Significance
RecQ DNA helicases are critical enzymes for the maintenance of genome integrity. Here, we determined the first DNA complex structures, to our knowledge, of the human RECQ1 helicase. These structures provide new insight into the RecQ helicase mechanism of DNA tracking, strand separation, strand annealing, and Holliday junction (HJ) branch migration. We identified a surface region in the winged-helix domain of RECQ1 that is important for both dsDNA recognition and HJ resolution, and we used a combination of biochemical, analytical ultracentrifugation, and EM experiments to begin elucidating the molecular basis of the distinct HJ resolution activities of human RecQ helicases.
Keywords: DNA helicases, RecQ, genome stability, Holliday junction, fork reversal
Abstract
RecQ helicases are a widely conserved family of ATP-dependent motors with diverse roles in nearly every aspect of bacterial and eukaryotic genome maintenance. However, the physical mechanisms by which RecQ helicases recognize and process specific DNA replication and repair intermediates are largely unknown. Here, we solved crystal structures of the human RECQ1 helicase in complexes with tailed-duplex DNA and ssDNA. The structures map the interactions of the ssDNA tail and the branch point along the helicase and Zn-binding domains, which, together with reported structures of other helicases, define the catalytic stages of helicase action. We also identify a strand-separating pin, which (uniquely in RECQ1) is buttressed by the protein dimer interface. A duplex DNA-binding surface on the C-terminal domain is shown to play a role in DNA unwinding, strand annealing, and Holliday junction (HJ) branch migration. We have combined EM and analytical ultracentrifugation approaches to show that RECQ1 can form what appears to be a flat, homotetrameric complex and propose that RECQ1 tetramers are involved in HJ recognition. This tetrameric arrangement suggests a platform for coordinated activity at the advancing and receding duplexes of an HJ during branch migration.
RecQ helicases are a family of ATP-dependent motor proteins that play central roles in maintaining genome stability. Defects in three of the five human RecQ homologs give rise to distinct genetic disorders associated with genomic instability, cancer predisposition, and premature aging (1–5). The unique clinical features of these disorders support the notion that the different RecQ helicases have nonoverlapping functions, but the molecular basis for their different enzymatic activities remains unclear. RecQ helicases catalyze ATP-dependent DNA unwinding in the 3′–5′ direction. Additionally, members of this helicase family have been shown to tackle an unparalleled breath of noncanonical DNA structures, such as fork DNA, G-quadruplexes, D-loops, and Holliday junction (HJ) structures (6–8). However, our understanding of the physical mechanisms by which RecQ helicases recognize and process their physiological substrates remains remarkably limited.
RECQ1 is the shortest of the human RecQ-family helicases, comprising the bipartite ATPase domain common to all superfamily 2 (SF2) helicases, the RecQ-specific C-terminal domain (RQC), and short extensions on the N and C termini. We recently discovered a specific function of RECQ1 in branch migration and restart of reversed DNA replication forks upon DNA topoisomerase I inhibition that is not shared by other human RecQ helicases, such as Werner (WRN) or Bloom (BLM) syndrome proteins (9). On the other hand, BLM is the sole human RecQ helicase member specifically able to resolve double-HJ junction structures in conjunction with DNA topoisomerase III alpha and the RMI1 and RMI2 accessory proteins (10–12). These findings lead us to hypothesize that the specialized functions of each human RecQ protein in HJ resolution and reversed fork restart arise from key structural properties that are unique to each protein.
In this work, we determined the first X-ray structures, to our knowledge, of two DNA-bound forms of human RECQ1. These structures complement earlier DNA-free forms that we determined of the RECQ1 catalytic core (2.0-Å resolution for the ADP-bound form) (13). Comparisons of these structures with new structures recently determined for both bacterial and human RecQ helicases in their DNA-bound and unbound forms reveal important insights into conformational changes linked to DNA binding, translocation, and unwinding. We also used biochemical, analytical ultracentrifugation, and EM tools to describe a tetrameric form of RECQ1, which has a role in binding an HJ. We posit that different quaternary structures might account for the specialized functions of each human RecQ protein in genome maintenance.
Results
Structure of a Complex of RECQ1 with a Tailed-Duplex DNA.
The human RECQ1 protein used in crystallization was a truncated form, containing amino acids 49–619 and lacking 48 and 23 amino acids at the N and C termini, respectively. The truncated RECQ1 protein, termed RECQ1T1, contains the conserved catalytic core of RecQ helicases and is fully active in unwinding forked DNA substrates. The protein was crystallized in complex with a pair of DNA oligonucleotides comprising a short (13 nt) oligo (“bottom”) fully base-paired to a longer (27 nt) oligo (“top”), with 7-nt extensions at the 5′ and 3′ ends. The final model, refined to a resolution of 2.9 Å, contains a RECQT1 dimer comprising residues 63–592 (residues 64–593 for chain B). Each chain also contains a zinc ion, two sulfates, and an ethylene glycol molecule. A single-stranded tailed duplex is associated with each chain, with the shorter oligo being fully resolved (13 bases) and the longer 27-nt strand being partially ordered (nucleotides 6–26 resolved). In addition, a nucleotide from the 5′ end of the longer strand of an adjacent molecule in the crystal [identified in the Protein Data Bank (PDB) file as residue G3 from chain O] interacts with the C21 at the β-hairpin junction of RECQ1, in a configuration that may resemble an extension of the chain (Fig. S1A).
