to 40% of total P in sponge tissue. At our sampling site, the concentration of P in the water column ranged from 0.1 to 0.5 μM, with a high nitrogen-to-P ratio, an indication of possible P limitation (13). It was recently reported in the oligotrophic Sargasso Sea that accumulation of polyP in phytoplankton enhanced the recycling of P in the local environment (14), and polyP metabolism can be important in the ocean (15). Considering the prevalence of sponges in the coral reef and the very low P concentration in the surrounding seawater, the amount of P sequestered within sponges is likely to be a significant proportion of the total P in the ecosystem.
Table 1.
PolyP and total P weight fraction in marine sponges (n = 3 for each sample)
| Sponge sample | PolyP ± SD, % dry weight | Total P ± SD, % dry weight | PolyP/total P, % |
| X. muta | |||
| Epidermal layer | 0.080 ± 0.010 | 0.325 ± 0.042 | 24.6 |
| Mesohyl layer | 0.075 ± 0.011 | 0.298 ± 0.034 | 25.2 |
| I. strobilina | 0.092 ± 0.012 | 0.342 ± 0.116 | 26.9 |
| Whole sponge | |||
| M. laxissima | 0.126 ± 0.015 | 0.314 ± 0.038 | 40.1 |
| Whole sponge |
PolyP is found in almost all organisms, ranging from bacteria to plants and animals (16). The biochemistry of polyP has been extensively studied in prokaryotes, where its major roles are considered to be storing P and serving as an energy reserve (16). The main polyP synthesis gene in prokaryotes is polyphosphate kinase (ppk). Polyphosphate kinase reversibly polymerizes phosphate to form high-energy bond polymers and is widespread in diverse prokaryote species. The gene responsible for the synthesis of polyP in eukaryotes was identified as the integral membrane vacuolar transporter chaperone complex (vtc4 gene) in yeast and diatoms (17). We searched the genome of the sponge Amphimedon queenslandica for potential ppk and vtc4 genes. BLAST results gave only one hit for the ppk gene. The sequence has no intron and is closely related to sequences from chemolithoautotrophic bacteria at around 60% amino acid similarity.
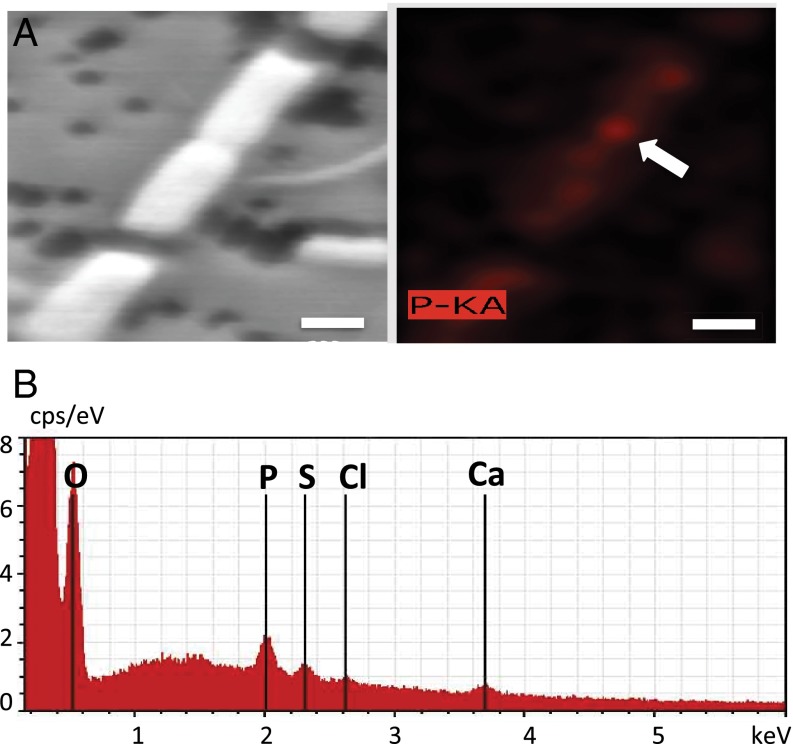
We separated the microbial community from host tissues by Percoll density gradient centrifugation and found a high proportion (38 ± 11%, n = 12) of microbial cells possessed intracellular polyP granules (Fig. 1B). Interestingly, we also obtained an enrichment culture of diazotrophic cyanobacteria from sponge tissue with the ability to accumulate polyP (Fig. 1C). EDS mapping showed P hot spots in bacterial cells, and spectra on the point of interest showed major Kα peaks for the elements P, oxygen (O), and Ca (Fig. 3). We were also able to amplify the ppk gene from the DNA of all three sponge species and from cDNA of X. muta and I. strobilina, indicating the existence and expression of the ppk gene in those samples. Amplicons of ppk genes from sponge I. strobilina were cloned. Sequence results showed that those genes were related to autotrophic nitrogen-fixing cyanobacteria, aerobic anoxygenic phototrophic bacteria, and sulfur cycle-related Proteobacteria (Table S1), suggesting that these microbial symbionts might be able to obtain energy through phototrophic or chemotrophic pathways, potentially providing the energy needed for polyP synthesis.
