Significance
Interferon γ-inducible protein 16 (IFI16) and cGMP-AMP synthase (cGAS) have both been proposed to directly detect herpesviral DNA in herpes simplex virus (HSV)-infected cells and initiate interferon regulatory factor-3 signaling, but it has been unclear how two DNA sensors could both be required. Both are required in human fibroblasts for detection of HSV and transfected DNAs. We found evidence that IFI16 plays a direct role in HSV DNA sensing, whereas cGAS produces low amounts of cGAMP and promotes the stability of IFI16. Our results demonstrate a new function for cGAS in the maintenance of normal levels of IFI16 and provide an explanation for the multiple proposed DNA sensors.
Keywords: protein–protein interactions, DNA sensing, innate immunity, virus–host interactions
Abstract
Interferon γ-inducible protein 16 (IFI16) and cGMP-AMP synthase (cGAS) have both been proposed to detect herpesviral DNA directly in herpes simplex virus (HSV)-infected cells and initiate interferon regulatory factor-3 signaling, but it has been unclear how two DNA sensors could both be required for this response. We therefore investigated their relative roles in human foreskin fibroblasts (HFFs) infected with HSV or transfected with plasmid DNA. siRNA depletion studies showed that both are required for the production of IFN in infected HFFs. We found that cGAS shows low production of cGMP-AMP in infected cells, but instead cGAS is partially nuclear in normal human fibroblasts and keratinocytes, interacts with IFI16 in fibroblasts, and promotes the stability of IFI16. IFI16 is associated with viral DNA and targets to viral genome complexes, consistent with it interacting directly with viral DNA. Our results demonstrate that IFI16 and cGAS cooperate in a novel way to sense nuclear herpesviral DNA and initiate innate signaling.
The innate immune response is a crucial component of host immunity and is the first line of defense against microbial pathogens, including bacteria and viruses. The initial events during infection of a host cell induce intracellular signaling pathways, resulting in the production of proinflammatory cytokines and IFNs. These effector molecules activate an antiviral state in neighboring cells and recruit immune cells to promote clearance of infection. Viral nucleic acids are potent activators of these signaling pathways and are recognized by a subset of host cell pattern recognition receptors (PRRs). These PRRs include the membrane-bound Toll-like receptors, the cytosolic RIG-I–like receptors, and a broad class of putative DNA sensors, which include both cytosolic and nuclear proteins (1).
Unlike viral RNAs, which are distinct from cellular RNAs and therefore recognized by intracellular PRRs, DNA genomes of viruses that replicate in the nucleus are thought to be chemically and structurally similar to host DNA (2–4). It was therefore generally accepted that viral DNA sensing was limited to the cytoplasm where aberrant DNA accumulation would be perceived as “foreign.” However, this dogma has recently been challenged by the identification of DNA-sensing pathways that are active in the nucleus. The Pyrin and HIN-containing interferon γ-inducible protein 16 (IFI16) protein, initially described as a cytosolic DNA sensor (5), has been demonstrated to be nuclear in many cells and to promote the activation of inflammasomes (6, 7) and production of IFNs (8, 9) in response to herpesvirus infection. These initial studies involved short-term siRNA depletion of IFI16; in addition, a recent study showed that long-term knockdown of IFI16 expression led to abrogated IFN responses to not only DNA viruses, such as herpes simplex virus (HSV), but also RNA viruses, such as Sendai virus (10).
cGMP-AMP synthase (cGAS) was also identified as a DNA sensor that recognizes cytosolic DNA and subsequently produces cGMP-AMP (cGAMP), a second messenger that induces stimulator of IFN genes (STING)-dependent activation of interferon regulatory factor-3 (IRF-3) signaling (11, 12). HSV-1 infection of murine L929 fibroblasts or human THP-1 cells led to synthesis of cGAMP and dimerization of IRF-3 (11, 12). Furthermore, fibroblasts from cgas−/− mice showed decreased induction of IFNβ in response HSV-1 infection, indicating a role for cGAS in the innate immune response to herpesviral infection (13).
A number of potential DNA sensors have been reported (14, 15), but IFI16 and cGAS appear most crucial for cellular detection of herpesvirus infections. The essentiality of two putative DNA sensors for innate responses to HSV infection raised a question, however, in that it was unclear how both IFI16 and cGAS could be required if serving as redundant DNA sensors. If the two sensors were completely redundant in one or more pathways, there would be no effect of depletion of either protein. Similarly, if one protein was sufficient as a sensor, both would not be required. Cooperativity between the two proteins could lead to a dual requirement. This analysis prompted us to evaluate whether cGAS and IFI16 cooperate to induce IFN expression in a single cell type.
In this study we demonstrate that both IFI16 and cGAS are necessary for the induction of IFNβ in primary human foreskin fibroblasts in response to transfected plasmid DNA and herpesvirus infection and that there are substantial differences in the responses to the two stimuli. We obtained evidence that IFI16 and cGAS cooperate in sensing HSV DNA by a mechanism in which IFI16 serves as the primary DNA sensor, and cGAS plays an indirect role in the response to incoming nuclear herpesviral DNA by interacting with IFI16 and promoting its stability rather than through the production of cGAMP.
Results
IFI16 and cGAS Are both Expressed in Normal Human Cells.
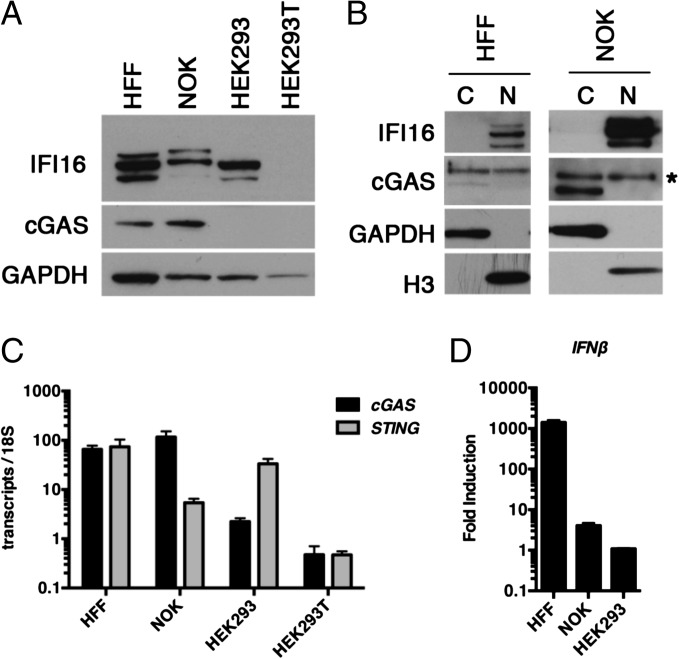
To define the relative roles of cGAS and IFI16 in innate sensing in a single cell type, we wished to define human cells that expressed both of these proteins. We therefore initially examined the basal levels of cGAS protein in normal human foreskin fibroblasts (HFFs), a normal oral keratinocyte (NOK) cell line immortalized with human telomerase reverse transcriptase (16), HEK293 cells, and HEK293T cells. cGAS protein was detectable in both HFF and NOK cells but not in HEK293 or HEK293T cells (Fig. 1A), the latter as reported previously (11, 12). Consistent with this observation, we detected markedly higher cGAS RNA levels in HFF and NOK cells compared with HEK293 or HEK293T cells (Fig. 1C). Surprisingly, although cGAS has been described as a cytosolic protein (11, 12), using cellular fractionation we found that human cGAS was localized in both the nucleus and the cytoplasm of HFF and NOK cells (Fig. 1B). The subcellular distribution of cGAS in HFF and NOK cells was confirmed by confocal immunofluoresence microscopy (Fig. S1A). Immunofluorescence analysis of cells depleted for cGAS or IFI16 showed reduced signal for the depleted proteins, showing the specificities of the antibodies (Fig. S1B). Interestingly, human cGAS appeared to be mainly nuclear upon stable ectopic expression in HEK293 cells (Fig. S1B). We found that IFI16 was also expressed in HFF and NOK cells, although the IFI16 isoforms in NOK cells appeared to run at a higher apparent molecular weight than those in HFF (Fig. 1A). Furthermore, the innate immune response to HSV-1 infection at 6 h postinfection (hpi) was greater in HFF cells than in NOKs or HEK293 cells (Fig. 1D). Based on the robust induction of IFNβ RNA in HSV-1–infected HFFs observed above and previous reports that HFFs are competent for IFI16 activity (7, 8, 17), we chose HFF cells to study the relative contributions of cGAS and IFI16 to the innate immune response to foreign DNA.
