Significance
The discovery that 5-methylcytosine (5mC) can be iteratively oxidized by mammalian ten-eleven translocation (TET) proteins marks a breakthrough in the field of epigenetics. To better understand the evolutionary and functional linkage of TET family members, we characterized NgTET1 from the protist Naegleria gruberi, which bears homology to both TET and base J-binding protein, a thymidine hydroxylase in trypanosomes. We show that NgTET1 performs iterative oxidation of both 5mC and thymidine (T) (minor activity) on various DNA forms, and that these activities can be modulated by mutagenesis. We also present evidence for the effect of sequence context on both 5mC- and T-oxygenase activities. Finally, we show the utility of NgTET1 at direct methylome profiling using single-molecule, real-time sequencing.
Keywords: TET proteins, NgTET1, 5-methylcytosine, SMRT sequencing, bacterial methylome
Abstract
Modified DNA bases in mammalian genomes, such as 5-methylcytosine (5mC) and its oxidized forms, are implicated in important epigenetic regulation processes. In human or mouse, successive enzymatic conversion of 5mC to its oxidized forms is carried out by the ten-eleven translocation (TET) proteins. Previously we reported the structure of a TET-like 5mC oxygenase (NgTET1) from Naegleria gruberi, a single-celled protist evolutionarily distant from vertebrates. Here we show that NgTET1 is a 5-methylpyrimidine oxygenase, with activity on both 5mC (major activity) and thymidine (T) (minor activity) in all DNA forms tested, and provide unprecedented evidence for the formation of 5-formyluridine (5fU) and 5-carboxyuridine (5caU) in vitro. Mutagenesis studies reveal a delicate balance between choice of 5mC or T as the preferred substrate. Furthermore, our results suggest substrate preference by NgTET1 to 5mCpG and TpG dinucleotide sites in DNA. Intriguingly, NgTET1 displays higher T-oxidation activity in vitro than mammalian TET1, supporting a closer evolutionary relationship between NgTET1 and the base J-binding proteins from trypanosomes. Finally, we demonstrate that NgTET1 can be readily used as a tool in 5mC sequencing technologies such as single molecule, real-time sequencing to map 5mC in bacterial genomes at base resolution.
Modified DNA bases exist in all forms of life, from viruses to mammals with many different biological roles. Accordingly, diverse mechanisms have evolved to “write,” “read,” and “erase” these modifications. In mammals, 5-methylcytosine (5mC) is the major form of DNA modification and is implicated in many crucial developmental processes. In human and mouse, 5mC can be successively oxidized into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) by the ten-eleven translocation (TET) family of oxygenases (1–4). The bases of 5fC and 5caC can be excised by thymine DNA glycosylase (4). The 5mC-oxidation–coupled base-excision repair pathway provides a plausible route for active demethylation in mammalian cells. Many other species, from simple to complex, maintain DNA methylation machinery throughout their life cycle that may contribute to epigenetic regulation. Therefore, an interesting perspective is to examine shared and distinct features of TET oxygenases in diverse eukaryotes (5, 6).
The human and mouse genomes encode three paralogous TET proteins, TET1, TET2, and TET3, which presumably carry out both redundant and distinct functions (7, 8). TET proteins belong to the diverse group of α-ketoglutarate (αKG) and Fe(II)-dependent oxygenases (5). Subgroup classification based on sequence similarity links the TET proteins to base J-binding proteins (JBP1 and JBP2), which are primarily present in trypanosomes and possess thymidine (T)-hydroxylation activity (1). Further bioinformatic analysis revealed eight paralogous TET/JBP-like genes in the genome of Naegleria gruberi, a single-celled amoeboflagellate protist that is a distant cousin of the parasitic trypanosomes, evolutionarily far removed from vertebrates (5, 9). Interestingly, genetic components for “writing” 5mC (i.e., homologs of mammalian DNA methyltransferases) are also present in the genome (9). These components may parallel the methylation/oxidation processes in mammalian cells.
We show here that new insights into the TET family of enzymes can be obtained by studying a representative from a protist that may have shared an ancestral TET enzyme with mammals, but then evolved separately from vertebrates for much of eukaryotic evolution. We have previously reported the in vitro biochemical activity and structure of an active N. gruberi TET/JBP-like protein, termed NgTET1 (10). We showed that like mammalian TETs, NgTET1 is capable of catalyzing the oxidation of 5mC to 5hmC, 5fC, and 5caC in vitro. The crystal structure of NgTET1 in complex with a symmetrically methylated oligonucleotide (oligo) reveals a base-flipping mechanism in which the DNA is bent and the flipped 5mC is positioned in the catalytic binding pocket (10). The hydrogen-bond networks between NgTET1 and substrate are specific to the flipped 5mCpG dinucleotide, and we reported a substrate preference for 5mCpG-containing oligo DNA (10). Here we extend this observation by reporting the activity of NgTET1 on various types of DNA containing different methylation motifs and in different conformations. Importantly, we show that it exhibits T-oxygenase activity, similar to JBP1 and JBP2, but can catalyze the formation of further oxidized T species, 5-formyluridine (5fU) and 5-carboxyluridine (5caU), in addition to 5-hydroxymethyluridine (5hmU). We compare the in vitro activities of NgTET1 and the catalytic domain of mouse TET1 (mTET1CD) on various substrates and show that the two enzymes exhibit similar 5mC-oxygenase activities but vary in the extent of their T-oxygenase activities, with NgTET1 displaying notably higher T-oxygenase activity than mTET1CD. Finally, we demonstrate the utility of NgTET1 in methylome sequencing applications, such as single molecule, real-time (SMRT) sequencing.