Fig. 3.
SEM image of a cyanobacterial enrichment culture from the sponge I. strobilina and energy dispersive X-ray elemental analysis on the corresponding area. (A) SEM image of the filamentous cyanobacterial Leptolyngbya sp. (Left) and a corresponding heat map of P signal (Right). (Scale bar: 500 nm.) (B) EDS spectra of the point of interest (white arrow in A) show a P signal, along with signature peaks from other elements, such as Ca, chlorine (Cl), sulfur (S), and O.
Sponge Hosts Provide a Suitable Redox Environment for PolyP Accumulation
The presence of polyP and Ca phosphate granules in sponge mesohyl and the polyP accumulation in their microbial symbionts suggest that there is a potential efficient P sequestration mechanism in the sponge holobiont. Phosphate minerals in marine sediment are a major sink for the removal of reactive phosphate from the water column (18). However, the mechanism responsible for the formation of those minerals is not fully characterized. One important factor for apatite formation is a high concentration of the component ions in the local environment. Research suggests that a pulse of phosphate accumulation in sedimentary pore water could trigger the formation of apatite (18). It is known that biogenic apatite formation is also promoted by coupling with iron chemistry and polyP cycling in suitable redox gradients (19). Previous studies suggested that giant sulfur bacteria in the upwelling environment (20, 21) and diatoms in the water column (22) significantly contribute to this process through polyP accumulation.
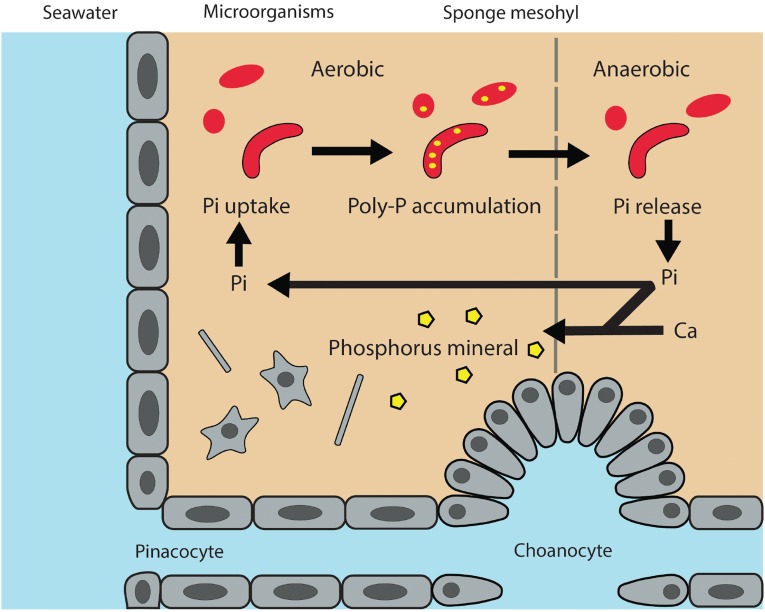
Combining our observations and known factors in P-containing mineral formation, we here propose a working model of P mineralization through the sponge–symbiont system in the benthic environment (Fig. 4). First, filter-feeding by sponges efficiently removes particles between 0.2 and 10 μm in the water column, a fraction dominated by marine picoplankton and nanoplankton (23). Limited nutrient flux studies compared dissolved phosphate content before and after water flow through sponges. Results suggested that sponges are a minor source of phosphate (24); a large proportion of particulate P is therefore likely retained by the sponge holobiont. With the high-affinity phosphate transporters and phosphatases that are commonly found in bacteria (25), sponge microbial symbionts may have the “luxury” of acquiring excess phosphate. Constant water flow facilitates O penetration in sponge tissues and creates a redox gradient. During periods of active pumping, the mesohyl is oxygenated and the bacterial symbionts can convert the excess phosphate to the polyP storage form. When pumping activities cease, the O level in the mesohyl can be quickly depleted (26), which could lead to polyP degradation for energy production, generating a pulse of phosphate release. The induction of polyP release as redox conditions shift from oxic to suboxic/anoxic conditions is well demonstrated in modern activated sludge sewage treatment plants, where the microbially mediated phosphate release and polyP accumulation are controlled by alternating aerobic and anaerobic phases (27). Phosphate released to the water column by sponge-associated microbes will be quickly diluted and consumed by P-starved microorganisms. In contrast, the porous matrix within the sponge might increase the chance of local ion accumulation. The transformation to apatite may be promoted in the porous microenvironment within the sponge mesohyl, where pulses of phosphate released from polyP degradation in anaerobic conditions may result in high concentrations of dissolved phosphate in tiny compartments, a scenario similar to the scenario in the sediment, where elevated phosphate concentration in pore water triggers mineral formation (28). Alternatively, high concentrations of polyP combined with Ca in the sponge mesohyl can act as a nucleating template to form apatite (29). This production of apatite from polyP in sponges may be one of the sources of the granules of biogenic apatite commonly observed in marine sediments (21, 22).