Fig. 1.
Expression and localization of IFI16 and cGAS in human cells. (A) Western blot of IFI16 and cGAS in normal and immortalized cell lines. Lysates from equal numbers of cells were loaded on a 4–12% SDS/PAGE gel and probed for cGAS and IFI16 protein expression. (B) Nuclear/cytoplasmic fractionation of human cells. GAPDH and H3 were evaluated to confirm fractionation efficiency. The asterisk marks the cGAS specific band lost during siRNA depletion. (C) Basal levels of cGAS and STING in human cells. RNA was harvested from cells and analyzed by qRT-PCR. Transcripts were normalized to 18S rRNA. (D) Induction of IFNβ RNA in response to HSV-1 infection. Cells were infected with HSV-1 d109 virus at an MOI of 10 and RNA was harvested 6 hpi. IFNβ RNAs were normalized to 18S rRNAs and then relative to mock-infected samples.
cGAS or IFI16 Depletion Reduces the Innate Immune Response to HSV-1 Infection in HFF Cells.
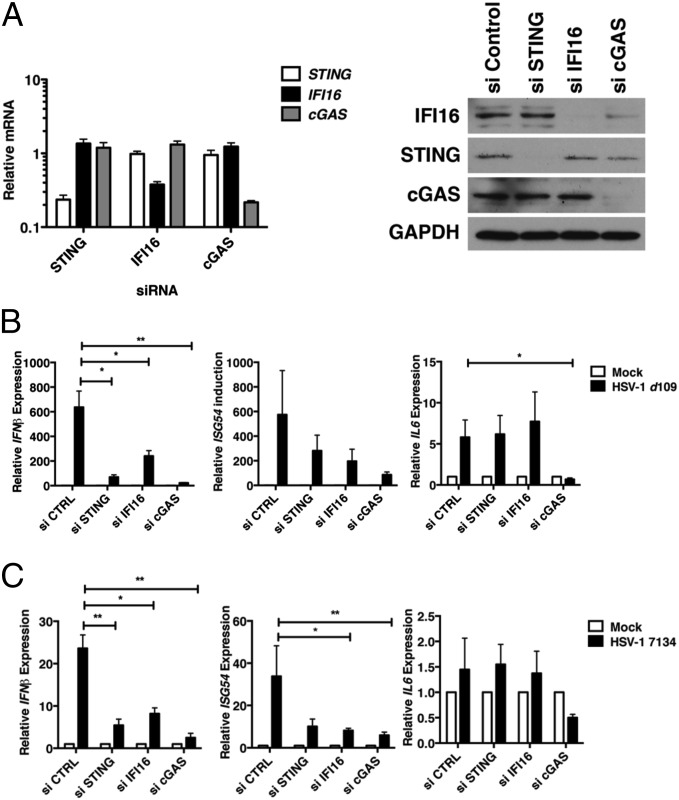
The partial localization of cGAS to the nucleus in HFFs suggested that the protein might sense herpesviral DNA at that site during productive infection. To test whether cGAS was involved in the innate immune response to HSV-1 in HFFs, we examined antiviral cytokine and IFN-stimiulated gene (ISG) expression by quantitative RT-PCR (qRT-PCR) in cells depleted for cGAS with siRNAs in parallel with cells transfected with siRNAs to deplete STING or IFI16. We observed a reduction in STING, IFI16, and cGAS RNAs (Fig. 2A, Left) and proteins (Fig. 2A, Right) with their cognate siRNAs at 72 h posttransfection (hpt). Surprisingly, we consistently observed that efficient cGAS depletion in HFF cells resulted in a reduction in basal IFI16 protein levels (Fig. 2A, Right). This finding was not the result of a reduction in basal IFI16 RNA levels because IFI16 transcripts appeared to be slightly elevated rather than reduced in the absence of cGAS (Fig. 2A, Left). These results indicated that cGAS may regulate IFI16 protein synthesis or stability.
Fig. 2.
Effects of depletion of STING, IFI16, or cGAS on innate immune responses to HSV-1 infection in normal human fibroblasts. (A) Efficiency of siRNA-mediated reduction of cellular gene expression at the RNA (Left) and protein (Right) level. RNA and protein were harvested from siRNA-transfected cells at 48 hpt. STING, IFI16, and cGAS transcripts were quantified by qRT-PCR and normalized to 18S rRNA. Whole-cell lysates were analyzed by Western blotting to determine the steady state protein levels of IFI16, STING, and cGAS. GAPDH was examined as a loading control. (B and C) Cytokine and IFN expression in HSV-1 d109 (B) or HSV-1 7134 (C) virus-infected human fibroblasts. siRNA-transfected cells were infected with the indicated HSV-1 recombinant viruses at an MOI of 10, and total RNA was harvested at 6 hpi. ISG54, IFNβ, and IL-6 expression were measured by qRT-PCR, normalized to 18S rRNA, and plotted as a fold-induction over mock-infected cells. *P < 0.05, **P < 0.01.
To examine whether cGAS depletion resulted in reduced innate immune signaling in response to HSV-1 infection, we infected STING-, IFI16-, or cGAS-depleted cells with one of two HSV-1 recombinant viruses, d109 (18) or 7134 (19), which show enhanced antiviral signaling because of the absence of the known viral inhibitors of the innate immune response (8, 20–22). We examined the induction of three cellular genes, ISG54, IFNβ, and IL-6, which are differentially regulated by specific transcription factors upon activation of innate signaling pathways. At 6 hpi, depletion of STING, IFI16, or cGAS resulted in a significant reduction in IFNβ transcripts in response to either the d109 or 7134 viruses (Fig. 2 B and C, Left). Depletion of IFI16 or cGAS and infection with d109 virus led to reduced, although not statistically significant, levels of ISG54 (Fig. 2B, Center), whereas depletion of STING, IFI16, or cGAS and infection with the 7134 ICP0− virus led to statistically significant reductions in levels of ISG54 RNA (Fig. 2C, Center).
Depletion of cGAS but not STING or IFI16 resulted in decreased IL-6 transcripts in response to d109 virus infection (Fig. 2B, Right). The 7134 virus did not induce detectable IL-6 RNA expression at 6 hpt (Fig. 2C, Right), but we consistently observed a reduction in IL-6 transcripts below control-treated cells when cGAS was depleted, consistent with transcript levels being a combination of induction of the innate immune response by HSV-1 and a reduction associated with the virion host shut-off function (21). Interestingly, STING depletion did not result in decreased IL-6 transcripts, indicating that this cGAS-dependent effect on IL-6 transcription may be independent of STING. Furthermore, these results argued that the effect of cGAS on IRF3-dependent signaling in response to HSV-1 may be caused, in part, by the decreased levels of IFI16 observed upon cGAS depletion, complicating the interpretation of these results.
cGAS Regulates IFI16 Protein Stability.