Results
NgTET1 Is an Fe(II)/αKG-Dependent 5-Methylpyrimidine Oxygenase.
5mC-oxygenase activity.
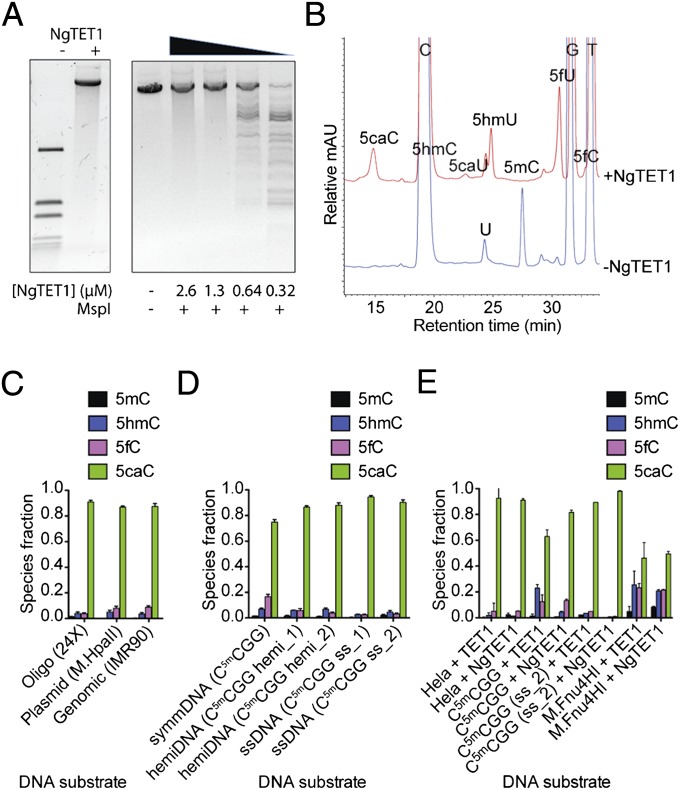
Full-length NgTET1 was expressed and purified to homogeneity and tested for activity on DNA containing 5mC. First, a restriction enzyme (RE)-based assay was used to test protection of pRS(M.HpaII), a linear plasmid in which all internal Cs in a CCGG recognition site are methylated by the endogenously expressed M.HpaII methyltransferase, upon treatment with NgTET1 (Fig. 1A). The plasmid is cleaved at the same recognition sequence (C5mCGG) by MspI for both 5mC and 5hmC sites. When oligo substrates contain 5caC or symmetrical 5fC, MspI cleavage does not occur (Fig. S1 and Table S1). Fig. 1A illustrates that the observed MspI protection is dependent on the concentration of NgTET1 used in a 30-min reaction at 34 °C, the optimal temperature for the NgTET1 reaction (Fig. S2). Full protection from MspI digestion is achieved at 0.01 µM plasmid DNA (equivalent to 0.3-µM 5mC sites) and an NgTET1 concentration of 1 µM and higher (Fig. 1A).
Fig. 1.
Enzymatic activity of NgTET1 on oligo, plasmid and gDNA. (A) RE-based assay showing protection of NgTET1-treated pRS(M.HpaII) plasmid against digestion with MspI at varying concentrations of NgTET1. (B) LC-MS (Agilent 1200)-based assay reflecting NgTET1 reaction species using mammalian gDNA IMR90. Reactions contained 1.5 μg sheared (1.5-kb) DNA and 4 μM NgTET1. (C–E) Quantification of NgTET1 reaction species as measured by LC-MS (Agilent 1200 for C and D; 6490 Triple Quad LC-MS for E) for different types of DNA. The error bars (in black) represent the SEM (n ≥ 3). (C) Two micromolars oligo (Table S1), 1.5 μg plasmid, and 1.5 μg gDNA were used with 4 µM NgTET1. (D) Four micromolars 5mC sites for symmDNA, hemiDNA and ssDNA (Table S1) were used with 8 μM NgTET1. (E) Two micromolars ds- or ss-oligo (Table S1) or sheared (1.5-kb) HeLa (0.5 μg) or M.Fnu4HI (0.2 μg) gDNA were used with 6.7 µM NgTET1 (in Mops buffer pH 6.9) or mTET1CD.
We also used a liquid chromatography-mass spectrometry (LC-MS)–based assay as a more sensitive method to detect and quantify each species in the oxidation reaction, as previously described (10). Fig. 1B shows a representative chromatogram from an LC-MS–based activity assay in the absence or presence of NgTET1 and genomic DNA (gDNA) from human cells (IMR90) as substrate. 5mC of IMR90 is completely converted to 5caC (major product) with small amounts of 5hmC and 5fC remaining after a 1-h incubation with NgTET1 at 34 °C. The amount of 5mC and its oxidized species present in the reaction is quantified and displayed in Fig. 1C for three different types of DNA: a 56-bp double-strand DNA (dsDNA) oligo substrate containing 24 5mCpGs (see Tables S1 and S2 for list of all substrates used in this study), pRS(M.HpaII) plasmid, and IMR90 gDNA. All three substrates contain 5mC methylation at multiple CpG sites on both strands of dsDNA. Nearly all of the 5mC (<1% unreacted) in each of these three substrates is converted to ≥87% 5caC, with small amounts of 5hmC and 5fC remaining (Fig. 1C and Table S3).