Fig. 4.
Conceptual diagram illustrating the uptake, accumulation, and release of phosphate (Pi), as well as potential P biomineralization through polyP granule formation.
We propose in our conceptual diagram (Fig. 4) that the cycling of P inside sponges might favor biomineralization over biological activities, divert phosphate from re-entering the water column, and increase the burial efficiency of P in the form of apatite, resulting in long-term geological sequestration. This P sequestration pathway raises several questions for future investigation: (i) Is the polyP pool in sponges transient with spikes in the dissolved P pulse being released in the sponge “effluent”? (ii) Are polyP granules released from sponges, thereby resulting in sequestration of particulate P in the surrounding sediments? (iii) If so, how significant is this removal by sponges of previously bioavailable P relative to other fluxes of particulate P to the sediment? (iv) Is apatite formation in sponges a significant pathway for marine P sequestration?
Marine primary production is thought to be constrained by nitrogen availability in the short term but subject to P limitation on longer geological time scales, based on the assumption that biological nitrogen fixation should be able to keep pace with the P supply in the long run. P in marine ecosystems is supplied by riverine input and is eventually lost to sedimentation in the ocean floor (30). With increasing P input from anthropogenic mining, bacterial polyP accumulation in sponges can create an important avenue that removes excessive P from the water column to marine sediment burial. Repeating oscillation between anoxic and oxic states in the sponge mesohyl might drive selection of polyP-accumulating organisms within the symbiotic community. These bacterial assemblages in sponges may serve as a new source to explore the diversity of phosphate-accumulating microorganisms and to discover novel ppk genes with potential for bioremediation applications (31).
As one of the most primitive animals on Earth (32), sponges can maintain growth under very low-O conditions (33), an environment that likely prevailed in the continental sea floor during the Early Neoproterozoic era. It has been proposed that marine sponges may have played a vital role in the removal of phosphate to marine sediment and the expansion of the oxic zone at the ocean bottom, eventually paving the way for animal evolution (34). Based on our findings, microbial symbionts in sponges may have made significant contributions to this benthic P cycle in early Earth history. The recently proposed concept of a “sponge loop” suggests that sponges can efficiently convert dissolved nutrients to particle forms through shedding cells, thus enhancing elemental cycles and retaining nutrients in the local environment (35, 36). Our study suggests that microbial symbionts can contribute to this process through polyP metabolism. With the wide distribution of sponges in the oceans and their increasing population in many coral reef sites (37), sponges and their diverse microbial communities play a significant role in mediating element cycling in the interface between the water column and marine sediment.