RECQ1T1 exhibits four structurally defined domains, identified by color coding in Fig. 1A. Two RecA-like domains (D1 and D2) contain the widely conserved helicase/ATPase motifs; the nucleotide-binding site is located between these two domains, as shown in crystal structures of RECQ1/ADP complex [PDB ID code 2V1X (13)], as well as in other SF1 and SF2 helicases [PDB ID codes 3PJR and 2DB3 (14, 15)]. In the present structure, a sulfate ion derived from the crystallization solution occupies a position equivalent to the position of the β-phosphate of the ADP (Fig. S1B). Further downstream is a region characteristic of RECQ-family helicases, termed the RQC. This region is composed of two separately folded domains, a Zn-binding domain and a winged-helix (WH) domain. A prominent β-hairpin motif, which forms a wing of the WH domain, has been shown to be critical for the helicase activity of RECQ1, but its deletion had no effect on ssDNA-dependent ATPase activity (13).
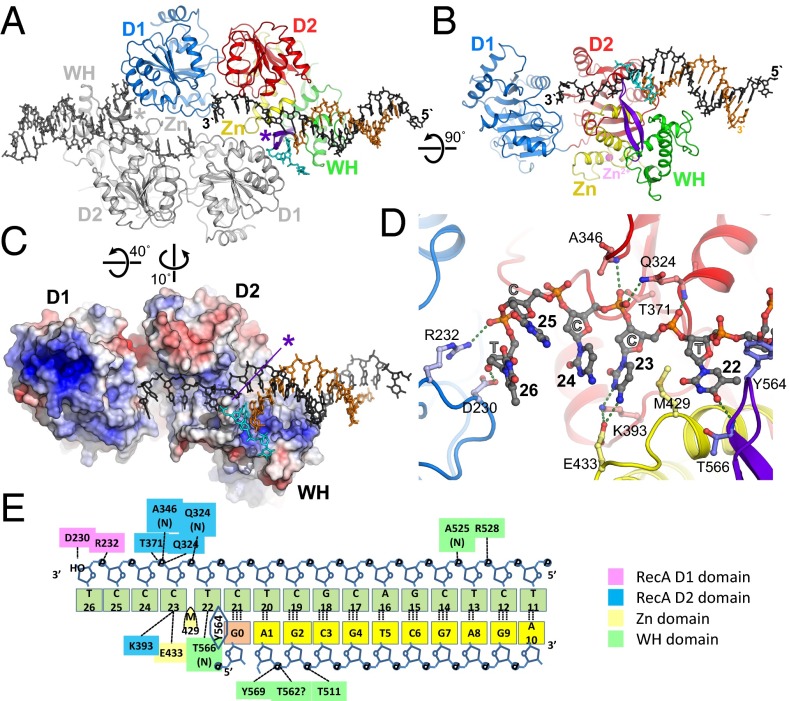
Fig. 1.
Overall structure of the RECQ1/DNA complex and trajectory of the ssDNA tail. (A) Schematic representation of the RECQ1 dimer bound to two-tailed DNA duplexes. One monomer is colored by domain: blue, first RecA domain (D1); red, second RecA domain (D2); yellow, Zn-binding domain (Zn); green, WH domain; purple, β-hairpin (also denoted by asterisk). The top and bottom strands of the tailed duplex are colored black and orange, respectively. A third ssDNA strand from an adjacent complex in the crystal, which base-pairs at the separation junction, is colored cyan. The second RECQ monomer and its bound tailed duplex are colored gray. (B) Perpendicular view of isolated monomer/DNA showing the trajectory of DNA. (C) Molecular surface of RECQ1 colored by electrostatic potential (+5kT/e− to −5kT/e−; blue, positive; red, negative). The β-hairpin is indicated by an asterisk. (D) Detailed view of the trajectory of the 3′ single-stranded tail. Bases 22–26 are shown, along with interacting protein residues. Domains are colored-coded as in A. (E) Summary of protein/DNA contacts. The top and bottom DNA strands are marked in green and yellow, respectively; G3 (pink) indicates a Gua residue from the DNA chain of an adjacent molecule in the crystal (details are provided in Fig. S1A).
A distinct characteristic of RECQ1 is its oligomeric structure. Full-length RECQ1 purifies as a mixture of tetramers and dimers (16). The truncated version of the protein used in this study (RECQ1T1) lacks the N terminus, which is required for tetramerization; the protein appears as a dimer both in solution and in crystals. The dimer is arranged head to tail, with the β-hairpin in each WH domain interacting with the N-terminal helicase domain (D1) of the opposite subunit (16) (Fig. 1A). Mutational analysis has demonstrated that the dimeric structure is stimulatory but not essential for DNA unwinding activity of RECQ1T1; this observation contrasts with the essential requirement for the β-hairpin structure (13, 16).
The present structure includes a dimer of RECQ1T1 in complex with two tailed-duplex DNA molecules. Interestingly, each DNA molecule contacts only one subunit of the dimer (Fig. 1B). This finding is compatible with the observation that the dimer is not essential for fork unwinding activity, and that dimer formation is not dependent on DNA (16). In the description below, we follow the trajectory of the DNA along a RECQ1 monomer, and explore the implications for the mechanism of helicase action. A summary of protein/DNA contacts is presented in Fig. 1E.
Trajectory of the DNA.