In addition to its oxygenase activity on dsDNA symmetrically methylated on both strands (symmDNA), NgTET1 oxidizes 5mC on hemimethylated (hemiDNA) and single-strand DNA (ssDNA) (Fig. 1D). After a 1-h incubation, the amounts of 5mC, 5hmC, 5fC, and 5caC as quantified by the LC-MS assay are comparable for all three types of substrates (Fig. 1D and Table S1). The ability of NgTET1 to catalyze oxidation of hemiDNA or ssDNA is consistent with the observation that NgTET1 forms hydrogen-bond contacts with 5mC on only one strand in the crystal structure of the enzyme, in complex with a symmetrically methylated dsDNA oligo substrate (10).
The relatively permissive substrate specificity of NgTET1, as indicated by its activity on these various substrates, raised the question of whether similar promiscuity is observed with the mammalian TET proteins. The activity of the C-terminal catalytic domain of mTET1CD on hemiDNA and ssDNA has been reported previously (11). Here we compare the activity of mTET1CD on ssDNA, dsDNA, and gDNA, using an LC-MS–based activity assay to measure the amount of 5mC, 5hmC, 5fC, and 5caC after a 1-h incubation. Indeed, we found that mTET1CD can convert 5mC to 5caC in all substrates tested, with similar efficiency as NgTET1 (Fig. 1E).
T-oxygenase activity.
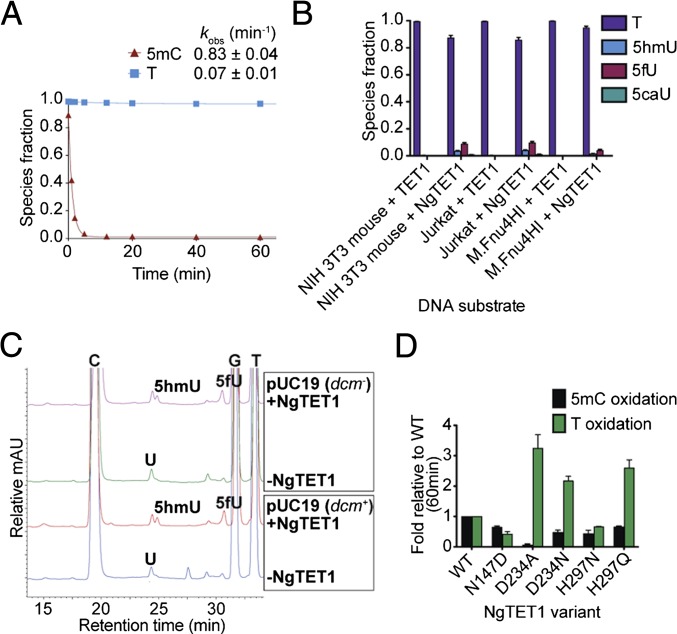
Bioinformatic analysis suggests an evolutionary linkage between the TET proteins and JBPs, which catalyze the hydroxylation of the methyl group in T to form 5hmU (5, 12). We detected LC-MS evidence for the formation of 5hmU, as well as the further oxidized species, 5fU and 5caU, in the reaction of NgTET1 on DNA (Fig. 1B). The formation of the oxidized T species is dependent on NgTET1, Fe(II), and αKG, and possibly follows a similar catalytic mechanism to that of 5mC oxidation. The decay of T is, however, significantly slower than the decay of 5mC, as observed for the C5mCGG oligo, for which less than 3% of the total number of Ts are oxidized, whereas nearly 100% of the total 5mC bases are oxidized after a 1-h reaction (Fig. 2A and Table S1). However, a direct kinetic comparison of 5mC- and T-oxygenase activity using this particular substrate is not possible, given the excess number of Ts (n = 20) compared with 5mC sites (n = 2). Attempts to perform a quantitative comparison using oligos with the same sequence bearing either a single T or 5mC site have been unsuccessful because of lack of detection of T oxidation. Nonetheless, we conclude that the T-oxygenase activity of NgTET1 is minor compared with its 5mC-oxygenase activity.
Fig. 2.
T-oxygenase activity of NgTET1. (A) Kinetic time course depicting the decay of 5mC or T for a reaction with 4 μM NgTET1 and 2 μM oligo C5mCGG. Reaction species were detected and quantified by LC-MS (Agilent 1200). The data are fit to a single exponential and the observed rate constants with SEM are provided. (B) Quantification of oxidized T reaction species using 6.7 μM mTET1CD or NgTET1 (in Mops buffer pH 6.9) as measured by LC-MS (6490 Triple Quad LC-MS) for 0.2 μg sheared (1.5-kb) gDNA substrates. The error bars (in black) represent the SE (SEM) (n ≥ 3). (C) LC-MS (Agilent 1200) traces comparing T-oxygenase activity by NgTET1 (10 μM) on methylated and unmethylated pUC19 plasmid DNA (2.5 μg). (D) LC-MS (Agilent 1200) quantification of 5mC or T after a 1-h reaction of 4 μM NgTET1 WT or variant proteins with 2 μM oligo C5mCGG. Error bars (in black) represent the SEM (n ≥ 3).