Materials and Methods
Sample Collection, Sponge Tissue Section, and Cell Separation.
Marine sponge samples of X. muta, I. strobilina, and M. laxissima were collected from Conch Reef in Key Largo, Florida (24° 57.11′ N, 80° 27.57′ W), in July 2011 by self-contained underwater breathing apparatus (SCUBA) diving. Sponge tissue samples were rinsed three times with artificial seawater, fixed with 4% (wt/vol) paraformaldehyde at room temperature for 2 h, rinsed with 20 mM Tris buffer (pH 7.0), and then transferred to 70% (vol/vol) ethanol at −20 °C until use. Fixed tissues were embedded in Cryomold (Sakura) with Tissue-Tek O.C.T medium (Sakura) and solidified on dry ice. Sponge sections of 6–10 μM were made on a cryostat microtome (Sakura Tissue-Tek Cryo3) under −30 °C and mounted on Superfrost Plus slides (Electron Microscopy Science). Percoll density gradient centrifugation was used to separate microbial cells from the sponge tissue. Paraformaldehyde-fixed sponge tissue preparations were loaded on top of discontinuous gradients containing 10%, 25%, 50%, and 100% (vol/vol) Percoll, respectively, and then centrifuged at 800 × g for 15 min at 4 °C. The gradient allowed the enrichment of microbial cells in the 10% and 25% fractions, and sponge cells and spicules were enriched in the 50% and 100% portions of the gradient tube.
Extraction and Measurement of Total PolyP from Sponge Samples.
The extraction and measurement of polyP followed the protocol described by Martin and Van Mooy as their “core” protocol (12). Lyophilized sponge tissue (100 mg) from three individuals of each species was finely ground, dissolved in 20 mM Tris buffer (pH 7.0), homogenized by vortexing, sonicated for 15 s, immersed in boiling water for 5 min, and then sonicated again for 15 s. The samples were treated with 40 units/mL DNase (New England BioLabs) and 400 units/mL RNase (New England BioLabs) at 37 °C for 10 min to remove DNA and RNA. Proteinase K (Fermentas) was added to a concentration of 20 mg/mL, and samples were incubated at 37 °C for 30 min and centrifuged at 16,100 × g for 1 min. Supernatants were collected, and the remaining pellets were subjected to four additional successive extractions following the same protocol (Fig. S1). Supernatants collected from the first four extractions were pooled for polyP fluorometric measurement. The last extraction was used to determine the background fluorescence generated by the interaction between DAPI and other substances in the extracts. The matrix effect was evaluated by adding 2 nmol of polyP standard into the extraction solution. This effect inflated the signal by 5–12%. Fluorescence data were adjusted accordingly, and polyP concentrations were calculated based on the equation in the supplemental information of the study by Martin and Van Mooy (12). Total P was determined using standard chemical techniques (38) by the Nutrient Analytical Service laboratory (Horn Point Laboratory, University of Maryland Center for Environmental Science).
The binding of polyP to DAPI shifts the peak DAPI emission wavelength from 475–550 nm under UV-light excitation, and the fluorescence intensity at this shifted wavelength is proportional to the concentration of polyP. Fluorescence measurements were carried out on a FluoroMax-4 spectrofluorometer (Horiba Scientific). The excitation wavelength was set to 415 nm with a 3-nm slit to minimize the interfering signal from free DAPI in the solution; emission wavelength was 550 nm with a 3-nm slit. Fluorescence emission spectra were recorded from 425–600 nm in 1-nm increments. PolyP standard was purchased from Sigma–Aldrich (S4379). Aliquots were dissolved in 20 mM Tris buffer (pH 7.0). The working solution for DAPI staining was 100 μM, and the solution was added to the samples at a 1:10 dilution to give a final DAPI concentration of 10 μM. Samples were mixed by vortex, incubated at room temperature for 5 min, mixed again by pipetting, and transferred to standard optical quartz cuvettes for fluorescence spectral recording. The quantification of polyP is based on the units of orthophosphate in solution; that is, the moles of polyP presented indicate the moles of orthophosphate regardless of the chain length of different polyP species (Fig. S2). The fluorescence signal is presented as thousands of counts per second in all related figures and tables.