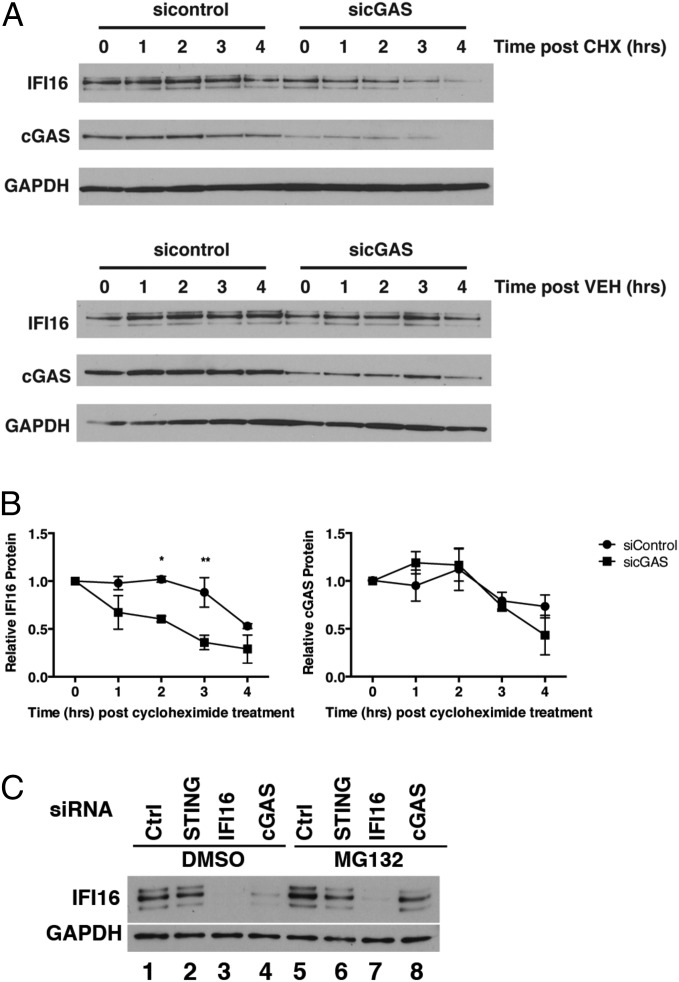
We had observed that cGAS depletion resulted in decreased IFI16 protein levels in normal HFF cells (Fig. 2A). To test whether cGAS regulates IFI16 protein levels by modulating protein stability, we determined the half-life of IFI16 in the presence or absence of cGAS. We treated HFFs with control or cGAS siRNAs and then incubated the cells with cycloheximide (CHX) or medium containing vehicle control for 4 h. IFI16 and cGAS levels were measured at 1-h intervals by Western blotting (Fig. 3A), and we observed that IFI16 shows a half-life of 1.9 h in the absence of cGAS compared with 4.2 h in the presence of cGAS (Fig. 3B). Therefore, IFI16 was less stable in the absence of cGAS.
Fig. 3.
Effects of cGAS on IFI16 protein stability. (A) HFF cells were treated with control or cGAS siRNAs for 48 h and then incubated in medium containing 100 μg/mL CHX or control medium for 0, 1, 2, 3, and 4 h. Total cellular lysates were harvested, and IFI16 protein levels were examined by Western blotting. (B) IFI16 and cGAS levels were quantified by ImageJ. Protein levels were set to 1 at time 0 h for each condition. IFI16 protein levels in sicGAS treated cells were initially 50% of the sicontrol-treated cells. (C) siRNA-depleted HFF cells were incubated with 1 μM MG132 or control medium for 8 h. Total cellular lysates were harvested, and IFI16 protein levels were examined by Western blotting. *P < 0.05, **P < 0.01.
We hypothesized that the difference in IFI16 stability might be a result of proteasomal degradation; therefore, we examined the effect of inhibition of proteasomal activity on IFI16 protein levels. HFFs depleted for cGAS were incubated with or without the MG132 proteasome inhibitor, and IFI16 protein levels were measured by Western blot at 8 h posttreatment. IFI16 protein levels were reduced again when cGAS was depleted (Fig. 3C, compare lanes 1 and 4), but when MG132 was added, we observed a restoration of IFI16 protein in cGAS-depleted cells to levels comparable with control siRNA-treated cells (Fig. 3C, compare lanes 4 and 8), consistent with reduced stability of IFI16 being due to proteasomal degradation.
cGAS and IFI16 Interact.
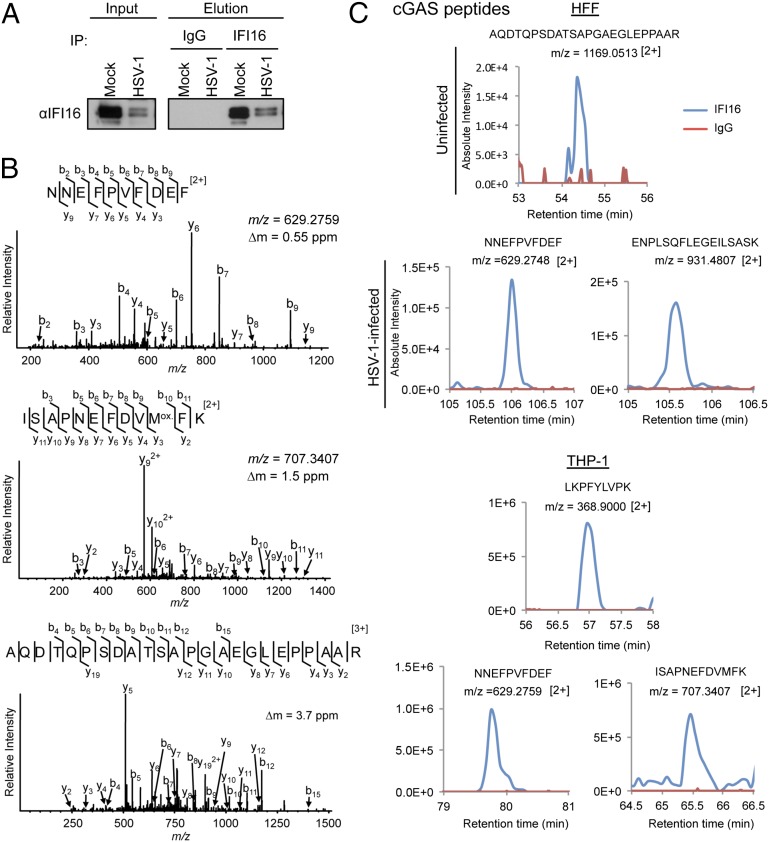
Because both IFI16 and a subset of cGAS showed nuclear localization, we hypothesized that they might interact. We therefore performed a targeted proteomic analysis of IFI16 in HFF cells. Endogenous IFI16 was immunoaffinity purified from uninfected cells, as well as at 3 hpi from cells infected with WT HSV-1 by using magnetic beads conjugated to monoclonal antibodies against IFI16 (Fig. 4A). The coisolated proteins were digested with trypsin, and targeted LC-MS/MS analyses were performed on an LTQ Orbitrap Velos instrument to specifically search for the possible presence of cGAS peptides. Indeed, cGAS peptides (Fig. 4B) were detected with IFI16 isolated from both uninfected and infected HFFs (Fig. 4C, blue lines), but were not detected in control IgG isolations (Fig. 4C, red lines). Similar results were observed with IFI16 immunopurified from differentiated monocytes (Fig. 4C).
Fig. 4.
IFI16 and cGAS interact during HSV-1 infection in HFF cells. (A) Western blotting analyses of HFF cells lysates (Input), and immunopurified (Elution) fractions demonstrating the isolation of endogenous IFI16 in uninfected (Mock) and cells infected with WT HSV-1 virus at an MOI of 10 for 3 h. (B) Confirmation of amino acid sequences of endogenous cGAS peptides identified in the IFI16 immunopurifications. Representative collision-induced dissociation MS/MS spectra are shown. (C) Representative extracted ion chromatograms of cGAS peptides, detected by LC-MS data and visualized using Skyline software. The peaks illustrate the detection of cGAS peptides in either IFI16 (blue) or IgG (red) isolations in uninfected HFF cells or HFF cells infected with WT HSV-1.
HSV-1 Infection Does Not Induce a Robust cGAMP Response.