Although T oxidation appears to be minor, this activity may have some physiological relevance as 5hmU formation through T oxidation was recently reported for the mammalian TET proteins in mouse embryonic stem cells (13). We compared the in vitro T-oxidation activity of NgTET1 and mTET1CD on various gDNA and oligo substrates (Figs. 2B and 3D). Intriguingly, significantly higher levels of 5hmU and 5fU formed in the reaction of NgTET1 compared with mTET1CD, and 5caU was detected only in the NgTET1 reaction. To further characterize this activity, we first tested whether T oxidation is dependent on 5mC methylation of the substrate DNA (i.e., if cytosine methylation is required for binding or recruitment of the DNA to the active site of NgTET1 before T oxidation). We compared T-oxygenase activity on pUC19 produced in a DNA cytosine-C5-methyltransferase+ (dcm+) Escherichia coli strain (methylated at C5mCWGG sites) compared with that from a dcm− strain (without cytosine methylation), isolated under identical conditions. As expected, LC-MS analysis of the reaction products shows peaks corresponding to 5hmC, 5fC, and 5caC for pUC19 (dcm+) but not pUC19 (dcm−) in the presence of NgTET1 (Fig. 2C). On the other hand, both substrates form comparable amounts of oxidized T products (5hmU and 5fU) in the presence of NgTET1 (Fig. 2C), suggesting that this activity is not dependent on the presence of 5mC in the DNA substrate.
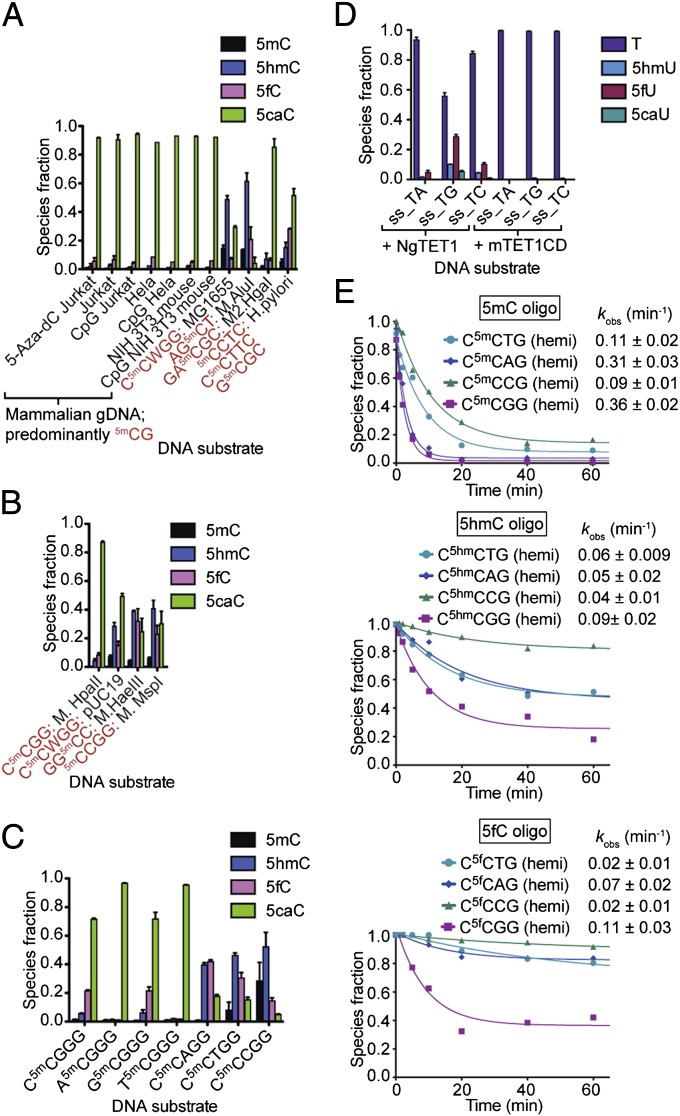
Fig. 3.
NgTET1 activity is dependent on nucleotide-sequence context. Distribution of NgTET1 reaction species: (A and B) As quantified by LC-MS (Agilent 1200), using excess enzyme (20 µM) with (A) genomic (2.5 μg, sheared to 1.5-kb) or (B) plasmid (2.5 μg) DNA containing different methylation sequences as indicated in red (Table S2); (C and D) As quantified by 6490 Triple Quad LC-MS for (C) 30-min reaction of 8 μM NgTET1 with 4 μM oligo DNA or (D) 6.7 μM NgTET1 (in Mops buffer pH 6.9) or mTET1CD with 1.6 μM oligo DNA. For A–D, error bars (in black) represent the SEM (n ≥ 3). (E) Kinetic traces, with species fraction determined by LC-MS (Agilent 1200), of NgTET1 with 5mC-, 5hmC-, or 5fC-containing oligos hemimethylated at a single CpX site (Table S1). Reactions were done in Mops buffer (pH 6.75) using 8 μM NgTET1 and 4 μM DNA. The data are fit to a single exponential and the observed rate constants with SEM are provided.