The trajectory of the DNA can be conceptually divided into three regions (Fig. 1B). The 3′ ssDNA tail lies across the RecA-like domains D2 and D1, forming extensive contacts with the D2 domain and some contacts of the extreme 3′ end with the D1 domain. The dsDNA region, which assumes a standard B-like conformation, lies along the C-terminal WH domain. The branch point, represented by the last base pair before the 3′ ssDNA tail, interacts with the β-hairpin, which interrupts the pattern of base pairing. The trajectory of the DNA is almost entirely lined by areas of positive surface potential contacting the phosphate backbone (Fig. 1C). This arrangement is in general agreement with models of helicase action: An ATPase-driven conformational motion between the RecA domains, coupled to cycles of tight and loose DNA binding, produces a tracking motion along the ssDNA tail, whereas a physical barrier, often a β-hairpin, couples the translational motion to DNA strand separation.
3′ ssDNA Tail.
In common with other SF2 helicases, the ssDNA tail runs across the two RecA-like domains. The two residues closest to the branch point (T22 and C23) are held tightly by a network of interactions (Fig. 1D). The phosphate between T22 and C23 interacts with the main-chain amide (NH) group of Q324; the phosphate between C23 and C24 interacts with the side chains of residues T371 and Q324 and the main-chain (NH) group of A346. These amino acid residues are within the conserved helicase motifs IV (Q324), IVa (A346), and V (T371), and they form homologous contacts with DNA/RNA in other SF2 helicases (summarized in Fig. S2). Interestingly, Q324 in RECQ1 replaces a widely conserved Arg at this position in other helicases, but both the main-chain and side-chain contacts are conserved.
In addition to the phosphate-binding interactions, the nucleotides immediately downstream of the branch point are held in place by protein/base interactions. Residue C23 is hydrogen-bonded to two residues, K393 and E433; the base of T22 is hydrogen-bonded to the main-chain amide of T566. M429 intercalates between bases T22 and C23, forming an extensive hydrophobic interaction with C23. Together, this configuration forms a tight grip of the ssDNA, which could act as a ratchet, preventing slippage of the DNA during a power stroke. A very similar arrangement is seen in crystal structures of the HEL308 helicase [PDB ID code 2P6R (17)], where an Arg side chain (R350) is positioned between nucleotide bases flanking the ssDNA/dsDNA junction (Fig. S2A). Structures of two viral NS3 helicases in complex with single-stranded oligos may have analogous features (Fig. S2 B and C): In hepatitis C virus NS3 helicase [PDB ID code 3KQH (18)], V432 is inserted between bases 1 and 2, whereas in structures of the NS3 helicase of Dengue virus 4 [PDB ID code 2JLV (19)], the 5′-terminal nucleotide abuts a hydrophobic wall consisting of L129, P431, and L443. These SF2 helicases are all capable of 3′–5′ translocation; interestingly, the DEAD-box helicase VASA [PDB ID code 2DB3 (14)], which is not thought to translocate, does not have an analogous feature (Fig. S2D).
The nucleotides at positions 24 and 25 span the gap between the two RecA domains and do not interact with the protein directly (Fig. 1D). The next DNA/protein contact is to the N-terminal RecA domain (D1): R232 forms a salt bridge with the phosphate 5′ to T26, and the 3′-OH can form a polar contact with D230. Unfortunately, the 3′ tail in the crystal structure is too short to follow the full trajectory of the DNA along the protein. Residues R232 and D230 are part of a region, the aromatic-rich loop (ARL), which is highly conserved in the RecQ family but not in other helicases. This region was found to be essential for helicase activity but not for ATPase activity in both Escherichia coli RecQ (20) and human RECQ1 (13). The aromatic loop is directly downstream of conserved motif II (DEVH), which includes the metal-binding E220. This proximity could provide a means of coupling ATPase activity or nucleotide occupancy to ssDNA binding. The interaction of the ARL with the DNA is also observed in structures of DNA complexes of both Cronobacter sakazakii RecQ [PDB ID code 4TMU (21)] and human BLM (PDB ID code 4CGZ). A structure-based sequence alignment of the RecA domains of SF2 helicases is shown in Fig. S3. The DNA-contacting residues of RECQ1 in motifs IV, IVa, and V have homologous counterparts in all SF2 proteins for which information is available. The contacts with the ARL are conserved uniquely in RecQ-family protein. DNA contacts with residues in motifs Ia and Ib are absent in our RECQ1 structure because the 3′ ssDNA does not extend into this region.
ssDNA/dsDNA Junction.