DNA/RNA Extraction, PCR Amplification, and Cloning of ppk Gene.
Total genomic DNA and RNA from three sponge species were extracted using a TissueLyser System (Qiagen) and an AllPrep DNA/RNA Mini Kit (Qiagen) with an RNase-free DNase (Qiagen) treatment step for RNA samples. RT of RNA was performed using RevertAid Reverse Transcriptase (Thermo Scientific) with random primers and following the manufacturer’s protocol. The ppk genes were amplified from genomic DNA and cDNA using primers designed by McMahon et al. (39), with the following conditions: 94 °C for 5 min; 35 cycles of 94 °C for 1 min, 50 °C for 45 s, and 72 °C for 2 min; and a final extension at 72 °C for 12 min. RNA samples without the RT step were included as a PCR template to check for residual DNA in the RNA samples. PCR products of about 1,300 bp in length were ligated into PCR-XL-TOPO vectors and transformed into ONEShot TOP10 chemically competent Escherichia coli cells using the TOPO XL PCR Cloning Kit (Invitrogen Life Technologies). Plasmid DNA was extracted from individual clones, purified by a Mini Prep Spin Kit (Qiagen), and sequenced by an ABI PRISM 3130XL genetic analyzer (Applied Biosystems). Sequences of the ppk gene obtained in this study were deposited in the GenBank database under accession nos. KP233195–KP233199.
Visualization of PolyP Granules by Fluorescence Microscopy.
The sponge tissue sections on Superfrost Plus slides (Electron Microscopy Science) were stained by 10 μM DAPI solution for 2 min, washed with MilliQ water (Millipore) three times, dehydrated by ethanol, and then imaged under a Zeiss LSM510 duo inverted confocal microscope equipped with a 40× Zeiss Plan Neofluar objective lens (1.3 N.A.) in the Department of Physiology, University of Maryland, Baltimore. Sample sections were excited by a 405-nm laser light source, and fluorescent emission signals were separated by a NFT515 (NebenFarbTeiler 515) filter into two channels. Channel 1 collected the emission wavelength from 420–515 nm, representing nucleotide-DAPI signals. Channel 2 was set up to collect the emission wavelength above 530 nm, representing a shifting peak of 550 nm for polyP-DAPI signals. Fig. 1A shows an image merged from both channels.
Bacterial cell fractions obtained from sponge tissue and cyanobacterial enrichment culture were stained with 10 μM DAPI solution for 2 min, filtered onto a 0.2-μm polycarbonate filter (Millipore), washed with MilliQ water three times, and imaged by an Axioplan universal microscope (Zeiss) equipped with a UV-G 365 filter set and an X-Cite series 120Q light source (Lumen Dynamic).
SEM-EDS.
The Percoll fraction enriched with polyP signal under epifluorescence microscopy was selected and filtered onto a 0.2-μm black polycarbonate filter. The filter samples were mounted on the graphite substrate, coated by carbon, and imaged under a JEOL 5700 Scanning Electron Microscope under voltage of 10 kV. Energy dispersive X-ray spectroscopy was used to determine element composition on a focus point. Cyanobacterial enrichment culture was chemically fixed as described above. A P signal heat map of cyanobacteria filaments and corresponding SEM images were obtained using an Hitachi SU-70 Ultra High Resolution Field Emission Scanning Electron Microscope under 10 kV.
Supplementary Material
Acknowledgments
We thank Jan Vicente and Jindong Zan for helping with sample collection in Key Largo, FL, and Joe Pawlik for providing a research cruise opportunity. We thank S. Nazar for sequencing support and J. Mauban (Department of Physiology, University of Maryland, Baltimore) for assisting with confocal microscopy. We acknowledge Dr. W. A. Chiou and J. P. Padilla (Maryland NanoCenter and NispLab) and Dr. L. Takacs (University of Maryland, Baltimore County) for their SEM-EDS analysis support. This research was funded by National Science Foundation Grant IOS-0919728 (to R.T.H.) and a Fulbright S&T fellowship (to F.Z.). This paper is University of Maryland Center for Environmental Science contribution no. 4985 and Institute of Marine and Environmental Technology contribution no. 15-142.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP233195–KP233199).