We next probed the role of specific amino acid residues in the recognition of 5mC and T in the active site of NgTET1. In the crystal structure, the flipped 5mC is situated in the active-site pocket and stabilized by hydrogen-bonding interactions with the side chains of three key residues: aspartic acid 234 (D234), histidine 297 (H297), and asparagine 147 (N147) (Fig. S3) (10). Here we describe both 5mC- and T-oxidation activities of several site-directed variants of these residues with a 56-bp dsDNA substrate C5mCGG (Table S1), plotting the ratio of the amount of 5mC and T remaining after a 1-h reaction compared with that of the WT NgTET1 reaction (Fig. 2D). We also performed a kinetic time-course analysis for selected variants to monitor the decay of both 5mC and T over time (Fig. S4). Alteration of any of these residues resulted in a decrease (relative to WT) in the total amount of 5mC converted after 1 h, ranging from 17-fold for D234A to 1.5-fold for H297Q (Fig. 2D). The observed effects of these alterations on NgTET1 activity are in agreement with our previously published results (10). Interestingly, T oxidation was decreased for some, but not all, of the variants. Alteration of D234 to A, most strikingly, greatly diminishes 5mC activity, but actually increases T activity by approximately threefold compared with WT. This pattern is also observed for D234N and H297Q, although to a lesser extent (Fig. 2D). The same trends are observed in the overall kinetic time courses of the NgTET1 variants (Fig. S4). D234 is proposed to make interactions specific for a C by interacting with the exocyclic amino group N4 of 5mC (Fig. S3), rather than T, which carries a carbonyl oxygen at the corresponding position. By disrupting this interaction and substituting D with the much smaller A, or to a lesser extent N, the active site pocket may more easily accommodate a T. However, the activity on T is still low (total conversion ∼7%) relative to the total amount of Ts present in the DNA molecule. It is yet unclear how the H297 to Q substitution may increase T oxidation, in the absence of any structural information for this variant.
The Extent of 5mC Oxidation Is Dependent on Sequence Context.
We reported previously that NgTET1 has a strong preference for 5mCpG sites in oligo substrates (10). Here we test the activity of NgTET1 on other types of DNA, both genomic and plasmid, and confirm that NgTET1 activity is dependent on the methylation sequence context (Fig. 3 A and B and Table S2). The amount of residual 5mC after a 1-h reaction varies greatly, with the most complete conversion of 5mC observed for substrates containing 5mCpG methylation (e.g., mammalian gDNA) and the least conversion for substrates with non-5mCpG methylation (e.g., MG1655) (Fig. 3 A and B and Table S2). A similar context preference is observed for both NgTET1 and mTET1CD on M.Fnu4HI gDNA, which is methylated at G5mCNGC sites (Fig. 1E).
To examine the effect of the methylation sequence context on the activity of NgTET1 more closely, we designed oligo substrates bearing an N15mCN2GG methylation motif, where N1 is maintained at C when N2 is A, T, C, or G, and N2 is maintained at G when N1 is A, T, C, or G (Table S1). Our results reflect that the nucleotide 5′ upstream of 5mC plays a minor role in the extent of 5mC oxidation by NgTET1, whereas the nucleotide 3′ downstream exhibits a much more pronounced effect on this activity with a substantial increase in the intermediate species (5hmC and 5fC) when N2 is not a G (Fig. 3C). The relative amount of 5caC formed after a 30-min reaction for non–5mCpG-containing substrates is only 6–20%, compared with 72% for the 5mCpG-containing substrate (Fig. 3C). These results are consistent with those of plasmid and genomic substrates (Fig. 3 B and C), as well as previous observation (10). We next tested substrate preference for NgTET1 T-oxygenase activity. A set of ssDNA oligo substrates containing nine Ts, each followed by either a G, A, or C was compared (Fig. 3D and Table S1), and the results reflect a TpG sequence preference.
Interestingly, each oxidative step of the reaction seems to be differentially sensitive to sequence context. For example, much lower amounts of 5caC are formed on G5mC substrates (M.AluI and M.HaeIII) than C5mCWGG substrates (MG1655 and pUC19), despite approximately the same amount of oxidized 5mC. These differences prompted us to further investigate the observed sequence context preference when the starting substrate contains 5hmC or 5fC, rather than 5mC. Fig. 3E shows the decay of 5mC, 5hmC, or 5fC over time for hemiDNA substrates containing a single modification. The data demonstrate the faster kinetics of the first step of the overall NgTET1 reaction (i.e., 5mC to 5hmC) compared with the two subsequent steps (Fig. 3E). In addition, the decay of 5mC proceeds to completion with nearly all of it gone after a 1-h reaction, whereas a considerable amount of starting material remains for the 5hmC- and 5fC-containing substrates (∼20% and ∼40%, respectively). Despite these differences in kinetics between each step of the reaction, the substrate preference for CpG sites is observed for the 5hmC- and 5fC-containing oligos as well, and CpC-modified oligos are clearly the poorest substrates (Fig. 3E). Overall, these results suggest that the reaction kinetics of each oxidative step by NgTET1 not only depends on the type of cytosine modification but also on the sequence context flanking the modified cytosine.
Mapping 5mC Using NgTET1 in SMRT Sequencing.
SMRT sequencing has been shown to readily detect most modifications in a DNA template, such as N6-methyl-adenine and N4-methyl-cytosine, but is limited in its detection of 5mC (14–16). In SMRT sequencing, the DNA polymerase kinetics are monitored by measuring the interpulse duration (IPD), the length of time between two successive nucleotide incorporation events (14, 15). The effect on the IPD ratio, a measurement of the IPD compared with an unmodified control template, is indicative of the modification of a specific base. By using NgTET1 to convert 5mC to 5caC, the newly modified base gives a stronger signal enabling better detection of 5mC (17).