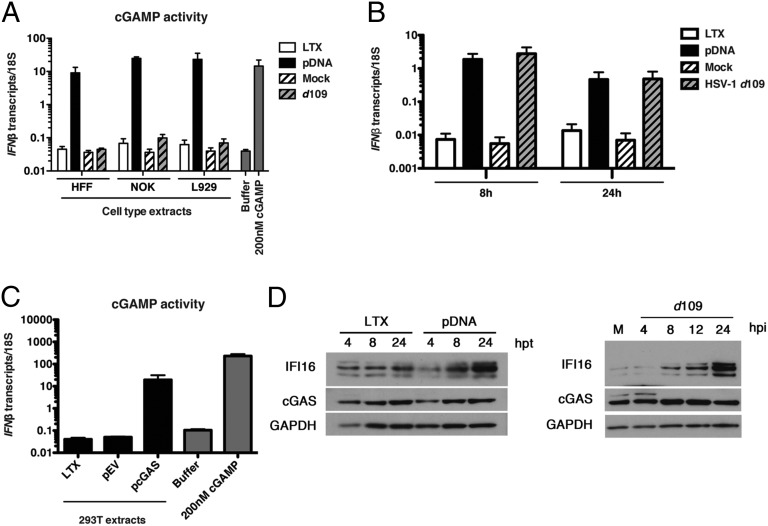
The results presented above describe a new function for cGAS in the stabilization of IFI16, but they did not show why IFI16 was also required to induce innate signaling. To address why cGAS was not sufficient, we examined the production of cGAMP—the product of cGAS—in HFFs, NOKs, and mouse L929 cells upon infection with HSV-1 d109 or following plasmid DNA transfection. cGAMP activity in cell extracts prepared as described in Materials and Methods was measured in a modified bioassay based on the original method of Gao et al. (23) by assaying the induction of IFNβ transcripts in a permeabilized secondary reporter cell (HFF) at 24 hpi or hpt. Transfection of plasmid DNA into the three types of cells resulted in robust cGAMP activity, whereas HSV-1 d109 infection resulted in low cGAMP activity in all cell lines examined (Fig. 5A). We were unable to detect cGAMP activity in HFF cells following d109 infection, and NOK and L929 cells produced a low but reproducible response to virus infection (Fig. 5A). Interestingly, when transcript levels were compared in the infected and transfected cells, we observed a similar fold-induction of IFNβ RNA in response to both stimuli (Fig. 5B), indicating that cGAMP production during viral infection did not correlate with the observed induction of the IFNβ transcripts.
Fig. 5.
cGAMP activity in plasmid DNA-transfected or HSV-infected HFF cells. (A) cGAMP production in response to DNA transfection or HSV-1 infection. HFFs, NOKs, and L929 cells were transfected with 200 ng of plasmid DNA or infected with HSV-1 d109 virus at an MOI of 1 for 24 h. Extracts of the transfected or infected cells were prepared, DNase- and heat-treated, and incubated with permeabilized HFF cells for 6 h. Reporter cells were treated with 200 nM cGAMP as a positive control. IFNβ RNA induction was analyzed by qRT-PCR and normalized to 18S rRNA. (B) Cellular gene expression in response to transfected or viral DNA. Cells were infected and transfected as in A. Total RNA was harvested at 8 and 24 h poststimulation and analyzed by qPCR. (C) 293T cells were transfected with 1 μg total DNA: pEV or pcGAS for 24 h. Permeabilized HFF cells were incubated for 30 min with DNase- and heat-treated cell extracts, harvested at 6 h, and IFNβ induction was analyzed by qRT-PCR. Reporter cells were treated with 200 nM cGAMP as a positive control. IFNβ transcripts were normalized to 18S rRNA. The data represent three independent experiments. (D) HFF cells were transfected with 200 ng of plasmid DNA (Left) or infected with HSV-1 d109 (MOI = 3) (Right). Cells were harvested at the indicated times, and cellular proteins were examined by Western blotting.
To eliminate the possibility that there was plasmid carryover in our cell extracts being used for cGAMP bioassays, we transfected plasmid DNA into HEK293T cells, which express no cGAS (Fig. 1), prepared a cellular extract, and exposed permeabilized HFF cells to this extract. We observed that there was very little or no induction of IFNβ RNA in cells treated with the extracts prepared from the empty plasmid vector-transfected HEK cells in comparison with extracts of HEK293T cells treated with only transfection reagent (LTX) (Fig. 5C). Therefore, this control experiment showed that there was no DNA carryover in the samples prepared from the transfected cells (Fig. 5C).
These results showed that although plasmid DNA transfection induced strong cGAMP production, infection with the HSV-1 d109 virus, which induced a strong IFN response, did not robustly activate cGAS production of cGAMP in normal human fibroblasts. We did not observe a reduction in cGAS protein levels in d109 virus-infected HFF cells over a 24-h time course (Fig. 5D), indicating that the absence of detectable cGAMP was not due to cGAS turnover during viral infection.
IFI16 but Not cGAS Is Recruited to the Site of Incoming Viral Genomes During Infection.
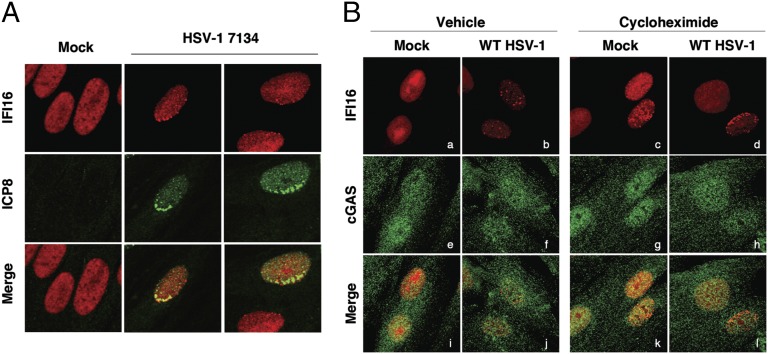
IFI16 has been shown to localize to intranuclear sites near incoming viral genomes, as marked by the HSV-infected cell protein (ICP)4 (24) or ICP8 protein (Fig. 6A). These results and the association of IFI16 with viral DNA, as shown by chromatin immunoprecipitation (9, 17), argue that IFI16 binds directly to viral DNA. To test if cGAS localized similarly to IFI16, we examined the localization of IFI16 and cGAS at 2.5 hpi in HFF cells infected at high multiplicity of infection (MOI) in the presence of CHX, which was used to prevent the synthesis of viral proteins that may inhibit these pathways. In control cells infected with WT HSV-1, IFI16 accumulated in foci within the nucleus (Fig. 6 B, b) as described previously (8), but we observed no redistribution of cGAS (Fig. 6 B, f). In the presence of CHX, IFI16 relocalized to sites near the periphery of many of the nuclei in these cells (Fig. 6 B, d) but cGAS showed no apparent relocalization (Fig. 6 B, h). Because IFI16 localized to viral genomes but cGAS did not, this was consistent with IFI16 being the primary sensor of HSV-1 DNA in the nucleus. Interestingly, CHX treatment also resulted in the intranuclear relocalization of IFI16 in ∼5% of mock-infected cells (Fig. 6 B, c), indicating that blocking host protein synthesis may activate nuclear IFI16 reorganization or filament formation.
Fig. 6.
Localization of IFI16 and cGAS in HSV-infected cells. (A) HFF cells were mock-infected or infected with HSV-1 7134 virus (MOI = 10). Cells were fixed at 24 hpi, and cells on the edge of viral plaques were imaged by IFI16 and ICP8 staining. (B) Human foreskin fibroblasts were mock-infected or infected with WT HSV-1 at an MOI of 100 in the presence or absence of 100 μg/mL CHX. Cells were fixed and stained at 2.5 hpi for IFI16 and cGAS. (Magnification: 63×.)