See Commentary on page 4191.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423768112/-/DCSupplemental.
References
- 1.Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71(2):295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster NS, et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12(8):2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Easson CG, Thacker RW. Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front Microbiol. 2014;5:532. doi: 10.3389/fmicb.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hentschel U, Piel J, Degnan SM, Taylor MW. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol. 2012;10(9):641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- 5.Fan L, et al. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci USA. 2012;109(27):E1878–E1887. doi: 10.1073/pnas.1203287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkinson CR. Net primary productivity in coral reef sponges. Science. 1983;219(4583):410–412. doi: 10.1126/science.219.4583.410. [DOI] [PubMed] [Google Scholar]
- 7.Deutsch C, Sarmiento JL, Sigman DM, Gruber N, Dunne JP. Spatial coupling of nitrogen inputs and losses in the ocean. Nature. 2007;445(7124):163–167. doi: 10.1038/nature05392. [DOI] [PubMed] [Google Scholar]
- 8.Björkman KM. Polyphosphate goes from pedestrian to prominent in the marine P-cycle. Proc Natl Acad Sci USA. 2014;111(22):7890–7891. doi: 10.1073/pnas.1407195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karl DM. Microbially mediated transformations of phosphorus in the sea: New views of an old cycle. Annu Rev Mar Sci. 2014;6(1):279–337. doi: 10.1146/annurev-marine-010213-135046. [DOI] [PubMed] [Google Scholar]
- 10.Aschar-Sobbi R, et al. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc. 2008;18(5):859–866. doi: 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- 11.Seki Y, Nitta K, Kaneko Y. Observation of polyphosphate bodies and DNA during the cell division cycle of Synechococcus elongatus PCC 7942. Plant Biol (Stuttg) 2013;16(1):258–263. doi: 10.1111/plb.12008. [DOI] [PubMed] [Google Scholar]
- 12.Martin P, Van Mooy BA. Fluorometric quantification of polyphosphate in environmental plankton samples: Extraction protocols, matrix effects, and nucleic acid interference. Appl Environ Microbiol. 2013;79(1):273–281. doi: 10.1128/AEM.02592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer JN, Briceño HO. 2010 Annual Rreport of the Water Quality Monitoring Project for the Water Quality Protection Program of the Florida Keys National Marine Sanctuary. US Fish and Wildlife Service; Miami, FL: 2011. [Google Scholar]
- 14.Martin P, Dyhrman ST, Lomas MW, Poulton NJ, Van Mooy BA. Accumulation and enhanced cycling of polyphosphate by Sargasso Sea plankton in response to low phosphorus. Proc Natl Acad Sci USA. 2014;111(22):8089–8094. doi: 10.1073/pnas.1321719111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Temperton B, Gilbert JA, Quinn JP, McGrath JW. Novel analysis of oceanic surface water metagenomes suggests importance of polyphosphate metabolism in oligotrophic environments. PLoS ONE. 2011;6(1):e16499. doi: 10.1371/journal.pone.0016499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornberg A, Rao NN, Ault-Riché D. Inorganic polyphosphate: A molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 17.Hothorn M, et al. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science. 2009;324(5926):513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- 18.Delaney ML. Phosphorus accumulation in marine sediments and the oceanic phosphorus cycle. Global Biogeochem Cycles. 1998;12(4):563–572. [Google Scholar]
- 19.Goldhammer T, Bruchert V, Ferdelman TG, Zabel M. Microbial sequestration of phosphorus in anoxic upwelling sediments. Nat Geosci. 2010;3(8):557–561. [Google Scholar]