We first used NgTET1 to treat pRS(M.HpaII), containing 15 C5mCGG recognition sites on each strand. Under our reaction conditions, all of the 5mC is reacted and 87% 5caC is generated (Fig. 1C). Fig. S5 shows the plasmid-wide view of IPD ratio data for pRS(M.HpaII) treated with NgTET1 and a representative IPD ratio profile in a 50-bp window. An increase in IPD ratio is detected at each predicted 5mC site, and the observed primary IPD ratio peak at the +2 position and a less prominent peak at the +6 position relative to the modification are consistent with the kinetic signature observed for a 5caC modification (17). As a result, all 30 5mCpG sites in pRS(M.HpaII) were readily detected (Fig. S5).
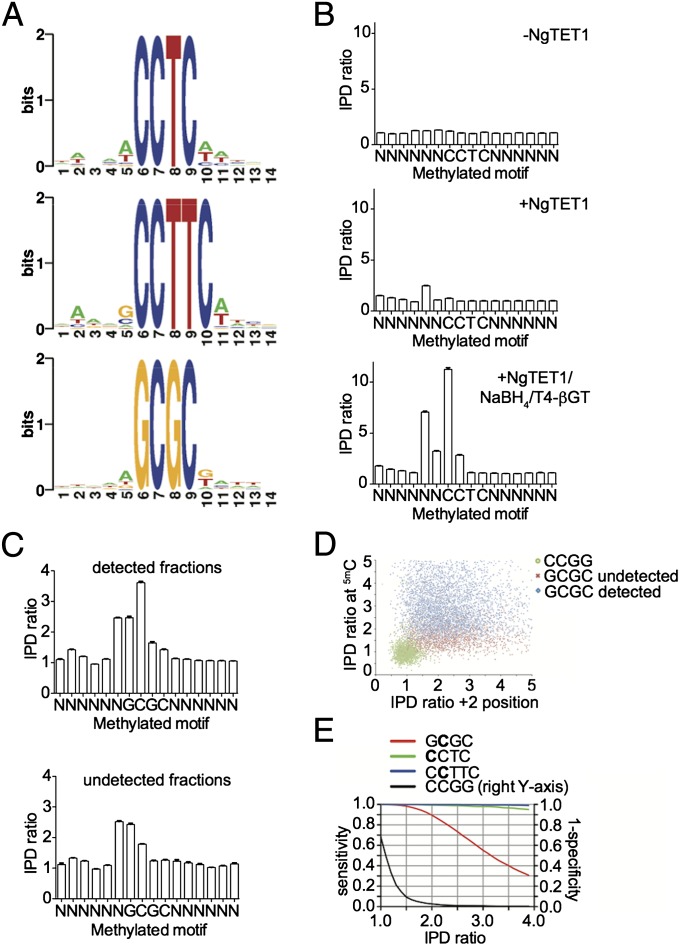
We then used NgTET1 to map the methylome of Helicobacter pylori strain 26695 gDNA, in which there are three known active cytosine-5-methylases with the specificities: G5mCGC, 5mCCTC, and C5mCTTC (18, 19). The reaction of H. pylori gDNA with NgTET1 results in an overall 5mC conversion of ∼95% but with only 52% 5caC formed (Fig. 3A). The relatively low conversion efficiency to 5caC may be because of the fact that two of the three known 5mC methylation motifs in H. pylori are in a non-CpG context. Nonetheless, all three methylated motifs were detected and the sequence contexts around the methylated sites show that there is no significant bias, suggesting that NgTET1 did not preferentially convert a subset of these sites (Fig. 4A).
Fig. 4.
SMRT sequencing of H. pylori gDNA using NgTET1. (A) Sequence logos for 5mCCTC, 5mCCTTC and G5mCGC motifs detected by SMRT sequencing of NgTET1-treated gDNA. (B) IPD ratio plots corresponding to the 5mCCTC motif in gDNA treated in the absence of NgTET1 (Top), with NgTET1 (Middle), or with NgTET1/NaBH4/T4-βGT (Bottom). (C) IPD ratio plots for the sequences detected (Upper) versus undetected (Lower) as belonging to the G5mCGC motif for gDNA treated with NgTET1/NaBH4/T4-βGT. For B and C, the error bars (in black) represent the SEM. (D) Scatter plot of IPD ratio values at the methylated and +2 positions for G5mCGC and CCGG sequences for gDNA treated with NgTET1/NaBH4/T4-βGT. (E) Plot of sensitivity and specificity as a function of IPD ratio for gDNA treated with NgTET1/NaBH4/T4-βGT.
Because of the high levels of 5hmC and 5fC formed in the reaction with H. pylori gDNA (Fig. 3A), we wanted to improve the detection of 5mC by using sodium borohydride (NaBH4) to reduce 5fC to 5hmC, and T4-β-glucosyltransferase (T4-βGT) to convert all 5hmC to β-glucosyl-oxy-5-methylcytosine (5gmC), a modification that would readily be detected by SMRT sequencing (20, 21). This protocol results in a product mixture of 5caC and 5gmC with negligible amounts of 5mC, 5hmC, or 5fC (Fig. S6). Using this approach followed by SMRT sequencing, we observed an improved signal in the genome-wide average IPD ratio profile, leading to a higher percentage detection of all three methylated motifs (Fig. 4B). Note that in addition to the IPD ratio increase in the +2 position, 5gmC increases the IPD ratio at the modified cytosine position.