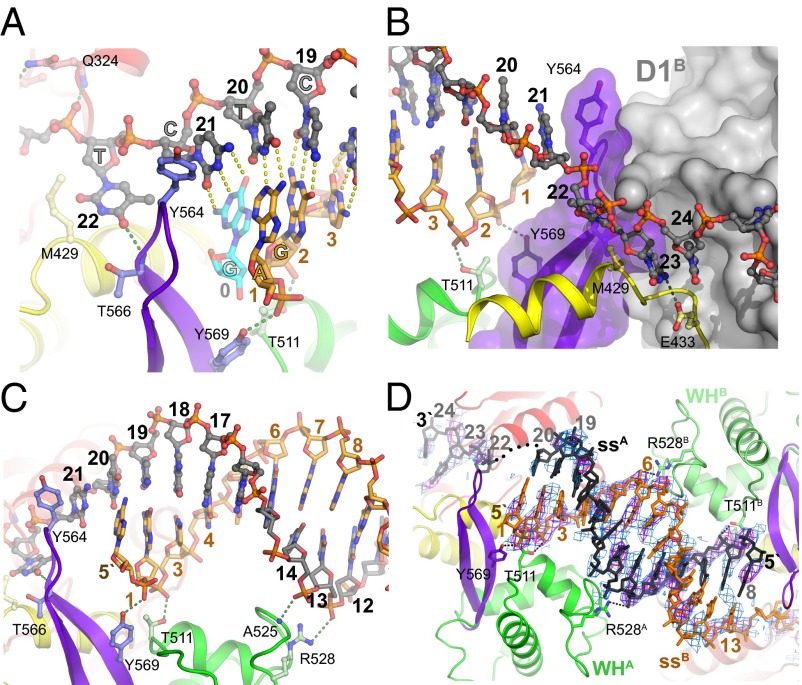
The last base pair of the duplex region in the crystal involves C21 of the top strand and a Gua base from the 5′ single-strand end of a DNA molecule from a neighboring molecule in the crystal (Fig. 2A and Fig. S1A). Although this arrangement is likely to be an artifact of the crystal structure, we believe this Gua base represents the location of the last base of the duplex in the context of the enzyme. The ultimate base pair interacts with Tyr-564, located at the tip of the β-hairpin (Fig. 2A). This arrangement is highly similar to that arrangement seen in a DNA complex with the isolated WH domain of WRN, in which the aromatic side chain (F1053) is similarly positioned [PDB ID code 3AAF (22)]. The aromatic wedge probably serves to compensate for the disrupted stacking interaction with the disrupted base pair. The top strand, which follows a normal helical trajectory up to this point, is sharply kinked so that the next base (T22) is unstacked and points away from the helix axis, into a wide groove that could accommodate either a purine or pyrimidine base. We have shown before that the β-hairpin, in addition to its role in DNA strand separation, also forms part of the contact surface of the RECQ1 dimer (16); mutations that selectively disrupt the protein/protein surface did not abolish DNA unwinding but reduced its efficiency by 60–80%. Fig. 2B shows the β-hairpin (purple) interacting with the D1 domain of the dimeric partner RECQ1 molecule (gray). The structure clearly shows that the dimerization and DNA-unwinding functions occur at opposite sides of the β-hairpin. Furthermore, it suggests the possibility that the observed dimeric arrangement enhances helicase activity by buttressing the hairpin in a favorable orientation for strand separation.
Fig. 2.
Interactions with the dsDNA and branch point. (A) Detailed view of the branch point region. The side chain of Y564 at the tip of the β-hairpin motif forms a physical barrier in front of the last base pair (Cyt21-Gua). Dotted lines represent hydrogen bonds (green, DNA/protein; yellow, DNA/DNA). The DNA strands are colored as in Fig. 1A. (B) Orientation of the β-hairpin is stabilized by the dimeric arrangement. The branch point is viewed looking from the D1 domain toward the adjacent RECQ1 molecule. The β-hairpin (purple), which forms a physical strand-separating barrier, is supported by the D1 RecA domain of the neighboring monomer (D1B, gray). (C) Interaction of the phosphate backbone of the duplex region with the WH domain. (D) Structure of RECQ1 cocrystallized with ssDNA. A noncomplementary pseudoduplex is formed between two adjacent WH domains, with the 3′ end extending between the D2/D1 domains. The modeled pseudoduplex is displayed, along with σ-a–weighted “omit” electron density maps calculated at 3.4 Å before the inclusion of the DNA in the refined model. The 2Fo–Fc electron density (blue mesh) and Fo–Fc electron density (magenta) maps are contoured at 1.2σ and 2.5σ, respectively.
Duplex DNA.
Following the DNA from the branch point toward the 5′ end of the top strand, the duplex extends from C21 to G8 as a well-defined B-form helix. The helix contacts the protein at two points in the WH domain: Phosphates C3 and G2 of the bottom strand form hydrogen bonds with T511 and Y569, respectively (Fig. 2 A and C). Phosphates C14 and T13 of the top strand contact the main-chain NH group of A525 and the side chain of R528. The interactions are restricted to the phosphate backbone, which is sequence-independent and presumably easy to slide.
Structure of a Complex of RECQ1 with an ssDNA Oligonucleotide.
In our screen for RECQ1/DNA cocrystallization, we obtained diffracting crystals from a complex of RECQ1T1 with an ssDNA oligonucleotide, 5′-GGATCTCGACGCTCTCCCTT. The crystals were twinned and diffracted to a lower resolution, but the bound DNA could be clearly visualized. The asymmetrical unit of the ssDNA complex contained two RECQ1 dimers, with the ssDNA oligonucleotide bridging two adjacent dimers (Fig. S4). Strikingly, the DNA bound in a double-stranded conformation (Fig. 2D), although there is no significant self-complementarity in the relevant region of the oligonucleotide. Accordingly, although the sugar-phosphate backbone conforms roughly to B-DNA conformation, the interior of the DNA is ill defined. The overall binding arrangement of the duplex to the protein is similar to the binding arrangement seen in the complex with the tailed-duplex DNA (Fig. S4B). In particular, each strand contacts the protein through the phosphates, interacting with residues T511 and R528. The DNA has the same interactions with neighboring protein molecules in the crystal.
Structural Basis for DNA Annealing.