- 20.Bailey JV, Joye SB, Kalanetra KM, Flood BE, Corsetti FA. Evidence of giant sulphur bacteria in Neoproterozoic phosphorites. Nature. 2007;445(7124):198–201. doi: 10.1038/nature05457. [DOI] [PubMed] [Google Scholar]
- 21.Schulz HN, Schulz HD. Large sulfur bacteria and the formation of phosphorite. Science. 2005;307(5708):416–418. doi: 10.1126/science.1103096. [DOI] [PubMed] [Google Scholar]
- 22.Diaz J, et al. Marine polyphosphate: A key player in geologic phosphorus sequestration. Science. 2008;320(5876):652–655. doi: 10.1126/science.1151751. [DOI] [PubMed] [Google Scholar]
- 23.Pile AJ, Patterson MR, Witman JD. In situ grazing on plankton <10 μm by the boreal sponge Mycale lingua. Mar Ecol Prog Ser. 1996;141:95–102. [Google Scholar]
- 24.Ribes M, et al. Functional convergence of microbes associated with temperate marine sponges. Environ Microbiol. 2012;14(5):1224–1239. doi: 10.1111/j.1462-2920.2012.02701.x. [DOI] [PubMed] [Google Scholar]
- 25.Wanner BL. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt C, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 1357–1381. [Google Scholar]
- 26.Hoffmann F, Larsen O, Tore Rapp H, Osinga R. Oxygen dynamics in choanosomal sponge explants. Mar Biol Res. 2005;1(2):160–163. [Google Scholar]
- 27.McMahon KD, Read EK. Microbial contributions to phosphorus cycling in eutrophic lakes and wastewater. Annu Rev Microbiol. 2013;67(1):199–219. doi: 10.1146/annurev-micro-092412-155713. [DOI] [PubMed] [Google Scholar]
- 28.Ruttenberg KC, Berner RA. Authigenic apatite formation and burial in sediments from non-upwelling, continental margin environments. Geochim Cosmochim Acta. 1993;57(5):991–1007. [Google Scholar]
- 29.Hirschler A, Lucas J, Hubert J-C. Bacterial involvement in apatite genesis. FEMS Microbiol Lett. 1990;73(3):211–220. [Google Scholar]
- 30.Paytan A, McLaughlin K. The oceanic phosphorus cycle. Chem Rev. 2007;107(2):563–576. doi: 10.1021/cr0503613. [DOI] [PubMed] [Google Scholar]
- 31.Imsiecke G, et al. Inorganic polyphosphates in the developing freshwater sponge Ephydatia muelleri: Effect of stress by polluted waters. Environ Toxicol Chem. 1996;15(8):1329–1334. [Google Scholar]
- 32.Li C-W, Chen J-Y, Hua T-E. Precambrian sponges with cellular structures. Science. 1998;279(5352):879–882. doi: 10.1126/science.279.5352.879. [DOI] [PubMed] [Google Scholar]
- 33.Mills DB, et al. Oxygen requirements of the earliest animals. Proc Natl Acad Sci USA. 2014;111(11):4168–4172. doi: 10.1073/pnas.1400547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenton TM, Boyle RA, Poulton SW, Shields-Zhou GA, Butterfield NJ. Co-evolution of eukaryotes and ocean oxygenation in the Neoproterozoic era. Nat Geosci. 2014;7(4):257–265. [Google Scholar]
- 35.de Goeij JM, et al. Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science. 2013;342(6154):108–110. doi: 10.1126/science.1241981. [DOI] [PubMed] [Google Scholar]
- 36.Richter C, Wunsch M, Rasheed M, Kötter I, Badran MI. Endoscopic exploration of Red Sea coral reefs reveals dense populations of cavity-dwelling sponges. Nature. 2001;413(6857):726–730. doi: 10.1038/35099547. [DOI] [PubMed] [Google Scholar]
- 37.Bell JJ, Davy SK, Jones T, Taylor MW, Webster NS. Could some coral reefs become sponge reefs as our climate changes? Glob Change Biol. 2013;19(9):2613–2624. doi: 10.1111/gcb.12212. [DOI] [PubMed] [Google Scholar]
- 38.Aspila KI, Agemian H, Chau ASY. A semi-automated method for the determination of inorganic, organic and total phosphate in sediments. Analyst (Lond) 1976;101(1200):187–197. doi: 10.1039/an9760100187. [DOI] [PubMed] [Google Scholar]
- 39.McMahon KD, Dojka MA, Pace NR, Jenkins D, Keasling JD. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl Environ Microbiol. 2002;68(10):4971–4978. doi: 10.1128/AEM.68.10.4971-4978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.