Using this method, 89%, 98%, and 57% of the sites genome-wide are reported as methylated for the 5mCCTC, C5mCTTC, and G5mCGC motifs, respectively (Table S4). These detection percentages are slightly improved from earlier results for H. pylori 26695 using mTET1CD (80%, 92%, and 50%) (19). However, the low detection for the G5mCGC motif is puzzling, especially because NgTET1 should not exhibit bias on this site (Fig. 4A). We therefore compared the IPD profiles between the GCGC sites detected as methylated and the sites detected as unmethylated, as reported by the Pacific Biosciences analysis pipeline, and noticed that the algorithm appears to ignore those sites with increased IPD ratio signals only at the +2 position (Fig. 4C). The same observation was made for the 5mCCTC motif (Fig. S7). Fig. 4D shows the scatter plot of the IPD ratio values at the methylated and +2 positions among the methylated G5mCGC (detected in blue and undetected in red) and the unmethylated CCGG motifs, as a comparison. It can be seen that in the 2D IPD ratio space by both methylated and +2 positions, the methylated “cloud,” which is more diffusive, can be separated from the unmethylated “cloud,” which is more bounded (Fig. 4D). There are a variety of machine-learning techniques that can incorporate these features into a classifier to detect modification. Here we explored a simple hard decision boundary for both methylated and +2 positions, and plotted the corresponding detection sensitivity, as well as specificity in Fig. 4E. Note that we used the genome-wide CCGG sites, which are known to be unmethylated in the genome, as the true negative set to calculate the detection specificity. It can be seen that by imposing the same IPD ratio cut-off (e.g., 1.75) at both modified and +2 positions, it is possible to dramatically increase the detection sensitivity to over 90% while maintaining high specificity (false-discovery rate < 5%) for all motifs (Fig. 4E). Overall, the above results demonstrate that NgTET1 can be applied to SMRT sequencing to assist in mapping bacterial methylomes, and that further optimization in the modification detection algorithm can be made to increase performance.
Discussion
Members of the TET/JBP family are distributed over a wide phylogenetic distance and often show patterns of lineage-specific expansion. Among them, JBP1 and JBP2 have been shown to possess T-hydroxylation activity, whereas multiple paralogous genes in mammals, mushroom (Coprinopsis cinerea), and honey bee (Apis mellifera) have been shown to possess 5mC-oxygenase activity (1, 6, 22, 23). Our results demonstrate that NgTET1 and the evolutionarily distant mTET1CD possess both 5mC- and T-oxygenase activities, which may initially have been shared by a single ancestral enzyme. Although 5mC-oxygenase activity in both enzymes is almost identical (Fig. 1E), mTET1CD T-oxygenase activity is significantly lower than that of NgTET1 (Figs. 2B and 3D). Indeed, the highest-level oxidation product, 5caU, is obtained only in the reaction of NgTET1. This is, to the best of our knowledge, the first in vitro evidence for oxidation of T on DNA to 5caU by any DNA modifying enzyme. Based on these observations, we hypothesize that unlike the heterolobosean NgTET1, and the evolutionarily close kinetoplastid JBP1/2, the mammalian TETs may have gradually lost most of their T-oxygenase activity, possibly to accommodate the emergence of multiple 5hmU glycosylases in the mammalian genome. More work is required to elucidate whether the oxidized Ts are epigenetically relevant or merely a promiscuous activity of these enzymes.
Interestingly, the mutant D234A (and H297Q, to a lesser extent) of NgTET1 appears to have reversed its substrate preference with almost no 5mC-oxygenase activity and increased T-oxygenase activity (Fig. 2D). Structure-based sequence alignments reveal that the residues D234 and H297 are conserved in seven or six of the eight NgTET homologs, respectively, whereas a pairwise comparison between NgTET1 and the mammalian TET counterparts shows conservation of H297 but an N at the D234 position (10). Indeed, the crystal structure of the TET2 catalytic domain reveals analogous roles for N1387 and H1904 in 5mC binding (24). Equivalent residues in JBP1/JBP2 are D218/D396 and R287/R463, respectively (24), which further demonstrates the importance of these two residues in 5-methylpyrimidine recognition and the delicate balance between these two activities.
Both NgTET1 and mammalian TETs exhibit a substrate preference for G immediately 3′ downstream of 5mC (10, 24). Here we show that replacement of this G with any of the three other nucleotides severely slows down 5fC conversion to 5caC, whereas a modest decrease is also observed for the 5mC and 5hmC decay rates in NgTET1 reaction (Fig. 3E). These observations could suggest marked structural variations in the NgTET1 active-site pocket for the 5fC substrate compared with 5mC and 5hmC substrates. In addition, the same sequence preference is exhibited when there is a T, rather than a 5mC, being oxidized (Fig. 3D). Structural studies with a T bound in the active site may shed additional light on the differences between these two catalytic activities. The biochemical and structural information on hand also raises further questions as to how NgTET1 or other TET analogs target their substrates on the genome for oxidation activity. More in vivo studies are required to elucidate any epigenetic significance to these observations in both organisms.
The catalytic properties of TET enzymes render them powerful tools in methylome sequencing applications. One potential disadvantage is that variable detection sensitivity for different methylation or hydroxymethylation motifs could be encountered, mainly in bacterial methylome sequencing as a result of the inherent substrate preference of TETs. Nonetheless, NgTET1 exhibits nearly complete oxidation activity of 5mC for all mammalian genomic DNA tested, and a full conversion of 5mC to any combination of its oxidized products is the only essential criterion for the accurate mapping of 5mC using our NgTET1/NaBH4/T4-βGT SMRT sequencing method. This method is distinct from other methylome sequencing methods, such as TET-assisted bisulfite sequencing (TAB-seq) (25) or oxidative-bisulfite sequencing (oxBS-seq) (26), in the fact that it does not include the relatively harsh bisulfite treatment that could result in DNA degradation (27–29). Although this method does not currently distinguish between 5mC and 5hmC (both TAB-seq and oxBS-seq distinguish between these two modifications), NgTET1 could be potentially used in a manner analogous to that of mammalian TET1 in TAB-seq to map 5hmC.