The presence of a noncomplementary DNA double helix formed at the interface with the WH domain prompted us to ask whether this interaction could drive the annealing of cDNA strands in solution, possibly by forming a structural and electrostatic template for the formation of the initial, energetically unfavorable base pairs. DNA annealing activity has been observed with the full-length RECQ1 protein (23, 24) and some other RecQ family members [BLM, RECQ5, and WRN (25–27)], as well as with proteins unrelated to RecQ (28). We decided to investigate the contribution of the WH domain to DNA annealing and unwinding by testing purified mutant proteins.
Several different elements were targeted: mutation of Y564 at the tip of the β-hairpin; progressive deletions of the β-hairpin; mutations of the dsDNA binding residues T511, K514, and R528 in the WH domain; an isolated WH domain (amino acids 481–624); and a larger protein fragment, including the D2, Zn, and WH domains (amino acids 282–624). The results are summarized in Table S1, together with a summary of mutagenesis results from earlier work (16). Strikingly, we found that the WH domain in isolation is as effective as the full-length tetrameric protein in DNA annealing (Fig. S5A). Unexpectedly, a longer protein fragment spanning the D2-Zn-WH domains completely lacked annealing activity (Fig. S5B). This finding suggests that the contribution of the WH domain to strand annealing is strongly dependent on its conformational state.
Second, in the context of the RECQ1T1 protein, a point mutation of Y564 at the tip of the β-hairpin, as well as deletion of the β-hairpin, regenerates the DNA annealing activity, which is not detected when the hairpin is present (Table S1). It is possible that the strand-annealing activity of the hairpin-mutated RECQ1T1 is unrelated to the annealing activity of the full-length protein and, instead, is a consequence of the defect in strand separation activity. However, an intriguing possibility is that the hairpin may play an important role in the regulation of the strand-annealing activity of full-length RECQ1 (RECQ1FL). If such is the case, it is tempting to speculate that protein oligomerization may be required to reorient Y564 and create a conformation able to promote the annealing reaction in the context of the full-length protein. Lastly, mutations of T511, K514, and R528 in the full-length protein significantly impair DNA helicase activity but have little impact on the annealing activity, suggesting that other structural features contribute to the mechanisms of annealing (Fig. S5 D–G and Table S1). These results provide the first glimpse, to our knowledge, of the RECQ1 mechanism of strand annealing. If the energy barrier to spontaneous annealing at room temperature is electrostatic repulsion and the loss of entropy when bringing two strands to proximity, the protein may serve as a structural template to counteract the electrostatic repulsion and to nucleate a double-helical structure, which can then propagate with little additional energetic cost.
HJs Are Preferentially Bound by a Flat RECQ1 Tetramer.
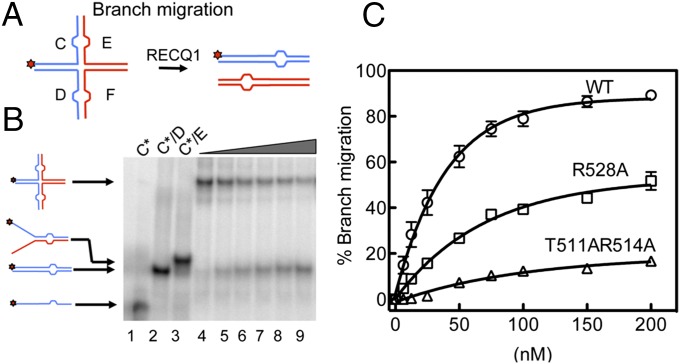
RECQ1FL protein possesses HJ branch migration activity (29, 30). To gain insight into the mechanism of HJ recognition, we tested if the T511, K514, and R528 mutations in the WH domain affect the branch migration activity of RECQ1. Our results showed that all these mutants have a reduced branch migration activity, suggesting that there are indeed unique structural features that account for the HJ resolution activities of the five human RecQ proteins (Fig. 3 and Fig. S5H).
Fig. 3.
Analysis of the branch migration activity of RECQ1FL and mutants R528A and T511A/K514A. (A) Schematic of the HJ substrate with a heterology region of one base used to study branch migration. (B) Branch migration assays as a function of protein concentration. Lanes 1–3: DNA migration markers. Lanes 4–9: branch migration assays performed using increasing RECQ1 concentrations [(0, 25, 35, 50, 100, and 200 nM) and a fixed concentration of HJ (2 nM)]. (C) Plot of the branch migration activity as a function of protein concentration. The data points represent the mean of three independent experiments. Error bars indicate SEM.
RECQ1 can form two different assembly states: a dimeric form that is proficient in DNA unwinding, in agreement with the structural data, and a tetrameric form that might be required for more “specialized activities,” such as HJ resolution (16). This notion is supported by earlier work, where we showed that the form of RECQ1 lacking the N-terminal 48 residues (RECQ1T1), which associates into dimers but not tetramers, is devoid of branch migration activity (30). To measure the number of RECQ1 motors loaded on a four-way junction structure precisely, we used a combination of sedimentation velocity and equilibrium analytical ultracentrifugation experiments (Fig. S6 A–C). The sedimentation velocity experiments showed a sedimentation coefficient s20,W = 13.8 S, which is consistent with the molecular mass of a tetramer bound to the HJ (Fig. S6A). The sedimentation equilibrium profiles collected at two different rotor speeds confirmed these findings and yielded an apparent molecular mass of 394 ± 12 kDa for the RECQ1/HJ complex, which is very close to the estimated molecular mass for a tetrameric RECQ1/HJ complex (372 kDa) (Fig. S6C). Furthermore, our fluorescence anisotropy studies confirmed that RECQ1 has higher affinity for HJ substrates (80.3 ± 3.5 nM) relative to linear dsDNA duplexes (Kd > 250 nM) suggesting that RECQ1 specifically recognizes the four-way junction of the HJ substrate (Fig. S6D).