Comparison of the Mechanisms of Action of cGAS and IFI16 in Infected Cells with Transfected Cells.
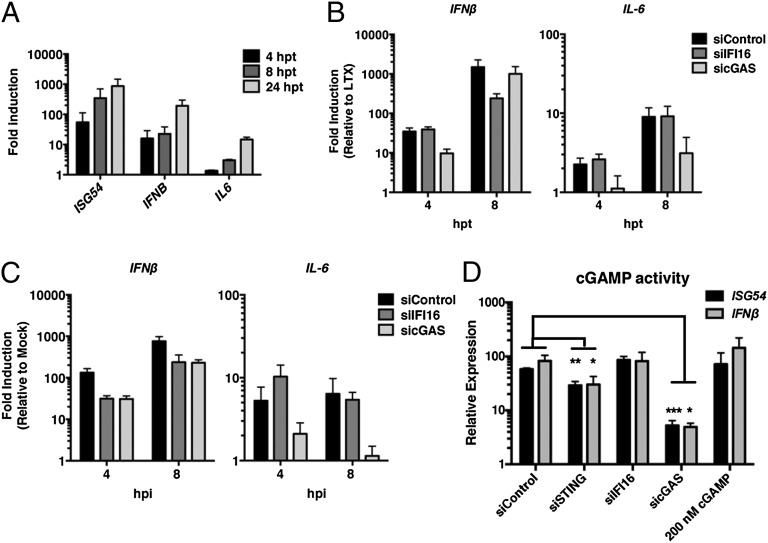
Our studies on cGAMP production indicated that the mechanisms of DNA sensing are different in infected versus transfected human cells. To further characterize the relationship between cGAS and IFI16, we examined the kinetics of their activity in HFF cells in response to plasmid DNA transfection. ISG54, IFNβ, and IL-6 mRNAs accumulated post-DNA transfection over a 24-h time course (Fig. 7A). At 4 hpt we observed that depletion of cGAS, but not IFI16, resulted in decreased expression of innate immune genes compared with control siRNA-treated cells (Fig. 7B). In contrast, at 8 hpt we observed that depletion of IFI16 resulted in reduced IFNβ transcripts, corresponding to a decreased necessity for cGAS in the induction of this gene. Although the exact kinetics varied between experiments, the requirement for cGAS always preceded that of IFI16 during DNA transfection. Therefore, IFI16 is required for the larger induction of IFNβ that occurs at later times after transfection. We did not observe a similar transition in the requirement for cGAS and IFI16 during HSV-1 infection in that both were required through 8 hpi (Fig. 7C), further highlighting the differences between the responses to transfection and DNA virus infection. Together these results indicated that the requirement for cGAS precedes the requirement for IFI16 in sensing transfected DNA, but not HSV-1 DNA in infected cells.
Fig. 7.
Contribution of cellular signaling components to innate immune responses to transfected DNA. (A) Kinetics of innate transcriptional responses to transfected DNA. HFF cells were transfected with 200 ng of plasmid DNA, and total RNA was harvested and analyzed by qRT-PCR at the indicated time points. Values were normalized to cellular GAPDH and are plotted as a fold-induction over a DNA-negative Lipofectamine LTX control. (B and C) Kinetics of innate transcriptional responses in cGAS- or IFI16-depleted cells transfected with plasmid DNA (200 ng) (B) or infected with HSV-1 d109 (MOI = 3) (C). (D) cGAMP activity. siRNA-depleted cells were transfected as in B, and cGAMP was harvested at 24 hpt and assayed on reporter HFF cells. ISG54 and IFNβ mRNA induction were measured in reporter cells at 6 h posttreatment by qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001.
Because both IFI16 and cGAS appeared to be functional in transfected HFF cells, we asked whether or not IFI16 affects cGAS activity. HFF cells were transiently depleted for cGAS, IFI16, or STING, transfected with plasmid DNA, and whole-cell extracts were harvested at 24 hpt to assay for cGAMP activity. Although depletion of cGAS resulted in a significant reduction in the ability of cellular extracts to induce IFNβ and ISG54 transcripts in a reporter cell line (Fig. 7D), depletion of IFI16 had no effect on cGAS-dependent cGAMP activity. In contrast, STING depletion resulted in a slight but significant reduction in cGAMP activity (Fig. 7D). These results indicated that IFI16 does not affect the enzymatic activity of cGAS and that IFI16 and cGAS act through distinct signaling pathways in response to transfected DNA.
cGAS and IFI16 Are both Recruited to Cytoplasmic and Nuclear Foci in Cells Transfected with Plasmid DNA.
To further characterize the response of the two sensors to transfected DNA, we examined the localization of cGAS and IFI16 by immunofluorescence following transfection. At 4 hpt, we detected cGAS foci in the cytoplasm of transfected cells colocalizing with Cy3-labeled plasmid DNA (Fig. S2A, 4 hpt), consistent with previous reports (11). Interestingly, at 8 and 24 hpt we observed IFI16 filaments in both the cytoplasm and nucleus of transfected cells (Fig. S2A). These filaments, which have been reported to be associated with IFI16 oligomerization along foreign DNA (25), were directly adjacent to or colocalized with cGAS foci and plasmid DNA. Furthermore, intranuclear cGAS foci were also observed in the nuclei of a portion of transfected cells. When we counted numbers of foci scoring for the presence of cGAS, IFI16, or both proteins, we observed a different kinetics of recruitment of the two proteins; that is, cGAS foci appeared early during transfection whereas IFI16 was recruited at later time-points (Fig. S2B). Together with the above siRNA experiments, these results argued that cGAS and IFI16 act sequentially to fully activate the response to plasmid DNA. This finding seems to differ from the host response to HSV-1 infection, where both proteins appear to be required simultaneously through at least 8 hpi for downstream antiviral gene expression (Figs. 2 B and C and 5C).
In a previous study we did not observe IFI16 accumulation in the cytoplasm during HSV-1 infection of HFF cells (8), but here we detected both nuclear and cytoplasmic IFI16 filaments following DNA transfection. Consistent with transfected DNA being a potent activator of IFNβ, we observed increased levels of IFI16 protein following DNA transfection (Fig. 4B). Therefore, the accumulation of IFI16 in the cytoplasm upon plasmid DNA transfection may be because of newly synthesized IFI16 binding to cytoplasmic plasmid DNA during its transit to the nucleus. Alternatively, this accumulation could be due to movement or leakage of IFI16 from the nucleus into the cytoplasm.
Discussion
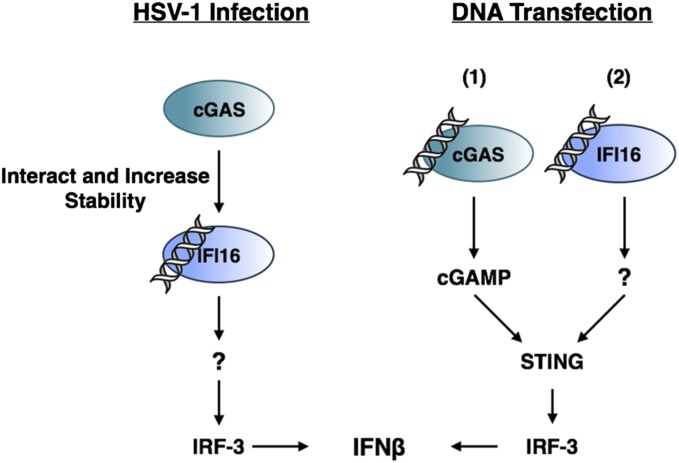
Reports of a large number of DNA sensors have prompted speculation that these sensors may function differentially in different cell types, species, or in response to different viruses. Alternatively, multiple sensors may be necessary in the same cell type and species to mediate the full response to foreign DNA (14, 15, 26). In particular, two DNA sensors, IFI16 (5) and cGAS (11, 13), have both been shown to be required for IRF-3 signaling in response to HSV infection, and it has been unclear how two DNA sensors could both be essential as primary DNA detectors. When we examined the relative contributions of two DNA sensors, cGAS and IFI16, in the host response to both nuclear and cytoplasmic foreign DNA in a single cell type, HFFs, we obtained evidence that although both are required for optimal responses to both stimuli, the mechanisms of sensing are different in HSV-infected cells from transfected cells (Fig. 8). In HSV-infected cells, cGAS is relatively inactive for production of cGAMP, compared with plasmid DNA-transfected cells, but interacts with IFI16 and stabilizes it so that IFI16 can localize and interact with incoming viral genomes. In transfected cells, cGAS initiates the initial response to plasmid DNA, likely binding it in the cytoplasm, producing cGAMP, and stimulating IFNβ production and IFI16 expression. IFI16 is responsible for the major part of the later IFNβ expression, with the newly made IFI16 potentially binding plasmid DNA in both the cytoplasm and nucleus. Our results document a previously unidentified mechanism in HSV-infected cells by which IFI16 and cGAS cooperate to induce innate signaling through the interaction of cGAS with IFI16 and the resulting stabilization of IFI16 (Fig. 8).