Using NgTET1 coupled with SMRT sequencing, we show comprehensive characterization of 5mC motifs in both plasmid and gDNA. As previously reported for SMRT sequencing of cytosine modifications (17), a prominent increase in signal at the +2 position is observed along with an increase at the modification position (0 position). Our analysis indicates that improvement in sensitivity can be made by incorporating both the methylated and +2 position into the 5mC calling algorithm (90% detection compared with 57% for the G5mCGC motif), while ensuring high specificity (Fig. 4E). It is important to note that NgTET1 efficiency, in its oxidation of 5mC on various substrates as well as its effectiveness as a tool for comprehensive genome-wide mapping of 5mC modification using SMRT sequencing, matches that of mTET1CD (19). The smaller size of NgTET1 renders it an ideal enzyme for production, sequencing applications, and future engineering efforts for the purposes of relaxing substrate specificity and enhancing 5mC conversion in all types of gDNA. We envision the use of NgTET1 in a variety of methylome sequencing applications for the critical understanding of the epigenomic function of 5mC and its oxidized forms.
Materials and Methods
Protein purification and preparation of DNA substrates are described in detail in SI Materials and Methods. Detailed descriptions of DNA substrates used are provided in Tables S1 and S2. See Table S5 for a list of ds-oligo substrates containing the MspI site, used in Fig. S1.
The NgTET1 reaction conditions and LC-MS–based activity assay were performed as described previously (10) and are described in detail in SI Materials and Methods. For the RE-based NgTET1 activity assay, purified DNA (300 ng) from each NgTET1 reaction was digested with 20 units (U) of BamHI (New England Biolabs) (to linearize the plasmid) and 50 U of MspI (New England Biolabs) in New England Biolabs CutSmart buffer (pH 7.9) for 1 h at 37 °C in 20-μL total volume. The reaction products were resolved on a 1.8% agarose gel.
Preparation of NgTET1 and NgTET1/NaBH4/T4-βGT reaction samples for SMRT sequencing and preparation of SMRTbell template libraries, sequencing, and analysis are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Derrick Xu and Meg Mabuchi for initial work on the project; Rick Morgan and Yvette Luyten for providing the pRS(M.HpaII) plasmid and M.Fnu4HI gDNA; Chandler Fulton and Elaine Lai for helpful discussions; and Bill Jack for critical review of the manuscript. This work is supported by New England Biolabs, and by National Institutes of Health Grants GM105132 (to Y.Z.) and GM049245-21 (to X.C.). Funding for the open access charge is from New England Biolabs.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417939112/-/DCSupplemental.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8(11):1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavez L, et al. Simultaneous sequencing of oxidized methylcytosines produced by TET/JBP dioxygenases in Coprinopsis cinerea. Proc Natl Acad Sci USA. 2014;111(48):E5149–E5158. doi: 10.1073/pnas.1419513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piccolo FM, et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol Cell. 2013;49(6):1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(4):1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritz-Laylin LK, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140(5):631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto H, et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506(7488):391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Yu M, He C. Mouse Tet1 protein can oxidize 5mC to 5hmC and 5caC on single-stranded DNA. Acta Chimi Sin. 2012;70(20):2123–2126. [Google Scholar]
- 12.Iyer LM, Zhang D, Burroughs AM, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41(16):7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaffeneder T, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014;10(7):574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 14.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 15.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark TA, et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40(4):e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark TA, et al. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 2013;11:4. doi: 10.1186/1741-7007-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Q, Morgan RD, Roberts RJ, Blaser MJ. Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc Natl Acad Sci USA. 2000;97(17):9671–9676. doi: 10.1073/pnas.97.17.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krebes J, et al. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 2014;42(4):2415–2432. doi: 10.1093/nar/gkt1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song CX, et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods. 2012;9(1):75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terragni J, Bitinaite J, Zheng Y, Pradhan S. Biochemical characterization of recombinant β-glucosyltransferase and analysis of global 5-hydroxymethylcytosine in unique genomes. Biochemistry. 2012;51(5):1009–1019. doi: 10.1021/bi2014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, et al. A TET homologue protein from Coprinopsis cinerea (CcTET) that biochemically converts 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine. J Am Chem Soc. 2014;136(13):4801–4804. doi: 10.1021/ja500979k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojciechowski M, et al. Insights into DNA hydroxymethylation in the honeybee from in-depth analyses of TET dioxygenase. Open Biol. 2014;4(8):140110–140118. doi: 10.1098/rsob.140110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu L, et al. Crystal structure of TET2-DNA complex: Insight into TET-mediated 5mC oxidation. Cell. 2013;155(7):1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Yu M, et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012;7(12):2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Booth MJ, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. 2013;8(10):1841–1851. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29(13):E65-5. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 2007;35(5):e29. doi: 10.1093/nar/gkl1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura F, Enomoto Y, Dairiki R, Ito T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012;40(17):e136. doi: 10.1093/nar/gks454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.