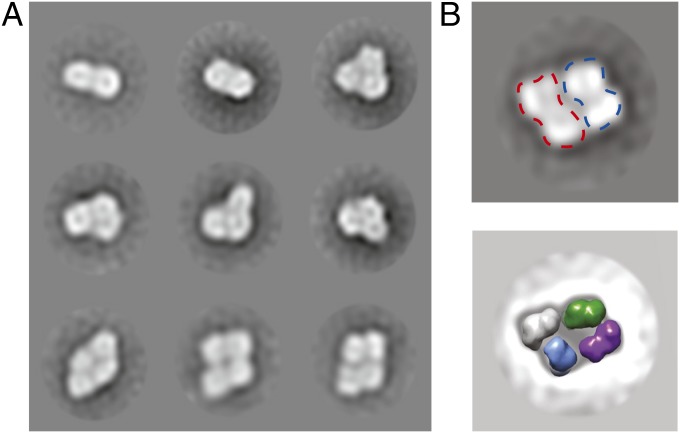
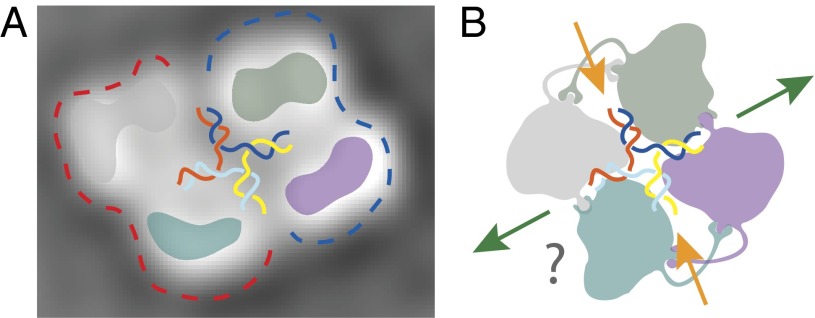
To gain insights into the mode of HJ binding by RECQ1, we used EM combined with single-particle 2D averaging to describe the architecture of the RECQ1 tetramer. A gel filtration fraction corresponding to the RECQ1 tetramer was further processed by means of a glycerol and glutaraldehyde gradient [GraFix (31)] to stabilize fragile, existing protein/protein interfaces gently. The GraFix treatment was essential to obtain negative-stain EM preparations tractable by single-particle analysis, whereas a non–cross-linked sample resulted in unresolvable heterogeneity. This sample was then used for negative-stain EM imaging. Reference-free 2D averages confirmed previous observations indicating that RECQ1 particles contain a variety of different stoichiometries (23), highlighting the presence of a mixed population of particles, including apparent dimeric and tetrameric forms (as well as apparent trimers that might be broken tetramers or 2D views of a tilted tetramer; Fig. 4A, Fig. S7, and Movie S1). RECQ1 tetramers mostly appear as rhomboid particles, which can accommodate four copies of monomeric RECQ1 atomic models (Fig. 4B). A rhomboid assembly is incompatible with a fourfold rotational (C4) symmetry in RECQ1 and more compatible with a dimer-of-dimers configuration, where two distinct dimerization interfaces (N-terminal appendix and C-terminal WH) would mediate tetramer formation, as described above. Importantly, our class averages provide compelling evidence for a flat configuration of the RECQ1 tetramer (with four RecQ1 protomers lying on a plane), which appears to contain two distinct twofold symmetry axes. A flat configuration of a four-membered RECQ1 motor assembly might provide the duplex unwinding and receding functions instrumental in HJ migration (Fig. 5), although further studies will be necessary to validate this model.
Fig. 4.
EM analysis of RECQ1 homo-oligomers. (A) Representative reference-free 2D averages of negatively stained RECQ1 particles. RECQ1 exists as a mixture of apparent dimeric, trimeric, and tetrameric assemblies. (B) Rhomboid, four-membered class average provides strong evidence for a flat configuration for the RECQ1 tetramer. (Top) Complex reveals an apparent dimer-of-dimers configuration (blue and red silhouettes), and can contain four copies of the RECQ1 monomer. (Bottom) Electron-density maps (25 Å; colored in purple, gray, blue, and green) were calculated from a monomeric atomic structure, isolated from the PDB entry (PDB ID code 2WWY). Box size, 168 pixels or 459 Å.
Fig. 5.
Speculative model of the RECQ1/HJ complex. (A) Two-dimensional class average of a flat tetrameric RECQ1 superposed to a cartoon HJ shown to scale. (B) Speculative cartoon representation of a tetrameric RECQ1 bound to an HJ. Tetramer formation is mediated by distinct interfaces, likely resulting in a dimer-of-dimer, rhomboid conformation. In other systems, HJ branch migration depends on two unwinding (orange arrow) and two annealing (green arrow) sites. A flat RecQ1 tetramer could provide these functions.