Fig. 8.
Model of the distinct roles of IFI16 and cGAS in sensing nuclear herpesviral or transfected DNA. Following HSV-1 infection (Left), IFI16 senses HSV-1 viral DNA in the nucleus and promotes downstream signaling to activate IFNβ expression. cGAS indirectly activates signaling by promoting IFI16 stability. During DNA transfection (Right), cGAS initially senses transfected DNA (1) followed by IFI16-dependent sensing (2).
cGAS Localizes Partially in the Nucleus, Interacts with IFI16, and Enhances Its Stability in Human Fibroblasts.
We observed that human cGAS localizes to both the nucleus and cytoplasm of human fibroblasts and keratinocytes cells using confocal microscopy and cell fractionation, a different localization pattern than reported for mouse cells and human THP-1 cells (11). Analysis of the human and murine cGAS sequence showed that the human gene product contains two potential nuclear localization signals, whereas the murine gene product does not contain these sequences (Fig. S3). This provides a possible basis for the nuclear localization of IFI16 in human cells, and it furthermore raises caution about assuming that the properties of cGAS are similar in murine and human cells.
We also observed that depletion of cGAS greatly reduced the levels of IFI16 protein. This effect might be exerted by cGAS inducing IFNβ, which increases IFI16 gene transcription, but IFI16 transcript levels were not reduced under these conditions, arguing for an effect on protein synthesis or stability. Measurement of the half-life under cGAS depletion conditions showed a twofold reduced half-life for IFI16 when cGAS was depleted. The reduced levels of IFI16 were restored when a proteasomal inhibitor was added, arguing for proteasomal degradation of IFI16 in the absence of cGAS. The interaction of cGAS with IFI16 could affect its conformation, prevent binding of an E3 ligase or adaptor molecule, or promote binding of a deubiquitinase so as to increase the stability of IFI16. The cellular ubiquitin ligases or deubiquitinases that basally or inducibly regulate IFI16 protein stability have not been described, so the identification of such proteins will likely shed light on the mechanism by which cGAS affects IFI16 stability.
DNA Sensing in HSV-1 Infected Cells.
The levels of cGAMP induced in HSV-1 infected cells are much lower than in transfected cells, arguing that cGAS enzymatic activity is low in HSV-1–infected HFF cells. Because there are approximately equal levels of IFNβ RNA induced in infected versus transfected cells, this indicates that the major induction of IFNβ in infected cells is not via cGAMP generated by cGAS. Furthermore, IFI16 is relocalized to HSV genomic complexes, whereas—in contrast to transfected cells—there is no apparent relocalization of cGAS in infected cells at these times of infection. Together these results bolster the hypothesis that IFI16 is the major DNA sensor in HSV-1–infected HFF cells.
In contrast to the supporting role played by cGAS, there is evidence from the literature that IFI16 is the primary DNA sensor in HSV-infected cells. IFI16 has been reported to bind to herpesviral DNA in HFF cells (9, 17). Furthermore, IFI16 localizes to intranuclear sites near the periphery of the nucleus near genome complexes (24) and within viral replication compartments during productive infection (27). Finally, HSV-1 ICP0 promotes the degradation of IFI16 in HFFs and prevents IRF-3 signaling (8). Therefore, the sum of these results argues that the major DNA sensor in HSV-infected HFF cells is IFI16, and that cGAS serves a novel function in maintaining nuclear levels of IFI16 (Fig. 8). The low levels of cGAMP induced in cells infected with the HSV-1 d109, which expresses no viral proteins de novo (18), argue that either low levels of viral DNA are exposed to cGAS in these conditions or that a viral inhibitor of cGAS is delivered to the infected cell by the virion.
Similar to our study, both cGAS and IFI16 have been reported to be involved in the response to HIV in phorbol myristate acetate (PMA)-differentiated human THP1 cells (28). It will be important to determine the specific mechanisms of action of each of these proteins under these conditions as well.
Roles for both IFI16 and cGAS in Innate Responses in Transfected HFF Cells.
In cells transfected with DNA, cGAS is required initially for IFNβ expression followed by a requirement for IFI16 in the later enhanced response. This finding is consistent with cGAS sensing the transfected DNA as it first enters the cytoplasm followed by IFI16 sensing it as it moves into the nucleus or as IFI16 moves to or accumulates in the cytoplasm upon de novo synthesis. In support of this theory, we observed an early sequential reorganization of cGAS followed by IFI16 filament formation, indicative of IFI16 activation (17, 25). Others have reported a later role for IFI16 in other cell types in response to transfected DNA. Zhang et al. (29) reported that the DDX41 DNA sensor was important earlier in the response to transfected DNA in dendritic cells whereas IFI16, which was up-regulated by the initial DDX41-dependent response, was subsequently involved in IFNβ expression. Unlike the phenotype reported for DDX41 (29), cGAS activity in HFF cells was transient and did not temporally overlap with IFI16 function. These results indicate that cGAS may act as a “sentinel” sensor in the cytoplasm, whereas IFI16 plays a later, apparently more central role in the response to plasmid DNA either upon the accumulation of DNA in the nucleus or upon accumulation of de novo synthesized IFI16 in the cytoplasm. The cellular responses that negatively regulate cGAS or result in a switch from cGAS-dependent to IFI16-dependent signaling are unknown. Unlike other signaling molecules, which are degraded upon activation [e.g., STING (30) and IRF-3 (31)], we do not observe a loss of cGAS upon DNA stimulation, indicating that the mechanism does not involve turnover of cGAS itself. It is possible that cGAS signaling results in the production of a factor that negatively regulates cGAS while promoting IFI16 activity, or that the IFI16 response gradually overtakes the cGAS response.
Roles in Other Cell Types and in Different Species.
It is possible that cGAS plays a more prominent role in directly detecting HSV-1 infection in other cell types, and additional studies examining multiple primary cell types might clarify such a role. It is also important to study primary cells because transformed cell lines and tumor-derived cell lines may not encode or express functional IFI16 (8, 9, 27), because IFI16 can act as a cell growth inhibitor (32). DNA sensors, such as IFI16 and cGAS, may also play different roles detecting and responding to related herpesvirus infections in different species. IFI16 is noted for rapid evolution in primates and other mammalian lineages (33) and has undergone repeated duplications and diversification, which is especially evident among rodents (34). Although cGAS appears to be a single-copy gene among mammals, it has also undergone rapid evolution in primates, similar to the evolution of several innate defense genes (35). Therefore, consideration of the context of infection, whether specific to cell type or species, will be crucial to further understanding the functional relationships between IFI16 and cGAS.
The importance of the cGAS/IFI16-IRF-3 pathway in vivo remains to be defined more fully. The closest murine homolog of IFI16, p204, has not been knocked out yet. irf3−/− mice show no defect in peripheral control of HSV-1 administered by the intravenous route (36) or the corneal route (37), but irf3−/− mice showed more central nervous system disease and viral replication than WT mice (37). Two independent reports have characterized two lines of cgas−/− mice to determine its role in vivo: Li et al. (13) reported that cgas−/− mice showed decreased IFN responses and were more susceptible to HSV-1 infection by the intraperitoneal route, and Schoggins et al. (38) demonstrated that cgas−/− mutations resulted in a down-regulation of basal IFN and resulted in increased RNA and DNA virus replication, including murine herpesvirus 68, indicating that the antiviral activity of cGAS is not specifically tied to DNA virus infection. Because there are no data on peripheral HSV clearance in the cgas−/− mice, more studies are needed to determine if IRF-3 and cGAS knock-out mice are consistent in their phenotypes with regard to HSV-1 pathogenesis.
With regard to possible pleiotropic effects of cGAS, cGAS depletion in our study did not result in a decrease in basal ISG (i.e., IFI16) transcription. This may represent a difference between transient and sustained depletion of cGAS or differences between mouse and human cells. However, given that cGAS stabilizes IFI16 at the protein level, it will be important to examine the effect of cGAS depletion or knockout on IFI16 levels in future studies. There may be indirect effects, however, when stable knockdown cells are studied. Thompson et al. recently reported that in long-term depletion experiments, loss of IFI16 leads to a decrease in IFN responses to multiple agents, including DNA and RNA viruses (10). Therefore, further studies are needed to fully define the functions of cGAS and IFI16.
In summary, our results show that both IFI16 and cGAS play essential roles in sensing of HSV DNA in infected human fibroblasts in culture. In contrast to the previously defined role for cGAS in synthesizing cGAMP, which activates STING, low levels of cGAMP, below the limit of detection, are produced by HSV-infected cells. Instead, in these cells cGAS binds to IFI16 and promotes its stability, allowing the accumulation of sufficient IFI16 to bind to HSV-1 DNA and initiate innate signaling. This finding illustrates a novel mechanism by which two DNA sensors can act together to provide the host cell with the ability to sense nuclear HSV-1 DNA.
Materials and Methods
Cells and Viruses.
HFF cells (American Type Culture Collection Hs27 ATCC CRL-1634) were cultured in DMEM supplemented with 10% (vol/vol) FBS and 2 mM glutamine and used at fewer than 10 passages. NOKs immortalized with human telomerase reverse transcriptase (16) were kindly provided by K. Munger (Tufts University School of Medicine, Boston, MA) and cultured in Keratinocyte-SFM (Gibco) supplemented with gentamicin and pen/strep. L929 cells were kindly proved by M. Nibert (Harvard Medical School, Boston, MA) and cultured in Joklik’s modified minimal medium supplemented with 4% (vol/vol) FBS, 2 mM glutamine, and pen/strep. HEK29293 and HEK293T cells were cultured in DMEM supplemented with 5% (vol/vol) FBS and 5% (vol/vol) bovine calf serum. cGAS-expressing HEK293 cells were constructed by transfection of a human cGAS expression plasmid or control vector plasmid into HEK293 cells. At 48 hpt, cells were cultured in 10 μg/mL blasticidin and blasticidin-resistant cells were pooled after 2 wk of selection and used for the described experiments.
The HSV-1 d109 virus (18) was grown and titrated as previously described (8). The ICP0-null 7134 virus was grown and titrated as described (19). WT HSV-1 KOS strain was propagated and titrated on Vero cells.
Plasmids and DNA Transfection.
HFF cells were plated at a density of 1 × 105 cells per 2.2-cm well at 24 h before transfection. Cells were transfected with 200 ng of plasmid DNA (pCDNA3.1) or LabelIT Cy3 plasmid delivery control (Mirus) with Lipofectamine LTX with Plus reagent (Life Technolgies) according to the manufacturer’s instructions. Human N-terminal FLAG cGAS (pcGAS) was constructed using the pSPORT CMV C6ORF150 clone (Open Biosystems) as a template for PCR to generate a NotI (FLAG)-Bstz17I fragment, which was subsequently ligated into the NotI-PmeI sites in pcDNA6 (Invitrogen) using the following primers: cGAS FLAG (5′-TTTTTTGCGGCCGCcagccATGGATTACAAGGATGACGACGAT AAGcagccttggcacgga-3′) and cGAS Rev (5′-TTTTTGTATACtcaaaattcatcaaaaactggaaact-3′), with the underlined sequences defining the added restriction sites, the italicized sequence defining an added kozak sequence, and the lowercase sequences defining the cellular gene.
siRNA Depletion.
HFF cells were plated at a density of 1 × 105 cells per well (12-well) 24 h pre-siRNA transfection. Cells were transfected with 50 nM of Control, IFI16, STING (On-TARGETplus) or cGAS siRNAs (11) (Thermo Scientific) using DharmaFECT 2 (Thermo Scientific) according to the manufacturer’s instructions. Overlay media was replaced at 24 hpt and depletion efficiency was determined at 72 hpt.
Western Blot and Antibodies.
Cells were harvested in NuPAGE LDS sample buffer (Life Technologies) or fractionated using NE-PER Nuclear Cytoplasmic Extraction kit (Thermo Scientific) and run on NuPAGE NOVEX 4–12% Bis-Tris Protein Gels (Life Technologies). Proteins were transferred to PVDF membranes overnight at 4 °C, and the membranes were blocked in 5% (wt/vol) milk (PBS) before incubation in primary antibody. Membranes were washed 3× with PBS containing 0.05% Tween-20 (PBST), incubated in secondary antibody for 1 h at room temperature, washed twice with PBST, and once with PBS. Membranes were incubated in SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and exposed to film. The antibodies used in this study were anti-IFI16 (IG7, 1:2,000; Santa Cruz), anti-MBD21(cGAS) (1:1,000; Sigma), anti-STING (1:1,000; Cell Signaling), anti-GAPDH (1:5,000; Abcam). Secondary goat anti-mouse and goat anti-rabbit antibodies were from Santa Cruz and used at a 1:5,000 dilution.
qRT-PCR.
RNA was harvested from infected or transfected cells using the RNeasy kit (Qiagen). An aliquot of 1 μg of RNA was DNase treated (DNA-free; Ambion) for 1 h at 37 °C. DNase-treated RNAs were reverse-transcribed (High-Capacity cDNA Reverse Transcription Kit; Life Technologies), and cDNAs were analyzed by real-time PCR (Step One Plus; Applied Biosystems) using Power SYBR green or Fast SYBR green master mixes. Values from cells infected with HSV-1 were normalized to 18S RNA levels, and values from cells transfected with plasmid DNA were normalized to 18S or GAPDH RNAs. The primers used in this study were: Human IFNβ: 5′-AAACTCATGAGCAGTCTGCA-3′, 5′-AGGAGATCTTCAGTTTCGGAGG-3′; Human ISG54: 5′-ACGGTATGCTTGGAACGATTG-3′, 5′-AACCCAGAGTGTGGCTGATG-3′; Human IL-6: 5′-ACCGGGAACGAAAGAGAAGC-3′, 5′-CTGGCAGTTCCAGGGCTAAG-3′; 18S RNA: 5′-GCATTCGTATTGCGCCGCTA-3′ 5′-AGCTGCCCGGCGGGTC-3′; Human STING 5′-CCTGAGCAGAACAACTGC-3′, 5′-GGTCTTCAAGCTGCCCACAGTA-3′; Human cGAS: 5′-GGGAGCCCTGCTGTAACACTTCTTAT-3′; 5′-CCTTTGCATGCTTGGGTACAAGGT-3′; Human IFI16; 5′-ACTGAGTACAACAAAGCCATTTGA-3′, 5′-TTGTGACATTGTCCTGTCCCCAC-3′; and GAPDH: 5′-TTCGACAGTCAGCCGCATCTTCTT-3′, 5′-CAGGCGCCCAATACGACCAAATC-3′.