Discussion
The present work describes the first crystal structures, to our knowledge, of human RECQ1 in complex with DNA. The structures, in combination with other published helicase structures, provide detailed insights on the RecQ helicase mechanisms of DNA tracking, strand separation, and strand annealing and on the molecular basis of their specialized enzymatic activities. A number of structures of RecQ helicases have now been solved, allowing a tentative reconstruction of structural transitions occurring during the ATPase catalytic cycle. The current structure of RECQ1 (PDB ID code 2WWY) is characterized by a tight interaction of the D2 domain with the 5′ part of the ssDNA, a 1-nt gap, and a weak interaction of the ARL with the next nucleotide, with no apparent interaction with motifs I and Ia of domain D1. The conformation of the protein is very similar to a conformation of RECQ1 bound to ADP [PDB ID code 2V1X (13)], so we assume this structure represents a posthydrolysis, postpower stroke state. The cleft between the domains is at its most open state, which may allow the release of the hydrolyzed nucleotide (and exchange for ATP). A second conformation, in which both the D1 and the D2 domains are tightly attached to the DNA at adjacent sites, may be represented by a structure of a DNA-bound RecQ protein from the bacterium C. sakazakii (PDB ID code 4TMU) (21). That structure maintains the highly conserved ratchet structure around the ssDNA close to the branch point: Specifically, residues R324, A346, T371, and K393 in RECQ1 are matched by the equivalent R246, A268, T293, and R315 in C. sakazakii RecQ (CsRecQ). In addition, the intercalating M429 is replaced by W424 (one α-helical turn away) in the bacterial structure. Crucially, the ssDNA chain binds tightly to the ARL immediately next to the D2 contact, followed by tight binding to helicase motifs Ia and Ib. Although this structure does not contain ATP, Manthei et al. (21) argue that it represents a conformation poised for ATP hydrolysis. Thus, it seems that the two structures represent two extreme states of the catalytic cycle. The translocation steps remain to be elucidated, as does the timing of ATP hydrolysis and release.
A recurrent feature of DNA helicases is the presence of a β-hairpin structure located at the junction of dsDNA and ssDNA; these structures have been shown previously, by mutational (13) and structural analysis (17, 22), to be essential for coupling ATPase activity to strand separation. As predicted, the current structure shows a similar arrangement, whereby the β-hairpin of the WH domain is in a position to affect strand separation both as a physical barrier and by providing an aromatic residue that may compensate for the loss of stacking at the last base pair of the DNA duplex. The WH domain (and the β-hairpin) is highly conserved structurally, although the sequences and charge distribution vary considerably among different RecQ helicases (Fig. S8). This sequence variation, in addition to changes in the spatial relationship between the WH domain and the remainder of the protein, may have significant implications on substrate recognition and activity. Here, we show that a surface region that includes residues T511, K514, and R528 in the WH domain of RECQ1 is important for HJ branch migration. These results provide the first structural insights, to our knowledge, into the molecular basis of the distinct HJ processing activities of human RecQ helicases.
The crystal structure of RECQ1 in complex with a single-stranded oligonucleotide has revealed an unusual arrangement of a DNA duplex of noncomplementary strands; to our knowledge, this arrangement is the only such representation in the PDB. The duplex is stabilized by binding to the WH domains and by the crystal lattice. However, our experiments showing that the isolated WH domain can catalyze DNA strand annealing suggest that this arrangement is not entirely a crystallographic artifact. Rather, the structure and charge distribution of the WH may provide a template that favors strand annealing. Thus, the interaction of the WH with the duplex DNA may play a dual role in promoting strand separation or strand annealing, depending on the context and the conformation of the whole protein. However, the question of the functional role of the annealing activity of RECQ1 remains open. We showed that mutations in the WH domain that have an impact on both unwinding and annealing result in a reduced branch migration activity. Based on these results, a tantalizing scenario is that HJ branch migration requires the transient opening of a few base pairs at the junction, followed by the annealing of the two complementary strands upon junction movement. However, further studies will be necessary to confirm this model, possibly by identifying new separation of function mutants that retain the ability to anneal DNA, but lack unwinding activity, in the context of the full-length protein.
RECQ1 is unusual among RecQ helicases in its quaternary structure, which can be dimeric or tetrameric. We have shown earlier that the RECQ1 tetramer possesses activities (HJ branch migration and DNA strand annealing) that are not shared with the homodimer, which has only 3′–5′ fork-unwinding activity (16). Here, our combination of EM and analytical ultracentrifugation experiments supports the notion that RECQ1 tetramers are involved in HJ recognition, reinforcing the idea that unique structural features and quaternary structures account for the specific role of human RECQ1 in HJ branch migration and reversed replication forks restart. Our EM findings indicate that RECQ1 tetramers are arranged to form an unexpected flat structure. As discussed above, HJ branch migration might result from coordinated unwinding and reannealing occurring at two pairs of opposing branches (Fig. 5B). The arrangement of two DNA forks on the RECQ1 dimer seen in our crystal structure (Fig. 1A) suggests how one pair of HJ branches may be simultaneously pulled inward and unwound. The two additional RECQ1 molecules in the tetramer may provide a template for reannealing of the two outward-driven DNA branches. The apparent asymmetry of the tetrameric arrangement would be compatible with such a mechanism, in which the WH domains may act in strand separation or in strand annealing, depending on their precise conformation. Further studies will help establish whether HJ binding induces a reconfiguration of the RECQ1 tetramer, instrumental for branch migration and strand annealing.