Immunofluorescence.
Transfected or infected cells were fixed with 2% (vol/vol) formaldehyde, permeabilized with 0.5% Nonidet P-40, and blocked in 5% (vol/vol) normal goat serum overnight at 4 °C. Fixed cells were incubated with primary antibodies for 30 min at 37 °C and washed two times with PBS containing 0.05% Tween 20 followed by one wash with PBS. Alexa Fluor 488- and 594-conjugated secondary antibodies (1:500; Jackson Immunoresearch) were incubated with cells for 2 h at 25 °C. The coverslips were washed as above and mounted on slides with ProLong Gold antifade reagent (Invitrogen). Images were acquired using an Olympus Fluoview Confocal Microscope with a 63× objective. Primary antibodies used for IF were: anti-MBD21(cGAS) (1:50; Sigma), anti-IFI16 (1:200; Abcam), anti-Lamin B1 (1:200; Abcam), anti-Lamin B2 (1:200; Abcam).
IFI16 Immunoaffinity Purifications and Targeted MS/MS Analysis on cGAS.
HFFs cultured as above were either uninfected (Mock) or infected for 3 h with WT HSV-1 at an MOI of 10, washed and scraped in PBS, resuspended in 20 mM Na-Hepes, 1.2% polyvinylpyrrolidone (wt/vol), pH 7.4, and frozen as pellets in liquid nitrogen, and processed, as described previously (39). Briefly, frozen cells were cryogenically lysed using a Retch MM301 Mixer Mill (Retch), and the frozen powder was suspended in 14 mL of 20 mM K-Hepes, pH 7.4, 0.11 M KOAc, 2 mM MgCl2, 0.1% Tween-20 (vol/vol), 1 µM ZnCl2, 1 µM CaCl2, 0.6% Triton X-100, 200 mM NaCl, 100 U/mL benzonase (Pierce), 1/100 protease inhibitor mixture (PIC; Sigma), 1/100 phosphate inhibitor mixtures 2 and 3 (PHIC; Sigma). The lysates were placed for 10 min on a roller at room temperature for endonuclease digestion, then homogenized for 30 s using a PT 10-35 GT Polytron (Kinematica), and subjected to centrifugation at 7,000 × g for 10 min at 4 °C. Immunoaffinity purifications were performed by mixing the supernatant with magnetic beads (7 mg per isolation), which have been conjugated with either anti-IFI16 antibodies (1:1 mixture of ab50004 and ab55328; Abcam) or with IgG for control isolations, as described previously (40). Coisolated proteins were digested with 12.5 ng/µL trypsin (Promega) in 50 mM ammonium bicarbonate overnight at 37 °C, and prepared for nLC-MS/MS analyses on a Dionex Ultimate 3000 nanoRSLC system (Dionex) coupled online to an ESI-LTQ-Orbitrap Velos (Thermo Fisher Scientific), as described previously (41). Peptides were separated by reverse-phase chromatography (Acclaim PepMap RSLC, 1.8 µm × 75 µm × 25 cm) over a 90-min discontinuous gradient of acetonitrile. To specifically search for putative cGAS peptides, data were imported into Skyline software (v2.6), and precursor extracted ion chromatograms (XICs) were obtained using the following settings: Isotope Count, 3; precursor mass analyzer, Orbitrap; Resolving power, 60,000 at 400 Th. The peak integration boundaries for the cGAS peptides in the IFI16 and IgG IPs were inspected manually. The identity of the cGAS peptides was confirmed by collision induced dissociation MS/MS fragmentation in the linear ion trap.
cGAMP Activity Assay.
Infected or transfected cells were trypsinized and washed with PBS. Pelleted cells were resuspended in the indicated volume of PBS and lysed by freezing at −80 °C. Cell extracts were nuclease and heat-treated as previously described (23) with modifications. Briefly, cell extracts were incubated with ∼1 U/μL Benzonase (Sigma-Aldrich) for 30 min at 37 °C. Cell extracts were then heated at 95 °C for 5 min and centrifuged for 5 min at maximum speed (16,100 × g) in an Eppendorf microcentrifuge. HFF were used as a reporter cell to measure cGAMP production. HFFs were permeabilized as described previously (42) with modifications. Briefly, media was aspirated from the HFFs, and digitonin permeabilization solution (50 mM Hepes pH 7.0, 100 mM KCl, 85 mM sucrose, 3 mM MgCl2, 0.2% BSA, 1 mM ATP, 0.1 mM DTT, 10 μg/mL digitonin) was added with 25 μL per well-treated cell extracts or 3′-5′-cGAMP (Invivogen #tlrl-cga-s) as a positive control. HFF reporter cells were incubated with extracts for 30 min at 37 °C and then replaced with supplemented media. RNA was harvested 6 h after the initial addition of extracts and qRT-PCR was performed, as described above.
Supplementary Material
Acknowledgments
These studies were supported by National Institutes of Health Grants AI106934 and DE023909 (to D.M.K.), DP1DA026192 and R21AI102187 (to I.M.C.), T32 AI007245 postdoctoral Training Grant (to N.M.B.), and American Heart Association predoctoral Fellowship 14PRE18890044 (to B.A.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424637112/-/DCSupplemental.
References
- 1.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brencicova E, Diebold SS. Nucleic acids and endosomal pattern recognition: How to tell friend from foe? Front Cell Infect Microbiol. 2013;3(37):1–14. doi: 10.3389/fcimb.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knipe DM, et al. Snapshots: Chromatin control of viral infection. Virology. 2013;435(1):141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 5.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013;87(9):5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA. 2012;109(44):E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci USA. 2012;109(26):10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson MR, et al. Interferon γ-inducible protein (IFI) 16 transcriptionally regulates type I interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J Biol Chem. 2014;289(34):23568–23581. doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unterholzner L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology. 2013;218(11):1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Orzalli MH, Knipe DM. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piboonniyom SO, et al. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 2003;63(2):476–483. [PubMed] [Google Scholar]
- 17.Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013;14(5):591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samaniego LA, Neiderhiser L, DeLuca NA. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72(4):3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai W, Schaffer PA. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66(5):2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eidson KM, Hobbs WE, Manning BJ, Carlson P, DeLuca NA. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J Virol. 2002;76(5):2180–2191. doi: 10.1128/jvi.76.5.2180-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol. 2004;78(4):1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melroe GT, DeLuca NA, Knipe DM. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol. 2004;78(16):8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341(6148):903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuchet-Lourenço D, Anderson G, Sloan E, Orr A, Everett RD. The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J Virol. 2013;87(24):13422–13432. doi: 10.1128/JVI.02474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrone SR, et al. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci USA. 2014;111(1):E62–E71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connolly DJ, Bowie AG. The emerging role of human PYHIN proteins in innate immunity: Implications for health and disease. Biochem Pharmacol. 2014;92(3):405–414. doi: 10.1016/j.bcp.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 27.Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci USA. 2013;110(47):E4492–E4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsen MR, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci USA. 2013;110(48):E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155(3):688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18(5):2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22(31):4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 33.Cagliani R, et al. Ancient and recent selective pressures shaped genetic diversity at AIM2-like nucleic acid sensors. Genome Biol Evol. 2014;6(4):830–845. doi: 10.1093/gbe/evu066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunette RL, et al. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209(11):1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daugherty MD, Malik HS. Rules of engagement: Molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 36.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 37.Menachery VD, Pasieka TJ, Leib DA. Interferon regulatory factor 3-dependent pathways are critical for control of herpes simplex virus type 1 central nervous system infection. J Virol. 2010;84(19):9685–9694. doi: 10.1128/JVI.00706-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoggins JW, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowles DL, Terhune SS, Cristea IM. Discovery of host-viral protein complexes during infection. Methods Mol Biol. 2013;1064:43–70. doi: 10.1007/978-1-62703-601-6_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4(12):1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Joshi P, et al. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328(5986):1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.