Analogous studies need to be extended to other human RecQ helicases to clarify how some of these enzymes are uniquely adapted to process potentially recombinogenic DNA structures that arise upon replication stress. Because most chemotherapeutic drugs interfere with DNA synthesis, these studies will also have a broad impact on our understanding of several chemotherapeutic modalities based on agents that inhibit DNA replication.
Materials and Methods
Detailed materials and methods are described in SI Materials and Methods.
RECQ1FL proteins (WT or mutated) were expressed and purified from baculovirus-infected Sf9 insect cells as described (16). RECQ1T1 proteins (spanning amino acids 49–619) were expressed and purified from recombinant E. coli as described (13).
Crystallization.
Diffracting crystals were obtained using the combination of top: 5′-CGGTATTGGATCTCGACGCTCTCCCTT-3′ and bottom: 5′-AGCGTCGAGATCC-3′. The two oligonucleotides were annealed, mixed with the protein at a 1.2:1 molar ratio, concentrated, and set for crystallization by vapor diffusion in sitting drops. Complexes of RECQ1T1 with single-stranded oligonucleotide 5′-GGATCTCGACGCTCTCCCTTT-3′ were prepared and crystallized in the same manner.
Data Collection/Structure Determination.
For dsDNA cocrystals, diffraction data were collected to a resolution of 2.9 Å on beamline I04 at the Diamond Light Source (Harwell, United Kingdom) from a crystal vitrified in reservoir solution supplemented with 25% (vol/vol) ethylene glycol. Crystals of the dsDNA complex belong to space group P21 and contain a single RECQ1 dimer associated with two DNA duplexes. The structure was solved by molecular replacement using the coordinates of the unbound enzyme (PDB ID code 2V1X) as a search model using the program PHASER. The complex has been refined with PHENIX using appropriate restraints (translation/libration/screw, grouped B, noncrystallographic symmetry). For ssDNA cocrystals, data were collected to 3.4 Å. Crystals belong to space group P21 and contain four RECQ1 molecules (two dimers) and four DNA molecules in the asymmetrical unit. Data collection and refinement statistics are summarized in Table S2.
DNA Unwinding, Strand Annealing, and Branch Migration Assays.
All enzymatic assays were performed as described using the oligonucleotides listed in Table S3 (more details are provided in SI Materials and Methods) (9, 16).
Single-Particle EM and Image Processing.
An S200 gel filtration fraction corresponding to RECQ1 tetramers was further processed by GraFix sedimentation (31). Negatively stained samples were imaged on a JEOL2100 (JEOL) microscope operated at 120 kV, and 2D averages from contrast transfer function-corrected (32) particles were calculated with RELION (more details are provided in SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank K. MacLellan-Gibson, R. Carzaniga, and L. Collinson for access to the EM. This work was carried out with the support of the Diamond Light Source beamline I04. The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, the Canada Foundation for Innovation, Genome Canada, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust (Grant 092809/Z/10/Z). This work was supported by Cancer Research UK funds (to A.C.), by NIH Grant R01GM108648 (to A.V.), by startup funding from the Doisy Department of Biochemistry and Molecular Biology and from the Saint Louis University Cancer Center (to A.V.), by grants from the President’s Research Fund of Saint Louis University, and by the GLIOMA-Interreg (Slovenian-Italian Cooperation 2007–2013) project (to A.V.). F.M. was funded, in part, by a Royal Society International Joint Project grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.H. is a guest editor invited by the Editorial Board.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2WWY and 4U7D).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417594112/-/DCSupplemental.
References
- 1.Ellis NA, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 2.Kitao S, et al. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22(1):82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 3.Siitonen HA, et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12(21):2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 4.Van Maldergem L, et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43(2):148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu CE, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996;272(5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 6.Bachrati CZ, Hickson ID. RecQ helicases: Guardian angels of the DNA replication fork. Chromosoma. 2008;117(3):219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 7.Bohr VA. Rising from the RecQ-age: The role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33(12):609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophys Chem. 2010;149(3):67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Berti M, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol. 2013;20(3):347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22(20):2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 12.Xu D, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22(20):2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pike AC, et al. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci USA. 2009;106(4):1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 15.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97(1):75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 16.Lucic B, et al. A prominent β-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 2011;39(5):1703–1717. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14(7):647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 18.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107(2):521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo D, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27(23):3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: Role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33(22):6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manthei KA, Hill MC, Burke JE, Butcher SE, Keck JL. Structural mechanisms of DNA binding and unwinding in bacterial RecQ helicases. Proc Natl Acad Sci USA. 2015;112:4292–4297. doi: 10.1073/pnas.1416746112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18(2):177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Muzzolini L, et al. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5(2):e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S, et al. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280(30):28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 25.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33(12):3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23(14):2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280(24):23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 28.Sugiman-Marangos SN, Peel JK, Weiss YM, Ghirlando R, Junop MS. Crystal structure of the DdrB/ssDNA complex from Deinococcus radiodurans reveals a DNA binding surface involving higher-order oligomeric states. Nucleic Acids Res. 2013;41(21):9934–9944. doi: 10.1093/nar/gkt759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazina OM, Rossi MJ, Deakyne JS, Huang F, Mazin AV. Polarity and bypass of DNA heterology during branch migration of Holliday junctions by human RAD54, BLM, and RECQ1 proteins. J Biol Chem. 2012;287(15):11820–11832. doi: 10.1074/jbc.M112.341347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popuri V, et al. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283(26):17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 31.Kastner B, et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5(1):53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 32.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.